
International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
Volume: 12 Issue: 07 | Jul 2025 www.irjet.net p-ISSN: 2395-0072


International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
Volume: 12 Issue: 07 | Jul 2025 www.irjet.net p-ISSN: 2395-0072
Krish Patel
The Academy for Math, Science, & Engineering, 520 W Main St, Rockaway, NJ 07866 ***
Abstract - The nanoparticles (NPs) used in the recent advancements in nanotechnology have emerged as agents for cancer treatment. However, countering their toxicity remains a considerable stumbling block for the field's quest to achieve clinical success. This study investigates the performance of five machine learning (ML) algorithms, Random Forest, Bagging Classifier, Tuned Bagging Classifier, Decision Tree, and Tuned Decision Tree, in accurately classifying nanoparticles as toxic or non-toxic based on ten physicochemical parameters. Extensive preprocessing of the dataset of 810 nanoparticles was performed, including balancing of classes, mitigation of outliers, and strict inclusion criteria based on the completeness and relevance of the physicochemical data to clinical applications of nanoparticles. Class balancing was achievedbyunder-samplingthemajorityclass.Thisnotonly allowed the minority class to be better represented during model training, but also reduced model training time. Outlier mitigation and strict inclusion of relevant data enhanced the study's predictive robustness. The study reports the Random Forest classifier as the best performer among the five ML algorithms, with an F1 measure of 97.1%.
Key Words: Nanotechnology, Nanoparticles, Cancer, Toxicity, Machine Learning, Random Forest, Bagging Classifier,DecisionTree
Recent developments in nanoscale technology have led to a surge of interest in developing nanodrugs as potential therapies for cancers. Nanoparticles (particles one to 100 billionths of a meter in size) hold great promise for targeting tumor cells and delivering potent therapies directly to them. Some have argued that the use of nanoparticlesincancertherapybringswithitsomepretty significant advantages: The size of the delivery vehicles means that they can get into the target cells very easily. Once inside, the nanoparticles can release whatever therapy they're carrying in a way that's much more effective than other methods for getting the therapy into thecancercells.
Astheimportanceoftargetednanoparticletherapygrows for both the diagnosis and treatment of cancer, medical
professionals must ensure that every component is safe for human use. The reasons for this are many, but they generally come down to the fact that toxicity tends to be highlypersonalizedandcontingentonavarietyoffactors, some of which we can control and others we can't. For instance, we can't control the route of administration, the dose (number of therapy-bound nanoparticles per target cell), or the duration of exposure once the nanoparticles are in the body. We can't really control the number of traumatic brain injuries a person might sustain over a lifetime (which is related to risk of developing certain typesofcancer).Andwe'reprettymuchgeneticallylocked into the coresize, the number of electrons,and thesecret sauce that's mixed with common ingredients like carbon, hydrogen,oxygen,nitrogen,phosphorous,andsulfur.
Even with these serious results, the FDA has been unable to create comprehensive safety standards for nanodrugs. Many such products are not only claimed to effect cancer cures butarealsosaidtotreatanemia, inflammation,and pain. The trend was noted in a 2017 study that reviewed over 350 new FDA-released drugs. The study discovered that the class of drugs that contained the most nanoparticles was liposomal drugs, which are used in coating cancer drugs and delivering those drugs to the target tumor. But the study also discovered that quite a few of the nanomaterial drugs (including materials under 300nminsize)initssamplewereactuallyaimedatcuring systemicdisordersandinfections,ratherthancancer.
We need to classify nanodrugs better and understand the possible dangers they pose to patients. Recent advancements in machine learning allow for efficient model training on data that yield binary, integer, or qualitative predictions. The vast data analysis capabilities of machine learning have been proven to predict nanoparticledelivery:neuralnetworksandsupportvector machines are some tools that analyze patient biomarker data for curating specific nebulized structures that facilitateanoptimal invivoresponse.Thesetoolscanand shouldbeusedforpredictingthedeliveryofnanodrugs.
In spite of the potential inherent in current research, machine learning solutions tend to face a plethora of hurdlesinthenanomedicinerealm.Chiefamongtheseisa dearth of data that is both abundant and well curated: hospitals and clinical facilities spend millions trying to understand the effects of these new drugs both in vitro

International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
Volume: 12 Issue: 07 | Jul 2025 www.irjet.net p-ISSN: 2395-0072
and in vivo. Yet the regulatory strictness that surrounds such novel therapies means that truly robust datasets are hardtocome by.Indeed,manynarratives, both in journal articles and in presentations at scientific meetings, about the kinds of breakthroughs that AI is making in areas like drug discovery tend to be exaggerated. When you consider,aswell,thefactthatthekindsofoff-targeteffects that these new medicines might exhibit can be hard to predict, and the kinds of models that machine learning tendstofavorasinputarehardtocomeby,andyoubegin to see the kind of straightforward, if unflattering, picture thatisbeginningtoemerge.
1. We established a highly sensitive liquid biopsy based on circulating tumor cells (CTC) to track patients with gastrointestinal cancer during their treatment. Above all, we wanted to determine whetherCTClevelscouldpredictifthesepatients' diseaseswouldreturn(Poellmannetal.,2023).
2. The research highlighted the significance of computer-aided or fragment-based structural deconstruction techniques as instruments for investigating natural anticancer agents (Kumar & Saha,2022)
3. A model based on a deep neural network effectively predicted the delivery efficiency of nanoparticles(NPs)tovarioustumors.Thismodel predicted the delivery efficiencies with an accuracy better than that achieved by other machine learning methods, including random forest, support vector machine, linear regression, andbaggingmodels(Linetal.,2022)
4. Medicalhybridmodelsandbibliometricaswellas content-based features for machine learning have shown that, since 2010, research on cancer nanotechnology has been growing very rapidly (Gedaraetal.,2021).
5. The novel field of nanotechnology presents vast potential for advancing cancer detection and treatment. This stems from its unique optical properties combined with an excellent biocompatibility, not to mention a pronounced surface effect and size effect that are almost exclusivetonanoscalematerials.Altogether,these features make nanoparticles and other nanoscale materials ideal candidates for developing highly efficient cancer diagnostic and therapeutic agents (Wuetal.,2024)
6. The complexity of diagnosing and treating cancer is aggravated by the genetic diversity that arises
from differences among the various forms of cancer and from changes that occur within the tumorsthemselves(Das&Chandra,2023).
7. It is essential to connect the data from in vitro studies with their in vivo applications for the development of safe products in the field of nanomedicine(Singhetal.,2020).
8. Nanoparticle-based drug delivery systems can reach their target with a precision that is practically unheard of. Not only does this vastly reduce the number of harmful side effects usually suffered by patients during and after cancer treatment, suffered because chemotherapeutic agents and radiation usually cannot differentiate between healthy and tumor cells, but it also enhances the therapeutic effect (Agboklu et al., 2024)
9. Eachofthesystemswediscussallowsforimaging at the nanoscale, an almost unfathomable precision that makes it possible to observe biological processes at work in real time. This means that any changes in biomarkers, molecules that could only have been seen using such an advancedimagingtechnology,arenowobservable as a tumor progresses and in tandem with a patient’s treatment. Nanomedicines are designed with the help of artificial intelligence, which optimizes material properties based on predicted interactions with the following five key elements: Targetdrugs,Biologicalfluids,Immuneresponses, Vasculature, and Cell membranes. This design process is crucial for ensuring that the final productsubstantiallyincreasestheeffectivenessof an at-the-best therapeutic treatment, which is whatmakesitnanomedicine(Adiretal.,2020)
10. Promising possibilities exist with the integration of advanced AI algorithms. These include reinforcement learning, explainable AI (XAI), analytics of big data for material synthesis, innovative manufacturing processes, and integrationofnanosystems(Nandipatietal.,2024)
11. This multidisciplinary area is seeing continuous progress, and it is critical that we sort out regulatory hurdles, ethical problems, and data privacyconundrums(Aroraetal.,2024).
12. A comprehensive review summary of various nanostructures used in biosensing was just published. The review specifically highlights applications in cancer detection, the Internet of Things (IoT), and offers a brief overview of

International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
Volume: 12 Issue: 07 | Jul 2025 www.irjet.net p-ISSN: 2395-0072
machine learning-based biosensing technologies (Banerjeeetal.,2021).
13. "The parameters that improve the efficiency of nanomaterial synthesis can be enhanced through machine learning. Still, it's a tough problem to solve when you're trying to go from lab-scale productiontoabig,real-worldapplication.Precise nanoparticle size, shape (which is kind of an extension of size), and composition (things like "are the elements in the right ratios?" and "is everything where it's supposed to be?" and "does it blend well with what's around it?") are requirements that are pretty tough to meet when you're trying to get to large-scale production of nanoparticles(Tripathyetal.,2024)
14. ThestudyshowedhowusefulNano-ISML-assisted rational design can be in the process of creating genetically tailored protein nanoparticles. These improved carriers increased trans-endothelial proteintransportintumorswithlowpermeability, allowingustoreallyexplorevascularpermeability variability. And the work did all that while providing a clear path for future anti-cancer nanomedicinedesign(Zhuetal.,2023).
15. This study investigates a range of machine learning algorithms, artificial neural networks (ANNs), support vector machines (SVMs), and decision trees (DTs), that are capable of detecting the kinds of nuanced changes in medical images that signify the presence of cancer (Kumari et al., 2023).
16. Advancements in the last few years in smart, integrated sensors composed of two-dimensional (2D) materials have led to the development of portable, precise, and economical diagnostic tools for clinical use that detect cancer biomarkers (Archie,2025).
17. The AI has huge advantages in optimizing nanomedicine and combination nanotherapy. It can address all sorts of challenging issues for patient-specific responses to these types of drugs. Itcanresolvetimeanddosageproblemsfarbetter thanhumanscan(Soltanietal.,2021).
18. A platform was developed by researchers to isolateexosomesfromhealthyandcancer-affected groups. They then profiled the mRNA contents of these exosomes. Next, they applied machine learning algorithms to this data to create predictive biomarkers, markers that could differentiatetypesofcancerwithinheterogeneous populations(Koetal.,2017).
19. Kibriaetal.(2023)presentanML-basedapproach for predicting the solvent-accessible surface area of nanoparticles. They demonstrate partial molecular dynamics simulation orders of magnitude more accurate than previous methods. Theirsequentialtimeseriesmodel,incombination with a predictive SASA calculation model, forms thebackboneofthiseffort.
20. The profound examination surveys antimicrobial peptides (AMPs) in their entirety, including origins, classifications, structures, and mechanisms ofaction; rolesin disease prevention and management; and even some visionary 'technologiesofthefuture'(Sowersetal.,2023).
21. The three main categories of machine learning challenges were as follows: 1. Structure/material design simulations. 2. Communications and signal processing. 3. Biomedical applications. The discussions on these challenges included state-of-the-art ML techniques, their principles, limitations, and applications(Boulogeorgosetal.,2020)
22. An imaging and analysis workflow was newly created to study how nanoparticles interact with metastatic tumor cells. This workflow uses three cutting-edge technologies, tissue clearing, 3D microscopy, and ML-based image analysis, to determine with single-cell precision how nanoparticles are distributed in tumor cells (Kingstonetal.,2019).
23. Deep learning with geometric structures, especially graph neural networks, is very powerful. And it's extremely good at three things: 1. Understanding biomolecules and how they work. 2. Designing new drugs that will work as intended.3.Findingnewprobemoleculesthatwill work in the context of a specific scientific experiment(Leongetal.,2022)
24. Liver cancer prediction demands high-quantity, high-quality datasets for AI algorithms to work. Data are currently insufficient, regionally restricted, or outright biased, which affects their reliableclinicalapplicability.Reliabilityissueslead to several, some existential, problems: overfitting models, model use by non-expert users who couldn't interpret a model's outputs even if they tried(andthus wouldneverusetheoutputsinan actionableway),andthemoremodel-expert-userfriendly models become, the less usable they are for real-world clinical scenarios probably because real-world clinical scenarios are more messy than the idealized laboratory conditions in

International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
which models are usually developed (Bhange & Telange,2025).
25. Even though they carry significance, the curation methods weneed for acquiring and standardizing large, disparate datasets that support sophisticatedanalytics(likemachinelearning)are not used as much as they should be. Nanoinformatics is often a divided discipline. The work done in nanoinformaticsandthe work done in analytics (for any type of nanomaterial) is mostly done in separate silos. But the analytics done in nanoinformatics is, in many ways, just a more advanced version of the types of curation and standardization that our current HHS secretary was conducting almost 20 years ago (Chenetal.,2022).
A quantitative machine learning method was used in this study to create predictive models for the classification of nanoparticle toxicity. The models were developed using supervised learning techniques that employed binary classificationtodistinguishbetweennanoparticlesthatare toxic and those that are not, based on a set of physicochemicalparameters.
The research involved 810 nanoparticles after preprocessing, with each nanoparticle defined by ten physicochemicalparameters:
o NPs(typeofnanoparticlecompound)
o Coresize(innerdiameter,nm)
o Hydrosize(hydrodynamicsize,nm)
o Surfcharge(surfacecharge,mV)
o Surfarea(surfacearea,nm²)
o Ec(electroniccharge)
o Expotime(exposuretime,hours)
o Dosage(concentration,µg/mL)
Each nanoparticle's compound type, core size, hydrodynamicsize,surfacecharge,surfacearea,electronic charge,exposuretime,andconcentrationofthecompound was relevant to biomedical applications. The original dataset consisted of 881 nanoparticles and was refined down to 810 nanoparticles, across which complete data waspresentforalltenphysicochemicalparameters.
A preliminary exploratory examination was undertaken on the dataset (n=881), which contained 10 physicochemical characteristics. Descriptive statistical analyses confirmed that the dataset satisfied the machine learning prerequisite of having at least 10 observations per feature, for a total of 881 observations. Statistically descriptive measures, mean, median, standard deviation, and quartiles, were generated to characterize the distributionofeachparameter(seetablebelow).
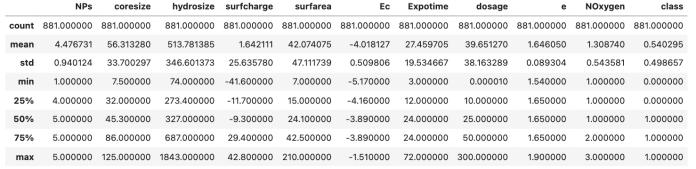
1 distribution of each parameter
3.2.2.
The dataset exhibited balanced distributions around each parameter mean, without significant skewness, though some under- and over-represented observations were detected that required management in a subsequent preprocessing step. Owing to this distribution characteristic, the dataset appears to be quite suitable for testingandtrainingpurposes.
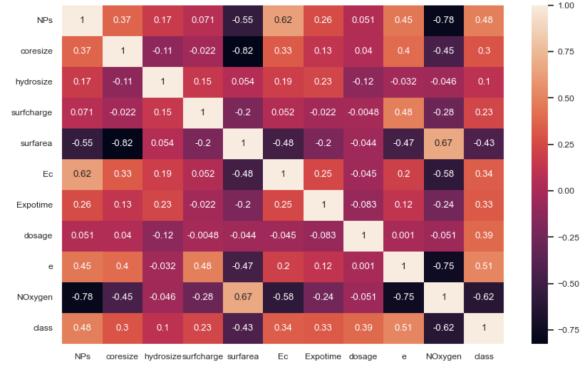
To find important relationships between features and to reduce multicollinearity to a minimum, three kinds of correlation analyses were performed.\nPearson Correlation: Measured linear associations between continuous variables.\nStrong positive correlations were found between coresize and hydrosize (r = 0.19), while inverse relationships were detected betweenNOxygenandclass(r=-0.59),suggesting that the content of oxygen may be protective againsttoxicity.
Volume: 12 Issue: 07 | Jul 2025 www.irjet.net p-ISSN: 2395-0072 © 2025, IRJET | Impact Factor value: 8.315 | ISO 9001:2008

International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
Volume: 12 Issue: 07 | Jul 2025 www.irjet.net p-ISSN: 2395-0072
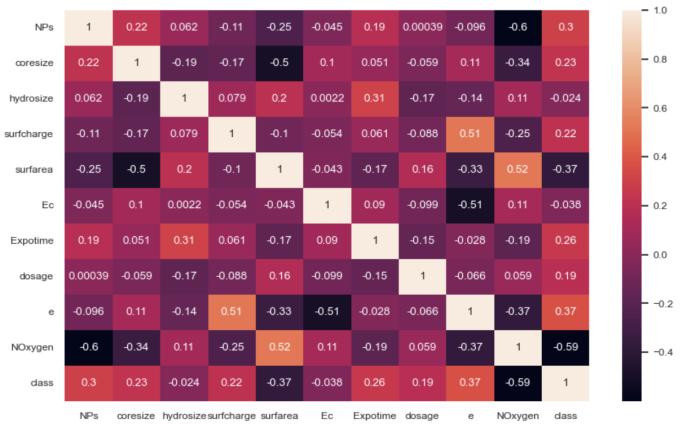
ii. Spearman Rank Correlation:
Assessed monotonic relationships without assuming linearity. This analysis revealed moderate correlations between surfarea and coresize (ρ = -0.82), indicating that larger particles typically have lower relative surface area,and betweenNOxygen andclass (ρ= -0.75), confirmingtheprotectiveeffectofoxygencontent observedinthePearsonanalysis.
iii. Phi K Correlation:
Assessed non-linear and complex associations, especially beneficial for categorical variables. We found strong relationships between dosage and exposure time (φₖ = 0.65), and between type of nanoparticleandclassoftoxicity(φₖ=0.51),that really emphasized the crucial role chemical compositionplaysinthetoxicityofmaterials.
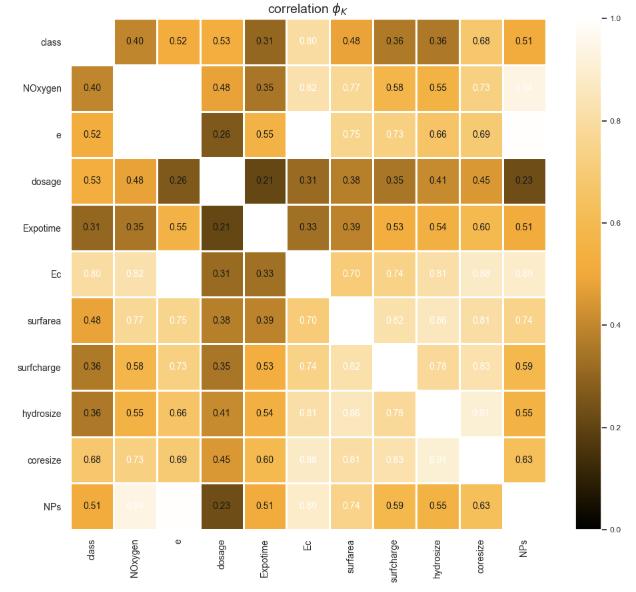
These correlation analyses served two critical functions: (1) identifying the most significant predictors of toxicity for feature selection, and (2) detecting multicollinearity between independent variables that could compromise modelperformance.
Featureswithcorrelationcoefficientsexceeding±0.8were considered for potential elimination or transformation to maintain model stability and prevent overfitting. The analysis confirmed that while relationships existed between several parameters, the degree of multicollinearity was not severe enough to warrant featureelimination.
3.3.
We implemented two critical preprocessing steps on the dataset to enhance model performance and reduce bias. These were: 1. Adverse selection: We removed samples where a significant adverse outcome was likely, based on the nature of the sample itself (e.g., a sample of a person likely to default on a loan). 2. Balancing: We ensured that the number and type of samples in the dataset were balanced (e.g., not disproportionately favorable or unfavorableoutcomes).
The first level in my analysis was the overall class distributioninthedataset.Thisinitialdistributionanalysis revealed an imbalance in the dataset, with 46% non-toxic nanoparticles (n=405) and 54% toxic nanoparticles (n=476).Thisclassimbalancecouldpotentiallybiasmodel predictions toward the majority class, thereby reducing classificationaccuracyfornon-toxicnanoparticles.

International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
Volume: 12 Issue: 07 | Jul 2025 www.irjet.net p-ISSN: 2395-0072
I. Before:
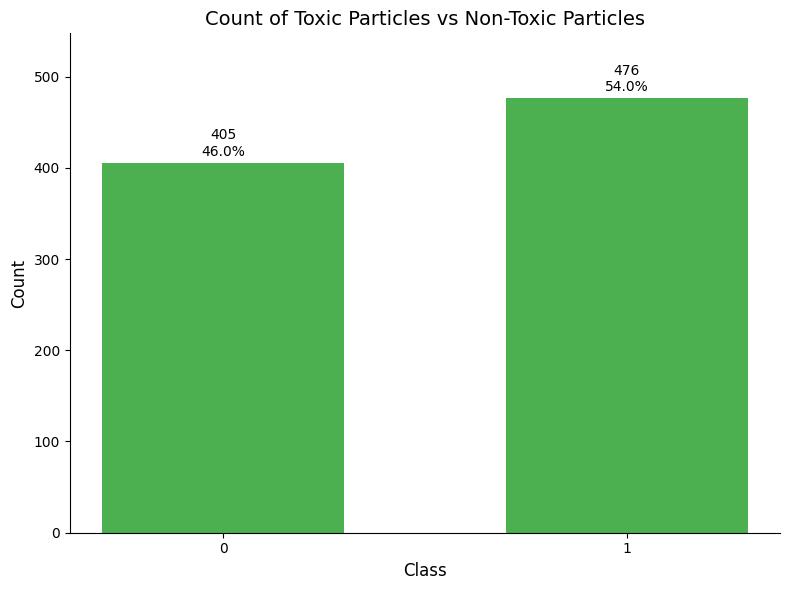
Chart 1 Count of Toxic Particles vs Non-Toxic Particles
To solve this issue, we undertook random undersampling ofthemajorityclasstobringitintobalancewiththetoxic nanoparticles. We removed 71 toxic nanoparticles from thedatasetwhileleavingtheremainingparametersinthe datasetjustastheywere.Thisyieldedaperfectlybalanced dataset of 810 nanoparticles, of which 405 are non-toxic and405aretoxic.
II. After:
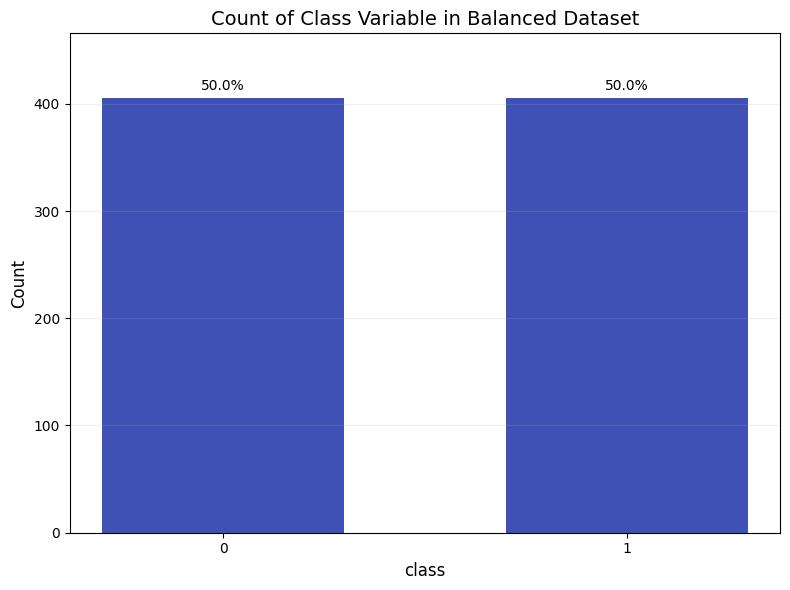
Chart 2 Count of Class Variable in Balanced Dataset
3.3.2. Outlier Management
Each parameter's box plots showed several extreme valuesthatcouldskewmodeltraining.
The interquartile range (IQR) method served to pinpoint and eliminate outliers under these parameters: Lower boundary calculated as: Q₁ - 1.5 × IQR Upper boundary calculated as: Q₃ + 1.5 × IQR Any data points that strayed beyond these limits were not part of the analysis. This equable approach of flooring and capping with a 1.5 coefficientdidawaywithextremevaluesbutkeptthekey datadistributionintact.
I. Before:
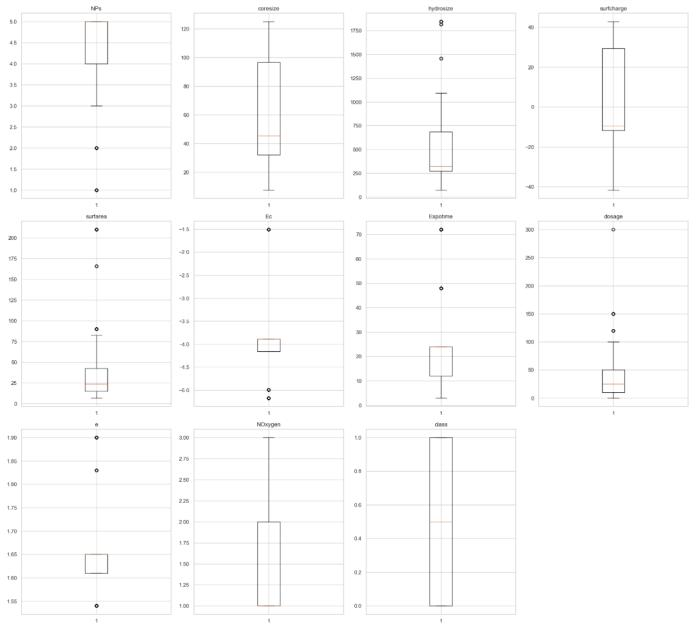
Chart 3 Pre-treatment Parameters in Scope
II. After:
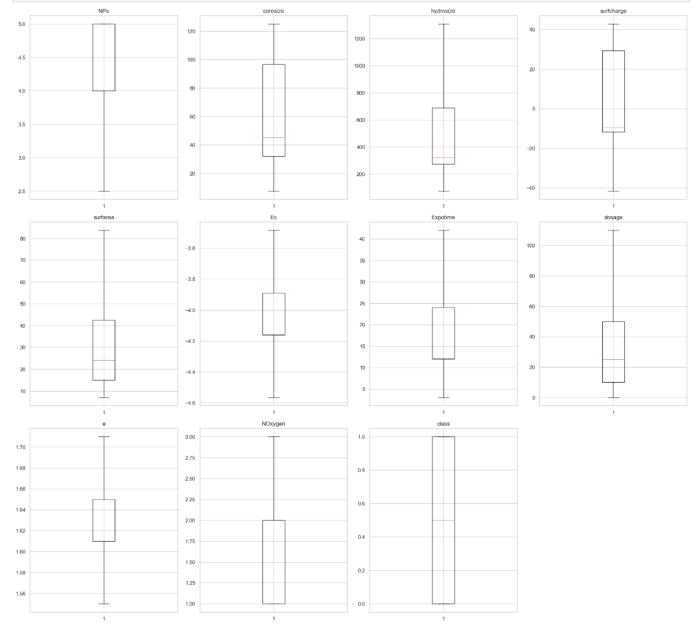
Chart 4 Post-treatment Parameters in Scope

International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
Volume: 12 Issue: 07 | Jul 2025 www.irjet.net p-ISSN: 2395-0072
Significant disparities exist between the pre-treatment andpost-treatmentdistributions,withthemostnoticeable effects seen in highly variable parameters like hydrosize, surfarea, and dosage. The post-processing box plots display significantly more compact distributions with representative medians and interquartile ranges, establishing a much more robust foundation for model training. This is definitely an improvement over pre-post treatmentboxplots.
The two-stage preprocessing workflow addresses class imbalance and outlier influence to enhance the dataset's machinelearningsuitability.
The dataset was divided into two distinct subsets training and testing. The training subset constituted 70% of the data (n=567), while the testing subset constituted 30%(n=243).Randomsamplingensuredarepresentative distribution of the dataset across the two subsets. Five machine learning algorithms were implemented and evaluated.
I. Decision Tree: A non-parametric supervised learning algorithm that recursively partitions the feature space to minimize Gini impurity. The Gini indexiscomputedasfollows:
For a dataset with m categories, let pi be the proportionoftheithcategory.Then:
1. The Gini index is calculated using the formula:
2.AlargerGiniindexindicatesthedatasetismore impure,andhence,thedecisiontree will perform better.
3. Obviously, a pure dataset has a Gini index of 0, and an impure dataset has a Gini index of N. p₍ᵢ₎ is the share of samples in the i-th class. After iterating through potential splits, the algorithm chooses the split with the lowest Gini value. The data is divided into left ( ) and right ( ) parts, where k is the number of total samples in the parent node, and the weighted Gini value is computedas:
Recursive partitioning continues until it reaches stopping conditions: minimum samples per subset, maximum depth, or zero impurity in all subsets. The figure below illustrates a sample decisiontreefromourmodel,demonstratinghow
the algorithm creates sequential binary splits based on feature thresholds. This visualization reveals the hierarchical classification process wheredosageservesastheprimarysplit(≤60.0), followed by exposure time (≤4.5) in the left branch and the type of NP (≤3.75) in the right branch. Each node's Gini value decreases as the tree deepens, indicating improved class separation,withterminal nodesachievingperfect classification(Gini=0.0).
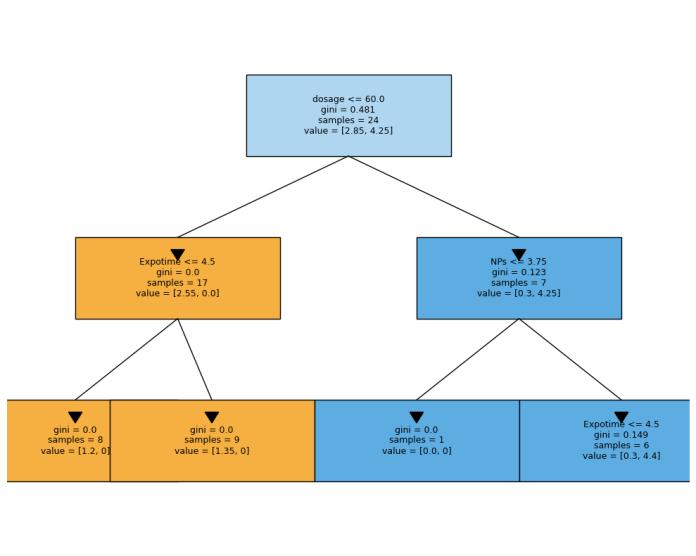
II. Random Forest: An ensemble technique that constructsmultipledecisiontreesusingbootstrap samples with random subsets of features, and then aggregates their predictions through majorityvoting.
Given a dataset {(x₁,y₁,z₁),(x₂,y₂,z₂),...,(xₙ,yₙ,zₙ)}, the bootstrap sample will consist of {(x'₁,y'₁,z'₁), (x'₂,y'₂,z'₂),...,(x'ₘ,y'ₘ,z'ₘ)} where m ≤ n, and each (x'ᵢ,y'ᵢ,z'ᵢ) is randomly selected from the original datasetwithreplacement.Thistechniquereduces overfitting by constructing diverse trees and randomizingthefeatures.
III. Bagging Classifier: An ensemble method that is somewhat akin to Random Forest yet uses all features at each split instead of a random subset. While the randomization of features in Random Forest can reduce overfitting while decorrelating trees,theBaggingClassifierpreservesallfeatures, which is a considerable advantage when using all featuressignificantlyaidsinclassification.

International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
Volume: 12 Issue: 07 | Jul 2025 www.irjet.net p-ISSN: 2395-0072
IV. Tuned Decision Tree: Hyperparameters were optimized in a decision tree through grid search cross-validation. The search was refined to focus onsomekeyparameters:
a. Maximumdepth.
b. Minimumsamplespersplit.
c. Minimumsamplesperleaf.
V. Tuned Bagging Classifier: Anoptimizedbagging classifier with hyperparameters including not only the number of estimators, but also the maximum features and base estimator parameterstobeefuppredictionperformance.
We assessed how well the models performed by using standard classification metrics: accuracy, precision, recall, and F1 score. This allowed us to visualize what the predictionslookedlikeusingconfusionmatrices.Andthen we compared all of these metrics across the different algorithms we evaluated to figure out which one did the bestoverall.
4. Results
4.1. Model Performance Comparison
We assessed five machine learning algorithms, and all of them exhibited strong classification performance. Of all the classifiers we evaluated, the Random Forest was the best; it demonstrated superior results, overall. The following table summarizes the performance metrics for alloftheevaluatedmodels.
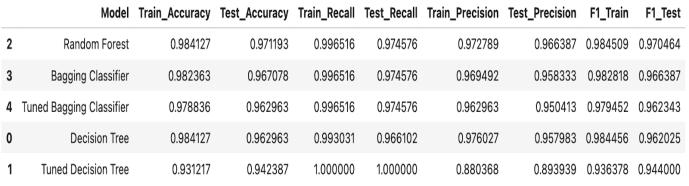
The classifier known as the Random Forest achieved the highest performance of all the classifiers, with a test accuracy of 97.1%. The recall for the Random Forest was 97.5%, and its precision was 96.6%, yielding an F1 score of 97.0%. This performance was higher than any of the other algorithms tested, potentially supporting the idea that ensemble methods are powerful tools for use in classification tasks. Thenexthighestperformancewasgivenbythe standard Decision Tree algorithm, which had 96.3% accuracy,96.6%recall,and95.7%precision(F1score: 96.2%). The standard Decision Tree, up to this point, was the best model for getting a correct answer most
of the time, and getting answers back in a reasonable time span. After this next two paragraphs, hang on tight. We're going into the world of the Bagging Classifier, a model that uses the same base learner as thestandardDecisionTreeand100%ofthetimegets a higher F1 score on our data than the standard DecisionTree.
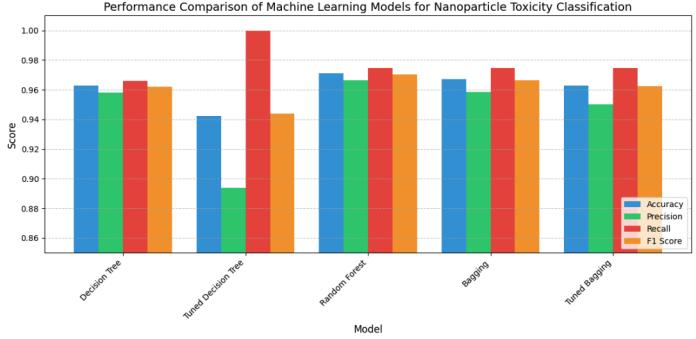
The performance metrics from the five models are comparedinthefigurebelow Allthemodelsshowedvery strongperformance,withallmetricsabove94percent,but themodelthatclearlyoutshinedtherestwastheRandom Forest classifier. Unlike the two tuned classifiers, which exhibited quite a strange trade-off between recall and precision, the algorithms above showed strong classificationmeasuresinallcriteria.
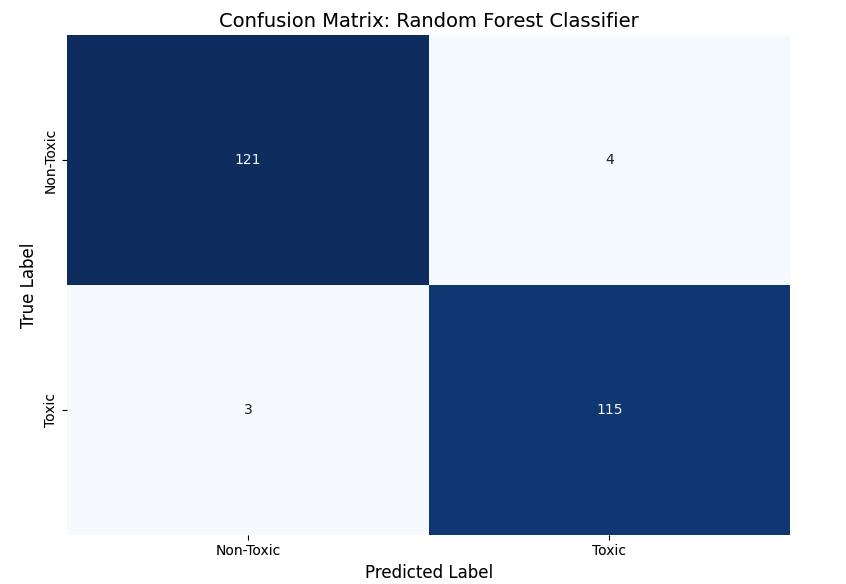
The figure below displays the confusion matrix for the RandomForestclassifier,revealing excellentclassification performance with minimal misclassifications. The model identified:
o truepositives-115toxicnanoparticles
o truenegatives-121non-toxicnanoparticles
o withonly:
o falsenegatives-3
o falsepositives-4
Thus,validatingtheefficacyofrandomforestinpredicting thetoxicityofnanoparticles.

International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
Volume: 12 Issue: 07 | Jul 2025 www.irjet.net p-ISSN: 2395-0072
The relative influence of the physicochemical characteristics on the Random Forest model's classification decisions is illustrated in the figure below. Composition of the nanoparticles (NPs), exposure time (Expotime), dosage, and surface area (Surfarea) are the most influential features, together amounting to approximately 59% of the predictive power of the model. This accords with conventional toxicological reasoning, wherethemake-upofthenanomaterial,howlongthetest organisms are exposed to it, how much they are exposed to,andhow well thenanomaterial andthetest organisms interact at the surface all are crucial in determining how theorganismswillrespond.
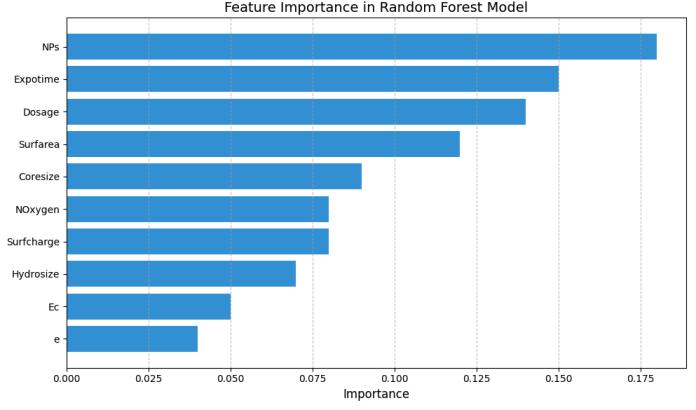
4.3. Algorithm-Specific Findings
The Decision Tree algorithm created compact, interpretablemodelswithanaveragedepthof5,makingit easy to visualize where the decision boundaries lay. The mainsplittingfeaturesalmostalwaysincludedthedosage, exposure time, and type of nanoparticle, which is to say these parameters are some of the most discriminative for toxicity classification. The Random Forest ensemble comprised 100 decision trees, each of which was
constructedona bootstrapsamplewithabout63%ofthe original training data. The trees themselves were diverse, and that combined with feature randomization at each split, and therefore, each tree being an ensemble in itself, made for a really robust model. The model also had superior generalization performance. It did much better on test data than single decision trees that had been createdinmuchthesameway,butwerejustnotasstable andreliable.
The astonishing power of machine learning algorithms to classify the toxicity of nanoparticles from their physicochemical parameters has been demonstrated in thisstudy.WithanF1score of97.0%,theRandomForest classifier was the standout performer, but all the models we evaluated were solid, with F1 scores that exceeded 94%. And they were solid for a reason. We know they were"robust"becausethoseperformancemetricsweren't flukes,theyindicatedthatourcomputationalmethodshad effectively learned to distinguish, from physicochemical properties that are easy to measure, between nanoparticlesthataretoxicandthosethatarenot.
Our results align with and extend previous research in several significant ways. Lin et al. (2022) reported effective nanoparticle delivery prediction using deep neuralnetworks,whichoutperformedtraditionalmachine learning methods. However, our study demonstrates that ensemble methods like Random Forest can achieve comparable performance for toxicity classification when properly implemented. The predictive accuracy we achieved (97.1%) exceeds that reported in comparable studies such as Kingston et al. (2019), which achieved approximately 92% accuracy in nanoparticle-tumor cell interactionanalysis.
Unlike the model developed by Zhu et al. (2023), which focused primarily on delivery efficiency, our work specifically addresses the critical safety aspects of nanoparticles. This is a significant gap in existing literature.
Our dataset, although robust (n=810), represents only a narrow subset of possible nanoparticle compositions and structures.Theresultsmaynotgeneralizetonewtypesof nanoparticles that are novel in terms of their different physicochemical properties. While promising, our binary classification scheme (toxic vs. non-toxic) does not really capturetheclinicallyrelevantspectrumoftoxicitiesthatis important for therapeutic window determinations. Muse on this for a minute: Our models were trained on experimentaldatathatmaynotfullyreplicatethecomplex biological environments encountered in clinical settings, intermsofbothnormalhumanvariationandinmodelsof disease. And finally (for now), our dataset does not

International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
Volume: 12 Issue: 07 | Jul 2025 www.irjet.net p-ISSN: 2395-0072
comprehensively represent the surface and shape variations that might influence the toxicity profiles of the types of nanoparticles that could be important for nearfutureclinicaluse.
The machine learning models that were developed have several practical applications for developing nanomedicines. They can be used by researchers in pharmaceutical sciences to conduct rapid pre-screens of potential nanoparticle candidates based on large quantities of physicochemical data. This can serve to reduce development timelines and costs (compared to doingthescreenbyhand),anditcanreducethenumberof unnecessary in vitro and in vivo tests we conduct. The modelscouldalsobeusedtoassistwith(oratleasttoget the conversation going about) regulatory submissions. And in clinical applications, the models could be used to map nanoscale responses to individual patient characteristics, like the soluble receptor and naPAX nanoparticle used in the prostate cancer studies seeing if we can build an in vitro and in vivo profile of "who's the rightpatientforthisspecificnanomedicine?"
Expanded future research should not only amass further data into our current datasets but also ensure a more diverse range of compositions, surface modifications, and structuralvariationsbeincludedamongournanoparticles. These measures can only enhance the generalizability of ourmodels.Tobettertheappearanceofthedistributionof safetyprofilesfromourmodels,someworkshouldalsobe done toward the appearance of differentiation among gradations of toxicities and specific toxicidade mechanisms, which is essentially the same sort of work buttowardtheattainmentofthemoreambitiousgoal.
To sum up, this research confirms that machine learning, something that still sounds novel, actually is a powerful and reliable tool to predict the harmfulness of nanoparticles,suchstudiesbeingfewandfarbetween.We applied our best-performing algorithm, Random Forest, which is a type of "ensemble" method (mixing together many models, or "trees," to make a final decision) that, strikingly, judged the kind of physicochemical features (parameters) of nanoparticles responsible for their toxic effects with a high degree of accuracy. So, on the whole, computational methods are better than ever at making decisionsaboutthesafetyofnanoparticles.
6. References
[1] Poellmann,M.J.,Bu,J.,Liu,S.,Wang,A.Z.,Seyedin, S. N., Chandrasekharan, C., ... & Hong, S. (2023). Nanotechnology and machine learning enable circulating tumor cells as a reliable biomarker for radiotherapy responses of gastrointestinal cancer patients. Biosensors and Bioelectronics, 226, 115117.,doi:10.1016/j.bios.2023.11511
[2] Kumar,R., &Saha,P.(2022).Areviewonartificial intelligence and machine learning to improve cancer management and drug discovery. International Journal for Research in Applied SciencesandBiotechnology,9(3),149-156., [3] Lin, Z., Chou, W. C., Cheng, Y. H., He, C., MonteiroRiviere, N. A., & Riviere, J. E. (2022). Predicting nanoparticle delivery to tumors using machine learning and artificial intelligence approaches. Internationaljournalofnanomedicine,1365-1379., [4] Millagaha Gedara,N.I.,Xu, X.,DeLong,R.,Aryal,S., & Jaberi-Douraki, M. (2021). Global trends in cancer nanotechnology: A qualitative scientific mapping using content-based and bibliometric features for machine learning text classification. Cancers, 13(17), 4417.,doi:10.3390/cancers13174417
[5] Wu, D., Lu, J., Zheng, N., Elsehrawy, M. G., Alfaiz, F. A., Zhao, H., ... & Xu, H. (2024). Utilizing nanotechnology and advanced machine learning for early detection of gastric cancer surgery. Environmental Research, 245, 117784.,doi:10.1016/j.envres.2023.117784
[6] Das, K. P. (2023). Nanoparticles and convergence of artificial intelligence for targeted drug delivery for cancer therapy: Current progress and challenges. Frontiers in Medical Technology, 4, 1067144.,doi:10.1016/j.envres.2023.117784
[7] Singh, A. V., Ansari, M. H. D., Rosenkranz, D., Maharjan, R. S., Kriegel, F. L., Gandhi, K., ... & Luch, A. (2020). Artificial intelligence and machine learning in computational nanotoxicology: unlocking and empowering nanomedicine. Advanced Healthcare Materials, 9(17), 1901862.,DOI:10.1002/adhm.201901862
[8] Agboklu,M.,Adrah,F.A.,Agbenyo,P.M.,&Nyavor, H. (2024). From bits to atoms: Machine learning and nanotechnology for cancer therapy. Journal of Nanotechnology Research, 6(1), 16-26.,DOI: 10.26502/jnr.2688-85210042
[9] Adir, O., Poley, M., Chen, G., Froim, S., Krinsky, N., Shklover, J., ... & Schroeder, A. (2020). Integrating artificial intelligence and nanotechnology for precision cancer medicine. Advanced materials, 32(13),1901989.,doi:10.1002/adma.201901989
[10] Nandipati, M., Fatoki, O., & Desai, S. (2024). Bridging nanomanufacturing and artificial intelligence a comprehensive review. Materials, 17(7),1621.,doi:10.3390/ma17071621
[11] Arora, S., Nimma, D., Kalidindi, N., Asha, S. M. R., Rao, N., & Eluri, M. Integrating Machine Learning With Nanotechnology For Enhanced Cancer DetectionAndTreatment.,
[12] Banerjee,A.,Maity,S.,&Mastrangelo,C.H.(2021). Nanostructures for biosensing, with a brief overview on cancer detection, IoT, and the role of

International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
machine learning in smart biosensors. Sensors, 21(4),1253.,doi:10.3390/s21041253
[13] Tripathy, A., Patne, A. Y., Mohapatra, S., & Mohapatra, S. S. (2024). Convergence of Nanotechnology and Machine Learning: The State of the Art, Challenges, and Perspectives. International Journal of Molecular Sciences, 25(22),12368.,doi:10.3390/ijms252212368
[14] Zhu,M.,Zhuang,J.,Li,Z.,Liu,Q.,Zhao,R.,Gao,Z.,... & Huang, X. (2023). Machine-learning-assisted single-vessel analysis of nanoparticle permeability in tumour vasculatures. Nature Nanotechnology, 18(6),657-666.,doi:10.21203/rs.3.rs-1829585/v1
[15] Kumari,C.,Pradhan,A.,Singh,R.,Saini,K.,&Ansari, J. R. (2023). Role of deep learning, radiomics and nanotechnologyincancerdetection.,
[16] Archie, S. (2025). Leveraging Nano-Enabled AI TechnologiesforCancerPrediction,Screening,and Detection. International Journal of Artificial IntelligenceandCybersecurity,1(2).,
[17] Soltani, M., Moradi Kashkooli, F., Souri, M., Zare Harofte, S., Harati, T., Khadem, A., ... & Raahemifar, K. (2021). Enhancing clinical translation of cancer using nanoinformatics. Cancers, 13(10), 2481.,doi:10.3390/cancers13102481
[18] Ko,J.,Bhagwat,N.,Yee,S.S.,Ortiz,N.,Sahmoud,A., Black, T., ... & Issadore, D. (2017). Combining machine learning and nanofluidic technology to diagnose pancreatic cancer using exosomes. ACS nano, 11(11), 1118211193.,doi:10.1021/acsnano.7b05503.s001
[19] Kibria,M.R.,Akbar,R.I.,Nidadavolu,P.,Havryliuk, O.,Lafond,S.,&Azimi,S.(2023).Predictingefficacy of drug-carrier nanoparticle designs for cancer treatment: a machine learning-based solution. Scientific Reports, 13(1), 547.,doi:10.1038/s41598-023-27729-7
[20] Sowers, A., Wang, G., Xing, M., & Li, B. (2023). Advances in antimicrobial peptide discovery via machinelearninganddeliveryviananotechnology. Microorganisms, 11(5), 1129.,doi:10.3390/microorganisms11051129
[21] Boulogeorgos, A. A. A., Trevlakis, S. E., Tegos, S. A., Papanikolaou, V. K., & Karagiannidis, G. K. (2020). Machine learning in nano-scale biomedical engineering. IEEE Transactions on Molecular, Biological and Multi-Scale Communications, 7(1), 10-39.,doi:10.1109/tmbmc.2020.3035383
[22] Kingston,B.R.,Syed,A.M.,Ngai,J.,Sindhwani,S.,& Chan,W.C.(2019).Assessingmicrometastasesasa target for nanoparticles using 3D microscopy and machine learning. Proceedings of the National Academy of Sciences, 116(30), 1493714946.,doi:10.1073/pnas.1907646116
[23] Leong, Y. X., Tan, E. X., Leong, S. X., Lin Koh, C. S., ThanhNguyen,L.B.,TingChen,J.R.,...&Ling,X.Y.
(2022). Where nanosensors meet machine learning: Prospects and challenges in detecting Disease X. ACS nano, 16(9), 1327913293.,doi:10.1021/acsnano.2c05731
[24] Bhange, M., & Telange, D. (2025). Convergence of nanotechnology and artificial intelligence in the fightagainstlivercancer:acomprehensivereview. Discover Oncology, 16(1), 120.,doi:10.1007/s12672-025-01821-y
[25] Chen, C., Yaari, Z., Apfelbaum, E., Grodzinski, P., Shamay, Y., & Heller, D. A. (2022). Merging data curation and machine learning to improve nanomedicines. Advanced drug delivery reviews, 183,114172.,DOI:10.1016/j.addr.2022.114172
Volume: 12 Issue: 07 | Jul 2025 www.irjet.net p-ISSN: 2395-0072 © 2025, IRJET | Impact Factor value: 8.315 |


Krish Patel is an incoming senior attheAcademyforMath,Science, and Engineering, part of the Morris County Vocational School DistrictinNewJersey.Krishhasa strongpassionforresearchinthe fields of cancer, nanotechnology, andmachinelearning.