




The new benchmark in grossing
• Discover next generation grossing
• You dreamt it, we built it
USER SAFETY
Sliding safety shield, backdraft and downdraft ventilation.
Bio-decontamination system to disinfect all the surfaces.
MACRO DIGITAL DOCUMENTATION
Built-in MacroPATH system for documenting the dissection procedure and creating image-enhance reports.



OPTIMAL ERGONOMICS
Adjustable working height. Mobility through sturdy industry casters. Compliance to anthropometric requirements (EN ISO 14738).
TAILOR-MADE MODULARITY
UltraGROSS counts on stainless-steel AISI 316 modules to be placed across the large basin, to match any working set-up, even with two-operators.
SMART USER INTERFACE
8” touchscreen terminal with all functions at reach. Capability to upload external files, like AAPA guidelines and training videos.
Editor
Lisa Cambridge, NZCS DipQA B.ApplManagement, MNZIMLS, NZIMLS, Rangiora
Deputy Editors
Michael Legge, PhD MRSB FIBMS FNZIMLS FFSc(RCPA), University of Otago, Dunedin
Holly Perry, DipMLS MAppSc(Hons) PhD MNZIMLS, University of Otago
Emeritus Editor
Rob Siebers, PGCertPH FNZIC FNZIMLS FRSB HonFNZAP, Wellington
Editorial Board
Paul Austin, MSc(Hons) DipMLT MNZIMLS, LabPlus, Auckland
Jillian Broadbent, FNZIMLS, NZIMLS, Rangiora
Julie Creighton, DipMLS, FNZIMLS, Canterbury Health Laboratories, Christchurch
Sujata Hemmady, PGDipMLSc, MMLSc, MNZIMLS, LabPlus, Auckland
Chris Kendrick, GradDipSci MSc MNZIMLS, Massey University, Palmerston North
Craig Mabbett, BMLSc PGDipHSM, LabCare Pathology, New Plymouth
Holly Perry, DipMLS MAppSc(Hons) PhD MNZIMLS, University of Otago
Mohd. Shahid, MBBS MD PhD FNZIMLS, PGDipHSM, Arabian Gulf University, Bahrain
Terry Taylor, BSc DipMLS MNZIMLS, Southern Community Laboratories, Dunedin
Sharon Tozer, DipBis Stud, AT CAANZ, NZIMLS, Rangiora
Robyn Wells, BApllSci(MT) GradCert Haem, Milton, Australia
Formatting
Sharon Tozer, AT DipBusStud, Executive Office NZIMLS, Rangiora
About the Journal
The New Zealand Journal of Medical Laboratory Science (the Journal) is the official publication of the New Zealand Institute of Medical Laboratory Science (NZIMLS). The Journal is peer reviewed and publishes original and review articles, case studies, technical communications, and letters to the Editor on all subjects pertaining to the practice of medical laboratory science. The Journal is open access (www.nzimls. org.nz/nzimls-journal) and is published three times per year in March, July, and November. Hard copies are circulated to all NZIMLS members and universities and research units in New Zealand and overseas. Current circulation is about 2,800 copies per issue. Printing is by Blueprint Ltd, Christchurch on environmentally responsible paper using elemental chlorine free third party certified pulp sourced from well managed and legally harvested forests and manufactured under the strict ISO14001 Environmental Management System. The Journal is indexed by CINAHL, EMBASE, SCOPUS, Informit, Thomson Gale, EBSCO and Biosis Citation Index, and the Journal Editors are members of the World Association of Medical Editors (www.wame.org).
Brief instructions to authors
The Journal accepts original submissions from anyone and anywhere. Comprehensive instructions can be found on the NZIMLS website (www.nzimls.org.nz/instructions-to-authors. html). All submissions will undergo single-blind peer review and possibly plagiarism checking with iThenticate™ software. If accepted for publication, copyright is vested in the author(s) under terms of the Creative Commons Attribution License (www. creativecommons.org/licenses/by/2.5/legalcode). The authors are responsible for the scientific content and views. Opinions expressed in the Journal are not necessarily those of the Editors, Editorial Board, or Council of the NZIMLS.
Advertising and subscription
Advertisement bookings and enquiries should be addressed to the NZIMLS Executive Officer, Sharon Tozer: sharon@nzimls.org.nz. Phone +64 3 313 4761.
Journal cover © Joseph Manieda, Te Whatu Ora Hawke’s Bay

Editorial Climate change, heath and pathology. Michael Legge 107
TH Pullar Address
Building resilience: keeping afloat during turbulent times. Angela Brounts 110-112
Original articles
Optimisation of sample volume for the 25% sulfosalicylic acid method for quantitative evaluation of proteinuria.
Nimesha D Ranawaka, Rajika G, Kalani B, Indika D Siriwardhana 114-117
Correlation of IL-6 with D-dimer, LDH, S-ferritin biomarkers, comorbidities and clinical outcomes of COVID-19 patients admitted in a tertiary hospital in Coimbatore, India – a retrospective study based on 2,569 patients.
Lavanya Sriramajayam, Murali Alagesan, Sudha Ramalingam, Karthikeyan Ramaraju, Jayavardhana Arumugam, Roshini Rajasekaran, Dhanushya Etraj, Sankarganesh Jeyaraj 119-124
Association of smoking with serum electrolytes, liver enzymes, and haematology parameters: a single-centre experience from Jordan.
Saad Al-Fawaeir 126-130
Immunological disturbances associated with Prader Willi syndrome in Egyptian patients.
Haiam Abdel Raouf, Rania Fawzy Mahmoud Abdelkawy, Hala T. El-Bassyouni, Shams Kholoussi, Mohammed M. Sayed-Ahmed, Azza E. Abd-Elnaby, Assem M. Abo-Shanab 131-135
The relationship between adiponectin and insulin sensitivity in obese patients with non-alcoholic fatty liver disease.
Dina Morsy A. Mohamed and Raghda M. Ghorab 136-141 (Supplementary Tables at https://mix.nzimls.org.nz/journalsrecent.html)
Case Studies
Para-Bombay Ah phenotype: case series from a tertiary care hospital in Malaysia.
Kaalpana Jayakumar, Rabeya Yousuf, Nur Afifah Suhemi, Nor Fadzliana Abdullah Thalith, Suria Abdul Aziz, Lailatul Hadziyah Mohd Pauzy and Qhasmira Abu Hazir 142-144
BMLSc and BMLSc(Hons) student research projects
Michael Legge asks us to consider the impact of climate change on human health and health care services with his editorial, describing how changes in climate will results in significant changes in environmental organisms, including viruses and pathogens on human health and to the pathology sector in research and diagnoses of diseases, previously only seen in tropical latitudes and how our laboratories now need to be prepared.
Each year the NZIMLS invites a prominent New Zealand medical laboratory scientist or pathologist to deliver the TH Pullar Address. This year Angela Brounts, a recently retired Operations Manager at ESR Wellington, presented her address titled; “Building resilience: keeping afloat during turbulent times” at the NZIMLS ASM 2024 in Christchurch and it is reprinted in this issue.
Evaluation of proteinuria is considered as an important marker in the diagnosis, and management of many diseases affecting the kidneys and some other systemic diseases. Siriwardhana and colleagues from the University of Ruhana, Sri Lanka evaluated a 25% sulfosalicylic acid turbidimetric assay for the quality evaluation of proteinuria and diagnosis of kidney diseases. This nationally patented method utilises 2mL of sample volume for urine protein quantitation and this study aimed to optimise sample volume and evaluate the potential to reduce the sample for subsequent automation. Results determined that the sample volumes of 1mL and 0.5mL were not significant compared to the validated 2mL sample and concluded that the methods would be a reliable and cost-effective alternative to the highly sophisticated pyrogallol red (PGR) dye-binding method at the lower ranges of proteinuria (0.03-0.45g/L) and feasible for automation.
COVID-19 disease is associated with an excessive host immune response leading to multiorgan failures and immunerelated manifestations. Clinical outcomes are influenced by disease severity, age, immune response, and comorbidity factors. In a retrospective study, Jeyaraj and colleagues at the PSG Institute of Medical Science and Research in India evaluated the correlation of interleukin-6 (IL-6) with D-dimer, LDH, serum-ferritin, disease severity, and comorbidities of COVID-19 patients. Their results showed higher respiratory distress in COVID-19 patients with IL-6 >7pg/mL compared to patients with IL-6<7pg/mL. IL-6 levels were positively correlated to biomarkers D-dimer, LDH and serum ferritin, duration of stay in ICU, high oxygen requirements and comorbidities including hypertension and diabetes in a study population of 2569 patients. Research presented by Saad Al-Fawaeir from Jadara University in Jordan examined the association between cigarette smoking and liver enzymes aspartate aminotransferase (AST), alanine aminotransferase (ALT), serum γ- glutamyl transferase (γ-GT) and alanine aminotransferase (ALT), total bilirubin and electrolytes (PO4-3, Ca+2, K+, Cl-, Na+, Fe+2), in 220 male participants (180 smokers, 40 non-smokers). Data showed cigarette smoking was correlated with deranged liver enzymes with a significant mean difference in serum γ-GT, ALT, ALP, AST, and total bilirubin in participants who had smoked for between 1 and 20 years compared with non-smokers. In addition, electrolytes levels of Ca+2, PO4-3, and Fe+2 were statistically significant (p=<0.05) comparted to non-smokers. The study provided insight into the potential hazards of smoking, which may contribute to increased mortality and morbidity rates. However, it is important to note that this study identified correlations but did not establish causation. Sayed-Ahmed and researchers at the National Research Centre in Cairo, Egypt, evaluated children with Prader–Willi Syndrome (PWS), a rare complex genetic disorder. The loss of expression of paternal genes in the PWS critical region of chromosome 15 q11-q13 affects multiple body systems and present major manifestations including; hypotonia, mild mental retardation, hypogonadism, growth hormone insufficiency and short stature. The study included eleven children with PWS diagnosed clinically and confirmed by FISH and 25 non-
PWS controls. Immune cell-count results revealed statistically significant higher total lymphocyte count and lower absolute cytotoxic T lymphocyte (CD8) count in PWS patients compared to controls (p= 0.0241 and 0.0134 respectively). Comparing the immunoglobulin results of PWS patients with control subjects, showed statistically significant elevation in IgG and IgM (p=0.0099 and 0.0040 respectively). IL-33 level was significantly increased while procalcitonin was significantly decreased in PWS patients in comparison to controls (p= 0.0009 and 0.0006 respectively). KREC expression showed statistically significant elevation in PWS patients in comparison to the healthy controls (p= 0.0107). This study showed the importance of the implementation of flow cytometric measurement of lymphocyte subsets, cytokines evaluation, and immunoglobulins quantification to elucidate the immunological disturbances in PWS patients.
"Non-alcoholic fatty liver disease" (NAFLD) occurs due to fat deposition and infiltration in the liver in absence of other factors causing steatosis like alcohol consumption, hereditary disorders or use of steatogenic medications. Mohamed and Ghorab from the National Research Centre in Cairo, Egypt explored the relationship between adiponectin levels and insulin resistance in obese patients with simple steatosis versus those who developed non-alcoholic steatohepatitis (NASH) Non-alcoholic liver disease. Serum adiponectin and insulin levels were measured in 60 obese patients, 30 with NASH and 30 with simple steatosis. Their results showed that the increase in ALT and Albumin had an independent effect on increasing Insulin level; with significant statistical difference (p<0.05). The increase in ALT also had an independent effect on increasing HOMA-IR; with significant statistical difference (p<0.05). The study concluded there is an important relationship between serum adiponectin and insulin resistance in obese patients with NASH in comparison to simple steatosis patients, which reflect the importance of serum adiponectin as a potential therapeutic target in treatment of obesity complicated with NASH.
ABO is a clinically significant blood group system, especially in pre-transfusion testing. The rare Para-Bombay phenotype may remain undetectable on ABO grouping due to its weak or absent H antigen. Para-Bombay individuals are expressed as Ah, Bh and ABh respectively based on the presence of A and B genes. Abu Hazir and colleagues at Hospital Canselor Tuanku Muhriz and the National University of Malaysia present a case series illustrating the findings of the para-Bombay Ah phenotype. All these three patients were confirmed as para-Bombay Ah blood group after further tests were performed to resolve the ABO discrepancy. Cases presented here help to demonstrate how prompt detection of ABO discrepancy in forward and reverse grouping, as well as comparison with previous records can identify a series of para-Bombay cases. For future transfusion H antigen negative blood is highly recommended to be transfused unless there is no availability.
As well as our regular features; Science Digest, Recent Reviews and the Pacific Way, we publish a conference report from our Barrie Edward - Rod Kennedy Scholarship winner, the HSIG Report, Otago BMLSc and BMLSc(Hons) student research project abstracts from Semester 1, 2024 and the 3rd annual Journal Christmas Quiz.
Retirements from the profession include a special mention to Michael Small, from Medlab Wanganui who has recently retired after more than 34 years in the profession and some words from Sue Little from the NZ Blood Service. We wish them all the best and thank them for their hard work and contribution.
On behalf of the NZIMLS and the Editorial Office, I wish you all a safe and happy holiday season and wish you luck and fortitude for whatever 2025 may throw at us.
“All great changes are preceded by chaos!” – Deepak Chopra
Lisa Cambridge Editor
Michael Legge
We tend to consider climate change with significant weather events, rising sea levels, extreme heat, wildfires and flooding. However, climate change represents a fundamental threat to human health and ultimately health care services. The changing patterns of the physical conditions associated with climate change possess a series of compounding influences on human health as well as both animal and environmental health. Globally the likelihood of the increasing impact of infectious diseases especially vector and water borne disease will increase. Associated with increasing temperature is the tolerance of humans to heat which impacts particularly on the very young and aging populations. A report by the Ministry for the Environment in 2020 (1) makes no mention of the impacts of climate changes and human health. Equally, it does not consider the significant changes which may result in environmental organisms such as bacteria and fungi (2). Changes in temperature and carbon dioxide will impact on bacterial growth and adaptability in the environment. Viral diseases favouring humidity and temperature will survive longer when there are suitably favourable conditions such as influenza, covid and yellow fever (3). In addition, changes in climate will favour common diseases such as measles and polio (3). There is increasing concern about the increase of antimicrobial resistance of some human pathogens. Data from the USA indicates that consistent daily minimum temperature of 10oC will lead to an increase in antibiotic resistance for Escherichia coli, Klebsiella pneumoniae and Staphylococcus aureus (2). Research has shown that over half of known human pathogenic diseases can be aggravated by climate change mitigated by the physical changes in weather (3). The simultaneous emergence of Candida auris on three continents demonstrates adaption of fungi to new, alternative, temperatures attributed to global warming (4). Added to these concerns is the increasing transmission of animal viruses to humans, often via a secondary host such as Nipah virus and Ebola. Of particular concern are the vector-based diseases such as malaria, dengue fever, West Nile virus and Zika virus. Rising temperatures and the frequency of international travel make conditions for the respective arthropod vectors more favourable and have the potential to create new animal reservoirs. For example, West Nile virus is now associated with over-wintering mosquitos in birds, horses and other mammals in Europe and has emerged as a disease not normally associated with European countries.
The increase is due primarily to summer heat waves, placing previously unaffected populations at risk due to climate change. Where does pathology fit with the effects of climate change? New Zealand has been relatively isolated from most of the diseases normally found in warmer climates. However, increasing temperatures and humidity has the potential to create favourable conditions for many of the previously diseases not normally identified in this country. A constant consideration for epidemiology research to assess unknown diseases and their aetiologies is essential. This would also include transfusion and transplantation services. There is a need for a better understanding and identification of vector-borne and other diseases and for clinicians to be alert to unusual signs and symptoms. Pathology will provide the evidence for accurate diagnosis and data on the disease epidemiology, but to be effective there is a need for upskilling the workforce to understand and recognise results that vary from the norm. A range of diseases not normally associated with the northern hemisphere are starting to emerge and it is essential that the pathology services in New Zealand are prepared for the potential for this to occur in this country.
Michael Legge, PhD, MRSB,FIBMS, FNZIMLS, FFSC (RCPA), University of Otago, NZIMLS. Email: mike.legge@nzimls.org.nz
1. National Climate Change Risk Assessment for New Zealand: Main Report Ministry for the Environment 2020: ISBN: 9781988579931.
2. Cavicchioli R, Ripple WJ, Timmis KN, et al. Scientists’ waring to humanity: microorganisms and climate change. Nat Rev Microbiol 2019; 17(9): 569-586. https://doi. org/10.1038/41579-019-0222-5
3. Mora C, McKenzie T, Gaw IM, et al. Over half of known human pathogenic diseases can be aggravated by climate change. Nat Clim Chang 2022; 12: 869-876.
4. Casadevall A, Kontoyiannis DP, Robert V. On the emergence of Candida auris: climate change, azoles, swamps and birds. mBio 2109; 10(4): e01397-19.
Advertise your company in the NZIMLS Journal of Medical Laboratory Science
The Journal is distributed free to all members of the NZIMLS, which at present numbers approximately 3,500. This means that the Journal is read by most medical laboratory scientists and technicians and enters all hospital and community medical laboratories in New Zealand. It is also sent to members employed in commercial and veterinary laboratories as well as government establishments, and members overseas.
For prospective advertising enquiries please contact: sharon@nzimls.org.nz
Advertising rates are available at: https://www.nzimls.org.nz/advertising-rates-pre-press-specifications
Angela Brounts
Tēnā koutou, tēnā koutou, tēnā koutou katoa
Ko Angela Brounts tōku ingoa
Nō Otautahi ahau
Kei te noho au ki Te Whanganui-a-Tara
Ko Ngāti Pākeha tōku iwi
Nō te whenua o Ingarangi tōku Whānau.
My name is Angela Brounts, I whakapapa back to Otautahi, Christchurch and I currently live in Whanganui-a-Tara, Wellington. I am a pakeha New Zealander whose family came from England. Today I will be talking about change, and some ideas to improve resilience in an ever-changing laboratory landscape. I don’t have all the answers but I’m going to suggest a few things worth thinking about.
First of all, I wonder what Thomas Pullar would make of me introducing myself in Te Reo. Even ten years ago I wouldn’t have considered it myself, but now it’s just the way I do things. Many organisations train their staff to introduce themselves in Te Reo now and my last employer the Institute of Environmental Science and Research or ESR is one. The Māori word for science is putaiao and the tagline for ESR, is ‘He Putaiao, He Tangata’, literally ‘The science, The people’ expressed as ‘Science for Communities’. Even if your organisation doesn’t provide Te Reo classes I bet you have a bit in your charter about honouring Te Tiriti and possibly a question about it as part of your recruitment or interviewing process. If you get the opportunity to learn some Te Reo, I encourage you to take it. I have found it immensely satisfying and it could be really helpful the next time you are pursuing research funding or looking for a job.
Many of you will work for Te Whatu Ora, the centralised organisation brought in by the last Government to replace individual District Health Boards or DHB’s. Publicly funded medical laboratories have had several different iterations in the last 40 odd years I’ve been working in the sector. Like me, you may have even been round long enough to remember the District Health Offices and Hospital Boards that in the 80’s became Area Health Boards. In the early 90’s these were restructured to Crown Health Enterprises when the chief executives were referred to as the Big CHE’s (cheese). In the mid 90’s there was another renaming to Hospital Health Services, and by the late 90’s we had 21 District Health Boards. The names and how they have sliced and diced the health sector have changed many times and no doubt will again in the future.
I remember private labs Pearson’s and Godfrey’s back in the 1980’s here in Otautahi Christchurch, and Grey Street labs in Palmerston North during a time when many of the private laboratories were owned by individual or groups of pathologists. I’m sure there were others. I’m not sure any of these exist anymore, the last might have been a Pathlab but don’t quote me. Before Awanui there was Southern Community Laboratories, Medlab and who remembers Cardinal? I worked for them for a short while returning from maternity leave. They agreed to employ me for three hours three mornings a week in haematology. The only other option I was given at another Otautahi lab was 20 hours a week, half days, five days a week. This was considered the only part time option with no flexibility. Only nine hours a week was groundbreaking stuff. How things have changed. Some of this is because operating hours have changed. Most labs used to be open 8-5 then someone was on call until 8am the next morning. All very rigid. Now many of you will be required to work rostered shifts as well as weekends. Until you have worked a midnight till 8 shift you have no idea what that is like and how difficult it is. To those of you who work those difficult shifts, thank you. If you are also looking after children during the day, you are a legend!
My lab career started in 1981, when I was only 16 years old. Can you imagine it? Straight out of the sixth form at high school into a medical laboratory science apprenticeship. These don’t exist anymore. Looking back, I was just a child and played some pranks on colleagues that would get your employment terminated these days, even though they weren’t particularly dangerous. Things like blowing up plastic dropper bottles with dry ice. It was a very different environment. There were six of us who went to Polytech on day release and night classes to get a New Zealand Certificate in Science: Medical Science after 3 years. Some other parts of the country did things differently. I know Whanganuia-Tara, Wellington had block courses. After 3 years of general on the job training in all disciplines and completing the NZ Certificate, we spent a further two years specialising in a chosen field called ‘O’ and ‘A’ levels, very English terminology, and pass some exams to gain registration as a technologist, equivalent to today’s scientist. You could do either two “O” levels or an “O” and an “A”. I did two “O”’s in haematology and biochemistry making me versatile as I had a husband in the Air Force and we moved around. Ironically my first scientist job after registration was in a microbiology lab, and so was my last role at ESR. The path to registration is very different now but interesting to note that many of my peers at the time are still working in medical laboratories around the country, so the ‘old way’ of training has served us well.
What is the point of all this nostalgia? The only constant all my working career has been change…and we haven’t even talked about the science yet. As things continue to change, we need to keep asking ourselves, what does this mean for us?
What do these changes mean for us as individuals, for the patients and referrers we serve, and what does this mean for us a profession?
The theme for this year’s conference of Navigating the Great Unknown is very timely. Darwin once said in a now famous quote “It’s not the strongest of the species who survive, it’s those who are most responsive to change”. Those of us who have worked in the medical laboratory science for any length of time have seen plenty of change and you’ll be very aware there is more coming. I feel for our colleagues in the public service who have recently been restructured again.
How do we become resilient to these constant changes?
The first thing I will say is keep calm. The health of New Zealanders is not going to suddenly improve and we will always be needed. We already know that most of all diagnoses rely on laboratory results. So, what are our challenges? Resilience is seeing a challenge as an opportunity so let’s look at things both ways.
The reality is we have an aging workforce. Terry Taylor disseminated some great information about this in his outgoing Presidents national tour of labs. Many of your colleagues are already retirement age and most labs have a predominance of people over 50 years old. The bad news is these people are going to retire. Some good news is they don’t have to, and many continue to perform valuable work past 65.
You may have noticed an issue keeping younger staff. They have different expectations about learning and promotion opportunities than my generation did. They are often looking for more flexibility, work life balance, and a clear career path. They don’t want to stay doing the same thing for decades. Managers need to work out how to meet these needs and encourage younger people into the workforce. If you work with a young person with potential and ambition, give them opportunities to
lead. Encourage them into some management training and give them the time to study. Offer flexible hours where you can. The easier you make it for people to stay the less likely they are to leave.
Thinking about opportunities does your lab have a succession plan? Does your laboratory team have a plan for when your senior people are about to retire? Are there designated people ready to step up… does there need to be? All things to think about. Take a minute now to think about all the people you work with who might retire in the next 5 years. So, what does this mean for you? Change is also opportunity for those able to step up.
The elephant in the room is usually pay, and to keep younger staff this needs to be addressed. Te Whatu Ora aside our laboratories are mostly owned by multinational companies who are motivated to some extent by making a profit for their shareholders. This is just a reality. The work we do is essential and while I’m not suggesting anything, it’s clear the only way we have any bargaining power is if we all work together.
Let’s also consider disruptive technology
More of which is always just around the corner. What skills do we need to respond to rapid advances in scientific understanding and changes, although not always necessarily improvements, in technology? I used to make up biochemistry reagents for the new SMAC machine wearing an apron, face shield and the same style of white gumboots my Dad wore at the freezing works. Now we buy our chemistry reagents in nice tidy little barcoded bottles and put them on a machine that achieves far more accuracy and quality in the results than we could dream about back in the early 1980’s. We buy in stains, media, primers and probes, antibodies, you name it. Everything comes in a kit.
Our job is now much more often focussed on quality control. While the machines are faster and more accurate than we ever were, our job is to check they are calibrated and working properly. A biochemist nowadays needs to understand more about statistics and possibly less about classical biochemistry. I certainly never used the citric acid cycle after I’d graduated, but I learnt, and used, a lot of knowledge about normal ranges, standard deviation, Levy-Jennings charts and uncertainty of error. We have also become an important part of the maintenance crew. Troubleshooting issues and understanding when a part needs changing is a key skill in most areas. When turnaround time is important, knowing how to change a probe or sensor so you don’t have to wait for the repair technician is crucial. We also need IT skills as we rely on computing to analyse and report our results. Rule number one as told to me many times by my colleague Andrew Crooke, “Turn it off, wait 30 seconds, then turn it back on again”. Isn’t it amazing how often that works.
So, is the scientist still needed?… of course we are. We must get the right sample onto a well performing machine, make sure the controls are in range, and set the parameters for automatic validation and reporting. We often only eyeball the out-of-range results now.That’s not a bad thing. We can now put through much higher volumes of work in much shorter times. That’s efficiency. Technology helps us concentrate on the things the machines can’t do. I don’t miss making reagents, and the endless filing that used to be part of my role back in the early 1980’s.
So be the best statistician and maintenance engineer you can be. Pursue opportunities to skill up in these areas. They make you valuable in your workplace.
I’d like to step sideways here for a minute to give a shout out to all the phlebotomists. This has to be the most underrated skilled job in our profession. Anyone who has ever had a blood test (and if that’s not you yet it will be,) knows how important a skilled phlebotomist is. They are often the only interaction members of the public get with our services. They are our face, and they do a great job. It is true you can’t get a good result out of a badly taken sample. Phlebotomists deserve our admiration so take a bow… Thank you.
So, what is next?
There are new technological advances all the time. Some are small and some are radical. I remember when molecular science was new, running in-house made gels, then polymerase chain reaction or PCR tests. “Molecular Science” had its own department. Now microbiology, where I have primarily been working for the last 15 years, is all about molecular testing, and genome sequencing particularly next generation sequencing is the big new thing. Labs have provided Sanger sequencing, 16S and MaldiToF for a while, but it’s getting easier and cheaper to sequence the whole genome of viruses and bacteria. And to look for the marker genes that can highlight susceptibility to diseases like cancer, or genetic abnormalities in humans. From pinpointing a single gene, we can now look at the whole genome with so much more information. This means we need, as a health professional, to consider important ethical concepts such as the sovereignty of that information and who has access to it.
What does this mean for patients and for you as scientists?
How do we prepare for this move to genomics?
The analysis of sequencing is all about statistical algorithms so if you are thinking about doing some further training, I’d recommend learning one of the statistical programmes that is commonly used. At ESR they start people off with a programme called R, which is something that may be taught at your local high school. It’s not especially difficult, more an extension of Excel. Don’t be scared, you can do it. There are plenty of other programmes available with cool nerdy sounding names like Python, ChewBBACA, Kraken, Pangolin, porechop, Krocus and Guppy. Some of them have courses available online if you are interested but get some advice first so you choose one that will be relevant to your future. More on data science in a minute. How long will it be before we are all having our genome sequenced to try and predict our human susceptibilities and frailties? Some people already buy a kit online, take a buccal swab and send it to a lab for this type of analysis.
Is it possible other providers than New Zealand registered medical laboratories will be offering testing services in the future?
Think about the use of rapid antigen or RAT tests for COVID-19. While they continue to be an important tool, they are not all equal and not necessarily the same quality as a laboratory test, overseen by a trained medical science professional. How many people got an incorrect result because they didn’t administer the test correctly? We’ll never know. I had a COVID RAT test administered by a nurse overseas who had clearly flooded the immunochromatography slide so it didn’t work and had to be repeated. I had no confidence in either of the results she obtained.
A good thing is that Medical Laboratory Science is a registered profession and only people registered with the Medical Sciences Council of New Zealand are allowed to call themselves Medical Laboratory Scientists, Medical Laboratory Technicians or PreAnalytical Technicians. These are protected titles and nonregistered people using them can be prosecuted. This is very important for our profession and something it is in our best interests to protect. If this protection is ever challenged, we need to fight for it so be prepared to stand up for your profession and the professional standards that are protected by the Medical Sciences Council. Read their emails, respond to requests for feedback on new policies and consider standing for a role on the Council. What the Medical Sciences Council does matters.
And that goes for the New Zealand Institute of Medical Laboratory Science who we know as the NZIMLS as well. They are our professional body. They represent us, set exams for people wanting registration as Technicians, and provide us with education and Continuing Professional Development via their Journal and scientific meetings like this one. It’s easy to take the mahi they do for granted but without the Institute it would be harder for the Medical Sciences Council to protect our
profession. To support them we need to write papers the Journal can publish, stand as a local representative, or volunteer for a scientific meeting organising committee. All these things teach you valuable skills, introduce you to a new network of people, and look great on your CV. It’s a real buzz to see your name on a paper.
Protecting our status as professional scientists could well be one of the most important battles we face in the future. Because of the next challenge I’m going to talk about.
Preventative diagnostics
Back in the 1980’s we used to mostly do testing on people who were unwell. The tests were expensive and the volumes we could process were limited. People who were sick got priority. Now a much larger percentage of our testing is routine health checks on people who feel perfectly healthy. A lot of us do and should have a regular check-up with our GP to check our liver and renal functions, glucose, lipids. Prostate checks are now a blood test, to the relief of a lot of people. Those over 60 can participate in a bowel screening programme. These are all preventative tests.
This is why a lot of the tests we do are in the normal range and can be auto reported if they pass the relevant QC checks. This is what makes our test volumes rise much faster than our population. We are not sicker; we are just checking we aren’t sick. This is not just being the worried well, it’s diagnosing an issue before it becomes a problem. Just like car maintenance and warrants of fitness the sooner you can determine if you have a problem the sooner you can get to a solution. So, what is going to happen when we all want to check our gut biome to determine what supplements or genetic therapies we might need to consider for better health? This is already huge overseas. Just something to think about.
We are going to need a lot more computing power for a start to deal with all the data. And who is going to do all the analysis? How much data science is being taught in the Bachelor of Medical Laboratory Science courses. I don’t know the answer but if it’s not already significant it needs to be.
So, are data scientists really medical laboratory scientists? Should they be registered as Medical Laboratory Scientists if they sit a computer and never enter the wet lab or touch a patient sample? They do produce ‘results’.
This is a question the Medical Sciences Council have been pondering for the last couple of years after a paper submitted with the suggestion they have a new scope of limited registration for data scientists. I believe that has to be a ‘Yes’. A couple of my colleagues who work in this area delivered the Council Chief Executive and Registrar a presentation about next generation sequencing to help them understand the issue. Watch this space.
Another key question… So, who checks these algorithms used to determine genetic code used for diagnostic testing and checks their validity for the diverse ethnic populations of New Zealand? We know that to date most clinical testing has been performed on Caucasian men. What does that mean for us?
Maybe we need to take a good look at where our normal ranges originated, and a study looking at diversity among different populations could really benefit us as a nation. If you’ve noticed something interesting in a population or have an idea regarding something worth investigating, put together a proposal and talk to your pathologist or manager about whether they could support a project. It may not cost much except your time to do the data analysis. Don’t forget to get ethics approval from the Manatu Hauora, Ministry of Health and Disability Ethics Committees if you are using patient data, even if it’s anonymised.
What about pharmacokinetics?
If you haven’t heard this term before it’s what the body does to a drug and refers to the movement of drug into, through, and out of the body, considering its absorption, bioavailability, distribution, metabolism and excretion. Scientists are using genetic markers to predict how an individual might metabolise drugs.
In tandem with kidney function this could be a powerful tool to help determine an individual’s optimum doses for antibiotics, chemotherapy and other drugs. Thereby minimising unintended and unwanted consequences such as organ damage and saving money by giving the necessary dose and no more. And the great news is once you have sequenced a human individual’s genome, it shouldn’t change. You only have to do it once!
If you sequence the whole genome all the information you need is right there, you just need to know how to interpret it. The body of knowledge about which genes code for what expressions of phenotype is expected to grow exponentially. I started writing this 6 months ago and things have already changed hugely. What does that mean for us? Where does AI fit in?
There’s that big data again
We’d better start growing our computing power now to handle all the gazillions of terabytes of processing we are going to need it. As our population ages it is also likely to get sicker. It’s an unfortunate fact that more functions in the body break down as you get older. Don’t I know it!
This is going to be the future, and how we get there will not necessarily be navigated by us, as the service provider. To use the waka analogy the policy makers and funders will negotiate with the technology providers to decide the destination and speed of craft, but we who are doing the putaiao mahi, the scientists, will be ones maintaining the vessel, optimising economies, maintaining the course and paddling as fast, skilfully and efficiently as we can.
In a nutshell, to be an effective paddler in this waka and ride out the turbulent waters ahead we need to upskill in statistics, data science, machine maintenance, information technology, and strive to protect our profession by encouraging succession planning and professional development via support for the Medical Sciences Council of New Zealand and the New Zealand Institute of Medical Laboratory Science.
To quote the often used whakatauki or Māori proverb; “He waka eke noa”. We are all in this together. And common sense tells us we are much more likely to be successful if we are all paddling in the same direction.
Nō reira, e āku hoa, e ngā kaiwhakahaere o te kaupapa nei
Kei te mihi, Kei te mihi, Kei te mihi.
To my friends, colleagues and the organisers of this conference, thank you.
To Willow Grace-Morton for her help with my pepeha, and Helena Woods and Jacqueline Wright who peer checked this paper for me.
Angela Brounts, BMLSC, BSc, MBA, (formally) Operations Manager, ESR, Wellington, New Zealand
Correspondence: angelabrounts@gmail.com

Copyright: © 2024 The author(s). This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.


Specialty Immunoassay Plus Controls are the next generation of independent specialty IA controls, now in liquid configurations. These controls are automation ready and fully integrated with Unity QC data management to give you confidence in every run.
Features:
• Consolidated menu of 14 specialty IA analytes, including 6 NEW ones: PCT, IL-6, Active Vitamin B12, Fructosamine, TRAb, and TSI
• Enhanced Vitamin D range for Vitamin D Standardization Program (VDSP) compliant assays
• Improved open-vial stability for PTH (intact)
• InteliQ and Liquichek formats
Streamline your laboratory processes and improve turnaround time with Bio-Rad Specialty Immunoassay Plus Controls. Learn more: www.bio-rad.com/iacontrols
Nimesha D Ranawaka, Rajika G Jinadasa, Kalani B Gunawardana, Indika D Siriwardhana
ABSTRACT
Objectives: Evaluation of proteinuria is important for the diagnosis and management of kidney diseases. Recently, 25% sulfosalicylic acid (SSA) turbidimetric assay has been validated for the quantitative evaluation of proteinuria in 0.03 - 0.5 g/L range. This nationally patented method utilises a 2.00 mL sample volume for urinary protein quantification. The current study aims to optimise the sample volume for the 25% SSA assay, streamlining the method for automation. Methods: Urine samples collected from 30 healthy volunteers and 32 patients with the established diagnosis of chronic kidney disease were used for volume optimisation of the method comparing 2.00mL of original sample volume with 1.0mL and 0.50mL volumes. The data were analysed by general linear model for repeated measures to find the minimum possible sample volume. Results: Data showed no significant differences between the mean protein concentrations between 2.00 mL and 1.00 mL (0.25 vs 0.20g/L respectively, p=0.677) and 2.00mL and 0.50mL volumes (0.25 vs 0.26g/L respectively, p=1). Spearman’ correlation between urinary protein to creatinine ratio (PCR) with 25% SSA and pyrogallol red (PGR) methods in healthy individuals was 0.642 (p<0.01). Bland-Altman plot gave a bias of 0.72mg/mmol for PCR. Pairwise comparison of volume optimisation data indicated an insignificant volume effect for 25% SSA protein concentration as measured using 2.00mL, 1.00mL and 0.50mL volumes thereby potentiating the use of reduced sample volumes enabling automation of the method. Conclusions: The volume effect for 25% SSA protein concentration as measured using 2.00mL, 1.00mL and 0.50mL volumes was not significant and the use of reduced sample volumes enabling automation of the method was confirmed. Keywords: proteinuria, quantitation, turbidimetry, sulfosalicylic acid, chronic kidney disease
NZ J Med Lab Sci 2024; 78(3): 114:117
INTRODUCTION
Evaluation of proteinuria is considered as an important marker in the diagnosis, and management of many diseases affecting the kidneys and some other systemic diseases (1). Further, the presence of any degree of proteinuria is a major risk factor for the future development of kidney and cardiovascular diseases (2). Proteinuria can be defined as the presence of abnormally elevated amounts of protein in urine (3). It may occur because of both renal and certain incidental extra-renal causes. As normal urinary protein excretion is <0.15g/day, urinary total protein values ≥ 0.15g/day indicate presence of proteinuria (4).
Total protein measurement in urine is a conventional and costeffective test used in clinical decision making (5). Detection and evaluation of urine total protein involve a wide variety of methods ranging from point of care testing (POCT) to highly sophisticated laboratory analysis (6). Among them, pyrogallol red (PGR) dye-binding method is one of the commonly used methods. Turbidimetric assays such as the sulfosalicylic acid (SSA) method by precipitation of proteins are commonly used as qualitative methods (7). The use of varying concentrations of SSA to quantitate urine protein which has been evaluated previously, investigated for the quantitation of low-grade proteinuria (8). Previously, the 25% SSA turbidimetric method has been validated as a cost-effective method for the quantitative evaluation of proteinuria in lower range (0.03 – 0.5g/L) (8). This method has been patented for its novelty by the National Intellectual Property Office (NIPO) of Sri Lanka in 2023. In this method, the volume of urine sample used is 2.00mL in a turbidimetric assay utilising a spectrophotometer. However, to facilitate automation of this method, sample volume needs to be further reduced below 2.00mL.
The 24-hour urine collection has been recognized as the gold standard for the quantification of proteinuria. However, due to its impracticability in certain circumstances, assessment of protein to creatinine ratio (PCR) in a spot urine sample is mostly used in clinical practice, which is justified by the observation that protein and creatinine excretion remain constant during the day, in a person with stable kidney function (9). The previously validated 25% SSA method has shown PCR results comparable to PGR PCR (r=0.913, p<0.0001) in the method evaluation phase (8). Therefore, urinary 25% SSA PCR merits further investigation as an alternative for expensive methods such as urinary albumin to creatinine ratio (ACR) and PGR PCR which makes PCR affordable in resource scarce settings.
In the current investigations, the aim was to determine the minimum possible sample volume for the 25% SSA method utilising urine samples obtained from healthy individuals as well as from patients with chronic kidney disease (CKD). As the original study used the sample volume of 2.00mL, if the method is to be automated its applicability with lesser sample volumes needs to be evaluated within the applicable range of proteinuria of the assay (0.03 – 0.5g/L). This would enhance the clinical application of the method as a screening test for low grades of proteinuria.
Ethics Approval
Ethical approval for the study was obtained from the Ethics Review Committee of Faculty of Allied Health Sciences, University of Ruhuna, Galle, Sri Lanka (14.10.2020.005).
Sample Collection
The aim of this study was to determine the minimum possible sample volume for the 25% sulfosalicylic acid (SSA) method using urine samples from healthy volunteers and diagnosed patients with chronic kidney disease (CKD) (Figure 1).
A total of 62 samples were collected from 30 healthy volunteers between 18 to 65 years of age in Godakanda Public Health Midwife area, Karapitiya, Galle, Sri Lanka and 32 patients attending Nephrology Clinic, Teaching Hospital Karapitiya, Galle, Sri Lanka using convenient sampling method. Collected urine samples were transported to the laboratory in a cool box at 4.0±1.0⁰C. The samples were then centrifuged at 2500rpm for 5 minutes and the supernatant was divided into 4.00mL (for 25% SSA protein assay) and 5.00mL (for PGR protein and creatinine assays) aliquots. Using all the samples, SSA protein analysis was carried out on the same day of sample collection and the 5.00mL aliquots were stored at -80.0±0.5⁰C in tightly capped plain tubes. A stock solution of albumin at a concentration of 1.8g/L was prepared by dissolving bovine serum albumin (Albumin Bovine Fraction -V Qualikems, India) in distilled water. This stock solution was used to generate a dependent dilution series of albumin, with concentrations of 1.8, 0.9, 0.45, 0.225, 0.112, 0.056, 0.028g/L following a doubling dilution method.
Preparation of the standard curves for 2.00 mL, 1.00 mL and 0.50mL sample volumes
In this method, 2.00mL of each albumin standard was mixed with New Zealand Journal of Medical Laboratory Science
50µL of 1% TCA (trichloroacetic acid) followed by the addition of 100µL of 25% SSA and exactly 3 minutes after the acidification, absorbance values were measured at 600nm against the reagent blank using a double beam spectrophotometer (SHIMADSU 1800 UV-VIS, Japan). The reagent blank was prepared by adding 50µL of 1% trichloroacetic acid (TCA) and 100µL of 25% SSA to 2mL of distilled water. Each assay was in triplicate and the mean absorbance values for each concentration were obtained. Data tabulation and initial calculations were done by Microsoft Excel 2010 and the standard curves were constructed using Minitab 2016 software. Next standard curves for 1.00mL and 0.5mL volumes were prepared by the procedure described under the preparation of the standard curve for 2.00mL volume varying only the sample volume as 1.00mL and 0.50mL respectively without changing the TCA and 25% SSA levels accordingly. The standard curves were validated by running two levels of in-house Quality Control (QC) materials (0.1g/L and 0.3g/L) albumin spiked urine samples and QC values derived from the standard curves were compared with the QC values obtained from the PGR method.
of
The 2.0mL, 1.00mL and 0.50mL volumes of the supernatant of 30 urine samples from healthy individuals were analysed by the 25% SSA method following the procedure described under preparation of standard curves for each volume. As patient urine samples contained higher concentrations of protein which were not within the linear range of the standard curves for each sample volume, they were double diluted by adding 2.00mL of distilled water to 2.00mL of supernatant of each sample. Then, 2.00mL, 1.00mL and 0.50mL volumes of each dilution were analysed by the same procedure by which the samples from healthy individuals were analysed. Their protein concentrations were obtained using the equations generated by the relevant standard curve. The data were analysed by general linear model
for repeated measures in IBM SPSS version 25.0 software to find the optimum sample volume.
Precision verification using QC materials
In-house urine QC material for 25% SSA method was prepared using a normal urine sample in which PGR urine protein concentration is <0.03g/L and it was spiked with 2.0g/L albumin solution to prepare two levels of albumin concentrations: 0.1g/L as the low level and 0.3g/L as the high level. Each spiked sample was tested 20 times using 25% SSA method to evaluate intraassay precision. To evaluate inter-assay precision, each sample was tested in duplicate before analysing patient samples in 25% SSA method daily up to ten consecutive days. Commercially available, lyophilized QC materials were used for the batch analysis of creatinine and PGR protein assays, and they were reconstituted before use according to manufacturers’ instructions.
Evaluation of urinary 25% SSA PCR vs PGR PCR in a group of healthy individuals
Urinary protein analysis of the 30 healthy individuals by 25% SSA method was carried out following the procedure described above using 2.00mL of the supernatant of urine samples and the protein concentrations were calculated from the 2.00mL standard curve using measured absorbances. After thawing the refrigerated urine samples in a 37±2⁰C water bath, protein levels were determined by PGR dye binding method using BioSystems BA 400 fully automated analyser (Biosystems S.A., Spain). Urinary creatinine values by Jaffe colorimetric method were obtained without deproteinization at 37±2⁰C and a wavelength of 510nm with a path length of 1cm using BioSystems BA 400 analyser. Both PGR PCR and 25%SSA PCR values for all the 30 samples were calculated. Clinical applicability of urinary 25% SSA PCR vs PGR PCR in a group of healthy individuals was evaluated using Spearman’s correlation and agreement of the two methods was assessed by Bland-Altman analysis in SPSS version 25.0.
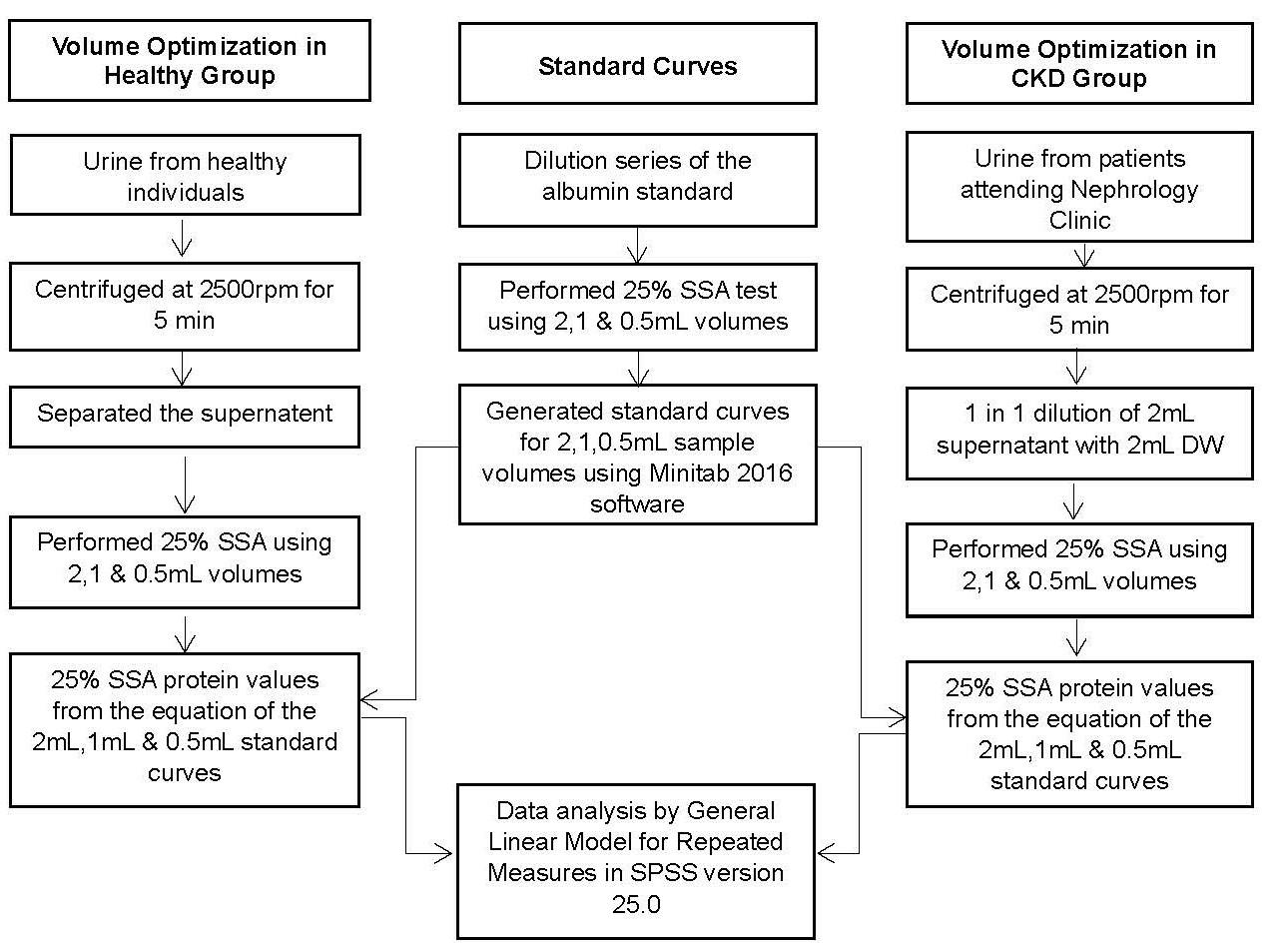
1. Flow diagram of the study. SSA: sulfosalicylic acid; DW: distilled water; rpm: rounds per minute
Standard curves for 2.00mL, 1.00mL and 0.50mL sample volumes
Standard curve for each 2.00mL, 1.00mL and 0.50mL sample volumes were plotted using the dilution series of the albumin standard. Three standard curves for the respective volumes were linear in the range of 0-0.45g/L range. The regression equations for 2.00mL, 1.00mL and 0.50mL sample volumes are given in Table 1.
Optimisation of the sample volume for the 25% SSA method
All the 62 samples whose PGR protein values ranged from 0.004 to 0.9g/L were analysed by 25% SSA method varying the volume of the supernatant of the sample in the order of 1.00mL and 0.50mL and their 25% SSA protein concentrations were derived from the relevant standard curve and the values compared with that of the original method of 2.00mL volume.
Table 1. Regression equations of the standard curves
Volume Regression equations of the standard curves
2mL Absorbance = 0.0144 (Concentration)- 0.0045
1mL Absorbance = 0.0278 (Concentration) -0.0211
0.5mL Absorbance = 0.0310 (Concentration) -0.0557
Tests of within-subjects effects determined that mean 25% SSA protein concentration did not differ significantly across the three sample volumes (F=1.844, p=0.163). A post hoc pairwise comparison using the Bonferoni corrections showed a decreased concentration between 2.00mL and 1.00mL volumes (0.25g/L vs 0.20g/L) which was not statistically significant (p=0.677). There was an increased concentration between 2.00mL and 0.50mL volumes (0.25g/L vs 0.26g/L) respectively which also was not statistically significant (p=1). Therefore, the results indicate that there’s no significant volume effect for 25% SSA method for urine protein concentration as measured using 1.00mL and 0.50mL volumes compared to volume 2.00mL.
Precision Verification
Results of intra-assay precision with coefficient of variation (CV%) for level 1 (0.1g/L) and level 2 (0.3g/L) QC were 5.2% and 2.9%, respectively whereas inter-assay precision results (CV%)
Table 2. Validation of standard curves
for level 1 (0.1g/L) and level 2 (0.3g/L) QC were 4.3% and 2.0% respectively (Table 2). Values for commercially available QC materials for urine total protein and creatinine were 0.75g/L (0.61 –0.91g/L) and 8.19 mmol/L (5.75-8.22mmol/L) respectively and they were within their ranges.
Evaluation of the clinical applicability of urinary 25% SSA PCR vs PGR PCR in a group of healthy individuals
The healthy individuals (14 males and 16 females) were between 18 to 60 years of age and had blood pressure ranging from 107/76 - 122/84mmHg. The urine samples collected were negative for glucose and protein on qualitative dipstick testing. Standard curve for 2.00mL sample volume was used to derive 25% SSA protein concentrations of the urine samples from healthy individuals which varied within 0.005 - 0.041g/L.
PGR protein concentration of the 30 healthy individuals varied within 0.004-0.058g/L range. As the values obtained for both SSA PCR and PGR PCR showed a non-parametric distribution, Spearman’s correlation was used to assess the degree of association between 25% SSA PCR and PGR PCR. According to the results, the significant Spearman’s correlation coefficient value of 0.642 which is within the (0.60-0.79) range confirms that there’s a strong, positive, monotonic relationship between 25% SSA PCR and PGR PCR (N=30, p<0.001). The Bland Altman plot gave a bias of 0.72mg/mmol, a lower limit of agreement (LOA) of -2.52mg/mmol and an upper LOA of 3.97mg/mmol (Figure 2).
SSA: sulfosalicylic acid; PGR: pyrogallol red.

ratio.
SSA: sulfosalicylic acid; PGR: pyrogallol red; PCR: protein to creatinine ratio; LOA: limit of agreement
DISCUSSION
The aim of this study was to determine the minimum possible sample volume for the 25% SSA method. We first evaluated the applicability of the 25% SSA method in measuring urinary protein in healthy individuals which has not been addressed in the original study. The SSA PCR which was nonparametrically distributed showed a significant positive correlation with PGR PCR as evidenced by Spearman correlation coefficient of 0.642 (P<0.0001).
In the original study, the volume of sample used for the 25% SSA method was 2.00 mL, which restricts its utilisation in semiautomated and automated analytical platforms. Therefore, we evaluated the method for the use of lower sample volumes. The current study evaluated the comparability of 1.00mL and 0.50mL sample volumes against the original method using 2mL. General linear model for repeated measures and the post hoc pairwise comparison using the Bonferoni corrections denoted
that the variation of the mean 25% SSA protein concentrations across the 2.00mL, 1.00mL and 0.50mL sample volumes were not statistically significant. Moreover, values derived from 0.50 mL volume displayed a closer proximity to that of 2.00mL volume compared to 1.00mL. Therefore, the lack of a significant volume effect for the protein concentration strongly favours the use of reduced sample volumes, enabling the automation of the 25% SSA method.
Besides the simplicity, and the feasibility of the method, the potential for using low sample volumes enabling automation may further enhance the potential use of this method. As per the previous cost evaluations, cost per test of 25% SSA method is about 0.27 LKR, (less than NZD 0.1 cent) while, it is about 25.00 LKR (NZD 0.13 cents) and 168.00 LKR (NZD 0.91 cents) respectively, for urinary protein estimation by PGR and for albumin (microalbumin) methods (8) Therefore, possible reagent volume reduction and the reduction of the use of other utilities which are associated with the reduced sample volume may result in further cost reduction. This may help reduce the cost per test for the detection of proteinuria and merit its use as a screening tool for low-grade proteinuria and the earlier diagnosis of kidney diseases and other associated disease conditions. Despite the benefits, several limitations are associated with this study. The method generates accurate results only with fresh urine samples. Higher protein concentrations require dilution since turbidimetry is used as the analytical method. Although bovine serum albumin factor (V) was used as the protein standard in this study, a stable total protein standard would be a more reliable substitute. The non-parametric distribution of the results for the evaluation of the clinical applicability of urinary 25% SSA PCR vs PGR PCR in a group of healthy individuals may have been due to the relatively small sample size, (n=30). Therefore, further studies with a larger sample size would generate more accurate results.
The urinary 25% SSA PCR showed a strong and significant positive correlation with PGR PCR for the samples obtained from healthy volunteers ensuring the clinical applicability of the 25% SSA method in health which has not been evaluated previously. Pairwise comparison of the volume optimisation data showed that both 1.00mL and 0.50mL sample volumes do not have a significant difference compared to 2.00mL which has been previously validated. Thereby, it can be concluded that the 25% SSA method is a reliable and cost-effective alternative for the PGR protein assay specially in the lower range of proteinuria (0.03 – 0.45g/L) and shows feasibility for automation with lower sample volumes.
The authors are thankful to the participants for their voluntary participation and kind co-operation; Prof. Bilesha Perera, Faculty of Medicine, University of Ruhuna for the assistance in data analysis; Director of Teaching Hospital Karapitiya and Dr. Dharshi Anuruddhika for granting permission to collect urine samples and details from patients; District Director of Health Services, Galle and Medical Officer of Health of Bope Poddala MOH area for granting permission to collect urine samples from healthy individuals; Dr.Manjula Dissanayake, Mr. Aruna Jayasooriya, Mr. Gayan Thebuwana and Nawaloka Laboratory, Galle for their co-operation and technical support to perform PGR protein and
creatinine assays using BioSystems BA400 analyser; Department of Medical Laboratory Science, Faculty of Allied Health Sciences, University of Ruhuna for providing the funding for the project and their collaboration throughout the research project.
Nimesha D Ranawaka, BSc Hons (MLS), MSc Student 1, 3
Rajika G Jinadasa, BSc Hon (MLS), Research Assistant 1, 4
Kalani B Gunawardana, PhD, MSc, BSc, Senior Lecturer 1
Indika D Siriwardhana, MBBS, MD (Chem Path), Senior Lecturer 2, 5
1 Department of Medical Laboratory Science, Faculty of Allied Health Sciences, University of Ruhuna, Galle, Sri Lanka
2 Department of Pathology, Faculty of Medicine, University of Ruhuna, Galle, Sri Lanka
3 University of Western Australia, Perth, Australia
4 University of Sri Jayewardenepura, Sri Lanka
5 Department of Biochemistry and Clinical Chemistry, Faculty of Medicine, University of Moratuwa, Moratuwa, Sri Lanka
Correspondence: Indika D Siriwardhana, Department of Biochemistry and Clinical Chemistry, Faculty of Medicine, University of Moratuwa, Sri Lanka
Email: indikads@uom.lk
1. Kashif W, Siddiqi N, Dincer AP, et al. Proteinuria: how to evaluate an important finding. Cleve Clin J Med 2003 70(6): 535-537.
2. Carter JL, Tomson CRV, Stevens PE, Lamb EJ. Does urinary tract infection cause proteinuria or microalbuminuria? a systematic review. Nephrol Dial Transplant 2006; 21(11): 3031–3037.
3. Beetham R, Cattell WR. Proteinuria: pathophysiology, significance and recommendations for measurement in clinical practice. Ann Clin Biochem 1993; 30(Pt 5): 425–434.
4. Marshall WJ, Lapsley M DA. Clinical Chemistry 2020; 9th edition, Elsevier, Missouri, USA.
5. Lamb EJ, MacKenzie F, Stevens PE. How should proteinuria be detected and measured? Ann Clin Biochem 2009; 46(Pt 3): 205-217.
6. Oni MO, Oguntibeju OO. Clinical and diagnostic importance of proteinuria: a review. Afr J Biotechnol 2008; 7(18): 316672.
7. Cheung CK, Mak YT, Swaminathan R. Automated trichloroacetic acid precipitation method for urine total protein. Ann Clin Biochem 1987; 24(2): 140-144.
8. Jinadasa A, Srimantha L, Siriwardhana ID, et al. Optimization of 25 % sulfosalicylic acid protein-to-creatinine ratio for screening of low-grade proteinuria. Int L Anal Chem 2021; 6688941.
9. Price CP, Newall RG, Boyd JC. Use of protein : creatinine ratio measurements on random urine samples for prediction of significant proteinuria: a systematic review. Clin Chem 2005; 51(9):1577-1586.
Copyright: © 2024 The author(s). This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
The Board of Examiners (BoE) have investigated several issues relating to the QMLT examination preparation including the workload distribution during the year in the NZIMLS Office. In 2025, applications will open on 1 February, and the closing date for application to undertake the 2025 QMLT examinations will be 15 April. Please note that the new earlier dates differ from those used for this year’s QMLT examination application. The examination date will remain the same i.e. the first Saturday in October.


Cardiac Advance control is the newest addition to our cardiac assessment portfolio. This next generation control is optimized with Troponin targets near the limit of instrument detection adhering to the latest guidelines set by AACC and IFCC for greater confidence in your test results. Consolidated with the most tested cardiac analytes and offered in automation-ready configurations, your QC workflow can be streamlined to help improve efficiency saving time and resources.
•Optimized cardiac Troponin targets
•Ten of the most tested cardiac analytes
•Automation ready configurations available
•Integrates with Unity QC data management
Learn more about Bio-Rad Cardiac Quality Controls by visiting: www.bio-rad.com/cardiac
Correlation of IL-6 with D-dimer, LDH, S-ferritin biomarkers, comorbidities and clinical outcomes of COVID-19 patients admitted in a tertiary hospital in Coimbatore, India – a retrospective study based on 2,569 patients
Lavanya Sriramajayam, Murali Alagesan, Sudha Ramalingam, Karthikeyan Ramaraju, Jayavardhana Arumugam, Roshini Rajasekaran, Dhanushya Etraj, Sankarganesh Jeyaraj
Introduction: COVID-19 disease is associated with an excessive host immune response leading to multiorgan failures and immunerelated manifestations. Clinical outcomes are influenced by disease severity, age, immune response, and comorbidity factors. In this retrospective study, we evaluated the correlation of Interleukin-6 (IL-6) with D-dimer, lactate dehydrogenase (LDH), Serum-ferritin, disease severity, and comorbidities of (n=2569) COVID-19 patients. A retrospective analysis of data from COVID-19 patients has been evaluated and the correlation of IL-6 to biomarkers towards comorbidity factors, D-dimer, LDH, and S-Ferritin parameters was estimated.
Materials and methods: Data from 2,569 COVID-19 patients 1687 males and 882 females were analysed.
Results: Respiratory distress was higher in COVID-19 patients with IL-6 > 7pg/mL values (26.0% (453/1743) compared to patients with IL-6 ≤ 7 pg/mL(11.4% (94/826) with a p-value of <0.001. IL-6 levels were found to be positively correlated with other biomarkers such as D-dimer, LDH, and S-Ferritin. A positive correlation of IL-6 with oxygen requirement, high flow oxygen, non-invasive and mechanical ventilation, and treatment in ICU wards was identified. COVID-19 patients with higher IL-6 values (> 7pg/mL) were found to be associated with the presence of comorbidities such as hypertension and diabetes than patients with IL-6 values ≤ 7pg/ mL group.
Conclusion: IL-6 was found to be positively correlated with biomarkers such as D-dimer, LDH, S-ferritin and comorbidities such as hypertension and diabetes in the study population of 2,569 patients.
Keywords: COVID-19, ICU admission, IL-6, D-dimer, S-Ferritin, comorbidities, biomarkers
NZ J Med Lab Sci 2024; 78(3), 119-124
COVID-19 caused by the SARS-CoV-2 virus, is a global health threat where countries have reported 771 million cases and 6.9 million deaths since December 2019 (1). Clinical symptoms of COVID-19 include fever, muscle and joint pain, fatigue, gastrointestinal disorders, and respiratory illness which can lead to mortality in severe cases. Changes in WBC count, and low lymphocytes are typical characteristics that occur during COVID-19 disease (2). Many of the patients experienced asymptomatic or mild pneumonia, but in severe COVID-19 cases, they exhibit arterial oxygen saturation (SpO2) ≤ 93), respiratory rate with values greater than 30 beats per minute, and lung infiltration (3). Acute respiratory distress syndrome (ARDS) and multiple organ failure were acting as leading causative factors of death in severe COVID-19 patients who required mechanical ventilation and admission to Intensive Care Units (4,5). Multiple factors are associated with clinical outcomes and variations in host immune response against the disease lead to difficulties in predicting the outcome during the initial progression of the disease (6–9). Clinical biomarkers act as a powerful tool to predict clinical development and help in the decision-making for treatment (10). A high level of inflammatory cytokines through immune system deregulation in the host as a cytokine storm, could lead to cytokine release syndrome (CRS), which plays a crucial role in the pathogenesis of COVID-19 disease (11–13). Patients with a history of cardiovascular disease are more vulnerable to an over-activated inflammatory response and subsequent multi-organ dysfunction, resulting in poor clinical outcomes (14,15). Close monitoring is advisable for these patients. Previous studies have indicated the importance and role of clinical haematology measurements such as CBC, which play a vital role in the rapid diagnosis of viral and bacterial respiratory pathogens which may lead to earlier treatment with better recovery outcomes (16,17). C-reactive protein (CRP), Interleukin-6 (IL-6), and procalcitonin levels in COVID-19 patients were found to correlate with disease severity and ICU admissions (18). These biomarkers are used to determine the severity of lung injury and lesions due to COVID-19 and the severity of COVID-19 disease (19). Elevated levels of IL-2, IL7, IL-10, granulocyte-colony-stimulating factor, IP-10, monocyte chemo attractant protein-1 (MCP-1), macrophage inflammatory
protein-1A (MIP-1A) and tumour necrosis factor-α (TNF-α) play an important role in the interaction between different immune cell types of critically ill COVID-19 patients who were admitted to ICU units in the hospitals (20). IL-6 and CRP are found to be elevated in relation to the progress of COVID-19 severity, whereas TNFR levels were found to be reaching peak levels at the onset of disease severity (21). Hyper activation of the host immune system and failure to differentiate between self and foreign antigens including the secretion of detrimental interleukin 6, are critical mediators for respiratory failure, multi-organ failure, and death.
D-dimer is found to be present in higher amounts in patients with diseases leading to hyper coagulation and fibrinolytic activity (22–24). It has been also shown that D-dimer levels were found to positively correlated with elevated levels of severe inflammatory and coagulation parameters such as CRP, procalcitonin and IL-6 (25).
Lactate dehydrogenase (LDH) enzymes are present in all the cell types in the body, and the human host responds to the tissue damages through the enzyme. LDH is directly associated with the clinical severity of COVID-19 (26). LDH was strongly found to be associated with mortality in patients with communityacquired-pneumonia (27).
Ferritin is an iron storage protein molecule plays an important role in several inflammatory and infectious diseases. During COVID-19 disease the pathogen virus utilizes the host iron contents leading to the reduced iron levels in the body. The body responds to the condition to produce more amounts of ferritin levels. The increased ferritin levels lead to more replication of virus, mitochondria defects, increase in ROS levels and damage to tissue and organs. Increased ferritin levels were shown to be associated in H1NI infection (28). High levels of ferritin were found to be associated with haemorrhage and death in Ebola patients (29). Examination of the values of such biomarkers helped the clinicians around the world to tackle the severity of COVID-19 disease and treatment regimens.
In this study, we aim to identify the correlation of IL-6 against comorbidity factors such as hypertension and diabetes, biochemical markers such as D-dimer, LDH, S-Ferritin, O2, and mechanical ventilation requirements, and ICU admissions in a
large cohort of 2,569 patients admitted in a tertiary hospital in India.
Study Design and Patients
Study data was obtained from PSG Hospitals, a tertiary hospital located in Coimbatore, Tamil Nadu, India. The hospital was approved by the Indian Council of Medical Research (ICMR), India, for treating COVID-19 patients during the pandemic period. Data was extracted from the hospital COVID-19 database (2,569 COVID-19 patients who were hospitalized during the treatment for COVID-19). All the patients met the COVID-19 disease severity according to the interim guidelines from the World Health Organization. Patient admission guidelines, management, and treatment were followed according to the national guidelines from the Indian Council of Medical Research (ICMR) a unit of the Government of India and the Ministry of Health and Family Welfare (MoHFW). Treatment modalities were antibiotic therapy, administration of dexamethasone, and Oxygen ventilation. Severely ill patients were admitted to the ICU units with oxygen support (invasive or non-invasive ventilation). Length of stay in the hospital for cases varied according to the symptoms and was discharged according to the guidelines from the Indian Council of Medical Research, Delhi, India.
SARS-COV-2 pathogen detection
SARS-CoV-2 RT-PCR was performed in the NABL and NABH accredited molecular diagnostics unit in the Department of Microbiology, PSG Institute of Medical Sciences and Research, Coimbatore, India. The unit has a BSL-2 lab facility and is approved by ICMR India to perform COVID-19 RT-PCR diagnostic tests. The protocol was followed by guidelines from WHO and ICMR, India. Nasopharyngeal and oropharyngeal swab specimens were collected from the patients during the admission and treatment period followed by immediate storage in the viral transport medium and transported in cold chain. Samples reaching the molecular diagnostics unit were processed on the same day without delay using a standardized protocol. Extraction was performed using GENEAID nucleic acid extraction kit and RT PCR was performed using Real Star SARS-CoV-2 RT-PCR Kit 1.0 (Altona diagnostics, GmbH, Germany or TRUPCR SARSCoV-2 RT qPCR Kit (3B BlackBio Biotech India Ltd).
Patient blood sample collection and biomarkers measurement
Blood and serum samples of volume (3 mL) were collected from the patients during the time of admission and on any day during the treatment and recovery phase. Serum IL-6 was detected using electrochemiluminescence immunoassay in Roche Cobas e411 analyser (Roche Diagnostics, CH-6343 Rotkreuz, Switzerland) in the Department of Biochemistry, inside the tertiary hospital. The concentration of ferritin and LDH were evaluated by electro-chemiluminescent immunoassay using Cobas e411
analyser. D-dimer was measured within 24 hours using Immunoturbidimetry method in a Sysmex CS 2400 coagulation analyser machine (Sysmex Corporation, Kobe, Japan) in the diagnostic division of Department of pathology in the hospital
Ethics approval
Ethics approval was obtained from the PSG Institutional Human Ethics Committee, PSG Institute of Medical Sciences and Research, Coimbatore, India (Ref No. 21/031).
Data collection and analysis
This study was single-centred with retrospective data analysis from COVID-19 patients admitted to a tertiary care hospital located in Coimbatore, Tamil Nadu, India. A separate database was established using data from medical reports comprising values of age, gender, symptoms, biochemistry and pathology lab reports, and comorbidities. The study was conducted in accordance with the principles of the ICH GCP guidelines. Study participants were COVID-19 positive by SARS-COV-2 RT-PCR test, precedent, or during the hospitalization. The admitted patients were grouped into four categories namely: Patient demographics, symptoms, and biomarkers IL-6, D-Dimer, LDH, and S. Ferritin were included for the analysis. Data were presented as values with percentages, where statistical significance was calculated using Fisher’s Exact test. Mann-Whitney U test, X² test, or Fisher’s exact test were applied to compare continuous and categorical variables in the IBM SPSS platform.
A database containing clinical details of 2569 patients with COVID-19 disease was included in the study. The patient flow chart is explained in Figure 1. Baseline demographics for the study cohort are given in Table 1. A male preponderance was observed in the group with IL-6>7pg/mL (Table 1). IL-6>7pg/mL was associated with an older age group (55.68 ± 14.00) than the younger age group (44.55 ± 15.94), as represented in Table 1. A chi-squared test was used to explore the association between ‘IL-6’ and Comorbidities such as diabetes and hypertension. Respiratory distress was observed to be significantly higher in the patient group with IL-6>7pg/mL with a p-value of 0.001 (Table 2). Comparing co-morbidities to IL-6 values, it has been revealed that patients with any comorbidities had higher IL-6 values (IL-6>7pg/mL group) with a p-value of < 0.001 (Table 3). Patients with the most common comorbidities, such as diabetes, hypertension, or any heart disease were found to exhibit IL-6 values >7pg/mL. High IL-6 (>7pg/mL) was positively correlated with higher levels of other biomarkers such as LDH, D-dimer and serum ferritin with p values less than 0.001 (Table 4). Increased requirements for oxygen, high-flow oxygen, noninvasive ventilation, and mechanical ventilation were found to be significantly correlated with the patient group having IL-6>7pg/ mL. They also had a significantly longer duration of ICU stay and hospitalisation (Table 5).
Table 2. Presenting symptoms and correlation of IL-6 values against presentation parameters fever, cough and respiratory stress
(Yes)***
Presentation: Cough (Yes)***
Presentation: Respiratory Distress (Yes)***
***Significant at p<0.05, 1 Chi-Squared Test, p value - 0.001
Table 3. Comorbidities patterns of COVID-19 patients versus IL-6 values
Comorbidities
Comorbidities:
at p<0.05, 1Chi-Squared Test, 3Fisher's Exact Test
Table 4. Laboratory biomarkers D-dimer, S-ferritin and LDH values correlating with IL-6 groups
***Significant at p<0.05, 2Wilcoxon-Mann-Whitney U Test
Table 5 Correlation of COVID-19 patients oxygen requirements, ICU stay, High flow oxygen, and Mechanical ventilation against IL-6 groups
***Significant at (p<0.05), 1 (Chi-Squared Test), 2 (Wilcoxon-Mann-Whitney U Test)

Figure 1. Patient flow diagram representing the enrolment of patients, inclusion and exclusion criteria and the data analysed for the study.
In this study, we aimed to investigate the association of IL-6 values with COVID-19 patients’ demographics, laboratory biomarkers, clinical presentation, duration of ICU stay, oxygen requirements and mechanical ventilation in an Indian cohort with data from 2,569 patients. Notably, higher IL-6 values (> 7pg/mL) were found to be associated significantly with D-Dimer, S. Ferritin, LDH values, ICU stay, high flow oxygen, mechanical ventilation, and comorbidities such as diabetes and hypertension. COVID-19 has had a devastating impact on the social and economic life of people throughout the world. Optimal care and treatment depend on the early identification of biomarkers in COVID-19 patients. It has been shown that corona virus activates excessive and uncontrolled immune responses in the infected host, leading to the development of acute respiratory
distress syndrome (ARDS). Immunopathogenesis in the host caused by the viral pathogen is also associated with host organ dysfunction and acts as a leading factor causing disorders in metabolic functions and mortality (17). It has been shown that the inflammatory pathways mediated in the host play an important role in the progression toward severity of COVID-19 disease, where cytokine storm augments the systems and immunemediated pathogenesis in the host (12).
The identification of parameters indicating the severity of infection may help clinicians to predict the outcome of patients with a high risk of unfavourable progress of the disease (30). Recognition of at-risk patients could permit the early institution of aggressive intensive care and antiviral and immune treatment to reduce or prevent the consequences associated with this proinflammatory state (31).
IL-6 acts as a robust predictor of severity in hospitalized patients with COVID-19 (19). A high level of IL-6 in serum samples collected from COVID-19 patients has been shown to be significantly correlated with the severity of the disease and acting as a biomarker to indicate the role of severe pathogenesis. Elevated IL-6 levels in COVID-19 patients have been linked with Major Adverse Cardiovascular Events (MACE) and/or mortality. During the initial hours after admission to the hospital, patients could be stratified by measuring IL-6 levels. Between the time interval between admission and discharge from the hospital for treatment, Interleukin-16 levels were found to be correlated with the alterations in their ability to taste and smell (32). When the IL-6 levels in COVID-19 patients were decreased during recovery time, it led to significant improvement in the ability to smell (p-value < 0.05) and taste (p = 0.047) functions during swab RT-PCR COVID-19 negative results timeframe. This indicated the key role of interleukin-6 in the pathogenesis of chemo-sensitive disorders in COVID-19 patients. IL-6 levels can be used as a biomarker for escalation of treatment in patients with COVID-19 infection (18). Monoclonal antibodies such as tocilizumab and sarilumab which were specifically targeting IL-6 receptor molecules, were approved in many clinical trials to reduce the severity of clinical manifestations and improve treatment outcomes (15,16,33). The long-term benefits of IL-6 inhibitors depend on patient safety, the cost associated with the treatment, treatment efficacy in different healthcare systems, and coverage under insurance schemes (34). Administration of IL-6 inhibitors was found to be associated with reducing mortality rate, cardiovascular death, improving the earlier rate of discharge during treatment, and reducing severe respiratory complications, suggesting that it can be successfully used to prevent cytokine release syndrome and mortality (35).
Previous study showed that the LDH measurement gives accurate information regarding the severity of injury and lesion in the lungs during COVID-19 pneumonia (26). Kojima et al reported in a study that the levels of LDH indicate the progression of severity of the disease in patients with mild to moderate disease during the admission (36). In a pooled analysis study, where results from nine published studies were taken into account, LDH measurement at the time of admission predicted the development of severity (six-fold increased odds) and mortality (16-fold increased odds) in COVID-19 patients (37). In a multi-centre study, patients who advanced to severity conditions were having higher levels of LDH when compared to the stable group (38). LDH in the Covid CALL score model study, was found to be acting as an independent risk factor to predict the patients progressing to severity levels (39). Elevated levels S-Ferritin plays an important role in mediating cytokine storm and secondary hemophagocytic lymphohistiocytosis (sHLH) in COVID-19 patients admitted in intensive care units (22,40,41) It has been shown that the levels of ferritin were significantly associated with mortality when compared between survivors and non-survivors (42). COVID-19 patients with co-morbidities such as diabetes, obesity and thrombosis had higher levels of ferritin when compared with patients without morbidity factors (42,43) Ferritin levels were found to be higher in severe COVID-19 patients with livery injury.
Different inflammatory markers, such as, C-Reactive-Protein, procalcitonin, serum levels of ferritin, erythrocyte sedimentation rate, and TNF-α have been reported to be significantly associated with an increased risk of developing severe COVID-19 (44–46). However, these results remain controversial because some other studies have reported no significant difference in the serum levels of CRP between the two groups, and the role of inflammatory parameters in monitoring COVID-19 progression is still unclear.
Limitations of the study include; a) the data for this retrospective study was obtained from a single centre hospital, not representing the cohorts from a multi-centre study, b) Only RT-PCR positive patients were admitted to the hospital for the treatment whereas negative RT-PCR results would not confirm the absence for COVID-19 disease, c) patients were represented
mainly from the urban population in the study area whereas data from remote and rural regions would be missed, d) the patient population represented in the study is very much restricted to the specific geographic location in the southern parts of India, not representing the overall Indian population.
We are sincerely grateful to PSG Institutions Management for facilities and infrastructure.
Data are available from the corresponding author upon request.
Lavanya Sriramajayam, MD, Associate Professor1
Murali Alagesan, MD, Professor2
Sudha Ramalingam, MD, Professor3
Karthikeyan Ramaraju, MD, FCCP, Professor4
Jayavardhana Arumugam,MD, Professor 5
Roshini Rajasekaran, MBBS, Junior Resident2
Dhanushya Etraj, BSC, Physician Assistant2
Sankarganesh Jeyaraj, PhD, Associate Professor6,7
1Department of Microbiology, PSG Institute of Medical Sciences and Research, Coimbatore, India
2Department of General Medicine, PSG Institute of Medical Sciences and Research, Coimbatore, India
3Department of Community Medicine, PSG Institute of Medical Sciences and Research, Coimbatore, India
4Department of Respiratory Medicine, PSG Institute of Medical Sciences and Research, Coimbatore, India
5Department of Pediatrics, PSG Institute of Medical Sciences and Research, Coimbatore, India
6PSG Centre for Molecular Medicine and Therapeutics, PSG Institute of Medical Sciences and Research, Coimbatore, India
7PSG Centre for Genetics and Molecular Biology, Coimbatore, India
Correspondence: Sankarganesh Jeyaraj, PSG Center for Molecular Medicine and Therapeutics, India
Email: sankarganesh@psgimsr.ac.in
1. WHO Coronavirus (COVID-19) Dashboard [Internet]. [cited 2023 Oct 16]. Available from: https://covid19.who.int
2. Guan WJ, Ni ZY, Hu Y, Liang WH, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020; 382(18): 1708–1720.
3. Lichter Y, Topilsky Y, Taieb P, et al. Lung ultrasound predicts clinical course and outcomes in COVID-19 patients. Intensive Care Med 2020; 46(10): 1873–1883.
4. Gershengorn HB, Hu Y, Chen JT, et al. The impact of highflow nasal cannula use on patient mortality and the availability of mechanical ventilators in COVID-19. Ann Am Thorac Soc 2021; 18(4): 623–631.
5. Meyer NJ, Gattinoni L, Calfee CS. Acute respiratory distress syndrome. Lancet 2021; 398(10300): 622–637.
6. Cao X. COVID-19: immunopathology and its implications for therapy. Nat Rev Immunol 2020; 20(5): 269–270.
7. Ing AJ, Cocks C, Green JP. COVID-19: in the footsteps of Ernest Shackleton. Thorax 2020; 75(8): 693–694.
8. Yang R, Gui X, Xiong Y. Comparison of clinical characteristics of patients with asymptomatic vs symptomatic coronavirus disease 2019 in Wuhan, China. JAMA Netw Open 2020;3(5): e2010182.
9. Gajate-Arenas M, García-Pérez O, Chao-Pellicer J, et al. Differential expression of antiviral and immune-related genes in individuals with COVID-19 asymptomatic or with mild symptoms. Front Cell Infect Microbiol 2023; 13:1173213
10. Battaglini D, Lopes-Pacheco M, Castro-Faria-Neto HC, Pelosi P, Rocco PRM. Laboratory Biomarkers for Diagnosis and Prognosis in COVID-19. Front Immunol 2022; 27:13: 857573.
11. Leisman DE, Ronner L, Pinotti R, et al. Cytokine elevation in severe and critical COVID-19: a rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir Med 2020; 8(12): 1233–1244.
12. Liu B, Li M, Zhou Z, Guan X, Xiang Y. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)induced cytokine release syndrome (CRS)? J Autoimmun 2020; 111: 102452.
13. Ye Q, Wang B, Mao J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J Infect 2020; 80(6): 607–613.
14. Smadja DM, Mentzer SJ, Fontenay M, et al. COVID-19 is a systemic vascular hemopathy: insight for mechanistic and clinical aspects. Angiogenesis 2021; 24(4): 755–788.
15. Nguyen N, Nguyen H, Ukoha C, et al. Relation of interleukin-6 levels in COVID-19 patients with major adverse cardiac events. Proc (Bayl Univ Med Cent) 2022; 35(1): 6–9.
16. Coomes EA, Haghbayan H. Interleukin-6 in COVID-19: a systematic review and meta-analysis. Rev Med Virol 2020; 30(6): 1–9.
17. Melo AKG, Milby KM, Caparroz ALMA, et al. Biomarkers of cytokine storm as red flags for severe and fatal COVID-19 cases: a living systematic review and meta-analysis. PLoS One 2021; 16(6): e0253894.
18. Herold T, Jurinovic V, Arnreich C, et al. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J Allergy Clin Immunol 2020; 146(1): 128-136.e4,
19. Broman N, Rantasärkkä K, Feuth T, et al. IL-6 and other biomarkers as predictors of severity in COVID-19 . Ann Med 2021; 53(1): 410–412.
20. Ruan Q, Yang K, Wang W, et al. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med 2020; 46(5): 846–848.
21. Gohda T, Murakoshi M, Suzuki Y, et al. Circulating tumor necrosis factor receptors are associated with mortality and disease severity in COVID-19 patients. PLoS One 2022; 17(10): e0275745.
22. Zhou F, Yu T, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020; 28:395(10229): 1054–1062.
23. Yao Y, Cao J, Wang Q, et al. D-dimer as a biomarker for disease severity and mortality in COVID-19 patients: a case control study. J Intensive Care 2020; 8:49. A look back at ASM24: Exhibition


Copyright: © 2024 The author(s). This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.




















Saad Al-Fawaeir
ABSTRACT
Objectives: Smoking is a preventable risk factor for morbidity and mortality A remarkable increase has been observed in the context of the prevailing trend of past years regarding smoking in Jordan. The present research examines the association between cigarette smoking and liver enzymes and electrolytes.
Methods: 220 male participants were recruited in this study of which 180 were smokers. Participants attended the outpatient clinic at Tohama Medical Center in Zarqa City. The study was conducted from September 2019 to February 2021. Aspartate aminotransferase (AST), electrolytes (PO4 -3 , Ca+2 , K+ , Cl, Na+ , Fe+2),Alanine aminotransferase (ALT), serum γ- GlutamylTransferase (γ-GT), total bilirubin, Alanine aminotransferase (ALT) were examined between the non-smokers and smokers. An independent t-test was employed to conduct inferential statistics.
Results: It was observed thatALT, γ-GT levels and total bilirubin were raised in smokers and statistically significant (p-value <0.05). The significant mean difference of serum γ-GT, ALT, ALP, AST, and total bilirubin was found between participants who smoked for 1-5, 5-10, 10-20 and >20 years and non-smokers. Significant (p<0.05) change was shown in Ca+2 , PO4-3 , and Fe+2 levels in cigarette smokers as compared to non-smokers. Haematological parameters were compared between non-smokers and smokers. The mean haematocrit, red blood cells, haemoglobin, and mean corpuscular volume of smokers were significantly increased as compared to those who did not smoke. Deranged liver enzymes and serum electrolytes were associated with smoking.
Conclusions: Cigarette smoking was correlated with deranged liver enzymes and serum electrolytes. Haematological parameters were also found to be higher in smokers than in non-smokers. The current study gives an insight into the hazards of smoking causing a consistent increase in mortality and morbidity rates.
Keywords: Cigarette smoking, Electrolytes, Potassium, Sodium.
NZ J Med Lab Sci 2024; 78(3) 126:130
INTRODUCTION
Tobacco is responsible for over 8 million deaths annually causing a serious concern in public health across the world. Among them 7 million were tobacco users and 1.2 million were non-smokers exposed to second hand smoke (1). Tobacco is found in cigars, kreteks, pipe tobacco, bidis, and cigarillos, however cigarettes are more popular. All these products are considered dangerous to health (2,3). There are more than 4,000 chemicals in a single cigarette, 200 of them are poisonous and are responsible for above 80 different categories of cancer (4,5). Examples of these poisonous components include carbon monoxide, nicotine, nitrogen oxide, hydrogen cyanide, and free radicals (6). Smoking causes the production of carbon monoxide, which binds strongly to haemoglobin, causing diseases and impacting numerous organs, including; high blood pressure (7), hypoxia, anaemia (8), pancreatic cancer, colon cancer, liver cancer, kidney cancer, lung cancer, strokes, chronic obstructive pulmonary disease, and heart disease (9,10). For example, the liver performs the functions of metabolism, the storage of glycogen, and the elimination of toxic and hazardous drugs and components (11). According to Bishop (2020), alanine transferase (ALT), aspartate transferase (AST) and the enzyme γ-glutamyl transferase (γ-GT), are particularly important for liver function. These enzymes might be used to forecast various clinical outcomes for both patients and healthy populations, as well as to detect liver damage or other abnormalities (12)
Electrolyte concentrations are controlled within the body by metabolic systems and are prerequisites for performing functions of contractility via potassium, calcium, and magnesium. Moreover, co-factor in enzyme activation regulation including somatic and volume regulation via sodium, chloride, and potassium, myocardial rhythm, and regulation of adenosine triphosphate through acid-base balance, ATPase, bicarbonate, potassium, calcium-regulated blood coagulation, and chloride (13). Pochineni & Rondon-Berrios (2018), suggested that the disturbances in electrolytes can cause serious, even fatal, metabolic abnormalities including renal failure, coronary heart disease, lung infection, endocrine system issues and liver disease. The management and distribution of electrolytes by the kidneys are one of the ways smoking impairs kidney function. Smoking hardens the arteries that supply the kidneys, making them smaller and less effective (14).
Cigarette smoke contains nicotine, which inhibits bone repair and increases the risk of infections at implant insertion sites. Nazeer et al. (15) demonstrated that a longer duration and higher frequency of smoking are associated with a lower implant survival rate compared to nonsmokers. Additionally, Qiu et al. (16) found higher levels of inflammatory cells, such as neutrophils and lymphocytes in smokers than in nonsmokers. These differences are attributed to physiological dysfunction and subsequent changes in glucose levels, cholesterol and blood pressure. Hassan et al. (17) discussed that smokers are more prone to heart disease, diabetes and arteriosclerosis due to elevated total cholesterol and blood pressure levels compared to nonsmokers.
The prevalence of smoking and diseases associated with smoking are observed and are considered one of the significant public health concerns in Jordan. In addition, although international smoking data is readily available, data is limited in our part of the world. Therefore, this study aimed to examine the association of smoking status with electrolyte levels, liver enzymes, and haematological factors between non-smokers and smokers in our cohort.
An analytical cross-sectional study was conducted at Tohama Medical Center in Zarqa City. This study was conducted from September 2019 to February 2021 with a total number of 220 male participants, of which 180 were smokers and 40 were nonsmokers. Ethical approval was obtained from the Institutional Ethics Committee (PHAR-06/11/08/2019) and participants provided informed consent enrolling them in the study.
Inclusion and Exclusion Criteria
The smoker’s group of 180 participants was divided into four subgroups based on the duration of smoking: 1-5 years (45 participants), 5-10 years (45 participants), 10-20 years (45 participants), and >20 years (45 participants). Participants aged above 18 years, males and at least a one-year smoking history were included in the study. Participants having chronic liver disease, chronic kidney disease, and overt thyroid dysfunction individuals on corticosteroid therapy, along with other states correlated with transformed serum ferritins, for instance,
hemochromatosis, chronic alcoholics, bleeding disorders, and anaemia were excluded from this study. Individuals who reported repeated blood transfusions were also not part of this study.
Separated serum separator gel tubes were used to collect venous blood samples. They were left for 15 minutes for clotting at normal temperature. Further, they were centrifuged at 4000g for 10 minutes. Afterwards, they were directly examined to test for their liver functionality and electrolyte parameters. Blood samples were collected for haematology analysis in EDTA tubes was performed through collecting blood samples. All the tests were conducted at Teryaq Alrohh Medical Laboratory. The study used Cobas 111 auto-analyzer (Roche Diagnostics GmbH, Mannheim, Germany) for examining AST, Serum γ-GT, alkaline phosphatase (ALP), ALT, total bilirubin and electrolytes (Na+, K+, Fe+2, PO4-3, Cl-, Ca+2). All these parameters were measured using Roche Diagnostics kits. Sysmex k 1000 haematology analyzer (Tao Electronics, Japan) was employed for determining the complete blood count. While the normal range was used as the criterion for testing the liver function parameters (γ-GT: 0-38 U/L, ALT (0-40 U/L), AST (0-40 U/L), ALP (42-129 U/L), total bilirubin (3.4 - 20.5µmol/L), Na (135 - 153mmol/L), K (3.5 - 5.3mmol/L), Ca (2.10 - 2.55mmol/L), Fe (8.8 - 25.1µmol/L), Cl (98 - 108mmol/L) and PO4 (0.84 - 2.1mmol/L). Upon request, the author would make the study data available for review.
The data was entered into Microsoft Excel and then imported to the statistical package for social sciences (SPSS; SPSS Inc. Headquarters, Chicago, III., USA) version 23.0 for data analysis. Descriptive statistics were calculated as mean and standard deviations for quantitative variables and frequency and percentages for qualitative variables. An Independent t-test was employed to evaluate the mean difference between smokers and non-smokers and a p-value ≤ 0.05 was considered significant statistically.
This study recruited 220 participants among them 180 (81.8%) were smokers and 40 (18.2%) were non-smokers. The mean age in years of smokers and non-smokers was 49.5±13.2 and 42.8±11.2 years respectively. Biochemical analysis was done and compared between both the groups, and it is presented in Table 1.It was observed that ALT, γ-GT levels, and total bilirubin were raised among smokers in comparison with non-smokers which was reported significant statistically (p-value <0.05). However, AST and ALP were not found statistically significant. Table 2 displays a comparative view regarding liver enzyme activity in line with the total time of having the use of tobacco with controls. The significant mean-variance in AST, serum ALT, ALP, γ-GT, and total bilirubin was found between a participant who was smoking for 1-5, 5-10, 10-20 and >20 years and non-smokers. Table 3 encapsulates electrolytes (K+, Na+, Cl-, PO4-3, and Fe+2) serum levels of non-smokers and smokers that display no significant change statistically in serum K+, Na+, and Cl- levels. In contrast, a significant rise was reported (p<0.05) in Fe+2 levels among smokers. A significant change in the concentration of PO4-3 and Ca+2 was noted among smokers. Table 4 shows a comparison of serum electrolytes according to the duration of smoking with controls. The significant mean difference in Na+, K+, Cl-, Fe+2, Ca+2, and PO4-3 was found between a participant who had been smoking for 10-20 and >20 years and non-smokers. However, no significant difference was observed in participants who were smoking for 1-5 and 5-10 years. Haematological parameters were associated between both groups. It was identified that the mean haemoglobin (Hb) of smokers and non-smokers was 16.21g/L ± 2.90 and 143.5g/L ± 4.8 respectively and it was found statistically significant (p-value 0.018). The haematocrit (HCT) of smokers was 51.34 ± 1.3 in comparison to non-smokers (42.71±1.82) and this was also statistically significant (p-value 0.039). Red blood cells (RBC) were also found to increase among smokers in comparison with non-smokers and found significance (p-value <0.05). The mean corpuscular volume (MCV) of smokers was significantly increased (91.7±1.17) as compared to nonsmokers (86.39±3.87) and was found significant (p-value 0.041). However, white blood cells and platelets were found statistically non-significant (p-value >0.05).
*p-value ≤ 0.05 was considered statistically significant
p-value ≤ 0.05 was considered statistically significant
Table 3: Serum electrolyte levels among smokers and non-smokers Parameters
(mmol/L) (K+) (Mean ± SD)
(mmol/L) (Na+) (Mean ± SD)
(µmol/L) (Fe+2) (Mean ± SD)
Ca+2) (Mean ± SD)
(mmol/L) (Cl-) (Mean ± SD)
(PO4-3) (Mean ± SD)
*p-value ≤ 0.05 was considered statistically significant
± 0.095
± 1.21
±
±
0.95
± 0.25
Table 4: Association of smoking duration and deranged serum electrolytes
Duration of Smoking (in years) Sodium (mmol/L) (Na+) (Mean ± SD)
*p-value ≤ 0.05 was considered statistically significant DISCUSSION
Smoking is a significant contributor to morbidity and fatality (18)as discussed by Murray et al (2020), more than 200 million deaths have been caused by smoking tobacco (19). Smoker mortality risk is calculated using daily cigarette consumption, smoking duration, inhalation intensity, and age of starting (20). Smoking contains various oxidants, including volatile aldehydes and radicals, which are considered major contributors to the damage of biomolecules (21). Chemicals found in cigarettes include; carbon dioxide, carbon monoxide, total hydrocarbons and nitrogen oxides for burning Talaiekhozani and Amani (22) reported that a single cigarette burns 0.5mg of carbon dioxide, 0.13mg of carbon monoxide, 0.01mg of nitrogen oxides and emits 0.01mg of hydrocarbons.
Compared to non-smokers, smokers are at a higher risk of cardiovascular diseases, malignancies, respiratory issues, hepatotoxicity, gastroesophageal reflux disease, peptic ulcers, blindness, and loss of bone matrix (23). Cigarette smoking has injurious adverse effects on the liver as it contributing to nonalcoholic fatty liver disease, primary biliary cholangitis, alcoholic liver disease and chronic viral hepatitis (24).
In this present study, an increase in Red Blood Cell (RBC) levels was observed, which is attributed to the high partial pressures of CO and CO2 gases in the bloodstream. This condition decreases the binding capacity of O2 with haemoglobin (Hb). Consequently, RBCs are produced by triggering the stem cells inside the red bone marrow and the levels of HCT also increase among smokers. Comparing non-smokers, the findings revealed a substantial increase in serum ALT and AST activity among smokers. The potential reason for this rise could be nitrosative stress (NSS). This refers to a state of intensity in the body, neutralizing and eradicating reactive nitrogencontaining compounds like nitrous oxide. Nitrosative stress
±
causes nitrosylation reactions that alter protein structure and impair normal body processes. Notably, NSS represent a highly oxidative condition involved in the deterioration of organs so, it can cause an irreversible damage to cell membranes, proteins, enzyme activity, the endoplasmic reticulum and nucleic acids, leading to necrosis and cellular death (25).
In a study by Chan-Yeung et al. (26) it was revealed the high γ-GT levels in smokers and in concordance with our study. Whether smoking could impact AST and ALT activity was up for debate. Some researchers claimed that smoking increased ALT (27). Nguyen and Kurtz (28) claimed that smoking only affected γ –GT (28). However, in our study, liver enzymes were found to be deranged among smokers in comparison with non-smokers Oni ET et al. (29) discussed that the serum gamma-glutamyl transferase (GGT) is a marker of oxidative stress, associated with increased cardiovascular (CV) risk. The impact of smoking on oxidative stress may be aggravated in individuals with nonalcoholic fatty liver disease (NAFLD).
Sodium with potassium plays an important role in maintaining the balance of electrolytes and water in the human body (28). According to this study, no substantial variations were observed in serum K+, Na+, and Cl- among the smokers in comparison with non-smokers and agreed with previous research by Jorde et al. (30), who also discussed that the serum level Ca+2 in the smoker's group was significantly low as comparison with the non-smokers. The reason may be related to the control of electrolyte levels, found inside and outside the cells which brings the loss of Ca+2 excess in the urine. Thus, smoking might cause a reduction in the density of bone and immersion of Ca+2 and vitamin D3 (31). We found an increase in serum PO4-3 levels in the smoker’s group which may be explained by the process of complexes found with calcium or irregularity in the secretion of parathyroid hormone (PTH).
To conclude, the results showed that cigarette smoking was correlated with deranged liver enzymes and serum electrolytes. Haematological parameters were also found to be higher in smokers than in non-smokers. The current study provides insight into the potential hazards of smoking, which may contribute to increased mortality and morbidity rates. However, it is important to note that this study identifies correlations but does not establish causation.
There are several limitations to this study, including a crosssectional design that limits the ability to infer causality, potential selection bias, and the reliance on self-reported smoking data, which may be subject to inaccuracies. Additionally, confounding factors such as diet, physical activity, and socioeconomic status were not fully controlled for, which could influence the results. Future longitudinal studies with advanced sampling techniques, such as stratified sampling and the use of biomarkers for smoking exposure, are needed to better understand the causal relationships and underlying mechanisms. Advanced sampling techniques could improve the study by ensuring a more representative sample, reducing bias, and increasing the reliability and validity of the findings. Longitudinal studies would also allow for the observation of changes over time and the potential long-term effects of smoking on health parameters. Smoking is a preventable factor, and public health interventions can be enhanced by arranging awareness sessions among the community, and by implementing measures that restrict smoking at workplaces, public places, and educational institutes. Print and electronic media can play a vital role in the cessation of smoking by disseminating information and promoting anti-smoking campaigns.
ACKNOWLEDGEMENTS
The author is thankful to all the associated personnel who contributed to this study by any means.
FUNDING
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
AUTHOR INFORMATION
Saad Al-Fawaeir, PhD, Assistant Professor Department of Medical Laboratory Science/Pharmacy Faculty, Jadara University, Irbid, Jordan
Correspondence: Saad Al-Fawaeir Email: Alfaouri1@yahoo.com
REFERENCES
1. Ahluwalia IB, Arrazola RA, Zhao L, et al. Tobacco use and tobacco-related behaviors - 11 countries, 2008-2017. MMWR Morb Mortal Wkly Rep. 2019; 68(41): 928–933.
2. Malenica M, Prnjavorac B, Bego T, et al. Effect of cigarette smoking on haematological parameters in healthy population. Med Arch 2017; 71(2): 132–136.
3. St Claire S, Gouda H, Schotte K, et al. Lung health, tobacco, and related products: gaps, challenges, new threats, and suggested research. Am J Physiol Cell Mol Physiol 2020; 318(5): L1004– L1007.
4. Fowles J, Bates M, Noiton D. The chemical constituents in cigarettes and cigarette smoke: priorities for harm reduction. A report to the New Zealand Ministry of Health. March 2000; 1-65. [Available from: https://www.researchgate.net/ publication/265540805]
5. Sinha SK, Haider MN. Tobacco and health effects. TOBACCO & PUBLIC HEALTH: Tobacco and Community Well-being: A Worldwide Health Crisis. 2024; 101.
6. Jain D, Chaudhary P, Varshney N, et al. Tobacco smoking and liver cancer risk: potential avenues for carcinogenesis. J Oncol 2021; 2021: 5905357.
7. Noel Baxter. Why a carbon monoxide test is an essential part of a GP and practice nurse’s kit. Primary Care Respiratory Update. 2016; 3(3): 27–28.
8. Hamza DH, Naji WA. Study of the smoking effect on some liver functions, blood pressure and some haematological parameters in smokers in Al-Muthanna province. Indian J Forensic Med Toxicol 2020; 14(4): 1995.
9. Ream C, Sabitsky M, Huang R, et al. Association of smoking and respiratory disease history with pancreatic pathologies requiring surgical resection. Cancers (Basel) 2023; 15(11): 2935.
10. Shi C, de Wit S, Učambarlić E, et al. Multifactorial diseases of the heart, kidneys, lungs, and liver and incident cancer: epidemiology and shared mechanisms. Cancers (Basel) 2023; 15(3): 729.
11. Abdulkader RT, Kadhim SA. The effect of smoking on liver enzymes and its functions. J Pharm Negat Results 2022; 690–700.
12. Bishop ML. Clinical chemistry: principles, techniques, and correlations, 8th Edition: Principles, techniques, and correlations. (Jones & Bartlett Learning, USA) 2020. ISBN: 9781284238860
13. Shrimanker I, Bhattarai S. Electrolytes. StatPearls [Internet], Treasure Island, (FL); 2024.
14. Pochineni V, Rondon-Berrios H. Electrolyte and acid-base disorders in the renal transplant recipient. Front Med 2018; 5: 261.
15. Nazeer J, Singh R, Suri P, et al. Evaluation of marginal bone loss around dental implants in cigarette smokers and nonsmokers. A comparative study. J Fam Med Prim Care 2020; 9(2): 729–734.
16. Qiu F, Liang CL, Liu H, et al. Impacts of cigarette smoking on immune responsiveness: up and down or upside down? Oncotarget 2017;8 (1): 268.
17. Hassan OK, Qasim MK, Hossein AS, Agha MMM. The effect of smoking and its period on the concentrations of the metals (Zn, Mn, Cu, and Fe), and its relationship with changes in glucose levels, total cholesterol levels, and blood pressure when compared with non-smokers. Int J Enhanc Res Sci Technol Eng 2018; 7(4): 63-67
18. Olié V, Pasquereau A, Assogba FAG, et al. Changes in tobacco-related morbidity and mortality in French women: worrying trends. Eur J Public Health 2020; 30(2): 380–385.
19. Murray CJL, Aravkin AY, Zheng P, et al. Global burden of 87 risk factors in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020; 396(10258): 1223–1249.
20. Qin W, Magnussen CG, Li S, et al. Light cigarette smoking increases risk of all-cause and cause-specific mortality: findings from the NHIS cohort study. Int J Environ Res Public Health 2020; 17(14): 5122.
21. Benowitz NL, Hall SM, Stewart S, et al. Nicotine and carcinogen exposure with smoking of progressively reduced nicotine content cigarette. Cancer Epidemiol Biomarkers Prev 2007; 16(11): 2479-2485.
22. Talaiekhozani A, Amani AM. Preparing emission factors of carbon dioxide, carbon monoxide, hydrocarbons and nitrogen oxides for cigarettes. Journal Air Pollution and Health 2018; 3(4): 219-224.
23. Murkamilov IT, Gordeev IG, Kaliev RR. The role of renal anaemia and cardiovascular disease in the progression of chronic glomerulonephritis. Ter Arkh 2016; 88(12): 57–61.
24. Premkumar M, Anand AC. Tobacco, cigarettes, and the liver: the smoking gun. J Clin Exp Hepatol 2021; 11(6): 700-712.
25. Pérez-Torres I, Manzano-Pech L, Rubio-Ruíz ME, et al. Nitrosative stress and its association with cardiometabolic disorders. Molecules 2020; 25(11): 2555
26. Chan-Yeung M, Ferreira P, Frohlich J, Schulzer M, Tan F. The effects of age, smoking, and alcohol on routine laboratory tests. Am J Clin Pathol 1981; 75(3): 320–326.
27. Whitehead TP, Robinson D, Allaway SL. The effects of New Zealand Journal of
cigarette smoking and alcohol consumption on serum liver enzyme activities: a dose-related study in men. Ann Clin Biochem 1996; 33(6): 530–535.
28. Nguyen MK, Kurtz I. Determinants of plasma water sodium concentration as reflected in the Edelman equation: role of osmotic and gibbs-donnan equilibrium. Am J Physiol Physiol 2004; 286(5): F828–F837.
29.Oni ET, Figueredo V, Aneni E, et al. Non-alcoholic fatty liver disease modifies serum gamma-glutamyl transferase in cigarette smokers. J Clin Med Res 2020; 12(8): 472-482.


30. Jorde R, Saleh F, Figenschau Y, et al. Serum parathyroid hormone (PTH) levels in smokers and non-smokers. the fifth Tromsø study. Eur J Endocrinol 2005; 152(1): 39–45.
31. Nagao Y, Sata M. Serum albumin and mortality risk in a hyperendemic area of HCV infection in Japan. Virol J 2010; 7: 375.
Copyright: © 2024 The author(s). This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.




Save the date!
NZIMLS Annual Scientific Meeting Wednesday 27 - Friday 29 August 2025
Distinction Hotel, Hamilton contact: Andrea Lee andrea.lee@adhb.govt.nz
See you there!
Haiam Abdel Raouf, Rania Fawzy Mahmoud Abdelkawy, Hala T. El-Bassyouni, Shams Kholoussi, Mohammed M. Sayed-Ahmed, Azza E. Abd-Elnaby, Assem M. Abo-Shanab
Background: Prader–Willi Syndrome (PWS) is a rare complex genetic disorder resulting from the loss of expression of paternal genes in the PWS critical region on the chromosome 15 q11-q13. PWS affects multiple body systems. The most consistent major manifestations include hypotonia, mild mental retardation, hypogonadism, growth hormone insufficiency and short stature. Other manifestations involve early childhood onset of hyperphagia and obesity, characteristic craniofacial appearance, immunological, behavioural, and sometimes psychiatric disturbances.
Objectives: The aim of this study is to evaluate cell mediated as well as humoral immunity in children with PWS. Participants and methods: This study included eleven Egyptian children with PWS diagnosed clinically and confirmed by the FISH technique and 25 non-PWS controls matching age and sex. CD3, CD16, CD19, CD4, and CD8 were estimated using flow cytometry. Serum immunoglobulin levels were measured using immunonephelometry. Interleukin 33 (IL-33), Human Leukocyte Antigen G (HLA-G), Procalcitonin and Human Obestatin (OB) were assessed by ELISA. Measurements of T-cell receptor excision circles (TREC) and k-deleting recombination excision circles (KREC) were performed using real-time PCR technique.
Results: The karyotype analysis in all patients was normal, FISH technique showed 15q11.2 deletion in two patients in chromosome 15. The immune cell-count results revealed statistically significant higher total lymphocyte count and lower absolute cytotoxic T lymphocyte (CD8) count in PWS patients compared to controls (p= 0.02405 and 0.01343 respectively). Comparing the immunoglobulin results of the PWS patients with that of control subjects, disclosed statistically significant elevation in IgG and IgM (p=0.0099 and 0.0040 respectively). Furthermore, IL-33 level was significantly increased while procalcitonin was significantly decreased in PWS patients in comparison to controls (p= 0.0009 and 0.0006 respectively). In addition, KREC expression showed statistically significant elevation in PWS patients in comparison to the healthy controls (p= 0.0107).
Conclusion: The present work sheds light on the importance of the implementation of flow cytometric measurement of lymphocyte subsets, cytokines evaluation, and immunoglobulins quantification to elucidate the immunological disturbances in PWS patients. Keywords: Prader-Willi syndrome; children; FISH; cytokines; Immunoglobulins; immune cells.
NZ J Med Lab Sci 2024; 78(3): 131:135
INTRODUCTION
Prader–Willi syndrome (PWS; OMIM #176270) is a complex neurodevelopmental disorder first described in 1956 (1). It is a rare disorder affecting about 1 in 25,000 live births (2). The principal features during infancy are failure to thrive, feeding difficulties, hypogonadism, growth hormone deficiency, and characteristically paternal 15q11-q13 chromosome deletion (3, 4), which results in the loss of transcription of several genes and RNA transcripts as the maternal region of chromosome 15 is inactive due to imprinting (5).
The main clinical manifestations of PWS include characteristic facial features as narrow forehead, almond-shaped eyes and triangular mouth. Infants with PW have small hands and feet, short stature and intellectual impairment (6, 7, 8). Growth hormone insufficiency is common, and replacement therapy improves the child’s growth, body composition, and physical aspects (9). Additionally, hyperphagia leads to obesity that may be dangerous, if not controlled. PWS is accompanied with increased risk of cardiovascular morbidity and mortality. The immune system activation is directly related with endothelial dysfunction, increasing arterial stiffness and the elevated risk for cardiovascular diseases in PWS (2).
The genes encoding the immune-related proteins as cytokines may influence the neurological development and disorders including autism spectrum disorders (ASD) (10, 11), and the mechanisms underlying the association of psychiatric conditions and the activation of the innate immune system in PWS subjects require more studies. It is supposed that lack of expression of paternal alleles from chromosome 15 q11–q13 region has a role in the neurodevelopment of patients. This chromosome deletion principally affects the secretion of the cytokines implicated in cell migration, proliferation and inflammatory processes (12). T cell receptor excision circles (TRECs) and kappa-deleting recombination excision circles (KRECs) are markers for the T and B cell development respectively. They are useful tools to assess T and B lymphocytes function (13). The diseases that are considered having low levels of T and/or B cells show decreased levels of TRECs and KRECs (14).
The aim of this study is to assess cell-mediated and humoral immunity in patients with PWS.
Subjects
This case–control study was conducted on patients referred to the outpatient of the Clinical Genetics Clinic, Medical Research Centre of Excellence, National Research Centre, Cairo, Egypt. Their parents gave written informed consent. The study was approved by the Ethical Committee of the National Research Centre (No. 19267) by the Declaration of Helsinki protocols. The study included two groups: the PWS group (11 children clinically diagnosed as PWS, 5 girls and 6 boys, their mean age is 8.9±3.5), the control group (25 typically developing children matching age and sex, 11 boys and 14 girls, their mean age is 9.3±5.6). All children enrolled in the study were subjected to full history taking, three-generation pedigree construction, and a thorough clinical examination of all body systems to detect any system abnormality. Exclusion criteria for all the participants were type 1 diabetes, the presence of metabolic disease (confirmed by the use of drugs), and pulmonary diseases. Patients were clinically diagnosed and confirmed by routine cytogenetic and fluorescence in-situ hybridization analysis.
5mL of peripheral venous blood samples were aseptically withdrawn from each patient and control and were divided as follows: 3mL were collected in a sterile plain tube. Blood was left to clot and then centrifuged at 4000-5000 rpm for 10 minutes then serum was separated and used for measurement of immunoglobulins (Igs), IL-33, HLA-G, Procalcitonin, and Obestatin. 2mL were divided into two EDTA tubes at a final concentration of 1.5mg/mL as follows: 1mL of blood taken for CBC as well as flow cytometric analysis which was done within 24 hours of collection and the remaining amount was used for DNA extraction which was done simultaneously or on refrigerated samples stored at 2-8°C within one week of collection.
Cytogenetic studies
G-banding chromosomal analysis confirmed by FISH was performed on all patients. Conventional cytogenetics analysis for all patients was applied following the standard protocols (15). Twenty-five metaphases were evaluated for each patient and karyotyped according to the ISCN (ISCN, 2016). FISH was carried out using Prader Willi/Angelman (SNRPN) localized to 15q11.2 labelled in red and 15q26.3 recognized in green as a control probe to allow the clarification of chromosome 15, according to the manufacturer’s instructions.
Determination of Immunoglobulins (IgA, IgM and IgG)
Measurement of serum immunoglobulin was performed by the method of immunonephelometry (16) using (Minineph TM, the Binding Site Ltd, PO Box 11712, Birmingham, B14 4ZB, U.K) as the manufacturer’s protocol.
Flow cytometric assessment of the lymphocyte subsets
Lymphocyte subsets immunophenotyping was done using flow cytometry (BD Accuri™ C6 Cytometer, USA). Analysis of lymphocyte surface markers was done on lysed whole blood using CD3 FITC labelled MoAbs for T lymphocytes, CD16 labelled with PE for NK cells and FITC labelled CD19 for B lymphocytes. CD4 FITC and CD8 FITC (BD Biosciences, USA.) were used for further quantitation of T helper and T cytotoxic lymphocyte populations, respectively in most of the patient samples and all control samples.
Enzyme-linked immunosorbent assay (ELISA)
Serum IL-33, Obestatin, HLA-G, and Procalcitonin levels of all study subjects were determined using Human Procalcitonin ELISA kit (EIAab, Co., Ltd, East Lake Hi-Tech Development Zone, Wuhan, China.) and Human IL-33, Obestatin and HLA-G ELISA kits (NOVA, Beijing, China.) by following the manufacturer’s protocol.
Real-time PCR for TRECs and KRECs measurement
Genomic DNA was isolated from peripheral blood leukocytes by QIAamp DNA Mini Kit (50 preps), Catalogue Number: 51304, Germany (https://www.qiagen.com/eg/). The assay was run on 7500 Fast Real-Time PCR (Applied Biosystems) using the specific primers and probes for TRECs, KRECs, and β-actin. RTPCR was carried out using TaqMan Universal PCR Master Mix II.10μLPCR reaction mix was added to the 5μL DNA samples, 1μL forward primer (Applied Biosystems, USA.), 1μL reverse primer (Applied Biosystems, USA.), 1μLTaqman TAMRA probe and 2μL H2O in a sterile 48-well PCR plate using real-time cycler conditions of initial activation stage at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 15 seconds,
and a combined primer/probe annealing and elongation at 60°C for 1 min. TRECs, KRECs, and β-actin copy number has been obtained by extrapolating the respective sample quantities from the standard curve obtained by serial dilutions of human genomic DNA (Promega, USA.). The initial copy numbers of TRECs, KRECs, and β-actin were calculated from the following equation: "Copies of the gene of interest = mass of gDNA / mass of haploid genome". The number of TRECs and KRECs in each sample was calculated per 1μL of extracted DNA. β-actin copy number was used to judge the successful amplification of each sample.
Data were collected, revised, verified, then edited on a personal computer. Data were then analysed using IBM SPSS ver. 16.0 program. Statistical tests used in the study were: Description of qualitative data was carried out by using frequency and percentage. Description of quantitative data was carried out by using minimum, maximum, mean, and standard deviation for normally distributed results or median and range for skewed results (17). Comparison between two quantitative, nonparametric variables was carried out by using Mann-Whitney’s U-test.
This case–control study included 11 patients clinically diagnosed with suspected PWS and 25 healthy controls matching age and sex. The demographic data and clinical characteristics of PWS patients and healthy controls are presented in Table 1. G-banding showed normal karyotyping for patients and controls. The FISH technique recorded deletion in 2 patients in chromosome 15 in all analysed metaphases and interphases (Table 1) (Figure 1).
There was a significant increase in IgG and IgM serum levels in PWS patients compared to the healthy controls (IgG p=0.0099 and IgM p=0.0040). While no significant differences were found in IgA levels in PWS patients compared to the healthy controls (Table 2).
IL-33 was significantly increased in PWS patients compared to the control group, while procalcitonin was significantly decreased (p= 0.0009 and 0.0006 respectively) as shown in Table 3.
The total leucocytic count was significantly higher in PWS patients compared to control subjects. Whereas absolute cytotoxic T lymphocyte (CD3+CD8+) count was significantly lower in patients with PWS as compared to the controls group (TLC p= 0.0241 and Absolute CD8 p= 0.0134) as shown in Figure 2.
The median values of KREC expression were significantly elevated in PWS patients in comparison to the healthy controls (p= 0.0107). However, no statistical difference was noted in TREC expression between patients and controls (Figure 3).
Table 2. Immunoglobulins levels in patients with Prader Willi compared to controls
Patients
Variables
Results presented as Median (Min-Max), * P< 0.05 significant versus controls.
Table 3. Biomarkers in Prader-Willi patients and healthy controls Variables
Results presented as
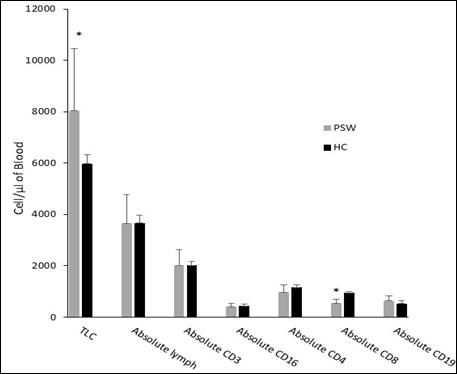
*P< 0.05

*P< 0.05 Significant versus controls.
Prader-Willi syndrome (PWS) is a rare neuro-developmental genomic imprinting disorder characterized by hypotonia, hyperphagia, and developmental delay. Prader-Willi syndrome is caused by a loss of expression for one or more paternally expressed genes in the 15q11.2-q13.1 region (the PWS/AS critical region) (18).
Obesity is a principal feature in patients with PWS arising in early childhood. It is often related with inflammatory processes and co-morbidities as diabetes mellitus, hypertension, cardiovascular disease, strokes and elevated C-reactive protein serum levels (19, 20). Furthermore, Hope et al. (2023) described an association between PWS and the innate immune system activation independent of the central obesity and insulin resistance (21).
Immunoglobulins are vital mediators of the humoral immune system via neutralization, opsonization, complement activation and phagocytosis of the pathogens (22). Many studies described an association between the decreased serum level of immunoglobulins and the disease severity in addition to the unfavourable outcomes in patients with sepsis (23).
In our study, there was a significant increase in IgG and IgM levels in PWS patients compared to the healthy controls (IgG p=0.0099 and IgM p=0.0040 respectively) (Table 2). In contrast, IgG and IgM antibody levels were found to be considerably lower in PWS patients than in a control group of healthy people in a previous investigation (24). Additionally, the study discovered that recurring infections, especially those of the respiratory system, were more common in individuals with PWS. However, Karaman et al. (2020) observed that PWS patients had normal levels of IgA, IgM and IgG (25). Moreover, in this report, no significant differences were found in IgA levels in PWS patients compared to healthy subjects.
Other factors relating to obesity in humans include several intercellular signalling proteins as cytokines which are released from numerous cells regulating the growth and proliferation of neuronal tissue and modulating the host responses to infections, inflammation, and chronic diseases (26, 27, 28).
Regarding the level of cytokines in PWS, the results of our study showed a significant increase of IL-33 in PWS patients as compared to the control group (Table 3). On the other hand (10) established that there were no significant differences in plasma cytokine levels in patients with PWS. Many psychotic events were detected in PWS patients with maternal disomy 15, during young adulthood. Najjar and Pearlman indicated that it may be related to high cytokine levels or neuroinflammatory mechanisms (29).
Previous studies focused on the patients with PWS revealed increased serum levels of inflammatory markers, independent of obesity (12, 30). Subclinical inflammation is frequently associated with several mental, psychotic and mood disorders as well as ASD that are highly prevalent in patients with PWS (12, 31, 32). Moreover, several studies showed that numerous immuneinflammatory markers, such as cytokines, acute phase proteins, and specific and nonspecific autoantibodies are elevated in the peripheral blood of patients with these mental disorders (33, 34). Muller stated that the diagnosis of PWS is a risk factor for several mental disorders and is associated with the activation of immune-inflammatory processes (35).
We measured three inflammatory markers, only procalcitonin showed a significant decrease in PWS patients as compared to the control group. Obestetin and HLA-g levels had no statistically significant differences in PWS patients compared to the control group (Table 3).
Karaman et al. (2020) observed that PWS patients had normal leukocyte counts, lymphocyte counts, and lymphocyte subgroups (CD3, CD4, CD8, CD19, natural killer cells) (25). In another study, Yahya Gul et al. (2022) detected lower percentages of CD16+ 56+ natural killer cells in PWS patients than in the control (24). Our findings indicated that the total leucocyte count was significantly higher in PWS patients compared with control subjects. Whereas absolute cytotoxic T lymphocyte (CD3+CD8+)
count was significantly lower in patients with PWS as compared to the control group (Figure 2). This is believed to be caused by increased apoptosis (cell death) and decreased synthesis of these cells in the bone marrow, which may contribute to their increased susceptibility to infections. There were no statistically significant differences in CD4, CD16, or CD19 in PWS patients than in the control group (Figure 2). Although other studies elucidated that they are lower in Prader Willi patients than in healthy individuals. The discrepancy between our results and others may be due to different sample sizes or different ethnic groups.
The elevated expression of KRECs in PWS patients may contribute to their increased susceptibility to infections. Our research showed that the median values of KREC expression were significantly elevated in PWS patients in comparison to the healthy controls (Figure 3). Xu et al. (2023) reported similar results (36). The reduction in KREC expression may contribute to the chromosomal instability and the dysregulation of gene expression. TRECs are used as a marker of T-cell development and function. In this study, no statistically significant difference was noted in TREC expression between patients and controls (Figure. 3). Butler et al. (2015) reported a decrease in TREC expression in Prader-Willi patients (12).
The present work sheds light on the importance of the establishing of flow cytometric measurements of lymphocyte subsets, cytokines evaluation, and immunoglobulins quantification to elucidate the immunological disturbances in PWS patients. Furthermore, the development of strategies to improve these patients' immune function is recommended.
Ethical Approval
This study was conducted in accordance with the principles outlined in the Declaration of Helsinki. The research protocol and procedures were approved by the Ethics Committee of the National Research Centre (NRC), Cairo, Egypt, under Ethics No. 19267. Informed consent was obtained from the parents of all participants for their inclusion in the study.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Conflicts of Interests
The authors declare there is no conflict of interest.
Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
Availability of Data and Materials
The datasets were generated and analysed during the current study. However, a deidentified version of the dataset may be available from the corresponding author upon reasonable request
Haiam Abdel Raouf, PhD, Professor of Immunogenetics, Head of Immunogenetics Department1
Rania Fawzy Mahmoud Abdelkawy, MD, Assistant Professor of Immunogenetics1
Hala T. El-Bassyouni, MD, PhD, Professor of Clinical Genetics 2 Shams Kholoussi, MD, Professor of Immunogenetics1
Mohammed M. Sayed-Ahmed, MD, Researcher of Clinical Genetics2
Azza E. Abd-Elnaby, PhD, Researcher of Human Cytogenetics3
Assem M. Abo-Shanab, PhD, Assistant Professor of Immunogenetics1
1Immunogenetics Department, Human Genetics and Genome Research Institute, National Research Centre, Cairo, Egypt.
2Clinical Genetics Department, Human Genetics and Genome Research Institute, National Research Centre, Cairo, Egypt.
3Cytogenetics Department, Human Genetics and Genome Research Institute, National Research Centre, Cairo, Egypt.
Correspondence: Mohammed M. Sayed-Ahmed, Clinical Genetics Department, Human Genetics and Genome Research Institute, National Research Centre, Cairo, Egypt.
Email: mohamdouh@hotmail.com
1. Prader A, Labhart A, Willi H. Ein syndrome von adipositas, kleinwuchs, kryptorchismus und oligophrenie nach myatonieartigem zustand im neugeborenenalter. Schweizerishe Med Wochen 1956; 86: 1260–1261. [German]
2. Viardot A, Sze L, Purtell L, et al. Prader–willi syndrome is associated with activation of the innate immune system independently of central adiposity and insulin resistance. J Clin Endocrinol Metab 2010; 95: 3392–3399.
3. Butler MG, Palmer CG. Parental origin of chromosome 15 deletion in prader–willi syndrome. Lancet 1983; 1(18336): 1285-1286.
4. Butler MG. Prader-willi syndrome and chromosome 15q11.2 BP1-BP2 Region: a review. Int J Mol Sci 2023; 24(5): 4271
5. Chung MS, Langouët M, Chamberlain SJ, Carmichael GC. Prader-willi syndrome: reflections on seminal studies and future therapies. Open Biol. 2020 Sep;10(9): 200195. doi: 10.1098/rsob.200195.
6. Driscoll DJ, Miller JL, Schwartz S, Cassidy SB. Praderwilli Syndrome. In: Adam MP (Eds.) et. al. GeneReviews® University of Washington, Seattle, 1993.
7. Cassidy SB. Prader-willi syndrome. J Med Genet 1997; 34: 917–923.
8. Ramsden SC, Clayton-Smith J, Birch R, Buiting K. Practice guidelines for the molecular analysis of prader–willi and angelman syndromes. BMC Med Genet 2010; 11:70. Available from: https://doi.org/10.1186/1471-2350-11-70.
9. Cassidy SB, Schwartz S., Miller JL, Driscoll DJ. Prader-willi syndrome. Genet Med 2012; 14(1): 10–26.
10. Butler MG, Lee PDK, Whitman BY. Management of prader–willi syndrome. 3rd Ed New York, NY: Springer-Verlag Publishers, 2006: 1–550.
11. Dykens EM, Lee E, Roof E. Prader–willi syndrome and autism spectrum disorders: an evolving story. J Neurodev Disord 2011; 3(3): 225–237.
12. Butler MG, Hossain W, Sulsona C, at al. Increased plasma chemokine levels in children with prader-willi syndrome. Am J Med Genet A 2015; 167A(3): 563–571.
13. Dasouki M, Jabr A, AlDakheel G, et al. TREC and KREC profiling as a representative of thymus and bone marrow output in patients with various inborn errors of immunity. Clin Exp Immunol 2020; 202(1): 60–71.
14. Thomas C, Durand-Zaleski I, Frenkiel J, et al. Clinical and economic aspects of newborn screening for severe combined immunodeficiency: DEPISTREC study results. Clin Immunol 2019; 202: 33–39.
15. Verma RS, Babu A. Human chromosomes principles and techniques. 2ed edn New York: McGraw-Hill, 1995.
16. Aksu G, Genel F, Koturoğlu G, et al. Serum immunoglobulin (IgG, IgM). Turk J Pediatr 2006; 48(1): 19-24.
17. Sottini A, Ghidini C, Zanotti C, et al. Simultaneous quantification of recent thymic T-cell and bone marrow B-cell emigrants in patients with primary immunodeficiency undergone to stem cell transplantation. Clin Immunol 2010; 136(2): 217-227.
18. Butler MG, Miller JL, Forster JL. Prader-Willi Syndromeclinical genetics, diagnosis and treatment approaches: an update. Curr Pediatr Rev 2019; 15(4): 207-244.
19. Butler MG, Bittel DC, Kibiryeva N, Garg U. C-reactive protein levels in subjects with prader–willi syndrome and obesity. Genet Med 2006; 8: 243–248.
20. El-Bassyouni HT, Hassan N, Mahfouz I, et al. Early detection and management of prader-willi syndrome in Egyptian patients. J Pediatr Genet 2019; 8:179–186.
21. Hope S, Nærland T, Kolset SO, et al. Systemic immune profile in prader-willi syndrome: elevated matrix metalloproteinase and myeloperoxidase and reduced macrophage inhibitory factor. Orphanet J Rare Dis 2023; 18(1): 185.
22. Kholoussi S, Kholoussi N, ElNady H, et al. Immunological markers in children with genetic disorders and recurrent respiratory tract infections. Open Access Maced J Med Sci 2020 Feb 05; 8(B):104-108.
23. Bermejo-Martin JF, Giamarellos-Bourboulis EJ. Endogenous immunoglobulins and sepsis: new perspectives or guiding replacement therapies. Int J Antimicrob Agents 2015; 46(Suppl1): S25-28.
24. Gul Y, Kapaklı H, Aytekin SE, et al. Evaluation of immunological abnormalities in patients with rare syndromes. Cent Eur J Immunol 2022; 47(4): 299-307.
25. Karaman S, Hazan F, Erdem SB, et al. Do microdeletions lead to immune deficiency? Cent Eur J Immunol 2020; 45(1): 69-72.
26. Skogstrand K, Thorsen P, Norgaard-Pedersen B, et al. Simultaneous measurement of 25 inflammatory markers and neurotrophins in neonatal dried blood spots by immunoassay with xMAP technology. Clin Chem 2005; 51:1854–1866.
27. Parmely MJ. USMLE Road Map: Immunology. NewYork, NY: Lange Medical Books/ McGraw-Hill Companies, 2006.
28. Salah EM, El-Bassyouni HT, Kholoussi S, et al. Behavioral problems, biochemical, and anthropometric characteristics of patients with Prader–Willi syndrome. Middle East J Med Genet 2015; 4: 63–69.
29. Najjar S, Pearlman DM. Neuroinflammation and white matter pathology in schizophrenia: systematic review. Schizophr Res 2015; 161(1): 1117–1118.
30. Thomas MM, Zaki ME, Youness E, et al. Measurement of serum chemerin, oxidized LDL, and vitamin D levels in prader–willi syndrome: a cross-sectional study in pediatric Egyptian patients. Journal of Child Science 2020; 10: e187–e195.
31. D’Acunto G, Nageye F, Zhang J, et al. Inflammatory cytokines in children and adolescents with depressive disorders: a systematic review and meta-analysis. J Child Adolesc Psychopharmacol 2019; 29(5): 362–369.
32. Saghazadeh A, Ataeinia B, Keynejad K, et al. A metaanalysis of proinflammatory cytokines in autism spectrum disorders: Effects of age, gender, and latitude. J Psychiatr Res 2019; 115: 90–102.
33. Misiak B, Frydecka D, Stanczykiewicz B, Samochowiec J. Editorial: peripheral markers of immune response in major psychiatric disorders: where are we now and where do we want to be? Front Psychiatry 2019; 10: 5. Available from: https://doi.org/10.3389/fpsyt.2019.00005.
34. Ormstad H, Bryn V, Saugstad OD, et al. Role of the immune system in autism spectrum disorders (ASD). CNS Neurol Disord Drug Targets 2018; 17(7): 489–495.
35. Muller N. COX-2 inhibitors, aspirin, and other potential anti-inflammatory treatments for psychiatric disorders. Front Psychiatry 2019; 10:375. Available from: https://doi. org/10.3389/fpsyt .2019.00375.
36. Xu Y, Hou X, Guo H, et al. CD16+ monocytes are involved in the hyper-inflammatory state of prader-willi syndrome by single-cell transcriptomic analysis. Front Immunol 2023; 14: 1153730.
Copyright: © 2024 The author(s). This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Dina Morsy A. Mohamed and Raghda M. Ghorab
Objectives:"Non-alcoholicfattyliverdisease"(NAFLD)isaveryimportantcommonhealthissue,causingobesityrelatedcomorbidity, the objective of this study was to explore the relation between adiponectin levels and insulin resistance in obese patients with simple steatosis versus those who developed non-alcoholic steatohepatitis (NASH).
Methods: Serum adiponectin and insulin levels were measured in 60 obese patients: 30 patients with non-alcoholic steatohepatitis (NASH) and 30 patients with simple steatosis. Their levels were compared to each other and correlated with clinical data, abdominal ultrasonography, fibrosis score, virology and immunological data.
Results: Patients with non-alcoholic steatohepatitis (NASH) showed highly significant increase in aspartate aminotransferase, (AST), alanine aminotransferase (ALT), total and direct bilirubin, ferritin, insulin level, and insulin resistance detected by Homeostatic Model Assessment for Insulin Resistance (HOMA-IR) compared to patients with simple steatosis (p<0.01). Fasting blood sugar (FBS) was highly significantly decreased in non-alcoholic steatohepatitis (NASH) in comparison to simple steatosis (p<0.01). Adiponectin was highly significantly decreased in non-alcoholic steatohepatitis (NASH) group in comparison to simple steatosis group (p<0.01).Asignificant positive correlation was observed between insulin level and Homeostatic ModelAssessment for Insulin Resistance (HOMA-IR) (p<0.01), while a significant negative correlation was found between adiponectin and insulin level (P=0.016). Furthermore, a highly significant negative correlation was observed between adiponectin and (HOMA-IR) (p<0.01).
Conclusion: There is an important relationship between serum adiponectin and insulin resistance in obese patients with NASH in comparison to simple steatosis patients, which reflects the importance of serum adiponectin as a marker for disease stratification and as potential therapeutic target in treatment of obesity complicated with NASH.
Keywords: Serum adiponectin level, insulin resistance, non-alcoholic steatohepatitis (NASH), non-alcoholic fatty liver disease (NAFLD), Homeostatic Model Assessment for Insulin Resistance (HOMA-IR), insulin level.
NZ J Med Lab Sci 2024; 78(3): 136:141
“Non-alcoholic fatty liver disease” (NAFLD) occurs due to fat deposition and infiltration in the liver in absence of other factors causing steatosis like alcohol consumption, hereditary disorders or use of steatogenic medications (1). The condition is often associated with metabolic risk factors such as obesity, diabetes, and dyslipidaemia (2). In defining simple steatosis, it refers to the presence of hepatic steatosis without indication of hepatocellular injury in the form of ballooning of the hepatocytes (3). Nonalcoholic steatohepatitis (NASH) is a form of NAFLD in which steatosis is associated with liver cell injury, fibrosis and tissue inflammation (1).
Obesity is considered a serious public health problem worldwide that is associated with substantial social and psychological consequences being recently increasing among younger age groups (4). It is frequently associated with insulin resistance, a condition that exacerbates liver inflammation, thereby accelerating the transition from simple steatosis to NASH (5). Several adipocytokines such as adiponectin, tumour necrosis factor-alpha (TNF-α), resistin and interleukins, were implicated in the pathogenesis of insulin resistance (6). Along with their effect on insulin sensitivity, adipokines have a significant influence on body fat mass and distribution, modulating body weight. They also affect appetite, blood pressure and several physiological processes as glucose homeostasis, inflammation, and liver function (7, 8). Consequently, various pathological conditions result from the imbalance in production of adipokines from adipose tissue such as hyperlipidaemia, hypertension, atherosclerosis, metabolic syndrome, and diabetes mellitus along with other disorders (9). These effects could be assessed through measurement of various analytes such as fasting blood sugar (FBS), lipid profile, liver enzymes and liver function tests (5, 10). Alternately, visceral fat accumulation also has an important impact on adipokine secretion profile (11).
Liver biopsy represents the gold standard for the definitive differentiation between simple steatosis and NASH (12). However, several non-invasive measures such as imaging techniques (e.g., FibroScan), serum biomarkers and scoring systems have emerged as reliable tools for diagnosing and staging NAFLD without the need for invasive procedures like liver biopsy (10, 13). These approaches offer a safer and more accessible means
of evaluating liver health, facilitating timely intervention and personalized management strategies for patients with NAFLD.
Scoring systems that have been developed to evaluate fibrosis include NAFLD fibrosis score (NFS score), fibrosis‐4 score (FIB-4), enhanced liver fibrosis score, BARD score, and European liver fibrosis, all aimed at predicting advanced fibrosis, thus minimizing the necessity for liver biopsy in most patients (14, 15). The regulatory role of adiponectin on the mechanism of insulin resistance, obesity and atherosclerosis has been previously discussed and investigated in many studies, however few studies have explored its relation to the pathogenesis and staging of NAFLD in obese patients.
The aim of this work was to study the relationship between adiponectin and insulin resistance in obese patients with simple steatosis versus those who developed NASH, as judged by fibrosis scores, and to investigate their role as markers for disease stratification and as potential therapeutic targets.
The present study included 60 obese patients who visited the outpatient clinic of Ain Shams University hospital whose age ranged from thirty to sixty years. Patients were categorized into two groups that included 30 patients in each. Group 1 included patients with simple steatosis diagnosed by abdominal ultrasonography with exclusion of fibrosis by FIB4 and NFS score. Group 2 had patients with NASH diagnosed by abdominal ultrasonography and elevated liver enzymes after exclusion of other causes that elevate liver enzymes. All patients underwent a complete medical history review, comprehensive clinical examination, abdominal ultrasonography, and laboratory investigations as part of the evaluation which included: liver enzymes (aspartate aminotransferase AST, alanine aminotransferase ALT), liver function tests (serum albumin, prothrombin time), serum bilirubin, total protein, alkaline phosphatase, serum creatinine, complete blood count, FBS, lipid profile, viral markers (HbsAg, HCV Ab), antinuclear antibody (ANA), anti-smooth muscle antibody (ASMA), serum ferritin, and serum ceruloplasmin. Serum adiponectin and serum insulin levels were determined by enzyme linked immunosorbent assay (ELISA) (Bioassay Technology Laboratory). “Homeostasis
model assessment for insulin resistance (HOMA-IR)” was done for all patients to assess insulin resistance (16). Patients with history of alcohol intake (above 20 g per day for females and 30 g per day for males), history of use of medications known to trigger steatohepatitis (e.g. valproate, amiodarone, or prednisone), viral hepatitis (hepatitis B, or C), metabolic causes of steatohepatitis e.g. haemochromatosis, Wilson disease and autoimmune liver disease were excluded from the study. Exclusion of advanced fibrosis in patients with simple steatosis was performed through two fibrosis scores: FIB4 and NFS. In addition, NFS score was calculated for all patients with NASH and accordingly, patients were classified as low fibrosis or intermediate/high fibrosis. The cut off used in NFS score for categorizing the likelihood of fibrosis was delineated as follows: less than -1.45 indicated a low probability, between -1.45 and 0.67 indicated an intermediate probability, and greater than 0.67 indicated a high probability (17).
All procedures performed were in accordance with the ethical standards of the Ethics Committee of Ain Shams University (approval number FMASU R 169/2023). Informed consent was obtained from all patients for being included in the study.
The data entry, processing, and statistical analysis were conducted using “MedCalc version 20 (MedCalc, Ostend, Belgium)”. Various statistical tests such as Mann-Whitney’s test, Chi-square test, multiple regression analysis, Spearman’s correlation and receiver operating characteristic curve (ROC curve) analysis were employed to determine significance. The presentation and analysis of data were tailored to the type of data found for each variable, whether parametric or non-parametric data. Statistical significance was defined as a p-value less than 0.05 (5%), while values below 0.01 were considered highly significant (HS). Parametric numerical data were expressed as mean, standard deviation (± SD) and range, whereas non-parametric numerical data were represented by median and interquartile range (IQR).
Frequency and percentage were employed for non-numerical data. Non-parametric quantitative variables were compared using Mann-Whitney’s test (U test), while qualitative variables were analysed using the Chi-square test. Correlation analysis employed Spearman’s method. Multiple linear regression was employed to assess the relationship between a quantitative variable and a set of independent variables. ROC curve evaluated the sensitivity and specificity of diagnostic measures categorizing cases into two groups.
This study included 60 obese patients who were categorized into 2 groups: 30 patients diagnosed with simple steatosis and comparably 30 patients with NASH. The patients had a mean age of (41 ± 8.8) years and a mean BMI of (34 ± 4.7). Regarding patients’ gender, most patients (63.3%) were males, while females accounted for (36.7%) of the total. Patients were matching for age and sex in both groups as indicated by lack of significant difference in those variables. BMI was highly significantly increased in NASH group compared to simple steatosis group (p<0.01) (Supplementary Table 1).
The laboratory parameters AST, ALT, ferritin, total and direct bilirubin showed highly significant increase in NASH vs. simple steatosis group (p<0.01). FBS was highly significantly decreased in NASH in comparison to simple steatosis (p<0.01) (Table 1). Comparative statistics for the tested markers showed that adiponectin was highly significantly decreased in NASH group when compared to simple steatosis group (p<0.01) (Table 1). Insulin level and HOMA-IR showed highly significant increase in NASH vs. simple steatosis (p<0.01) (Table 1). Serum ceruloplasmin levels were within the reference range for all patients, and ANA and ASMA tests were negative. Correlation studies showed a highly significant negative correlation between platelet count and FIB-4 score (p<0.001, r = -0.74), and between platelets and NFS score (p = <0.001, r = -0.6).
(μmol/L)
Tested Parameters
(μg/mL)
(71 –
(17 – 22)
(19 – 30)
(5.1 – 6.8)
(65 – 90)
(8 – 17)
(7 – 22)
(0.3 – 1)
Hb: haemoglobin, PLT: Platelet count, TLC: Total leucocyte count, T. Cholesterol: Total cholesterol, TGs: Triglycerides, FBS: Fasting blood sugar, Creat.: Serum creatinine, AST: Aspartate aminotransferase, ALT: Alanine aminotransferase, T. Bil.: Total bilirubin, D. Bil.: Direct bilirubin, Alb.: Serum albumin, INR: International normalised ratio, PT: Prothrombin time Statistical significance p-value < 0.05*, p-values < 0.01** were considered highly significant.
New Zealand Journal of Medical Laboratory Science
There were no significant correlations between adiponectin and fibrosis scores; either FIB-4 or NFS scores (Table 2). No significant correlation was found between baseline clinical factors namely age and sex with any of tested parameters (adiponectin, insulin level and HOMA-IR). Results of Spearman’s correlation analysis revealed a significant negative correlation between adiponectin and insulin level as well as HOMA-IR (p<0.01). Additionally, a significant positive correlation was observed between insulin level and HOMA-IR (p<0.01) (Table 2).
Multiple regression model for factors affecting adiponectin level showed that the decrease in ALT and direct bilirubin had an independent effect on increasing adiponectin level; with significant difference (p=0.027, p=0.009; respectively) (Supplementary Table 2). Multiple regression model for the factors affecting Insulin level showed that the increase in ALT and Albumin had an independent effect on increasing Insulin level; with significant
difference (p=0.012, p=0.01; respectively) (Supplementary Table 3). Multiple regression model for the factors affecting HOMAIR showed that the decrease in haemoglobin, platelets and FBS; had an independent effect on increasing HOMA-IR; with highly significant difference (p<0.01). The increase in ALT had also an independent effect on increasing HOMA-IR; with highly significant difference (p<0.01) (Supplementary Table 4).
According to ROC curve analysis, adiponectin demonstrated good accuracy (80%) in predicting patients with NASH using a cutoff point of ≤9.1, with a sensitivity of 90% and specificity of 70% (p<0.01). Insulin levels at a cutoff point of >3.8 showed fair accuracy (77%) in predicting NASH patients. Its sensitivity was 70% and specificity was 90% (p<0.01). Additionally, HOMA-IR, with a cutoff point of >1.1, displayed good accuracy (81%) in predicting NASH patients, with a sensitivity of 70% and specificity of 96% (p<0.01) (Table 3).
Table 2: Correlation analysis for adiponectin, insulin level, HOMA-IR and fibrosis scores
r: Spearman's rho (correlation coefficient), NFS score: NAFLD fibrosis score, PLTs: platelets. Statistical significance p-value < 0.05*, p-values < 0.01** were considered highly significant.
Table 3: ROC-curve of tested parameters to predict patients with NASH.
ROC (Receiver operating characteristic), AUC= Area under curve, SE= Standard Error. Statistical significance p-value < 0.05*, p-values < 0.01** were considered highly significant.
See Supplementary Tables at https://www.nzimls.org.nz/current-journal
NAFLD has emerged as a significant public health concern globally, affecting millions of individuals worldwide, becoming one of the most prevalent liver pathologies. The disease spectrum ranges from simple steatosis to NASH, which can further progress to cirrhosis and hepatocellular carcinoma (18). In developed countries, NAFLD affects up to 30% of adults and up to 10% of children (19). Its prevalence is rapidly increasing worldwide to the extent that it is predicted to emerge as the primary indication for liver transplantation in the future (20).
Insulin resistance is one of the hallmarks of NAFLD being fundamental in its pathogenesis as it is closely connected to obesity (21). Moreover, it is one of the multiple events determining the progress from simple steatosis to NASH (22). It describes the metabolic defect in the action of insulin hormone on target cells (e.g., muscle cells, hepatocytes, adipocytes, etc.) or at whole body (23). Additionally, overweight and obesity represent states of chronic low-grade inflammation which cause imbalance in the production of adipokines. Yet, the mechanisms underlying NAFLD remain to be fully explained. In this work, we conduct a comparative cross-sectional study on 60 obese patients, 30 patients with simple steatosis and 30 patients with NASH as judged by clinical, imaging, fibrosis scores and other laboratory parameters, with assessment of serum adiponectin, insulin level and HOMA-IR and correlation of data with clinical and laboratory findings. The patients had a mean age of (41±8.8) years and a mean BMI of (34±4.7). Most patients (63.3%) were males, while females accounted for (36.7%).
Analysis of the demographic data of the studied groups showed that there was no significant difference as regards age and sex between patients of the 2 studied groups. (p > 0.05).
A highly significant increase in BMI was seen in NASH group compared to simple steatosis group (p<0.01). This is consistent with Loomis et al. (2016) who observed significant and evident nearly linear correlation between BMI and the subsequent risk of developing simple steatosis/NASH, indicating a 5-10 times higher risk in obese individuals and a 10-14 times higher risk in severely obese individuals. This underscores the crucial role of preventing weight gain and implementing weight reduction strategies in the prevention and treatment of NAFLD (4).
In this study, BMI and gender did not have a significant correlation with either adiponectin or HOMA-IR (p > 0.05). This finding contrasts with previous studies that have reported a connection between the low levels of anti-inflammatory adipokines like adiponectin and obesity. Obesity initiates a persistent state of mild inflammation, which contributes to the onset of conditions such as insulin resistance state, type2 diabetes, atherosclerosis, hypertension, cardiovascular diseases, and certain types of cancer (24, 25). This discrepancy could be settled through studies with larger number of patients to confirm this association.
In the “Third National Health and Nutrition Examination Survey”, the prevalence of NAFLD, determined through ultrasound, was found to be approximately 4-8 times higher in obese individuals compared to those with normal weight across various ethnicities Moreover, this prevalence was observed to be higher in men than New Zealand
women and higher in individuals with diabetes (26). In this study, although most patients were males (63.3%); while (36.7%) were females, the correlations between sex and serum adiponectin level and HOMA-IR were non-significant (p > 0.05 respectively). Comparisons between laboratory parameters (Table 1) showed highly significant increases in AST, ALT, ferritin, total and direct bilirubin in NASH group in comparison to simple steatosis group (p<0.01). Further studies reported that fatty liver resulting either from alcohol consumption or NAFLD stands as the main cause of mild elevation in aminotransferases. As per data from the “National Health and Nutritional Survey”, approximately 23% of American adults exhibit this elevation in point-prevalence (27). Our findings regarding direct bilirubin are consistent with Chang et al. (2012) who observed the presence of an inverse correlation between the level of direct bilirubin and the incidence of NAFLD in a prospective cohort (28). Bilirubin levels were higher in NASH patients compared to simple steatosis patients. This was further confirmed by other studies which showed that bilirubin exhibited repressive effects on NAFLD development (29, 30).
The significant difference in ferritin levels among simple steatosis and NASH groups in this study was in consistent with many previous studies which proposed that hyperferritinemia can predict the risk of NASH with 92% sensitivity and 80% specificity as well as severe fibrosis (31, 32).
Our study revealed a highly significant decrease in FBS in NASH group when compared to simple steatosis group (p < 0.05) unlike previous study which did not find correlation between FBS and either total NAFLD activity scores or the staging of NASH. However, it was found that serum insulin level was correlated significantly with both (33).
Insulin level and HOMA-IR were significantly increased in NASH group in current study (p<0.01). This is consistent with findings of previous study which concluded that insulin resistance state has a critical role in the progression of NAFLD (34). Results of other studies had demonstrated that NAFLD has a strong significant correlation with insulin resistance as 70–80% of obese and diabetic patients included in these studies had NAFLD (35, 36).
Adiponectin is an adipokine with targeted secretion that plays a crucial role in regulating fatty acid oxidation and preventing lipid buildup in both adipose tissue and the liver (37). Additionally, it promotes glucose homeostasis by preserving insulin sensitivity in the liver (38). Our study revealed highly significant decrease in adiponectin, in NASH group compared to simple steatosis group (p < 0.01). This is consistent with previous studies which showed that serum adiponectin levels were lower in patients with NASH than those with simple steatosis (39). The same findings were documented by Bugianesi et al. (2005) and Pagano et al. (2005) who concluded that hypoadiponectinemia has a role in the pathogenesis of NAFLD and type 2 diabetes through impairing metabolism of fatty acids. It also promotes a state of chronic inflammation in the liver (40, 41). Another meta-analysis reported that total serum adiponectin was higher in controls than patients with hepatic steatosis or NASH (42). These data suggest that the maintenance of adiponectin levels may have a role in prevention of development of NASH by inhibiting the development of inflammation and fibrosis.
The current study demonstrated a highly significant negative correlation between platelet count and fibrosis scores either FIB-4 score or NFS score. These findings were consistent with previous studies which suggested that platelets have an important role in the process of liver fibrosis (43). Platelet count is included in many prognostic scores for liver fibrosis and cirrhosis (44). Regarding fibrosis scores, FIB-4 and NFS score, there was no significant correlation between adiponectin and fibrosis scores unlike other studies whose results showed a statistically significant negative correlation between adiponectin and grade of liver fibrosis (45, 46). However, our finding goes in line with Lucero et al. (2017) whose results did not show a
significant correlation between adiponectin and the degree of liver fibrosis (47).
Our study showed that the decrease in ALT and direct bilirubin had an independent effect on increasing adiponectin level; with significant statistical difference (p<0.05). A prior study which was done on healthy individuals showed the same results and found that adiponectin levels were negatively correlated with ALT and gamma-glutamyl transferase levels (48). On the other hand, another study did not find a significant correlation between serum adiponectin levels and ALT elevation in the study groups which included patients with hepatic steatosis, NASH and patients with hepatocellular carcinoma (49). Furthermore, results of another study showed that adiponectin was positively correlated with serum bilirubin levels in asymptomatic middle-aged people (50). This discrepancy among results may be because the later study was done in asymptomatic, non-smoking, middle-aged individuals with stable body weight, while our study was conducted on obese patients only. However, several other studies that were conducted in patients with liver cell injury such as cirrhosis and cholestasis affirmed our observed findings (50-52).
Results of the current study have shown that the increase in ALT and Albumin had an independent effect on increasing Insulin level; with significant statistical difference (p<0.05). The increase in ALT also had an independent effect on increasing HOMAIR; with significant statistical difference (p<0.05). In agreement with our finding regarding ALT, Sheng et al. (2018) had the same observation that showed that HOMA-IR increases as the levels of liver enzymes increase, and each enzyme had shown a significant association with HOMA-IR (33).The same results were previously found by Bonnet et al. (2011) who mentioned that increased ALT can be considered as a biomarker of both systemic and hepatic insulin resistance with associated increase in insulin secretion and decreased hepatic insulin clearance (53).
In the present study, serum adiponectin was found to be a promising biomarker target in the treatment of obesity complicated with NASH as there were highly significant negative correlations between serum adiponectin and each of Insulin level and HOMA-IR (p<0.01). Similarly, other studies demonstrated a negative association between adiponectin levels and HOMA-IR (54). A previous study concluded that adiponectin is the most common adipokine inversely correlated with insulin resistance, lipid accumulation, inflammation and NAFLD (55). Moreover, another study showed that adiponectin administration in humans and rodents had multiple effects like insulin-sensitization, anti-atherogenic, anti-inflammatory effects and in certain circumstances can decrease body weight. This could suggest that adiponectin replacement therapy in humans may have multiple potential therapeutic applications in the treatment of obesity, atherosclerosis insulin resistance/type 2 diabetes (56).
There is an important relationship between serum adiponectin and insulin resistance in obese patients with NASH in comparison to simple steatosis patients, which reflect the importance of serum adiponectin as a potential therapeutic target in treatment of obesity complicated with NASH.
ACKNOWLEDGEMENTS
The authors thank all patients who participated in this study.
Dina Morsy A. Mohamed, MD, MBBch, MSc, Lecturer 1 Raghda M. Ghorab, MD, MBBch, MSc, Researcher of Immunogenetics 2
1 Internal Medicine Department, Faculty of Medicine, Ain Shams University, E-mail: drdinam@yahoo.com
2 Immunogenetics Department, Human Genetics and Genome Research Institute, National Research Centre, Cairo, Egypt.
Correspondence: Raghda Mohammed Ghorab, Human Genetics and Genome Research Institute, National Research Centre, Cairo, Egypt.
E-mail: raghdaghorab@hotmail.com
1. Wilkins T, Tadkod A, Hepburn I, Schade RR. Non-alcoholic fatty liver disease: diagnosis and management. Am Fam Physician 2013; 88(1): 35-42.
2. Chalasani N, Younossi Z, Lavine JE, et al. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American association for the study of liver diseases, American college of gastroenterology, and the American gastroenterological association. Hepatology 2012; 55(6): 2005-2023.
3. Brunt EM, Janney CG, Di Bisceglie AM, et al. Non-alcoholic steatohepatitis: a proposal for grading and staging the histological lesions. Am J Gastroenterol 1999; 94(9): 24672474.
4. Loomis AK, Kabadi S, Preiss D, et al. Body mass index and risk of non-alcoholic fatty liver disease: two electronic health record prospective studies. J Clin Endocrinol Metab 2016; 101(3): 945-952.
5. Cetin EG, Demir N, Sen I. The relationship between insulin resistance and liver damage in non-alcoholic fatty liver patients. Sisli Etfal Hastan Tip Bul 2020; 54(4): 411-415.
6. Iwabu M, Yamauchi T, Okada-Iwabu M, et al. Adiponectin and AdipoR1 regulate PGC-1alpha and mitochondria by Ca(2+) and AMPK/SIRT1. Nature 2010; 464(7293): 13131319.
7. Mancuso P. The role of adipokines in chronic inflammation. Immunotargets Ther 2016; 5: 47-56.
8. Weschenfelder C, Schaan de Quadros A, Lorenzon Dos Santos J, et al. Adipokines and adipose tissue-related metabolites, nuts and cardiovascular disease. Metabolites 2020; 10(1): 32.
9. Salha T, Andrijevic D, Vrselja Z, et al. Chemerin blood levels are associated with MRI measured volumes of abdominal Adipose tissue compartments and lifestyle choices. Acta Clin Croat 2017; 56(4): 663-672.
10. Li G, Zhang X, Lin H, et al. Non-invasive tests of nonalcoholic fatty liver disease. Chin Med J (Engl) 2022; 135(5): 532-546.
11. Coelho M, Oliveira T, Fernandes R. Biochemistry of adipose tissue: an endocrine organ. Arch Med Sci 2013; 9(2): 191200.
12. Spengler EK, Loomba R. Recommendations for diagnosis, referral for liver biopsy, and treatment of nonalcoholic fatty liver disease and non-alcoholic steatohepatitis. Mayo Clin Proc 2015; 90(9): 1233-1246.
13. Piazzolla VA, Mangia A. Non-invasive diagnosis of NAFLD and NASH. Cells 2020; 9(4): 1005.
14. Zain SM, Tan HL, Mohamed Z, et al. Use of simple scoring systems for a public health approach in the management of non-alcoholic fatty liver disease patients. JGH Open 2020; 4(6): 1155-1161.
15. Cox B, Trasolini R, Galts C, et al. Comparing the performance of fibrosis-4 and non-alcoholic fatty liver disease fibrosis score with transient elastography scores of people with non-alcoholic fatty liver disease. Can Liver J 2021; 4(3): 275-282.
16. de Cassia da Silva C, Zambon MP, Vasques ACJ, et al. The threshold value for identifying insulin resistance (HOMAIR) in an admixed adolescent population: a hyperglycemic clamp validated study. Arch Endocrinol Metab 2023; 67(1): 119-125.
17. Angulo P, Hui JM, Marchesini G, et al. The NAFLD fibrosis score: a noninvasive system that identifies liver fibrosis in patients with NAFLD. Hepatology 2007; 45(4): 846-854.
18. Ahmed A, Wong RJ, Harrison SA. Non-alcoholic fatty liver
disease review: diagnosis, treatment, and outcomes. Clin Gastroenterol Hepatol 2015; 13(12): 2062-2070.
19. Tilg H, Moschen AR, Roden M. NAFLD and diabetes mellitus. Nat Rev Gastroenterol Hepatol 2017; 14(1): 3242.
20. Wong RJ, Aguilar M, Cheung R, et al. Non-alcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology 2015; 148(3): 547-555.
21. Manco M. Metabolic syndrome in childhood from impaired carbohydrate metabolism to non-alcoholic fatty liver disease. J Am Coll Nutr 2011; 30(5): 295-303.
22. Buzzetti E, Pinzani M, Tsochatzis EA. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD). Metabolism 2016; 65(8): 1038-1048.
23. Petersen MC, Shulman GI. Roles of diacylglycerols and ceramides in hepatic insulin resistance. Trends Pharmacol Sci 2017; 38(7): 649-665.
24. Liu C, Feng X, Li Q, et al. Adiponectin, TNF-alpha and inflammatory cytokines and risk of type 2 diabetes: A systematic review and meta-analysis. Cytokine 2016; 86: 100-109.
25. Hotamisligil GS. Foundations of immunometabolism and implications for metabolic health and disease. Immunity 2017; 47(3): 406-420.
26. Schneider AL, Lazo M, Selvin E, Clark JM. Racial differences in non-alcoholic fatty liver disease in the US population. Obesity (Silver Spring) 2014; 22(1): 292-299.
27. Harrison SA, Kadakia S, Lang KA, Schenker S. Nonalcoholic steatohepatitis: what we know in the new millennium. Am J Gastroenterol 2002; 97(11): 2714-2724.
28. Chang Y, Ryu S, Zhang Y, et al. A cohort study of serum bilirubin levels and incident non-alcoholic fatty liver disease in middle aged Korean workers. PLoS One 2012; 7(5): e37241.
29. Ucar F, Sezer S, Erdogan S, et al. The relationship between oxidative stress and non-alcoholic fatty liver disease: its effects on the development of non-alcoholic steatohepatitis. Redox Rep 2013; 18(4): 127-133.
30. Kwak MS, Kim D, Chung GE, et al. Serum bilirubin levels are inversely associated with non-alcoholic fatty liver disease. Clin Mol Hepatol 2012; 18(4): 383-390.
31. Ibrahim MA, Kelleni M, Geddawy A. Non-alcoholic fatty liver disease: current and potential therapies. Life Sci 2013; 92(2): 114-118.
32. Nakamura A, Yoneda M, Fujita K, et al. Impact of glucose tolerance on the severity of non-alcoholic steatohepatitis. J Diabetes Investig 2011; 2(6): 483-489.
33. Sheng X, Che H, Ji Q, et al. The Relationship Between Liver Enzymes and Insulin Resistance in Type 2 Diabetes Patients with non-alcoholic Fatty Liver Disease. Horm Metab Res 2018; 50(5): 397-402.
34. Marchesini G, Brizi M, Morselli-Labate AM, et al. Association of non-alcoholic fatty liver disease with insulin resistance. Am J Med. 1999;107(5):450-5.
35. Loomba R, Abraham M, Unalp A, et al. Association between diabetes, family history of diabetes, and risk of non-alcoholic steatohepatitis and fibrosis. Hepatology 2012; 56(3): 943-951.
36. Williams CD, Stengel J, Asike MI, et al. Prevalence of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology 2011; 140(1): 124-31.
37. Angulo P. NAFLD, obesity, and bariatric surgery. Gastroenterology 2006; 130(6): 1848-1852.
38. Berg AH, Combs TP, Du X, et al. The adipocyte-secreted protein Acrp30 enhances hepatic insulin action. Nat Med 2001; 7(8): 947-953.
39. Marques V, Afonso MB, Bierig N, et al. Adiponectin, leptin, and IGF-1 are useful diagnostic and stratification biomarkers
40. Bugianesi E, Pagotto U, Manini R, et al. Plasma adiponectin in non-alcoholic fatty liver is related to hepatic insulin resistance and hepatic fat content, not to liver disease severity. J Clin Endocrinol Metab 2005; 90(6): 3498-3504.
41. Pagano C, Soardo G, Esposito W, et al. Plasma adiponectin is decreased in non-alcoholic fatty liver disease. Eur J Endocrinol 2005; 152(1): 113-118.
42. Polyzos SA, Toulis KA, Goulis DG, et al. Serum total adiponectin in non-alcoholic fatty liver disease: a systematic review and meta-analysis. Metabolism 2011; 60(3): 313-326.
43. Fierbinteanu-Braticevici C, Dina I, Petrisor A, et al. Noninvasive investigations for n on-alcoholic fatty liver disease and liver fibrosis. World J Gastroenterol 2010; 16(38): 4784-4791.
44. Zijlstra MK, Gampa A, Joseph N, et al. Progressive changes in platelet counts and Fib-4 scores precede the diagnosis of advanced fibrosis in NASH patients. World J Hepatol 2023; 15(2): 225-236.
45. Mohamed MS, Youssef TM, Abdullah EE, Ahmed AE. Correlation between adiponectin level and the degree of fibrosis in patients with non-alcoholic fatty liver disease. Egypt Liver J 2021; 11(1): 78.
46. Nazal L, Riquelme A, Solis N, et al. Hypoadiponectinemia and its association with liver fibrosis in morbidly obese patients. Obes Surg 2010; 20(10): 1400-1407.
47. Lucero D, Miksztowicz V, Gualano G, et al. Non-alcoholic fatty liver disease associated with metabolic syndrome: Influence of liver fibrosis stages on characteristics of very low-density lipoproteins. Clin Chim Acta 2017; 473: 1-8.
48. Silva TE, Colombo G, Schiavon LL. Adiponectin: A multitasking player in the field of liver diseases. Diabetes Metab 2014; 40(2): 95-107.

49. Mavilia MG, Wu GY. Liver and serum adiponectin levels in non-alcoholic fatty liver disease. J Dig Dis 2021; 22(4): 214221.
50. Petelin A, Jurdana M, Jenko Praznikar Z, Ziberna L. Serum bilirubin correlates with serum adipokines in normal weight and overweight asymptomatic adults. Acta Clin Croat 2020; 59(1): 19-29.
51. Fantuzzi G. Adipose tissue, adipokines, and inflammation. J Allergy Clin Immunol 2005; 115(5): 911-919; quiz 20.
52. Kawagoe J, Ishikawa T, Iwakiri H, et al. Association between adiponectin production in coronary circulation and future cardiovascular events in patients with coronary artery disease. Int Heart J 2014; 55(3): 239-243.
53. Bonnet F, Ducluzeau PH, Gastaldelli A, et al. Liver enzymes are associated with hepatic insulin resistance, insulin secretion, and glucagon concentration in healthy men and women. Diabetes 2011; 60(6): 1660-1667.
54. Shabalala SC, Dludla PV, Mabasa L, et al. The effect of adiponectin in the pathogenesis of non-alcoholic fatty liver disease (NAFLD) and the potential role of polyphenols in the modulation of adiponectin signaling. Biomed Pharmacother 2020; 131: 110785.
55. Friedemann C, Heneghan C, Mahtani K, et al. Cardiovascular disease risk in healthy children and its association with body mass index: systematic review and meta-analysis. BMJ 2012; 345: e4759.
56. Li X, Zhang D, Vatner DF, et al. Mechanisms by which adiponectin reverses high fat diet-induced insulin resistance in mice. Proc Natl Acad Sci USA 2020; 117(51): 3258432593.
Copyright: © 2024 The author(s). This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited. of NAFLD. Front Med (Lausanne) 2021; 8: 683250.

Kaalpana Jayakumar, Rabeya Yousuf, Nur Afifah Suhemi, Nor Fadzliana Abdullah Thalith, Suria Abdul Aziz, Lailatul Hadziyah Mohd Pauzy and Qhasmira Abu Hazir
Para-Bombay phenotype may remain undetectable on ABO grouping due to its weak or absent H antigen. The discovery of paraBombay phenotype has also been reported after a discrepancy in blood grouping in previously known blood group individuals. This case series illustrates the findings of three para-Bombay Ah phenotypes in our tertiary centre. The first case was identified from a blood sample of a young female with underlying end stage renal failure who was electively admitted for left giant fistula excision. There was ABO discrepancy, and her blood group was further confirmed by extended tests. The second case was discovered in a previously known blood group. An elderly Malay female with a known blood group of A Rh D positive in our centre was admitted for bleeding right brachiocephalic fistula. ABO discrepancy was also seen, in which forward grouping was group O whereas the reverse grouping was consistent with group A. The third case was an elderly Chinese female who was admitted for severe iron deficiency anaemia. Similar to the second case, ABO discrepancy was observed in this blood sample. All these three patients were then confirmed as para-Bombay Ah blood group after further tests were performed to resolve the ABO discrepancy. This case series highlights the importance of resolving ABO discrepancy prior to transfusion as it can significantly affect patient management.
Keywords: Para-Bombay, blood transfusion, ABO discrepancy, blood grouping
NZ J Med Lab Sci 2024; 78(3) 142-144
ABO is a clinically significant blood group system, especially in pre-transfusion testing. Para-Bombay individuals are expressed as Ah, Bh and ABh respectively based on the presence of A and B genes. The para-Bombay phenotype includes two different genetic compositions. The first one includes a mutated weakfunctional FUT1 allele in the presence of a functional or nonfunctional FUT2 allele which encodes a critically low amount of H enzyme, producing very little A, B and H antigens on RBC. The second phenomena include a silent FUT1 gene with an active FUT2 gene, H deficient secretor status. Here, only FUT1 is silenced or non-functional, but FUT2 is functional and produces H, A and B type 1 antigens in the body fluid including plasma and there is some adsorption of these type 1 antigens from plasma to red cell membrane (1,2).
The para-Bombay phenotype is exceedingly rare with a ratio of 1:15 compared to the Bombay phenotype (3,4). Worldwide, the prevalence of para-Bombay phenotype is estimated to be around 1:1000, mostly reported in Eastern Asia with a frequency of 1:5000 among Thais, 1:8,000 among Taiwanese, and 1:15,620 in Hong Kong Chinese, and 1:12,000 among Chinese population (5,6). In Malaysia only few cases have been reported (4,7). The rare H-deficient secretor status is reported to be present in several of the ethnic population (8).
In this case series, we describe three cases of para-Bombay phenotype discovered at our tertiary centre, in Malaysia. The objective is to highlight the identification, test methods applied and importance of this rare phenotype in blood serology.
A 37-year-old female with underlying end-stage renal failure (ESRF) was admitted for excision of left giant brachiocephalic fistula. Her haemoglobin was 100g/L. Pre-transfusion investigations revealed ABO discrepancy between the forward and reverse grouping performed using a fully automated analyser (BioRad IH-500) and column agglutination technique (CAT). The forward grouping showed weak reaction (1+) with anti-A, and no reaction with anti-B while the reverse grouping showed no reaction with A1 cell and strong reaction (4+) with known B cell. However, there was a weak reaction (+) with known O cells. Rh D grouping showed strong positive reaction (4+).
Anti-H lectin test (Ulex europaeus) showed a negative result which signified absence of H antigen on the red cell and negative anti-A1 lectin (Dolichus biflorus). Para-Bombay Ah blood group was suspected. An extended test adsorption and elution test was performed and it showed weakened expression of A and H
antigen. Saliva study revealed presence of A and H substances in the saliva. Lewis phenotyping was also performed which was denoted as Le (a-b+), suggesting her as a secretor individual. All the above test was eventually confirmed her blood group as para-Bombay Ah (Table 2). Patient’s indirect coomb’s test was positive at screening cell III (Bio-Rad ID-DiaCell I-II-III Asia). Antibody identification using gel card method (Bio-Rad IDDiaPanel and ID-DiaPanel-P), showed presence of Anti- Mi(a) Direct antiglobulin test (DAT) was negative. Her probable red cells genotype was R1r (DCe/dce), MN, ss and Mi(a) negative. One unit of Mia-, Bombay (Oh) phenotype, Rh D positive, crossmatch compatible blood was obtained from the National Blood Centre (NBC). The surgery was otherwise uneventful, and blood transfusion was not required. She was discharged well with Hb of 102g/L.
A 71-year-old female with underlying ESRF admitted for bleeding of right brachiocephalic fistula. This was her second admission to our hospital. Her first admission was 6 months earlier, for urosepsis. She had a previous history of multiple admissions to other hospitals for underlying disease and she also had a history of red cell transfusion. No adverse transfusion reactions were reported.
During current admission, her haemoglobin was 67g/L. Two units of red cells were requested for transfusion during haemodialysis. On her previous admission at our hospital, her blood group was registered in the Laboratory Information System (LIS) as A Rh D positive with weakened A antigen expression.
On current admission, pre-transfusion testing by fully automated analyzer (BioRad IH-500) displayed ABO discrepancy. Forward grouping revealed a negative reaction with anti-A and anti-B while the reverse grouping showed a negative reaction with A cell and 4+ reaction with B cell. Reaction of the patient’s plasma with O cells was negative. Repeated ABO/Rh D grouping using the new sample also showed similar discrepancy (Table 1). Enhancement techniques were performed by prolonging the incubation at room temperature and 4°C as well as with enzyme treatment which revealed similar results. Tests with anti-A1 lectin (Dolichus biflorus) and anti-H lectin (Ulex europaeus) showed negative results. Adsorption and elution test revealed absence of A antigen. The ABO blood group was then confirmed as paraBombay Ah phenotype by the reference laboratory where the secretor study showed presence of A and H substances (Table 2). Blood was requested from NBC for transfusion and a total of
four units of red cells cross-matched compatible were supplied where three units were of Bombay and one unit was of paraBombay Ah blood group. Transfusion was uneventful and her haemoglobin raised to 113g/L Patient was discharged well on day 17 of admission.
Case 3
A 90-year-old female with underlying chronic kidney disease stage 4 and iron deficiency anaemia. She had previous history of admission to other hospital and history of uneventful blood transfusion with group O Rh D positive. Upon admission, her haemoglobin was 65g/L and required one unit of red cell transfusion. Pre-transfusion investigation from fully automated analyzer (BioRad IH-500) showed ABO discrepancy in which the forward grouping showed no reaction with anti-A and anti-B
Table 1: ABO grouping by BioRad IH-500 automated analyser
while reverse grouping showed strong reaction (3+) in B cell only and no reaction with A cell. There was no reaction with known O cells (Table 1). Enhancement technique for forward grouping with prolonged incubation at room temperature, 4°C and enzyme treatment showed positive (1+) reaction with anti-A. No enhancement was seen with anti-B. For reverse grouping, there was no reaction seen in A cell after enhancement technique. Tests with anti-H lectin and anti-A1 lectin showed negative results which further raised the suspicion of para-Bombay Ah. Adsorption and elution test revealed presence of A antigen. Secretor studies showed presence of A and H substance which confirmed paraBombay Ah phenotype (Table 2). Patient was transfused with one unit of deglycerolized para-Bombay Ah Rh D positive blood and her haemoglobin subsequently raised to 80g/L and the patient was then discharged, well.
Table 2: Summary of supplementary tests results
Patient Lectin
Anti-A1 lectin Anti-H lectin
Case - 1 0 0
Red cell
Adsorption and Elution
Weak A and H antigen (+)
Saliva secretor study
A and H substance detected
Case - 2 0 0
Case - 3 0 0
No A, B, H antigen detected
Presence of A antigen (1+)
Para-Bombay is a rare blood group variant with limited cases reported. This case series describe three cases of rare paraBombay Ah, detected in a tertiary care centre. In the paraBombay phenotype, FUT 1 is inactive or mutated with an active FUT 2 gene so that a trace of H/A/B antigen is present on the RBCs. The weak form of A and B antigens expressed on the RBCs of these individuals are serologically undetectable using routine ABO testing methods and require additional test of adsorption and elution technique for confirmation (1). Here, case 1 showed a weak reaction with anti-A in ABO grouping while the second and third case did not show any reaction in standard ABO grouping. Weak A antigen was detected by adsorption and elution test in cases 1 and 3, but not in case 2. A saliva test was able to detect secreted A and H substance in all three cases and confirm the grouping as Ah para-Bombay. All these tests are challenging, but are necessary to resolve any ABO discrepancy, to confirm the patient’s blood group.
It is noted that cases 2 and 3 were previously mislabelled as group A and group O and had received several transfusions
A and H substance detected
A and H substance detected
Other tests
DCT: Negative
Antibody screening: Positive
Cell 1:0
Cell II:0
Cell III: 2+
Antibody identification: Anti Mia
DCT: Positive (1+) with anti-IgG (1+).
Red cell elution study: Negative
Antibody screening: Negative
DCT: Negative
Antibody screening: Negative
without reported adverse transfusion reaction. The para-Bombay status was determined in subsequent admission. This kind of scenario is not the first to be reported in transfusion practice where the discovery of para-Bombay phenotype was only discovered after subsequent admissions or blood transfusion (4,9). The crossmatches for earlier transfusions in cases 2 and 3 were probably compatible because neither patient had developed anti-H (10). Usually, anti-H develops in Bombay individuals and reacts with H antigen on the O cell in reverse grouping or antibody screen. However, in para-Bombay, anti-H can be weak or absent as the red cell has some H antigen, demonstrable only at 4°C or by using adsorption and elution techniques (9,11,12). This leads to misidentification of the blood group as group O. In such cases, when normal ABO blood group red cells with compatible crossmatch at indirect anti-globulin test (IAT) are transfused, acute haemolytic transfusion reactions are rarely observed (11). The second and third cases demonstrated a good response following the previous transfusion and no prolonged hospitalizations or any adverse effects were reported. With regards to transfusion for these individuals, they should be
strictly transfused with H-deficient red cells if anti-H and/or antiIH is reacting up to 37°C. However, for patients with anti-H and/ or anti IH not reacting at 37°C, based on a case study in Taiwan, in case of unavailability of the Bombay or para-Bombay blood group, AHG compatible ABO identical red cells can be considered for transfusion (13,14). A few literature reviews concerning blood product transfusion particularly for para-Bombay phenotype has stated that group O red cells have been well transfused with no adverse transfusion reaction and with appropriate Hb increment and HCT response (4,6,15). We can relate this to our second and third patient who was previously grouped as O Rh D positive and had received O red cell transfusion. Besides that, we also have access to Bombay and paraBombay red cell as our National Blood Centre (NBC) successfully stored glycerolized frozen blood products of rare blood groups in their centre. In our practice, even though patients in cases 2 and 3 do not develop anti-H/anti-IH, since they were not in an emergency situation where blood was needed urgently, they had received deglycerolized Bombay (Oh) and para-Bombay Ah red cells in a timely manner. This is also the safest practice without jeopardizing patients’ care and lives after thorough discussion between clinician and transfusion medicine specialist. However, in the event of no access to Bombay or para-Bombay blood in an emergency situation, we support the option to transfuse para-Bombay individual with normal ABO blood group units, compatible by AHG (13,14,16). National Blood Centre also maintains the ‘Registry for Rare Phenotype Donors’ that enables prompt arrangement for donor recruitment. Patients with known blood group of Bombay/para-Bombay must be informed to Blood Bank prior to admission for any elective procedures or surgery so that arrangements can be made with National Blood Centre for their blood products and donor recruitments if necessary. Our cases highlight the importance of having national registry of donor with rare blood groups and role of frozen blood products to ensure quick and efficient blood supply.
Our para-Bombay cases demonstrated how prompt detection of ABO discrepancy in forward and reverse grouping, as well as comparison with previous records, enabled us to identify a series of para-Bombay cases. This highlights the importance of resolving ABO discrepancies, as urgently as possible since it may significantly affect transfusion management especially in the selection of blood products. Besides detailed serological tests, further confirmation with molecular genotyping is also recommended. For future transfusion H antigen negative blood is highly recommended to be transfused unless there is no availability. In such cases, a shared decision must be made by both clinician and transfusion medicine specialist upon weighing all the benefits and potential risks of transfusion of other blood groups to individuals with para-Bombay phenotypes.
ACKNOWLEDGEMENTS
We would like to thank all the pathologists, transfusion medicine specialists, medical officers, and immunohematology staff in NBC and Blood Bank Unit, Department of Diagnostic and Laboratory Services, HCTM who had contributed their precious time, effort and expertise in these cases.
AUTHOR INFORMATION
Kaalpana Jayakumar, MBBS1
Rabeya Yousuf, MBBS, MSc1
Nur Afifah Suhemi, MBBS1
Nor Fadzliana Abdullah Thalith, DipMLT1
Suria Abdul Aziz, BMedSci, MBBS, MPath (Hematology), Lecturer1,2
Lailatul Hadziyah Mohd Pauzy, BMedSci, BMBS, DrPath (Hematology)1
Qhasmira Abu Hazir, MBBS, DrPath (Hematology), Head of Unit, Blood Bank1
1Department of Diagnostic Laboratory Services, Hospital Canselor Tuanku Muhriz (HCTM), National University of Malaysia, Kuala Lumpur, Malaysia
2Department of Pathology, Faculty of Medicine, National University of Malaysia, Kuala Lumpur, Malaysia.
Correspondence Dr Qhasmira Binti Abu Hazir, Blood Bank Unit, Department of Diagnostic Laboratory Services, Hospital Canselor Tuanku Muhriz, National University of Malaysia, Kuala Lumpur, Malaysia
Email: qhasmira@hctm.ukm.edu.my
1. Harmening DM, Manning BL. The ABO Blood Group System. In: Harmening DM, ed, Modern Blood Banking and Transfusion Practices. 7th Edition, FA Davis Company, Philadelphia, USA. 2019: 119-148.
2. Soejima M, Koda Y. FUT1 variants responsible for Bombay or para-Bombay phenotypes in a database. Sci Rep 2023; 13(1): 17447.
3. Joshi SR, Vasantha K. A profile of rare bloods in India and its impact in blood transfusion service. Asian J Transfus Sci 2012; 6(1): 42–43.
4. Pei TP, Ahmad NH, Noor NHM. Para-Bombay phenotype of a pregnant mother in Malaysia: transfusion for an extremely premature baby. Oman Med J 2022; 37(1): e336.
5. Lei H, Shen Y, Wang Y, et al. A Para-Bombay blood group case associated with a novel FUT1 mutation c.361G>A. Transfus Med Hemother 2021; 48(4): 254-258.
6. Subramaniyan R. AB Para-Bombay phenotype: a rare blood group variant and its clinical significance. Hematol Transfuse Cell Ther. 2018; 40(1): 96–97.
7. Abdullah MR, Faizli AA, Noordin SS, et al. Transfusion practice blind spot in para-Bombay: a case report. Transfus Apher Sci 2021; 60(3): 103076.
8. Storry JR, Olsson ML. The ABO blood group system revisited: a review and update. Immunohematology 2009; 25(2): 48-59.
9. Chacko MP, Mathan A, Daniel D. Para-Bombay: A blind spot in blood grouping? Asian J Transfus Sci 2011; 5(2): 182-183.
10. Mohanan N, Henry N, Rafi AM, et al. An antibody worth mention. Asian J Transfus Sci 2016; 10(2): 152-154
11. Patel JN, Donta AB, Patel AC, et al. Para-Bombay phenotype: a case report from a tertiary care hospital from South Gujarat. Asian J Transfus Sci 2018; 12(2): 180-182.
12. Yashovardhan A, Chaitanya Kumar IS, Sreedhar Babu KV, et al. Para-Bombay phenotype - report of a rare blood group. J Clin Sci Res 2012; 3: 141-143.
13. Lin-Chu M, Broadberry RE. Blood transfusion in the paraBombay phenotype. Br J Haematol 1990; 75(4): 568–572.
14. Shih MC, Jen IM, Hwang DH, Lee MT, Shu CW. Blood transfusion in patients with the para-Bombay phenotype-2 case reports. Annual Meeting of Hematology Society ROC and Chinese Society of Blood Transfusion 1988: 31.
15. Bhatia HM, Sathe MS. Incidence of “Bombay” (Oh) phenotype and weaker variants of A and B antigen in Bombay (India). Vox Sang 1974; 27(6): 524-532.
16. Charlotte E, Stefen M, Young-LS, et al. Transfusion in a rare case of para-Bombay phenotype. 27th Regional Congress of the ISBT, June 17-21, 2017, Copenhagen, Denmark.
Copyright: © 2024 The author(s). This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Microplastics and human health
Contributed by Michael Legge
We live in a world of plastics. Current estimates of plastic production are around 400 million tonnes per year which is expected to double by 2040. Plastic disposal is problematic as when it breaks down in the environment, chemical ladened microplastic and nanoplastic particles are created and it is estimated that in the USA alone, plastic particles and residual chemicals are present in nearly all Americans. In a publication from an international collaboration between researchers in the USA and Italy (1). Clear evidence has been provided of the influence of micro and nanoplastics on human health. Using pyrolysis-gas chromatography-mass spectrometry, stable isotope analysis and electron microscopy the researchers analysed excised carotid artery plaques. Two groups of patients were investigated, those that micro and nanoplastics were present and those where they were not. Of those where the plastics were present, polyethylene was detected in the 150 of 257 patients and 31 had measurable amounts of polyvinyl chloride. Electron microscopy identified plastic particles among plaque macrophages. On long term follow-up those patients who had measurable micro and nanoplastics in their plaques had a higher risk of myocardial infarction, stroke or death at the 34 months follow-up than those where plastics were not detected.
How well do microwaves kill bacteria?
Microwaves have become an essential part of both domestic use and laboratory use and it is assumed that the energy is sufficient to provide safe food or not influence laboratory outcomes. There have been several studies investigating microbial populations on everyday items such a lift buttons, electronic devices etc, including coffee machine and dishwashers. A publication from Spain has investigated the bacterial population present in 30 microwaves from domestic use to laboratory use (2). Microwave energy is used to reduce microorganisms in food and extend the shelf life. Previous research has shown that the microwaves inactive a wide range of potentially pathogenic bacteria. However, the current authors maintain that no work has been undertaken to investigate microwaves as microbiological niches to determine microwave microbiomes. Using next generation sequencing and culture techniques, the microwave oven bacterial population was dominated by Proteobacteria, Firmicutes, Actinobacteria and Bacteroides, which the authors concluded were similar to human skin populations and that of kitchen surfaces. Laboratory microwaves had a higher population Nonomuraea species, Kocuria and Moraxella strains as well as those found in domestic microwaves. The authors conclude that these data reflect microwave radiation levels, food interactions and user use. They considered that the differences between domestic and laboratory use may reflect high-temperature resistant taxa and the result of selective pressure.
The popularity of tattoos has increased significantly during the last few decades with estimates between 20% and 30% of people getting tattoos, frequently starting at a relatively young age. Tattoo ink often contains carcinogenic chemicals, and the tattooing process invokes immune responses due to these inks. Coloured inks contain primary aromatic amines, black inks polycyclic aromatic hydrocarbons and metals such as arsenic, chromium, cobalt, lead and nickel are found in all inks. In addition, previous research has demonstrated that tattoo inks are deposited in the lymph nodes. However, despite this previous research the long-term health effects of the mobility of tattoo inks to lymph nodes has not been considered. However, there is a global rise in malignant lymphoma which has not been resolved. A Swedish research group has undertaken a casecontrolled study of all incident cases over a 10-year period of malignant lymphoma using data from the Total Population
Register as well as a questionnaire (3). The study had 1398 cases of lymphoma and 4193 controls. Tattoo assessment was made relating to participant tattoo colours and tattooed body surface area. The results from this research indicated that there was a 21% increase of tattooed individuals developing malignant lymphoma relative to the non-tattooed. Two other findings were that the tattooed body surface area did not affect susceptibility to lymphoma and that laser treatment to remove tattoos was associated with a higher risk of developing malignant lymphoma due to chemical reactions of the dyes with the laser treatment.
advances ‘biological age’ but birth reverses the clock
Previous research has demonstrated that pregnancy in mice changed their ‘biological age’ and was unrelated to chronological age. These research findings have been followed up by a collaboration in the USA to investigate whether the earlier research could be replicated in humans (4). The group investigated 119 pregnant women with low-risk pregnancies and collected blood samples at three timed points during pregnancy plus a sample three months after giving birth. The authors used a range of specific DNA methylation markers that are known to correlate with biological age and specific DNA methylation computer analysis systems to analyse the serial results. The results were consistent with those previously published for the earlier mouse research, confirming a significant positive association between the stage of pregnancy and biological aging. However, they also identified that after three months post-partum recovery the biological aging effects were reversed back to the non-pregnant state. The authors conclude that the changes identified may reflect a naturally occurring form of biological stressor associated with pregnancy and propose further research relating to maternal and fetal health because of the DNA methylation changes.
Red blood cells progressively age when stored developing a series of storage lesions. These are, in part, due to oxidative stress, metabolic changes and changes in morphology. There are a wide variety of factors which impact on red cell storage survival including genetic as well as many environmental factors. Previous research to determine major contributions of redox to red cell demise as well as red cell metabolism has not been previously investigated using multiple blood donations from the same donor. In a recent collaboration between scientists from the USA and Canada the investigators used a metabolomic approach on 643 blood consecutive blood units from volunteers on two separate occasions (5). In addition, they used a previously published method for Transfusion microarray for 879,000 single nucleotide polymorphisms (SNPs). Data was analysed by specific software. Sequential analysis of L-carnitine indicated a significant decrease in stored blood L-carnitine to the point of depletion and was associated with elevated haemolysis. These data correlated with red cell survival rates. Analysis of the SNP data identified polymorphisms in genes encoding L-carnitine transporters and metabolism. Donors carrying 2 alleles of the rs12210538 SLC22A16 SBP exhibited the lowest L-carnitine levels and significant haemolysis on storage. Supplementation of stored mouse blood with L-carnitine boosted post-transfusion recovery and it was proposed that this strategy could improve RBC storage quality.
Pathology services are becoming aware of the carbon footprints being created by providing their services. Australian data indicates that around 7% of carbon dioxide equivalents is contributed by the health care system. In addition, data from Australia indicates that between 12 to 44% of ordered pathology tests were not clinically indicated. Two research publications have investigated
the carbon footprint of pathology testing. The first from Australia investigated the carbon footprint of full blood examination, urea, electrolytes, coagulation profile, C-reactive protein and blood gasses. Included in the research was all consumable and associated waste for venipuncture and laboratory analysis, plus the water and electricity consumed (6). Overall, the different tests had variable carbon footprints, but the highest carbon footprint was related to the sample collection and phlebotomy. The authors conclude that although the relative carbon footprints of their test scenario were small the millions of tests performed each year contributes to the carbon footprint in health care. The second publication was from France where the carbon footprint of a histological procedure was analysed. (7). Here they assessed the use of a biopsy specimen through all the stages of processing including frozen section and immunohistochemistry. The highest carbon footprints related to materials (packaging etc), staff travel and reagents with electricity consumption fourth. The authors conclude that inclusion of life-cycle analysis would also enhance the carbon footprint analysis and that further studies on entire Pathology Departments would reveal the full carbon footprint cost.
Previous research has indicated that clinical embryologists experience high levels of stress and burnout which has been a consistent finding from various international studies. This has been related to not only physical factors but also to the lack of room for error. The latter which could result in significant issues for patients and the potential for legal action. A recent collaborative research publication from the USA and UK has investigated the comparative analysis of stress, somatization and burnout in both the UK and the USA in embryologists (8). Overall, The UK embryologists had a higher female population, graduate education, permanent contracts and full-time employment compared to those in the USA. Working overtime was the highest score for both the UK and USA followed by stress to be called out in times of emergencies. Many of the USA employment conditions were significantly worse than those in
the UK. Both the USA and the UK reported significantly greater stress scores associated with inadequate staffing. Burnout and being on-call scored highly as stress and burnout associated factors. Cryostorage had a very high level of anxiety. The authors conclude that the stressful environments in the embryologist’s workplace impact significantly on the embryologist’s wellbeing which in turn may affect the quality of work.
1. Marfella R, Prattichixxo F, Sardu C et al. Microplastics and nanoplastics in atheroma’s and cardiovascular events. New Eng J Med 2024; 390(10): 900-910.
2. Iglesias A, Martinez L Torrent D et al. The microwave bacteriome: biodiversity of domestic and laboratory microwave ovens. Front Microbiol 2024; 8:15:1395751.
3. Nielsen C, Jerkerman M, Joud AS. Tattoos as a risk factor for malignant lymphoma: a population-based case-control study. Lancet, eClinicalMedicine 2024; 21: 72: 102649.
4. Pham H, Thomson-Feliox T, Czamara D et al. The effects of pregnancy, its progression, and its cessation on human (maternal) biological aging. Cell Metab 2024; 36(5): 877878.
5. Nemkov T, Key A, Stephenson D et al. Genetic regulation of carnitine metabolism controls lipid damage and repair and aging RBC hemoloysis in vivo and in vitro. Blood 2024; 143: 2517-2533.
6. McAlister S, Barratt AL, Bell KJL et al. The carbon footprint of pathology testing. Med J Aust 2024; 212(8): 377-382.
7. Trecot A, Cottinet P-J, Donzel M. et al. Carbon footprint evaluation of routine anatomic pathology practices using eco-audit: Current status and mitigation strategies. Annal Diag Pathol 2023; 67:152210.
8. Murphy A, Lapczynski MS, Proctor G. et al. Comparison of embryologist stress, somatization, and burnout reported by embryologists working in the UK HFEA- licensed ART/IVF clinics and USA ART/IVF clinics. Hum Reprod 2024; 39(10): 22997-2304.
A final word from Sue I commenced my training on 1st February 1977 in the Pathology Department, Christchurch Hospital. In those days we moved around the various Pathology Departments whilst attending The Christchurch Polytechnical Institute on Day release and Evening classes. After that a couple of years of inhouse training to attain two "O" levels or an "A" Specialist level in our chosen disciplines. All the while working on the bench and taking full part in the evening, weekend and on-call rosters. You gained practical experience alongside theoretical knowledge in all the disciplines and learnt how they interrelate, as well as finding friends and mentors- lifelong contacts in some cases. Those of you who work at the smaller laboratories will still experience this facet of the job which unfortunately in my opinion, has been lost with the move to the Degree Courses.
Looking back on my 47 years in Pathology and my chosen discipline of Transfusion Medicine I can happily say I have no regrets, I am ready to enjoy my retirement. Goodbye and good luck to you all

The NZIMLS Council and your colleagues wish you all the best in your retirement. Thank you for your support of NZIMLS and your hard work over the past 20 years.

Warm Pacific greetings to you all from the PPTC
Laboratory Quality Management Course, 8th July- 3rd August 2024 (4 Weeks)
In July of this year, the PPTC hosted a Laboratory Quality Management Course. Participants who attended included those from the North Pacific and Fiji. This training course was delivered over 4 weeks and was based at the Pacific Pathology Training Centre (PPTC) in Wellington, New Zealand. Four medical laboratory personnel from the Federated States of Micronesia (FSM) attended this course was aimed primarily to progress the laboratory services in the FSM to reach international accreditation standards of practice, and in doing so improve the Laboratory Quality Management Systems of the four states. Participants included:
1. Aralai Tuione- FSM Kosrae state laboratory
2. Danilee Giltug- FSM Yap State Laboratory
3. Catherine Gootinngin- FSM Yap State laboratory
4. Herbert Johnny- FSM Pohnpei State laboratory
5. Mun Reddy- Fiji- WHO Programme Officer
This training provided encompassed a comprehensive theoretical and practical application of Laboratory Quality Management according to the ISO 15189 Medical Laboratories Standard. The basis of the course was the teaching of ISO15189 accreditation requirements and the application of the 12 Quality System Essential’s (QSE’s) covering laboratory services being provided. Participants were also trained on the framework of LQMS, features of which included documentation control and the roles of laboratory personnel in the diagnostic environment. This included governance roles, operational roles and support roles in the laboratory. The students were very appreciative for the learning opportunity and acknowledged that the training provided was a most valuable experience for them all.
Knowledge and experience in Lab Quality Management was given by the PPTC to the 5 students and this assisted in their journey towards the next level of achievement in terms of accreditation alignment. By having the opportunity to visit and observe accredited Medical Laboratories as part of this course, is undoubtedly a valuable and inspirational experience for them and we hope they can now implement the principles learnt and observed with confidence in their home laboratory. It has been a privilege and absolute pleasure to have the students visit NZ and to host them through these 4 weeks. The participant from Fiji (Mun Reddy) was also part of an extended training programme on External Quality Assurance (EQA) facilitated by the PPTC.

Blood Transfusion Course, 9th August- 13th September 2024
Soon after the completion of the Laboratory Quality Management course, the PPTC hosted a Blood Transfusion Course. This training course was delivered over four weeks at the Pacific Pathology Training Centre, to medical laboratory personnel; specifically, staff working in Blood Bank laboratories in the Pacific.
There were 3 participants from the Pacific:
1. Arieta Batinivoka- Fiji- Colonial War Memorial Laboratory
2. Peter Tosul- Vanuatu- Vila Central Hospital Laboratory
3. Rosa Tapunuu- Samoa- Tupua Tamasese Meaole Hospital Laboratory.
This course provided students with the following skills, knowledge and information:
• Transfusion transmitted diseases (including HIV, Syphilis, Hepatitis B and C).
• Donor selection criteria and collection.
• Blood processing, Blood Transfusion practise, ABO, Rh and other blood group systems.
• Blood Group Genetics and basic Immunology.
• Preparation of Coombs control cells, technical methods.
• Antibody screen and identification.
• Compatibility testing.
• Haemolytic Diseases of the Newborn.
• Transfusion reaction investigations and Hemovigilance
• Blood bank quality management processes.
• Equipment maintenance.
• Organisation of a Blood Bank including emergency planning, major trauma cases, backup planning.
• Appropriate use of blood components in transfusion medicine.
• Referral laboratory network.
• Practical sessions are provided, focusing on correct technique and fundamental basic procedure.
• Tours of NZBS Crossmatch Laboratory and the Blood Donor Centre.
The PPTC wishes to acknowledge the valuable contributions made by the New Zealand Red Cross and the Norman Kirk Memorial Scholarship for sponsoring a student each from the Pacific to attend this course.


Foundations of Haematology, 23rd September – 1st November (6 Weeks) (In progress)
The PPTC has been tremendously busy with the various programmes we provide. One of which includes In-country support. This involves discipline related training in the relevant Medical Laboratory Sciences where PPTC Consultants travel to countries in the Pacific to provide direct hands-on and application teaching and training to Pacific Laboratory staff according to the ISO 15189 Medical Laboratory Accreditation Standards. The PPTC takes pride in the work that it does, and such support is always a rewarding experience. So far this year the PPTC has covered over 7 different Pacific Island laboratories for which includes Fiji, Samoa, Tonga, Kiribati, Niue, Cook Islands, Solomon Islands and Kiribati.
Two of our Consultants attended professional development training in Singapore provided by Cepheid on GeneXpert analysers. This training was considered most valuable considering the high number of GeneXpert analysers in the Pacific. The two PPTC ConsultantsEmmanuel Marshall and Angela Lewis successfully completed the course as certified Super Users of the GeneXpert systems. The Super User training encompassed both application and engineering components of GeneXpert analysers. This certification allows both our consultants to provide the much-needed support to Pacific laboratories as well as provide user and assay training for staff. The PPTC believes in supporting and promoting staff professional development and with great pleasure congratulates its two consultants for their achievements.


If any New Zealand medical laboratories have items of diagnostic instrumentation as mentioned above that have been recently upgraded or continue to be stored in the laboratory but are actually surplus to requirements, the PPTC would be most grateful if such items could be donated through its Centre to Pacific Island laboratories where there is an exceptional need. Pacific laboratories have very restricted budgets and often cannot afford to replace troublesome instrumentation that continues to breakdown and which is often discontinued because it is so outdated.
Please contact:
Phil Wakem Chief Executive Officer
Pacific
Pathology
Training Centre
Wellington, New Zealand
Email: pptc@pptc.org.nz or phil@pptc.org.nz
Tel: 64-4-389 6294 or 027 2305483


Michael Legge
For some time, the NZIMLS Council has been investigating possible options for ongoing education for members who have qualified as QMLT. Following discussion with the University of Otago this year, an agreement has been reached that the NZIMLS QMLT qualification will be accepted for entry into the University into their first year Diploma of Science course. This is a full-time on-campus course lasting a year. Once successfully completed, then students will be qualified to enter the second
year full-time BSc programmes being offered by the University. Currently there is no distance taught options for this route. Discussions are ongoing for access to other degree programmes using this new option for QMLT. Full information can be found at www.otago.ac.nz and search for Diploma of Science, University enrolments for 2025 has recently opened. This is a significant outcome and recognition for the QMLT programme and qualification offered by the NZIMLS.
The following reviews below can be accessed for their Abstracts and “Open Access” is indicated, where applicable. Unfortunately, the NZIMLS cannot provide full access to the articles due to copyright restrictions, but full access may be available through various institution arrangements. Any feedback on this can be sent to: editor@nzimls.org.nz.
1. Slikenstedt E, Salles G, Campo E, Dreyling M. B-cell nonHodgkin lymphomas. Lancet 2024; 403(10438): 1791-1807.
2. Ling X, Wang C, Li L, Huang C et al. Third-generation sequencing for genetic disease. Clin Chim Acta 2024; 551:117624.
3. Klotman ME, Haynes BF. The other pandemic: lessons from 40 years of HIV research. J Clin Invest 2024; 134(13): e183039. [Open Access]
4. Casadevall A. Pandemics past, present and future: progress and persistent risks. J Clin Invest 2024; 134(7): e179519. [Open Access]
5. Sanchorawala V. Systematic light chain amyloidosis. New Eng J Med 2024; 390: 2295-2307.
6. Hou H, Zhang R, Li J. Artificial intelligence in the clinical laboratory. Clin Chim Acta 2024; 1:1559:119724.
7. Bayoumy AB, Mulder CJJ, Mol JJ, Tushuizen MT. Gut fermentation syndrome: A systematic review of case reports. United European Gastroenterol J 2021; 9(3): 332342.[Open Access]
8. Casadevall A. The mRNA vaccine revolution is the dividend from decades of basic science research. J Clin Invest 2024; 131(19): e153721. [Open Access]
9. Wulandari DA, Harati YW, Ibrehim AU, Pitaloka DAE. Multidrug resistance tuberculosis. Clin Chim Acta 2024; 1:559:11970. https://doi.org.10.1016/jcca.2024.119701
10. Meya DB, Williamson PR. Cryptococcal disease in diverse hosts. New Eng J Med 2024; 290(17): 1597-1610.
11. Burns JC. The etiologies of Kawasaki disease. J Clin Invest 2024; 134(5): e176938. [Open Access]
12. Janssen H, Koekkoek LL, Swirski FK. Effects of lifestyle factors on leukocytes in cardiovascular health and disease. Nat Rev Cardiol 2024; 21(3): 157-169.
13. Yu X, Zhu Y, Yin G, Wang Y et al. Exploiting hosts and vectors: viral strategies for facilitating transmission. EMBO Rep 2024; 25(8): 3187-3201. [Open Access]
14. Kaminski HJ, Coronel I, Kusner L. Myasthenia gravis: the future is here. J Clin Invest 2024; 134(12): e179742. [Open Access]
The Barrie Edwards & Rod Kennedy scholarship is one of the most significant awards offered by the NZIMLS. The award provides support for attendance at an international or national scientific meeting up to a maximum value of $7,500.
Applications for this prestigious scholarship are invited from Fellows, Members and Associate Members of the NZIMLS. Applicants must be current financial members of the NZIMLS and have been a financial member for at least two concurrent years prior to application. To be eligible applicants must make an oral or poster presentation as 1st author at their nominated scientific meeting. All applications are considered by a panel consisting of senior medical laboratory scientists (who are ineligible to apply for the scholarships). All applications are judged on professional and academic merit and the authors participation in the profession in NZ.
Congratulations to Sunny Jamati, BMLSc, a Technical Specialist in Coagulation Haematology at Waikato Hospital and our recent recipient of this scholarship.
ISTH 2024 was the 32nd Congress of the International Society on Thrombosis and Haemostasis (ISTH), in Bangkok, Thailand, at the Queen Sirikit National Convention Centre. The conference dates ran from 22nd to the 26th June 2024. I was able to travel to this event thanks to NZIMLS using the Barrie Edwards & Rod Kennedy scholarship. This is the largest haemostasis and thrombosis conference with approximately 4000 attendees. ISTH allowed me to experience the latest in thrombosis and haemostasis and I was given the opportunity to interact with the global experts in the field and advance my knowledge.
My contribution to ISTH was a poster personation titled “Overcoming high optical densities in a chromogenic assay.” The poster entailed a detailed investigation on the BIOPHENTM FVIII variant kit setup on the STAR Max 2® and the issues around setting this kit up as there wasn’t a known protocol available. This poster was received very well with a number of attendees commenting and asking questions.
There were many topics discussed but the highlight for me was the Mim8, a novel factor VIIIa mimetic bispecific antibody by Novo nordisk. This a treatment for haemophiliac patients that is in the trial phase. Geoff Kershaw presented an excellent study that he personally conducted with the different bovine kits on the market and the cross reactions that can occur with some of the kits unlike the drug Hemlibra which is also a bispecific antibody but doesn’t cross react the same way that Mim8 does. I found this extremely beneficial as no doubt this drug will eventually having funding in New Zealand.
Correspondence: Sunny Jamati, BMLSc, Haematology Department, Waikato Hospital, Hamilton Email Sunny.Jamati@waikatodhb.health.nz

Comparison of direct and standardised antimicrobial disk susceptibility testing for Enterobacterales isolated from urine
Sophia Barlow1, Susan Taylor2 and Wendy Annan2
1University of Otago, Dunedin and 2Middlemore Hospital, Auckland
Objectives: This study evaluated the reliability of direct antimicrobial susceptibility testing (AST) compared to standard disk-diffusion methods for 101 positive urine samples at Middlemore Hospital's clinical microbiology laboratory. It also examined discrepancies between CLSI and EUCAST guidelines. The standardised method included preparing a 0.5 McFarland inoculum from isolated colonies on a blood agar plate.
Methods: Direct sensitivity plates were processed with zone diameters measured. Colonies from blood plates were used to create a 0.5 McFarland standard using the bioMérieux DensiCHEK, ensuring uniform organism concentration for both manual and automated (VITEK 2) AST testing. The same inoculum was used for the VITEK2 N311 card and to prepare standard disc diffusion susceptibility plates on Mueller-Hinton agar. Antibiotics were placed on the agar plates within 15 minutes and incubated overnight at 35°C in O2 conditions. Zone sizes were read after 24 hours, with breakpoints interpreted per CLSI M100 2023 and EUCAST guidelines. Discrepancies were categorized as major (susceptible to resistant or vice versa) and minor (susceptible or resistant to intermediate and vice versa).
Results: The agreement between the two methods under CLSI guidelines was 91.3% across 1,002 comparisons for 10 antibiotics commonly used, tested on gram negative isolates at Middlemore. There were 77 minor and 10 major discrepancies, with major discrepancies involving ceftazidime, amoxicillin clavulanate, ampicillin, and aztreonam. Reinterpretation according to EUCAST guidelines displayed a 51.7% decrease in discrepancies.
Conclusion: Direct AST testing of urine samples using standard EUCAST guidelines provides reliable results with reduced turnaround-time compared to standard testing from colonies. The reduced time to reporting for select antibiotics may allow for escalation of therapy when empiric antibiotics are unlikely to be active.
Method comparison analysis for lipaemic samples: blood gas analyser and Cobas ISE
Pallak Bhatnagar1 and Karen Glover2
1University Of Otago, Dunedin and 2 LabPLUS, Auckland
Objectives: A Lipaemic Index is used to gauge the degree of turbidity within a sample which is often caused by lipaemia. The main cause of lipaemia is lipoproteins, importantly chylomicrons. When a sample reaches threshold or is rejected by the analyser due to lipaemia, ultra centrifugation is required to separate lipids from the sample. This process can be avoided by directly running the sample on a blood gas analyser. Therefore, this method comparison study will follow the electrolyte (sodium, potassium, and chloride) results of lipaemic samples that have been run on the Roche Cobas ISE and blood gas analyser ABL 800. Methods: A total of 23 samples were used that had arrived at LabPLUS with an increased lipaemic index. The purpose of this study is to identify whether the current method at LabPLUS should be continued or replaced with the blood gas analyser. Results: Sodium is a reliable assay and therefore shows a consistent trend where there is a smaller percentage difference between the blood gas analyser and cleared samples with the blood gas analyser results being the largest result. Chloride
is more susceptible to interferences from other negatively charged ions whilst potassium has a much larger uncertainty of measurement in comparison to effects from the electrolyte exclusion effect.
Conclusion: The criteria for the usage of blood gas analyser instead of Cobas was met during sodium analysis. Our statistics clearly indicated that the blood gas analyser regularly yields the highest results. The findings of the assay for potassium and chloride exhibited greater fluctuation due to increased interference from other ions and exceeded uncertainty of measurement range.
Evaluating discrepancies between the von Clauss assay and PT-derived fibrinogen assay to identify interferences
Khrystine Carullo1, Rhonda Lucas2 and Katherine Christie2 1University of Otago, Dunedin and 2Awanui Labs, Dunedin
Objectives: The von Clauss assay is the gold standard for reporting plasma fibrinogen concentration. However, it is susceptible to interferences and unreliable results from dysfibrinogenemia, high bilirubin levels, fibrin degradation products and significant levels of heparin. The aim of the study was to evaluate whether discrepant plasma fibrinogen results between the von Clauss assay and PT-derived fibrinogen assay can indicate the presence of interference and/or unreliable result in the von Clauss assay that requires testing with an alternative method such as Stago.
Methods: Data from routine coagulation studies samples analysed at Awanui Labs, Dunedin, between February 2024 and April 2024, were reviewed. Samples with both von Clauss and PT-derived fibrinogen results within the reporting limits were selected, resulting in 1565 samples. Statistical analysis was performed using Analyse-it software with Passing-Bablok plot, Bland-Altman plot and Student’s t-test to evaluate the variation between the assays.
Results: The Passing-Bablok plot displayed agreement (y=0.05 +1.063x) between the von Clauss and PT-derived fibrinogen assays. The Bland-Altman plot showed that the PT-derived fibrinogen assay results were, on average, 0.31g/L higher. Furthermore, both plots showed data points outside the 25% allowable difference and 95% limits of agreement, suggesting potential interferences and unreliability in the von Clauss assay. The variation between the von Clauss and PT-derived fibrinogen assays was statistically significant (p <0.0001).
Conclusion: The study demonstrated that significant variation between the assays may signal interferences and unreliable results in the von Clauss assay. However, testing with Stago is only necessary for discrepant results with von Clauss fibrinogen concentration within the clinical action limits (<1.0g/L or >9.0 g/L).
Steroid profile result comparison between serum and EDTA/lithium heparin samples
Claire Chan1, Berit Jensen2, Danielle Chillingworth2 and Chris Leaver2
1University of Otago, Dunedin and 2Canterbury Health Laboratories, Christchurch
Objectives: To compare anticoagulant EDTA and lithium heparin samples with serum samples to identify if there is a statistically significant difference in results for the steroids 17-hydroxyprogesterone, androstenedione, testosterone, dihydrotestosterone and 11-deoxycortisol. The aim of this study was to inform whether samples taken in these anticoagulant tubes could be accepted for testing of the above steroids.
Methods: Three matched samples from twenty patients of both genders and various ages, who had serum, EDTA and lithium heparin samples taken at the same time were collected over a one-month period. The samples were leftover or spare samples and were anonymised for this study. They were centrifuged at 3500rpm for 5 minutes with the serum/plasma separated and frozen at 20°C immediately after. The samples were thawed and underwent an organic extraction process before being analysed on the liquid chromatography mass spectrometer.
Results: Using Analysis of Variance (ANOVA), the p value between the three groups for all five steroids are observed to be greater than 0.05 (the threshold of significance) and very close to 1. The Bland Altman plots showed that results from almost all samples were within a 15% difference, with only the occasional outlier.
Conclusion: The ANOVA results support the null hypothesis: that there is no significant difference among group means and that the alternative hypothesis is very unlikely. This is further supported by the Bland Altman plots. Therefore, EDTA and lithium heparin samples could be accepted for the analysis of the above steroids as alternatives to serum.
Tristan Cherrill1, Dan Gyles2 and Abby Clayton2
1University of Otago, Dunedin and 2New Zealand Blood Service, Wellington
Objectives: Microcolumn gel cards are a fast and reliable technology that have replaced the conventional tube technique in most laboratories. New Zealand Blood Service’s current standard operating procedure for room temperature antibody identification uses conventional tube technique. This project assessed if DG neutral gel microcolumn cards provide comparative comparable results for room temperature panels with conventional tube technique.
Methods: Samples were acquired by adding anti-M, anti-P1, anti-Lea, and anti-Leb antisera to plasma to induce different agglutination strengths in tube. These samples were tested in replicate against antibody screening cells by tube and card method, producing 30 individual results for anti-M and anti-P1 Only limited data was acquired for anti-Lea and anti-Leb
Results: The anti-M samples produced 8 results of equal strength in the card and tube methods, and 22 results one grade lower in card than in tube. One anti-P1 sample measured an equal grade in card and tube, two samples measured three grades weaker in card, 16 samples measured two grades weaker, while the last 11 were only one grade weaker in card than in tube. Seven P1 samples tested negative in card but positive in tube. Three antiLea samples tested negative by card by positive by tube. Two anti-Leb samples tested negative by card but positive by tube. Conclusions: A Wilcoxon Test identified a statistical difference between card and tube when detecting anti-M and anti-P1 both with a p-value <0.0001. Anti-Lea and anti-Leb seemed to follow this pattern but there was not enough data to confirm. DG neutral gel microcolumn cards did not produce comparative results with conventional tube technique in room temperature antibody identification for anti-M and anti-P1
Comparative analysis of a Roche enhanced biotin-tolerant parathyroid hormone (PTH) assay with the current PTH assay
Enoch Chin1, Sian Horan2 and Christian Christian2
1University of Otago, Dunedin and 2Awanui Labs, Dunedin
Objectives: Parathyroid hormone (PTH) is an essential biomarker for assessing parathyroid function, calcium regulation, and associated disorders. Due to increased consumption of biotin supplements, there is a greater chance of biotin interference with the existing Roche PTH immunoassay. This study aimed
to validate a new Roche PTH assay designed for increased biotin tolerance and to compare its performance with the current Roche assay.
Methods: PTH Samples (n=39) were tested on the Roche Cobas 8000 e602 unit using both the current and new biotintolerant Roche PTH assays. Intra-run and inter-run precision were evaluated using pooled samples and daily quality controls. Analyse-it for Microsoft Excel was used to run statistical analyses such as Passing-Bablok regression and Bland-Altman plotting. Results: Statistical analysis showed the current and new Roche PTH assays showed a mean difference of 1.94pmol/L and a median difference of 1.0pmol/L. Passing-Bablok regression produced a fit equation of y= -0.1348 + 1.113x, indicating a small systematic bias and a 11.3% positive proportional bias in the new assay. The Wilcoxon rank-sum test showed a statistically significant difference (P=<0.0001) between the assays.
Conclusion: The updated Roche PTH STAT assay demonstrated promising results with a minor positive bias of 11.6%. Although this bias was statistically significant, it was not clinically significant. Therefore, it is viable to replace the current assay without compromising patient results, whilst reducing risk of biotin interference.
Comparison of routinely reported complete blood count parameters before brain natriuretic peptide testing and after
Sooin Choi1 and Ramesh Tiwari2
1University of Otago, Dunedin and 2Canterbury Health Laboratories, Christchurch
Objectives: To identify if there is a significant difference in complete blood count (CBC) parameters of normal whole-blood EDTA before and after centrifugation and removal of plasma aliquots. This comparison is done to see if the samples analysed for BNP (brain natriuretic peptide) should continue to be rejected for CBC testing.
Methods: Fifty samples were organized into 4 categories; CBCcontrol (initial results), CBCc (centrifuged and 0uL plasma aliquoted), CBC150 (centrifuged and 150uL plasma aliquoted), and CBC300 (centrifuged and 300uL plasma aliquoted). The samples were analysed for CBC test in duplicate on Sysmex XN20 and centrifuged using an Eppendorf 5804 centrifuge. Each categorised result was compared with the CBCcontrol and was named Group 1 (CBCcontrol v CBCc), Group 2 (CBCcontrol v CBC150) and Group 3 (CBCcontrol v CBC300). Seven reported parameters of the CBC test were taken into review.
Results: The ANOVA (Analysis of Variance) displayed no significant difference in parameters (P values >0.05) of Group 1 but only mean cell volume and mean cell haemoglobin for Groups 2 and 3. The remaining reported parameters in groups 2 and 3 were reported to have P-value < 0.05 and F-statistic > Scheffe's critical value, demonstrating a significant difference in results. The Bland-Altman plot supported this as the group 2 and 3 differences were scattered outside of the practical limitation of validity as the mean difference sits further from y = 0.
Conclusion: Five out of seven reported parameters of CBC had a significant difference once the plasma had been aliquoted from the sample. Hence a sample is unreliable for testing after plasma aliquoting. Thus, EDTA samples centrifuged and aliquoted for BNP testing should continue to be rejected for CBC test addition.
Development of a multiplex assay for the detection of uniparental disomy of chromosome 7 (UPD7)
Francis Costas 1, Jeshyloria Flores 2 and Kylie Drake 2
1 University of Otago, Dunedin and 2 Canterbury Health Laboratories, Christchurch
Objectives: Uniparental disomy (UPD) is the inheritance of a chromosome pair from the same parent; potentially resulting
in imprinting disorders. UPD often arises from trisomy rescue, through mitotic loss of one of three trisomic chromosomes. One example, trisomy seven (T7), is the most frequent rare autosomal trisomy detected following non-invasive prenatal screening (NIPT). Non-mosaic T7 spontaneously aborts during the first trimester, therefore NIPT-positive T7 cases often involve mosaicism and warrant investigation for UPD7. Testing involves trio (child, mother, and father) segregation analysis using short tandem repeats (STRs), which is inefficient and labour intensive. Therefore, this project aimed to develop a multiplex PCR assay to streamline UPD7 testing.
Methods: Sixteen informative markers across chromosome 7 were selected and fluorescently tagged primers designed. Optimisation was performed using commercially available DNA controls, through combining primers by fluorophore and then mixing the different fluorophores together. PCR was conducted using the Qiagen MultiplexMastermix at an annealing temperature of 60°C using the manufacturers protocol. Size separation was done using an ABI3500 Genetic Analyser (Thermo Fisher Scientific) and GeneMapper software was used for fragment analysis.
Results: Multiplexing all 16 markers into a single PCR and then into two fluorophores was challenging due to amplicon dropout and uneven amplification. However, four multiplex PCR reactions with four primers, adequately amplified 14 out of 16 markers.
Conclusion: We demonstrated that multiplexing 16 markers into four PCR reactions for UPD7 testing is feasible; reducing the number of PCR reactions per trio from 66 to 12, thereby easing the workload. Additional optimisation to achieve a single tube is necessary, but beyond this project’s scope.
Human T-Lymphotropic virus (HTLV): Comparison between Murex HTLV 1+2 assay and LIAISON XL recHTLV-I/II assay
Maria Costas1, Rebecca Dew2 and Rodger Linton2
1University of Otago, Dunedin, and 2Canterbury Health Laboratories, Christchurch
Objectives: Canterbury Health Laboratories currently utilises the Grifols Triturus with the Murex HTLV 1+2 Assay for diagnosing HTLV-I/II antibodies. However, consistent quality control failures and diagnostic instrument malfunctions indicated a need for a new diagnostic method. This study aims to compare the existing method with the DiaSorin Liaison XL, utilising the Liaison XL recHTLV-I/II assay, to determine the best diagnostic assay for Canterbury Health Laboratories.
Methods: A total of 51 samples previously tested on the Grifols Triturus were selected and retested on the DiaSorin Liaison XL. These included true positives, true negatives, equivocal level and low optical density samples. Samples were thawed, vortexed, and centrifuged prior to testing on the DiaSorin Liaison XL. Log data was collected using a USB flash drive and compared with previous results from the Grifols Triturus, as recorded in the Canterbury Health Laboratories database.
Results: The DiaSorin Liaison XL, utilising the Liaison Murex recHTLV-I/II assay, exhibited 100% specificity and 100% sensitivity. Log data showed among the 51 samples, 16 produced true positive results, and 35 produced true negative results, consistent with previous results and expectations.
Conclusions: The DiaSorin Liaison XL, when used with its specific reagents, controls, and methodologies, offers improved turnaround times and operational ease for Medical Laboratory Scientists. The performance of the DiaSorin Liaison XL surpasses that of the Grifols Triturus, providing more repeatable and accurate results. This performance supports the recommendation to transition to the DiaSorin Liaison XL for HTLV-I/II testing.
Effects of amiloride and avastin on retinal pigmented epithelial cell fate pathways
Matthew Cresswell, Robert Walker, Francesc March de Ribot and Tania Slatter
University of Otago, Dunedin
Objectives: Wet age-related macular degeneration (AMD) is a retinal disease that causes progressive vision loss of central vision hallmarked by choroidal neovascularisation and fibrosis. This fibrosis consists of a cellular infiltrate of pericytes. Pericytic fibrosis is also seen in chronic interstitial fibrosis of the kidney, in which previous research suggests amiloride, a diuretic, has anti-fibrotic effects facilitated through increased tumour protein p53 (p53) function. The aim of this study was to assess the effect of amiloride on AMD, using an AMD-induced cell line model. The level of p53 and p53 related proteins were measured following amiloride treatment in comparison to the current standard AMD treatment of avastin (bevacizumab).
Methods: A retinal pigmented epithelial cell line (ARPE-19) was cultured under different conditions (serum control, serum-starved control, serum-starved amiloride, serum-starved avastin, and serum-starved amiloride and avastin) with serum-starved ARPE19 mimicking a cell under macular degenerative conditions. Colorimetric and fluorescent immunohistochemistry were used on cell clots prepared following cell culture to measure levels of p53, p21, pAKT, and p65/NF-κB. In-situ hybridisation TUNEL and Basescope assays were used to investigate apoptosis and p53 isoforms respectively.
Results: The percentage of p53 positive cells were increased with amiloride treatment. The percentage of p21 positive cells was decreased with amiloride but increased with avastin alone. Negligible amounts of pAKT were identified under all conditions, and p65/NF-κB decreased with amiloride but increased with avastin. Both drugs showed low levels of apoptosis, and p53 isoforms showed scattered positivity in the starved control and starved amiloride groups.
Conclusion: Preliminary results show that amiloride and avastin have anti-inflammatory, anti-fibrotic effects on AMD. However, further research and repeat analyses are required before a conclusion can be made.
Pilot study to examine if prophylactic anti-D may also reduce alloimmunisation to non-D RBC antigens
Alice Doney1, Dr Krishna Badami2 and Yohana Stella2 1University of Otago, Dunedin, 2Canterbury Hospital Blood Bank, Christchurch
Objectives: Investigate possible protective effects against alloimmunisation to non-D RBC antigens by antenatal and postpartum prophylactic RhD immunoglobulin.
Methods: The proportion in a random subset (n = 1941) of patients receiving prophylactic RhD immunoglobulin provided by New Zealand Blood Service Christchurch during January 2020 -March 2024 who were alloimmunised to non-D RBC antigens was compared with the proportion in the general population alloimmunised to such antigens.
Results: Of the 1941 individuals in the random subset of those who received prophylactic RhD immunoglobulin, 597 (31%) had an irregular RBC antibody. Of these, 97% (578/597) were in fact passive anti-D (due to prophylactic RhD immunoglobulin). Nineteen of the 1941 (0.9%) had antibodies to non-D antigens. Of these, anti-C was most frequent followed by anti-E, anti-Cw, anti-Fya, anti-S, anti-s, anti-Lea, and anti-P1.
Conclusion: Antibodies to non-D RBC antigens were found in 0.9% of our study group. In comparable pregnant populations, these are found in 0.3%- 3.6% of individuals. This is insufficient to support the hypothesis that prophylactic Anti-D has a protective effect against non-D RBC antigen alloimmunisation.
Optimisation of the anti-HLA-DR immunohistochemistry protocol for identification of MHC-II complexes to aid the histopathological diagnosis of idiopathic inflammatory myositis conditions
Jessica Evans1 and Amanda Fisher2
1University of Otago, Dunedin and 2Awanui Labs, Dunedin
Objectives: The Dunedin Histology department has continuously reported issues regarding the diagnosis of idiopathic inflammatory myositis conditions. These issues are underpinned by the substantial overlap in the clinical and pathological characteristics seen within this group of diseases, which the current immunohistochemistry panel fails to differentiate. Therefore, the aim of this study was optimising the anti-human leucocyte antigen-DR (anti-HLA-DR) immunohistochemistry protocol for identification of the Major Histocompatibility Complex class II (MHC-II). Furthermore, refining the diagnostic differentiation of suspected idiopathic inflammatory myositis conditions, improving the diagnostic information readily available to ultimately underpin improved patient care.
Methods: The Novolink immunohistochemistry polymer kit was used to perform a series of manual assays, undertaken at room temperature on frozen fixed skeletal muscle. Different assays explored different antibody dilutions, incubation times and blocking steps until the optimal protocol was established. This was determined by Dr Ming Yang, who assessed the sections under light microscope to ensure the staining quality reached an optimal level for diagnostic information.
Results: The anti-HLA-DR antibody immunohistochemistry protocol demonstrated optimal staining when the antibody dilution was 1:100 for 15 minutes. Additionally, the test was performed at room temperature and included both a peroxidase and protein blocking step.
Conclusion: Optimisation of the anti-HLA-DR antibody expanded the diagnostic information readily available within the Dunedin Laboratory, most beneficial when used alongside the current immunohistochemistry protocols. Thereby, providing an easy and cost-effective way to enhance patient diagnosis for decreased turn-around times and improved patient care.
Verification of BD MAX™ Enteric Parasite Panel for consideration of use in Pathlab Bay of Plenty molecular laboratory
Felicity George1 and Nicolas Zacchi2
1University of Otago, Dunedin and 2Pathlab Bay of Plenty, Tauranga
Objectives: Verification of the BD MAX™ Enteric Parasite Panel for consideration of use in Pathlab Bay of Plenty Molecular laboratory for identifying Giardia lamblia, Cryptosporidium hominis/Cryptosporidium parvum and Entamoeba histolytica.
Methods: Twenty-four samples were included in this verification process of BD MAX™ Enteric Parasite Panel. Samples were tested and prepared as per manufacturer instructions. This verification included 3 known positive controls provided by Royal College of Pathologists of Australasia Quality Assurance Programme and the remaining 21 unpreserved patient stool samples were selected based on results provided by other testing methods currently used; ELISA at Pathlab Waikato immunology department or Biomerieux BioFire FilmArray at Pathlab Bay of Plenty Molecular laboratory (7 positive samples for G. lamblia, 6 positive samples for C. hominis/C. parvum and 8 known negative samples).
Results: Results for 75% (18 samples) provided by BD MAX™ Enteric Parasite Panel were in concordance with immunology results, BioFire results or a combination of both. Concordance does not apply to 25% (6 samples) that provided an ‘unresulted’ or ‘indeterminate’ result on BD MAX™ System. No further test
panels were available at the time of verification to retest these samples as per manufacturer’s recommendations, which might have reduced the 25% failure rate.
Conclusion: The BD MAX™ Enteric Parasite Panel showed a good concordance rate with existing testing methods, supporting its potential use in Pathlab Bay of Plenty Molecular laboratory This panel is feasible for the Molecular laboratory to implement, as it is similar to other BD MAX™ System test panels currently in use. Its implementation would reduce turnaround time of results to clinicians at a reasonable cost.
Comparative analysis of TPPA kit and TPHA kits by BioRad and Fortress: efficacy and accuracy in diagnostic applications
Yirui Gong1 and Helen van der Loo2
1Otago University, Dunedin and 2Awanui Labs, Dunedin
Objective: Syphilis, caused by Treponema pallidum subspecies pallidum, progresses through various stages, each with distinct clinical features. Accurate laboratory diagnosis is essential for effective management and prevention of transmission. This project aimed to assess alternative kits for syphilis testing by comparing the test results obtained from the SERODIA-TP-PA (Treponema pallidum particle agglutination) kit with those from Treponema pallidum Haemagglutination Assay (TPHA) kits offered by BioRad and Fortress.
Methods: Serum samples from 70 patients with a pre-diagnosis of syphilis were analysed. Initial testing was conducted using enzyme immunoassay (EIA), followed by TP-PA and rapid plasma reagin (RPR) tests on fresh samples. Subsequently, the samples were stored frozen and later thawed for TPHA testing using kits from two different manufacturers. Prior to TPHA testing, samples were centrifuged at 10,000 x g for 10 minutes. TPHA tests were performed manually according to the manufacturers' instructions. The sensitivity, specificity, and kappa agreement of the various testing methods were assessed.
Results: The BioRad TPHA kit demonstrated high specificity (100%) but lower sensitivity (85.9%), resulting in a kappa agreement ranging from fair to moderate. The Fortress TPHA kit showed better performance with 100% specificity and 90.5% sensitivity, and kappa agreement ranging from moderate to good. Conclusion: The evaluation revealed limitations in the sensitivity of the BioRad TPHA kit, underscoring the need for more reliable testing methods. One promising alternative is the Treponema pallidum Latex Agglutination (TPLA) test, which has demonstrated better sensitivity in previous evaluations. Sending samples to Wellington, where the TPLA test is available, may provide more accurate and reliable results for syphilis diagnosis. This approach could enhance the overall effectiveness of syphilis testing and improve patient outcomes.
An attempt at validation of a 1-step reverse transcription Real-Time PCR assay for Enterovirus detection
Senumi Gunawickrama1 and Paul Ogbuigwe2
1University of Otago Dunedin, and 2Health NZ – Waikato, Hamilton
Objectives: A 2-step, reverse transcription (RT) real time qualitative polymerase chain reaction (PCR) method is utilised by the Waikato Molecular Biology laboratory for Enterovirus (ETV) detection. This method is laborious as two separate mastermixes for RT and amplification need to be prepared. The aim of this study was to determine if a 1-step RT-PCR method, using the Quantbio UltraPlex®1-step ToughMix® (4x), can be used as a replacement for the 2-step method while maintaining high sensitivity for ETV.
Methods: A RT qualitative real time PCR protocol was developed using TaqMan chemistry on the Roche Lightcycler 480 II analyser, which included a separate RT reaction followed by amplification
and signal detection using the in-house ETV primers and probes. Samples were tested using the reference 2-step RT-PCR and the UltraPlex®1-step based RT-PCR and compared. The sensitivity of both methods was compared by carrying out a ten-fold dilution series.
Results: UltraPlex®1-step ToughMix®(4x) based assays detected presence/absence of ETV reliably by providing expected results, with 86% of samples giving a lower cycle threshold value, in the 1-step method than the in-house. However, the sensitivity of the 1-step RT-PCR assay was lower than the reference 2-step RT-PCR assay.
Conclusion: UltraPlex®1-step ToughMix®(4x) can be used for ETV detection. However, due to the decreased sensitivity observed in this study, further research needs to be done and modifications made to the mastermix to increase the sensitivity of the assay.
Validating the Vitek 2 AST-N434 card as the primary card for urine antibiotic susceptibility testing
Olivia Hurnard1 and Debra Gordon2
1University of Otago, Dunedin and 2Te Whatu Ora Waikato, Hamilton
Objectives: Urinary tract infections are common infections. Gram negative bacteria are the most common causative agents. With increases in drug resistance, efficient treatment is vital and relies on accurate antimicrobial susceptibility testing (AST). The objective of this study was to validate the new AST-N434 card as the primary card for UTI susceptibility testing by comparing it to the current card in use in the Waikato Hospital Laboratory (AST-N311).
Methods: Fifty-one isolates were tested across 8 species of gramnegative bacteria from urine cultures. Antibiotic susceptibilities were run on the N434 with the N311 card as the comparison. Manual sensitivities were used to validate fosfomycin and cefalexin, as these antibiotics are not included on the N311.
Results: When compared with the results from the N311, the N434 showed an overall 97.45% categorical agreement (CA) with 0.85% minor errors, 0.57% major errors, and 1.13% very major errors (VME). Five of the 15 antibiotics tested displayed 100% CA, and the highest error rate was seen in amoxicillin/ clavulanate with 8% VME. A shortage in fosfomycin MICs meant that <50% of the isolates could be manually tested for fosfomycin, so the data may not be representative. Four of the 8 species tested showed no errors, with the greatest error rate occurring in Proteus mirabilis with 7.5% VME.
Conclusion: The overall results showed the AST-N434 to be satisfactory for validation. However, due to the high error rates found in individual species and antibiotics, the AST-N434 cannot yet be validated for use. Further investigations using manual susceptibilities, including adequate fosfomycin coverage and more isolates, are required to validate the N434 card.
Method Evaluation of the Roche Apolipoprotein B assay
Jan Janerol1, Sian Horan2 and Christian Christian2 1University of Otago, Dunedin and 2Awanui Labs, Dunedin
Objectives: Apolipoprotein B (ApoB) is a better biomarker for assessing atherosclerosis risk than lipid panels alone. The aim of this project was to evaluate the Roche ApoB assay on the Cobas c502 analyser for the purpose of adding this test to the IANZ scope of practice in the Dunedin Awanui Lab.
Methods: Thirty-three frozen specimens were tested on Roche Cobas8000 c502 analyser following the manufacturer's instructions. Results for the same samples were obtained from Canterbury Health Labs. Between-run and within-run data were generated by running quality control material daily for 9 days and simultaneously 10 times, respectively. Statistical analysis was performed via Excel’s Analyse-it, generating Bland-Altman plot,
Passing Bablok, and Student’s T test.
Results: Between-run and within-run analysis showed satisfactory precision with quality controls being within 2SD and had a co-efficient of variation range of 1.53- 2.03%. Between sites, limits of agreement (95%) in Bland Altman plot were -0.335 to -0.139g/L, with a mean difference of –0.245g/L. Passing Bablok regression generated a strong positive correlation slope equation of y= -0.03211 + 0.0831x showing 16.9% negative proportional bias. A p-value of <0.0001 was calculated for student’s T test.
Conclusion: Evaluation has concluded that results generated were deemed statistically significant by the p-value, and clinically significant as they fall out of the Royal College of Pathologists Australasia analytical performance specifications. Therefore, further investigations towards the potential cause of this significance such, as sample pre-analytics or quality assurance, should be carried out before adding ApoB test to the scope of practice in Awanui Dunedin.
Re-evaluation of afternoon quality control panels and recommendations for hardware troubleshooting panel for Cobas c503 and e801
Mameaw Jungsakul1 and Matthew Fawkner2
1University of Otago, Dunedin and 2Awanui Labs, Taranaki
Objectives: Re-evaluate chemistry and immunoassays risk of imprecision. Propose updated afternoon quality control panels and troubleshooting panels for suboptimal hardware performance.
Methods: Each of twenty-four chemistry assays and four immunoassays were tested in 21 consecutive repeats using BioRad Level1 Liquichek Chemistry Control and Level1 Liquichek Immunology Control, respectively. The assays were ranked in 3 categories: experimental, retrospective1 (7 February 2024-7 May 2024), and retrospective2 (26 October 2023-26 December 2023) using 2 parameters: coefficient of variations for imprecision, and capability scores for probability of clinical implication. The five most high-risk chemistry assays and the four most highrisk immunoassays across 3 categories with consideration for hardware troubleshooting capability were proposed as the new afternoon panel. Hardware troubleshooting panels were derived from a webinar by Roche Diagnostics.
Results: The five most high-risk chemistry assays were alanine aminotransferase, aspartate aminotransferase, calcium, cholesterol, and triglycerides. The four most high-risk immunoassays were vitamin B12, folate, oestradiol, and betahuman chorionic gonadotropin. The recommended hardware troubleshooting assays for Cobas c503 are aspartate transferase, cholesterol, creatinine, glucose, total protein, and triacylglycerol. The recommended hardware troubleshooting assays for Cobas e801 are beta-human chorionic gonadotropin, vitamin B12, and hepatitis B surface antigen.
Conclusion: The combination of experimental and retrospective data provided insight into changing assay’s risk for imprecision. The difference between current and resulting panels demonstrated the need for more frequent re-evaluation (<30 months). Further use of existing data could provide cost-effective and up-to-date assessment of assays risk status. An established automated process has the potential to assist laboratorians through evidence-based optimization of laboratory quality control with minimal additional workload.
Comparing picric acid solutions in Gram, Van Gieson, and Elastic-Van Gieson stains
Patrick Kane1 and Rowena Hunter2
1University of Otago, Dunedin and 2Awanui Laboratories, Christchurch
Objectives: Picric acid is used to produce Picro-Acetone
and Van Gieson solutions. These solutions can be made in the laboratory which requires handling and storing solid picric acid, which has explosive properties. This study compared the staining quality of laboratory and commercially made picric acid solutions in the Gram (Brown-Hopps), Van Gieson, and ElasticVan Gieson (Verhoeff’s) stains.
Methods: For each of the three stains, three methods were used. One was the method currently used at Awanui Christchurch. The second was the same method but using a commercially made picric acid-containing solution. The third was a new method using a commercially made solution. The staining quality was evaluated by a pathologist.
Results: The current Gram method using the laboratory-made solution and the alternative method using the commercial solution were both assessed as ‘good’, while the current method using the commercial solution had poor staining quality. The current Van Gieson method using the laboratory-made solution was assessed as ‘good’, while the same method using the commercial solution was ‘excellent’. The alternative method had poor staining quality. The current Elastic-Van Gieson method using the laboratory-made solution was assessed as ‘good’. Both the current method and the alternative method using the commercial solution were ‘excellent’.
Conclusion: The current and alternative gram stain methods were comparable, further investigation should occur before changing the method and solution. For the two Van Gieson stains the new solution with the current method is preferred, producing ‘excellent’ staining quality. A shift of solution is justified but not of method.
BACT/ALERT
Chongyuan Li1 and Alistair Fisher2
1University of Otago, Dunedin and 2Awanui Labs, Christchurch
Objectives: This project aimed to demonstrate adequate function of the new BACT/ALERT system for incubation of blood culture bottles and detection of microbial growth.
Methods: Four types of bottles were tested. There is a carbon dioxide sensor with a pH indicator at the bottom of each bottle. As organisms grow, they produce carbon dioxide which lowers the pH in the bottles. The decreased pH causes the sensor to permanently change colour from grey green to yellow, which reflects more light.
The reflected light is measured by photodiode as reflectance units, which are plotted every 10 minutes. Using clinical specimens for instrument validation is not feasible due to the large number of samples and sample size needed. Instead, this procedure was based on close replication of clinical circumstances using a selection of control organisms. 11 organisms from ATCC control cultures and wild-type samples were used. The suspensions were diluted to a certain concentration (10-100 Colony-forming Unit/mL, which imitates clinical specimens. Each organism suspension was inoculated into bottles and then incubated in the instrument. The positive bottles were unloaded and subcultured onto plates to check if it was the same strain as the control plate.
Results: Most organisms had expected results except for Haemophilus influenzae. The laboratory uses the system for incubation of tissue aspirate samples so failure in detection of Haemophilus influenzae growth is not critical because this organism is not a common pathogen from tissue samples, although further investigation is required to support this.
Conclusion: These validation results support the use of the BACT/ALERT system.
of K2EDTA 6mL, 4mL and 2mL tubes on selected complete blood count parameters
Elinor McAuley1, Philip Ibrahim2 and Emma-Jayne Hillson2 1University of Otago, Dunedin and 2Awanui Labs, Christchurch
Objectives: Complete blood counts (CBC) are routinely
completed using 4mL and 2mL K2EDTA tubes on Sysmex XN analysers at Awanui Labs, Christchurch. However, on occasion only a 6mL K2EDTA tube is available. The aim of this study was to compare the results of different K2EDTA tube sizes across selected CBC parameters; haematocrit, mean corpuscular volume, white blood cell count, and the platelet count. The stability of the 6mL tube was included in the analysis. Methods: There were 32 volunteers from Awanui Labs, each providing a 2mL, 4mL, and 6mL sample. CBC tests were conducted manually on an XN-20 analyser across a selection of time points after collection; 1.5, 4, 8, and 24 hours. Comparisons were made between each tube size at 1.5 hours for each parameter. To examine the stability of the 6mL sample, there were comparisons between the 1.5-hour measurement and each subsequent time point.
Results: There were statistically significant results with p-values <0.05 between some of the 2mL and 6mL comparisons at 1.5 hours, along with the 6mL comparisons over time. However, the differences in the coefficient of variation were acceptable in each comparison when biological variability was used as the allowable limits of variation. This proved a lack of clinical significance in any of the differences. Further analysis included assessment of correlation with R values between 0.997-0.960 for each comparison, and Bland-Altman plots assessed bias and agreement.
Conclusion: The K2EDTA 6mL tube provided similar results to the 2mL and 4mL tubes, as well as similar results over time; supported by a lack of clinical significance. A further study could be completed to conclusively state the validity of the 6mL tube.
Garima Panta1 and Regie Iñigo2
1University of Otago, Dunedin and 2LabPLUS, Auckland
Objectives: This research aimed to optimise the POU2F3 antibody at Labplus using the immunohistochemistry BOND III machine. POU2F3 can be used as a biomarker to diagnose a neuroendocrine (NE) negative subtype of small cell lung carcinoma (SCLC-P). The optimisation of the POU2F3 antibody would aid in the identification of the NE-negative SCLC subtype. Methods: A rabbit monoclonal POU2F3 antibody from Cell Signaling Technology (E5N2D) was optimised. Eight sections were taken from a known case of SCLC-P and different protocols were applied in the BOND III machine. The antibody dilution of 1:200 with 15 minutes primary antibody incubation was applied. Different antigen retrieval protocols - no retrieval, enzyme 1 (proteinase K) for 10 minutes, and Heat Induced Epitope Retrieval (HIER) with low pH (citrate) or high pH (EDTA) buffers for 20, 40, and 60 minutes were applied. Bond Polymer Refine Detection System was used to visualise a brown precipitate to confirm the presence of POU2F3 protein.
Results: The slides were reviewed by the pathologists at LabPLUS. No antigen retrieval and enzyme 1 for 10 minutes resulted in negative staining. HIER with high pH protocols produced intense nuclear staining with notable cytoplasmic background staining while HIER with low pH protocols demonstrated good nuclear staining. HIER (low pH) applied for 60 minutes demonstrated crisp nuclear staining with no background staining.
Conclusion: An optimal protocol for Cell Signalling POU2F3 in the BOND III Machine was analysed and established with the experiment. The primary antibody incubation time of 15 minutes and HIER for 60 minutes with citrate buffer provided ideal staining for POU2F3. The optimisation process determined that the desired protocol had specific and reproducible results making it reliable for future use.
Method evaluation of the HemosIL AcuStar ADAMTS13 activity assay for rapid diagnosis of thrombotic thrombocytopenic purpura.
Divya Patel
University of Otago, Dunedin
Objectives: Thrombotic thrombocytopenic purpura (TTP) is a rapidly fatal microangiopathic haemolytic anaemia that leads to a prothrombotic state due to reduced activity of the vonWillebrand factor-cleaving protease ADAMTS13 (below 10%). The aim of this project was to evaluate the analytical accuracy of the fully automated, chemiluminescent HemosIL AcuStar ADAMTS13 activity assay. The results may be used to establish this test in Awanui Labs, Wellington to improve patient outcomes by eliminating unnecessary pre-emptive plasma exchange. An evaluation was done by performing a method comparison of the HemosIL AcuStar ADAMTS13 activity assay against the gold-standard fluorescence resonance energy transfer assay, FRETS-VWF73.
Methods: Ten frozen plasma samples derived from seven patients, two external quality controls, and six internal quality controls were run on the Bio-Flash analyser using the HemosIL AcuStar ADAMTS13 activity assay and retrospectively compared to the in-house FRETS-VWF73 assay performed at Canterbury Health Laboratory.
Results: Overall, the HemosIL AcuStar ADAMTS13 activity assay gave highly agreeable results to the FRETS-VWF73 assay. The Pearson correlation coefficient value was r =0.971, the mean bias was -0.840 (95% confidence interval, -19.21 to 17.53), and no false negatives or false positives were recorded.
Conclusion: The HemosIL AcuStar ADAMTS13 activity assay produced largely concordant results when compared with the FRETS-VWF73 assay indicating its potential to rapidly aid thrombotic thrombocytopenic purpura diagnosis. However, a larger sample size with a broader range of patient conditions should be tested in a validation study to support these initial findings.
Acknowledgement: The author thanks staff at Awanui Labs, Wellington for supervision and support of this project.
A timeless art: Diagnostic cytopathology in Aotearoa, New Zealand
Kimberly Pinto1, Elizabeth Pringle3, Tamaki Inoue2, Mubarak Ali2, Ursula van den Heever3 and Nirup Kumar3
1University of Otago, Dunedin, 2LabPlus, Auckland and 3Community Anatomic Pathology Service, Auckland
Objectives: Highlight the role, advantages, and limitations of diagnostic cytopathology in the era of molecular testing in Aotearoa New Zealand.
Clinical Presentation: Case 1: Woman, early thirties, in second trimester pregnancy presenting with abnormal vaginal bleeding (PVB) with clots. Case 2: 42-year-old woman with PVB and stenosed, vascular cervix with “easy bleeding” white patch at 2 o’clock. Case 3: 71-year-old man with right lung lesion and multiple bilateral pulmonary nodules as incidental findings on computed tomography pulmonary angiography.
Discussion: Cytological findings of atypical epithelial cells with high nuclear to cytoplasmic (N:C) ratios and hyperchromatic, pleomorphic nuclei for case 1 provided more actionable information than molecular testing alone, building the diagnosis of squamous cell carcinoma. However, inaccuracies in reporting were imparted by inter-condition mimicry and bias. In case 2, cytological findings of small diffusely spread cells with high N/C ratios, scant cytoplasm, prominent nucleoli and occasional binucleation necessitated cytology for finding a rare diffuse large B cell lymphoma of the cervix, unidentifiable by molecular testing. Unfortunately, cytology could not be independently diagnostic in this case, again falling prey to mimicry. In case 3, this shortfall
was convincingly overshadowed by the value of cytology. Findings of abnormal epithelial cells with hyperchromatic, roundoval eccentrically situated nuclei with coarse chromatin and moderate amounts of delicate, finely vacuolated cytoplasm were critical. These helped subtype lung adenocarcinoma, permitting access to molecular testing. Cytology was further indispensable as it provided the sample for molecular testing in this case. Overall, cytology is a timeless art, filling a requisite niche in the era of molecular testing in Aotearoa.
Assessing the efficacy of using formalin-fixed, paraffinembedded (FFPE) samples for MS-MLPA Analysis
Karla Rades1, Malou Andres2 and Amanda Dixon-McIver2 1University of Otago, Dunedin and 2IGENZ, Auckland
Objectives: Lynch Syndrome is the most common hereditary cause of colorectal cancers. It is caused by a germline defect in at least one of the miss-match repair genes; MLH1, MSH2, MHS6, PMS2, or an EPCAM deletion. The laboratory currently utilises Agena Mass Array – MALDI-TOF to determine Lynch syndrome associated methylation status. Methylation Specific Multiplex Ligation-Dependent Probe Amplification (MS-MLPA) is a quicker and cheaper alternative method. Most MS-MLPA probe-mixes are not intended for FFPE extracted DNA, however, FFPE is the most frequently received sample type in the laboratory. This project will investigate the ability of FFPE extracted DNA be used for MS-MLPA methylation status analysis via the MRC Holland SALSA MLPA Probemix P003 MLH1/MSH2.
Methods: Six whole blood extracted DNA samples from healthy patients, along with 11 FFPE extracted DNA samples from oncology patients with known methylation status were processed following the MRC Holland MS-MLPA protocol. Fragments were separated and visualised via ABI-SeqStudio. Results were analysed using coffalyser.net
Results: Both whole blood extracted DNA and FFPE extracted DNA were able to provide amplified fragments by MS-MLPA. The quality of both sources was inadequate for full analysis with coffalyser.net. The ABI-SeqStudio electropherograms produced enabled crude analysis.
Conclusion: DNA quality is a limitation of the MS-MLPA assay. MS-MLPA may be used for FFPE extracted samples provided sufficient quality DNA is able to be extracted. Further experimentation with fixation and extraction methods and assay procedure tweaking is required.
Tegan Rutherford1 and Samantha Kerruish2
1 University of Otago, Dunedin and 2 Whangarei Hospital Blood Transfusion Department
Objectives: The objective of this investigation was to compare the analytical performance of Grifols and Bio-Rad enzyme panels. Additionally, their performance was analysed after swapping the panel cells, and Grifols panel cells were used in Bio-Rad cards, and Bio-Rad panel cells were used in Grifols cards.
Methods: Using the microcolumn technique, antibodies of different specificities were tested against the Grifols Identisera Diana and the Bio-Rad ID-DiaPanel panels, using the untreated red cells for the indirect antiglobulin test phases and the papainised red cells for the enzyme phases. Antisera and external quality assurance samples were used. Next, two additional panels were performed for each of the same antibodies that swapped the panel cells and the cards. One panel was performed using Grifols cards and Bio-Rad cells, and another using Bio-Rad cards and Grifols cells.
Results: All the panels showed reactivity that matched the expression of their corresponding antigen in their respective indirect antiglobulin phases, except for one, which showed reactivity in two antigen-negative cells. Some antibodies did not react as expected in the enzyme phases, while others did.
The reactivity of some antibodies was not negatively affected by swapping the brands of cells and cards used, while some showed undesirable or unexpected reactions.
Conclusion: The Grifols and Bio-Rad panels were both almost always able to identify the antibodies of interest. Although the reactivity of some of the antibodies was not negatively affected by swapping the cells and cards, their overall performance was more satisfactory when the panels were performed as intended.
Heart failure marker suitability comparison in the medical laboratory: replacing BNP with NT-proBNP
Amelia Scott1 and Heather Hughes2
1University of Otago, Dunedin and 2Awanui Labs, Christchurch
Objectives: At Awanui Labs Christchurch, the heart failure (HF) marker B-type Natriuretic Peptide (BNP) is set to be replaced by N-terminal BNP (NT-proBNP) later in 2024. This method comparison aimed to evaluate and compare the efficacy of NT-proBNP to the BNP assay in HF classification, along with comparing analyte stability to assess the suitability of NT-proBNP for routine use.
Methods: EDTA serum samples (n=48, mean age=78 years), previously tested for routine BNP on the Alinity i system at Awanui Labs were selected across a variety of low, medium, and high BNP results ranging from 3.0-913.9 pmol/L. Samples were retested for NT-proBNP results on the Alinity i system within six days of collection.
Results: BNP and NT-proBNP results were compared for qualitative agreement. Two cases were excluded due to NTproBNP exceeding assay limits. Weighted Cohen’s kappa (κw=0.59), calculated from 40 results (excluding six noncomparable values), supported moderate to substantial agreement between assays for HF classification. NT-proBNP demonstrated better analyte stability: six days at 2-8°C vs. 24 hours for BNP; three days at room temperature vs. 4 hours. The coefficient of correlation was 0.858 and the coefficient of determination was 0.736; a p value <0.001 indicated statistical significance. Analysis of viable results showed bias as results increased, including three outliers; a second analysis focusing on clinically relevant NT-proBNP results (<120 pmol/L) showed reduced bias, including one outlier.
Conclusion: BNP and NT-proBNP showed comparable agreement for HF classification for clinically relevant result levels, despite statistical significance emerging as results increased. NTproBNP’s superior stability to BNP supports Awanui’s adoption of the assay as the preferred HF marker later in 2024.
Validation of acridine orange for microbiological staining
Kirushni Suthakaran1 and Kerry Miller2 1University of Otago, Dunedin and 2Microbiology, Te Whatu Ora, Whangārei.
Objectives: Acridine orange is a fluorescent dye first extracted from coal tar in the late nineteenth century. Acridine orange produces shades of orange and green fluorescence when it binds to acidic structures such as nucleic acids, which makes the stain appealing in the scientific field. Microbiological staining with acridine orange is dependent on maintaining the pH at pH 3-4 which allows for the differentiation of microorganisms from background elements. Following acquisition of a fluorescence microscope, a validation study was undertaken to determine if staining with acridine orange could detect organisms in previously false negative Gram stains.
Methods: Eighteen samples were tested, consisting of 4 negative samples and 14 positive samples comprising blood cultures (real and simulated) and sterile fluids. These samples were selected by the supervising scientist. Cytospin slides were prepared and stained using Gram stain and acridine orange stain. The slides were examined, compared and results were checked by the
supervising scientist.
Results: Of the 18 samples, 14 Gram-stained slides were consistent with expected results, whereas all 18 acridine orangestained slides gave expected results.
Conclusion: The validation results of the acridine orange stain, coupled with the quick and easy staining method, indicate that it would be a useful stain to use on potentially false negative Gram stains of blood cultures and sterile fluids. Consequently, in the Microbiology department at Whangarei Hospital, acridine orange stain has been approved as a secondary stain for such samples.
Verification of the Biomerieux BacT/ALERT 3D Microbial Identification System for detection of bacteraemia in Awanui Labs Nelson and Wairau
Imogen Taylor1 and Tony Barnett2
1University of Otago, Dunedin and 2Awanui Labs, Nelson
Objectives: The aim of this study was to evaluate the function of the bioMérieux BacT/ALERT 3D Microbial Identification System for growth and identification of growth of organisms from blood culture samples in the clinical microbiology laboratory.
Methods: Function of the BacT/ALERT analyser (bioMérieux, Marcy l’Étoile, France) was tested using quality control strain organisms, or wild type organisms with confirmed identifications. Seven bacteraemia associated organisms were selected for use in the study: Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, Cutibacterium acnes, Candida albicans, Aspergillus fumigatus and Bacteroides fragilis. BacT/ ALERT media bottles were inoculated with 0.1 mL of 106 dilution of each of these organisms, except for C. albicans for which a 104 dilution was used. The bottles were supplemented with 9.9mL of sterile donor blood. Bottles were then incubated in the BacT/ALERT according to standard protocol.
Results: The BacT/ALERT was able to successfully culture and identify growth of S. aureus, E. coli and C. acnes in all bottle types. Growth of P. aeruginosa and C. albicans was identified in the FA Plus, PF Plus, SA and SN media bottles. B. fragilis growth was identified in the FN Plus and SN media bottles. The BacT/ ALERT did not successfully culture A. fumigatus
Conclusion: The BacT/ALERT showed sufficient sensitivity in growing and identifying organisms from blood culture samples and is appropriate for use in the diagnostic microbiology laboratory. Since the completion of this study, the Bact/ALERT has been in use in Awanui labs including in Nelson and Wairau.
Alan Uthup1 and Megan Smith2
1University of Otago, Dunedin and 2Awanui Labs, Dunedin
Objectives: Increasing rates of antimicrobial resistance drive the need for rapid antimicrobial susceptibility testing (RAST), especially for positive blood cultures (BCs). In response, the European Committee on Antimicrobial Susceptibility Testing (EUCAST) developed the RAST method, performed directly from positive BCs, to report finalised antimicrobial susceptibilities from 4 to 16-20 hours. This study evaluated the 2022 EUCAST RAST method for positive BCs while assessing the practicality of implementing RAST into the current laboratory workflow at Awanui Labs, Dunedin.
Methods: Data was collected from 46 patient BCs that signalled positive for any of the eight RAST-approved BC organisms. The EUCAST RAST methodology was performed, and the inhibition zone diameters of antimicrobial susceptibility plates were measured at 4, 6, 8 and 16-20 hours of incubation.
Results: For E. coli-positive BCs, 98% of all zones could be read after 6 hours. At 4, 6, 8, and 16-20 hours, the percentage of uncategorised zones changed from 14% to 17%, 9%, and 7%. The percentage of categorical errors decreased from 55%, 5%,
and 3% from 4 to 8 hours but increased to 7% at 16-20 hours. Conclusion: The 2022 RAST method performed best for E. colipositive BCs when plates were read at 8 and 16-20 hours. Further testing is required to evaluate RAST against more resistance mechanisms and organisms other than E. coli. Though a simple, inexpensive method that reports susceptibilities quicker, the incubation-sensitive nature of RAST makes it difficult to implement into the laboratory workflow for routine use. Therefore, RAST should be implemented at Awanui Labs, Dunedin, only as a supplementary tool to detect resistance mechanisms quicker and provide same-day susceptibilities for severe cases of sepsis and for commonly encountered BC organisms.
Simultaneous analysis of 7 antibiotics in urine by liquid chromatography triple quadruple tandem mass spectroscopy and solid phase extraction
Ziqi Wang and Natalie Hughes University of Otago, Dunedin
Objectives: In developing countries, antibiotics can be sourced outside the health-system, which may impact antimicrobial resistance. To assess the extent of this, new antibiotic detection methods are required. This study aimed to develop a qualitative method to identify prior exposure to an initial panel of antibiotics—flucloxacillin, cefixime, tetracycline, amoxicillin, phenoxymethylpenicillin, and benzylpenicillin—in urine sample analysis with Liquid Chromatography-Triple Quadruple Tandem Mass Spectrometry (LC-QqQ-MS/MS) and Solid Phase Extraction (SPE)
Methods: Mass spectroscopy (MS) conditions of the 7 antibiotics were optimized under positive electrospray ionization. Separation of antibiotics was achieved on the Kinetex C18 column (50 x 2.1 mm, 100A, 2.6 µm Phenomenex NZ). Optimal mobile phases were identified from combinations of 0.1% aqueous formic acid (A) with either acetonitrile, methanol, methanol with 10% Isopropanol, 0.1% formic acid in methanol and 0.1% formic acid in acetonitrile (B), assessed by retention times and peak quality. Antibiotic solution stability was investigated at 4°C, 25°C, and 40°C, with a pH of 4.05 and 5.61. Initial investigations of polymeric SPE using antibiotic standards were also conducted. Results: Optimal MS conditions were identified, and two product ions were used for analysis. Two successful LCMS methods were developed. Stability tests indicated that degradation followed first-order kinetics and increased with higher temperature and acidity, and responsible for abnormal peaks observed for phenoxymethylpenicillin and benzylpenicillin. An SPE trial showed reversed-phase SPE cartridge gave highest antibiotic recovery of 39.34% to 126.35% for the seven antibiotics.
Conclusion: Two LC-QqQ-MS/MS methods were developed, capable of simultaneously detecting seven antibiotics within 12 minutes. Initial investigations into SPE methods had shown promising results and require further development and application to urine samples.
Abbott Alinity c-series: Phosphorous and Phosphorous2 assay comparison
Anna Welch1 and Tim Aitken2
1University of Otago, Dunedin and 2Hawke’s Bay Hospital, Hastings
Objectives: The comparison aimed to determine the suitability of the Abbott Phosphorous2 reagent in replacing the current Abbott Phosphorous reagent in routine testing. Another aim was to extend the calibration stability of the Phosphorous2 assay. Phosphorous is a highly abundant mineral in the body and is responsible for vital functions such as muscle contractility, serum pH buffering and electrolyte transport. Phosphorous is absorbed from food or resorbed from bone stores, filtered through the glomerulus, and reabsorbed within the proximal
tubule with serum concentrations controlled by the parathyroid hormone (PTH) and vitamin D. Disorders affecting PTH, vitamin D, intestinal absorption, storage, or renal function greatly alter serum concentrations and lead to life-threatening consequences. Methods: Fifty patient serum samples with previous phosphorous results were reanalysed using both the reference assay (Phosphorous) and test assay (Phosphorous2). Results were compared to assess the correlation between the two assays. To investigate calibration stability, quality control material was run every morning for 30 days and the daily variation was recorded.
Results: The measured phosphorous concentration of the Phosphorous2 assay showed very slight overall negative bias (-0.76%) compared to the original Phosphorous assay. Any variation was within allowable limits; the assay showed good precision. Calibration was stable for at least 27 days, 13 days longer than recommended.
Conclusion: Phosphorous2 is an acceptable replacement for the current Phosphorous reagent. The calibration can be extended to at least 27 days to save materials and minimise testing delays.
Evaluating the use of CHROMagar™ mSuperCARBA™ for the detection of carbapenemase-producing organisms
Carly Williams1 and Murray Robinson2
1University of Otago, Dunedin and 2Pathlab, Tauranga
Objectives: This report was undertaken to assess how well CHROMagar™ mSuperCARBA™ agar cultures carbapenemaseproducing organisms with a range of minimum inhibitory concentrations (MICs), and at a low dilution (representative of clinical specimens).
Methods: Fourteen carbapenemase-producing organisms (confirmed by the Institute of Environmental Science and Research) were selected for the experiment. The meropenem MIC of each organism was recorded from a MIC strip on Mueller Hinton agar. A Klebsiella pneumoniae isolate producing NDM-1 was used to determine the dilution factor in which the smallest number of single colonies could grow on mSuperCARBA (a 1/106 dilution). All isolates were serially diluted to 1/106 from a 0.5 MacFarland standard. Fixed volumes of the 0.5 MacFarland suspension and 1/106 dilution were inoculated onto mSuperCarba plates. Each 0.5 MacFarland standard was also inoculated onto MacConkey agar to perform a meropenem disc diffusion test. Results: All carbapenemase-producing organisms grew equally well on mSuperCARBA at a 0.5 MacFarland standard. Ten out of fourteen isolates grew isolated colonies on mSuperCARBA at a 1/106 dilution. The isolates had a meropenem MIC range of 1mg/L to >32mg/L, and the meropenem zones of inhibition ranged from 6mm to 25mm. Susceptibility/resistance to meropenem was interpreted with EUCAST guidelines.
Conclusion: CHROMagar mSuperCARBA is a selective agar that can culture a variety of carbapenemase-producing organisms. A low level of detection was observed, but further study should be done to support this. A CHROMagar mSuperCARBA plate would be a beneficial addition to a laboratory’s carbapenemase screening protocol, as it can help warrant further testing on specimens containing carbapenemase-producing organisms that show susceptibility/reduced susceptibility to carbapenems in vitro via the standard methods.
Theresa Winckler1, Sue Melvin2 and Tracey Hollings2 1University of Otago, Dunedin and 2Awanui Labs
Objectives: The study compared chloride results obtained from the Abbot i-STAT Chem8+ as used in the rural laboratory and results obtained from a Cobas in the reference laboratory (Dunedin), as currently there is no patient comparison. This investigation also considered whether later add-ons performed
on serum obtained from the same heparinised samples, performed on the Abbot i-STAT Chem8+ are acceptable.
Methods: Heparinised non-PST blood samples were routinely obtained overnight by non-laboratory medical staff and were run on the i-STAT in the ward. If this included a Chem8+ the same samples were then centrifuged during laboratory hours and the plasma was run again on the same i-STAT before being sent to Dunedin for analysis on the Cobas. Serum sodium was added, as it is expected to be proportional to chloride and may highlight any measurement discrepancies. Forty-eight samples were run.
Results: Differences between whole blood (mean: 101.3, SD: 3.8) and serum chloride (mean: 101.0, SD: 4.3) were minimal. Chloride measurements obtained from the Cobas had a consistently lower bias, regardless of time between measurements. A large proportion were outside the allowable difference of +/- 3%. Sodium measurements on the Cobas were variable but centred around the same mean as the i-STAT. Most fell within the allowable difference of +/-3%.
Conclusion: Due to a significant difference in chloride results between the i-STAT and Cobas, the two methods have been deemed not comparable. Therefore, a patient comparison will not be adequate. Add-on requests for chloride are acceptable using plasma on the i-STAT and within 24 hours. However, requesters should be made aware of potential risks using nonstandard methods.
Ruoling Xiao1, Rosie Greenlees2 and Esther Lau2 1University of Otago, Dunedin and 2Cantebury Health Laboratories, Christchurch
Objectives: To compare the effectiveness and accuracy of three different methods for leukocyte cell counts in body fluid and to assess the performance of the cell count method against semiquantification by direct Gram stain, and semi-quantification by cytocentrifuge preparation (cytospin) on Gram stain methods. The feasibility of conducting all three methods on a single specimen will be explored for comparative analysis. This study aimed to replace semi-quantification by direct Gram stain method with the cytospin method in a routine laboratory.
Methods: Forty specimens were collected, and all tests were performed within five days of arrival. Pleural fluid, CAPD, and other aspirate specimens were included. For cell count, a haemocytometer was loaded with 10μL of specimen. All cell counts were performed manually, except for bloodstained pleural fluid samples, which were counted by a haematology analyser. For the direct Gram stain, one drop of the specimen was loaded onto a slide and Gram stained. Twenty fields were examined under low power magnification.
For the cytospin method, 80μL of the specimen was processed through the cytocentrifuge chamber. Gram stains were performed with cytospin slides. Five fields were examined under high power magnification.
Results: The results suggested there was a positive linear relationship between each method. Except for six outliers, all other sample data fell within the 95% confidence interval of the linear regression between direct stain and cytospin stain results. Conclusion: Statistically, the cytospin stain method could replace the direct stain method based on this study. More fields are required to improve the precision of the results, and further evaluations are needed for standardisation.
Validation of the BacT/ALERT 3D blood culture instrument
Altha Zamora1 and Grant Cook2
1University of Otago, Dunedin and 2Awanui Laboratory, Timaru
Objectives: The study aimed to verify the performance of the Bact/Alert 3D blood culture instrument.
Methods: Controlled seeded blood culture bottles (63) were loaded in the blood culture instrument for microbial detection.
The BioMerieux culture bottles used for the verification were standard aerobic, anaerobic, fastidious aerobic, fastidious anaerobic, and paediatric. In addition, the test inoculums were plated onto blood and chocolate agar plates to check purity and colony count to ensure the organisms were ideally in the 10100 CFU inoculum range. Each bacterial strain was inoculated to achieve a McFarland 1.0 Standard equivalent to approximately 3x108 CFU/mL Therefore, a three-serial dilution (106) resulted in a bacterial count of approximately 300 CFU/ml or 30 CFU/0.1mL. For Candida albicans, a McFarland 0.5 was used with the serial dilution adjusted to two-serial dilution (104). The final dilution was injected into the blood culture bottles and subcultured onto agar plates for purity. The bottles were loaded into the BacT/ ALERT 3D for a 5-day incubation period or until flagged positive. Unexpected results were Gram-stained.
Results: The BacT/ALERT 3D system detected all bacteria and fungi in the blood tested in this validation. Most bacteria and Candida albicans were detected within 27 hours of incubation except Bacteroides fragilis. Fifty four out of 63 returned positive and 3 of the negative cultures returned false negative. The unexpected positive bottle was likely contaminated with the environment or skin flora.
Conclusion: The study demonstrated the BacT/ALERT 3D blood culture system's capability to detect aerobic bacteria, anaerobic bacteria, and fungi which was further confirmed with the gram-staining method.
Alan Zhang1 and Samantha Hutton2
1University of Otago, Dunedin and 2Awanui Labs, Wellington Hospital
Objectives: The translocation t(15;17)q(24;21) results in the fusion of the promyelocytic leukaemia (PML) gene to the retinoic acid receptor alpha (RARα) gene. This translocation, known as the PML-RARα fusion, is crucial to detect because it causes acute promyelocytic leukaemia (APML). This study aimed to verify the PML-RARα fusion detection assay currently performed on the LightCycler 2.0 by comparing its data to that generated by the newer and more advanced LightCycler 480.
Methods: This study involved both a comparative analysis and an assessment of measurement uncertainty. The comparative analysis included two runs on the LightCycler 480 using past patient samples previously tested on the LightCycler 2.0. The measurement uncertainty assessment utilised specific target concentrations from the Roche LightCycler PML-RARα kit standard row. These target concentrations were tested across three LightCycler 480 machines in the Wellington Hospital Awanui Molecular Diagnostics Laboratory. Together, these studies evaluated the sensitivity, specificity, and precision of the LightCycler 480.
Results: In the comparative study, all samples within the linear detection range of the assays were identified as positive. The coefficient of variation was below 2 for all tested concentrations, indicating excellent inter-machine reliability.
Conclusion: Although the data from this study were insufficient to fully verify the LightCycler 480 against the LightCycler 2.0, the results demonstrated a high level of intra-run precision and significant inter-assay precision among the LightCycler 480 machines.
Verification of the BioFire FilmArray Meningitis/ Encephalitis (ME) panel for use at Medlab Central
Xintong Zhang1 and Rebecca Lucas-Roxburgh2
1University of Otago, Dunedin and 2Med Lab Central, Palmerston North
Objectives: Rapid diagnosis and management of patients with central nervous system (CNS) infections is crucial to reduce morbidity and mortality. The aim of this study was to verify the
BioFire FilmArray Meningitis/Encephalitis (ME) Panel for use at Medlab Central.
Methods: Eighteen samples collected between 1st June 2022 and 31st January 2024 were used. Fifteen were clinical samples previously tested by routine methods, which comprised 13 positive and two negative cerebrospinal fluid (CSF) specimens. Routine methods were bacterial culture performed at Medlab Central, and CSF CNS PCR Infectious screen performed at Canterbury Health Laboratories. Three were external quality assurance samples (simulated CSF specimens) from T-lab Gisborne Hospital. ME Panel results were compared with routine methods results or T-lab test results for calculations of performance measures.
Results: Escherichia coli K1, Haemophilus influenzae, Neisseria meningitidis, Streptococcus pneumoniae, Enterovirus (EV), Herpes simplex virus 1, Herpes simplex virus 2 (HSV-2),
Human herpesvirus 6 and Varicella zoster virus were detected. After discordant analysis, the overall agreement rate for bacterial analytes and viral analytes were 100% and 55.6% respectively. The positive percent agreement (PPA) for EV and HSV-2 were 0% and 50% respectively. The ME Panel demonstrated a PPA of 100% for seven other analytes detected. The negative percent agreement (NPA) was 100% for 11 out of 14 analytes. Conclusion: This study showed acceptability of the ME Panel at Medlab Central in principle, as it has high PPA and NPA for most analytes detected. The work was limited by inadequate target coverage and low sample numbers. Sample age and suboptimal storage likely explain the low detection rate for RNA targets. Therefore, further work is required before verification can be completed, and the test introduced.
HSIG with a Hemoglobinopathy twist!
This year the meeting had a specific focus, haemoglobinopathies
A vital and ambiguous area that requires standardization across New Zealand. This event was spearheaded by Alan Neal and convened by Sunny Jamati. As always Sharon Tozer did her magic for a seamless day with excellent catering. Our generous sponsors included Roche, Stago, Abacus and ESL biosciences. The event was held at Victoria University of Wellington with over 60 people attending including NZIMLS and non NZIMLS members. We had a total of twelve speakers over the day. The presenters were of an excellent calibre with a wide range of talks encompassing Haemoglobinopathies and Thalassaemia’s.
The judges had the most difficult job deciding the top two presentations. Best presentation went to Rachel O’Connor from Wellington Awanui and runner up prize went to Alan Neal from Pathlab. We had two spot prizes from Abacus and ESL biosciences that went to Jordyn Moore.
Report by: Sunny Jamati (Co-Convenor), Technical Specialist -Coagulation Haematology, Waikato Hospital, Hamilton.

09 Nov 24
Mortuary SIG, Waikato Hospital, Hamilton
09 Nov 24 Immunology SIG, Rydges Hotel, Wellington
16 Nov 24
Pre-Analytical SIG, Auckland
Anita.Worrall@waikatodhb.health.nz
Sarah.Burge@awanuilabs.co.nz
Ajesh.Joseph@nzimls.org.nz
22 Nov 24 Molecular Diagnostics SIG, Wellington Hospital, Wellington Clive.Felix@ccdhb.org.nz
23 Nov 24 Microbiology SIG, Wellington Hospital, Wellington
Leeanne.Olsen@esr.cri.nz
28-29 Nov 24 NZIMLS Council meeting, Christchurch sharon@nzimls.org.nz
06 Dec 24 Tentative date for release of QMLT results sharon@nzimls.org.nz
NZIMLS EXECUTIVE OFFICE WILL BE CLOSED FOR THE CHRISTMAS/NEW YEAR PERIOD FROM MONDAY 23 DECEMBER 2024 - RE-OPENING MONDAY 13 JANUARY 2025
Read the articles carefully as most questions require more than one answer. Answers are to be submitted through the NZIMLS website. Make sure you supply your correct email address and membership number, it is recommended that you write your answers in a word document and then cut and paste your answers on the website.
You are reminded that to claim valid CPD points for successfully completing the journal questionnaire you must submit an individual entry. It must not be part of a consultative or group process. In addition, members who have successfully completed the journal questionnaire cannot then claim additional CPD points for reading the articles from which the questions were derived.
The site will remain open until Friday the 14th February 2025. You must get a minimum of eight questions correct per questionnaire to obtain 5 CPD points.
The Editor sets the questions but the CPD Co-Ordinator, Jillian Broadbent, marks the answers. Direct any queries to her at cpd@ nzimls.org.nz.
1. Proteinuria is defined as the presence of abnormally elevated amounts of protein in urine, what is considered normal urine protein excretion and what values indicate presence of proteinuria? What is an example of a Turbidimetric assay and what did its method validation provide?
2. The 25% SSA method uses precipitation of proteins to quantitate urine protein, what 3 sample volumes were evaluated by pairwise comparison in the Siriwardhana study, what differences were found? What conclusions were drawn about the potential for the 25% SSA method to be automated?
3. What do C-reactive protein (CRP), Interleukin-6 and procalcitonin levels in COVD-19 patients correlate with? What can these biomarkers determine?
4. What have elevated IL-6 levels been linked to in one of COVID-19's common symptoms? What was observed in this symptom over time and in recovery?
5. There are more than 4,000 chemicals in a single cigarette, including carbon monoxide. What does carbon monoxide bind to in the body and what can this cause?
6. What principal features are exhibited by the rare neurodevelopmental disorder Prader-Willi Syndrome (PWS) in infancy? Where is the chromosome deletion? What are the physical manifestations/characteristics of this disorder?
7. What is a principal feature in patients with Prader-Willi Syndrome arising in early childhood? What inflammatory processes and co-morbidities are often related to this disease?
7. What is the estimated prevalence of the para-Bombay phenotype worldwide? Why do the two different genetic compositions seen in the para-Bombay phenotype result in poor detection of this phenotype on the ABO blood group system?
9. What adipocytokines are implicated in insulin resistance? What else do they influence?
10. Non-alcoholic fatty liver disease (NAFLD) is becoming one of the most prevalent liver pathologies globally. What is the percentage of adults and children affected by NAFLD in developed countries? What is predicted to emerge from this disease?
1. Electronic crossmatch (EXM) is an alternative to serological crossmatching methods, what does this alternative rely on to confirm ABO blood group compatibility between donor and recipient? What does EXM involve?
It relies on the electronic record of the patient’s blood group, laboratory automation, and a robust computer system in the laboratory. EXM involves antibody screening, blood group checks using two different reagents, and electronic issuing to minimise mistakes and reduce workload.
2. One of the advantages of EXM is the reduction in workload for the laboratory. Briefly list the types of activities that are reduced for the laboratory staff specifically?
Hands on time is reduced, lab staff are less exposed to biohazardous material, scientists can allocate expertise to other tasks, less physical and mental stress
3. What is Haemophilia A? What can patients develop when receiving therapy? And what can they become more susceptible to?
Haemophilia A is an acquired or inherited deficiency of clotting factor VIII, patients can develop an immune response from neutralising alloantibodies against Factor FVIII making patients more susceptible to bleeding symptoms and higher risk of morbidity and mortality.
4. What are Factor VIII inhibitors? How do they inhibit?
Factor VIII inhibitors are Polyclonal IgG antibodies known as coagulation factor inhibitors. They bind to and then neutralize pro-coagulant plasma proteins, inhibiting a variety of haemostasis assays depending on the specific factor affected.
5. What is the reference method for antinuclear antibody (ANA) detection for patients with suspected connective tissue disease, autoimmune hepatitis and juvenile idiopathic arthritis? What are the problems associated with this method?
Indirect immunofluorescent assay using human epithelial cells (Hep-2 IFA) method. Manual interpretation is subject to both intra and inter-laboratory reader variation, affecting pattern recognition and endpoint titre determination.
6. What limitation was demonstrated in multiple Pathlab laboratories using NOVA View SWT? What is required for automated ANA readers?
Limitation was the variation between the same instruments at different laboratories and differences in manufacturers’ reagents. Harmonisation between different manufacturers reagent and instruments, an inter-manufacturer standardisation for automation ANA readers.
7. Familial Mediterranean Fever (FMF) is an autosomal recessive disorder and the most common genetic autoinflammatory disease. What is used as the current management of this disease? What is its role in treatment? Treatment is based on Colchicine which suppresses excessive monocyte activation and useful for reducing the severity and duration of symptoms, prevent acute attacks and the development of complications of amyloidosis
8. What was did Khalil et al to be a reliable inflammatory marker for FMF? And how could this help with the diagnosis of subclinical inflammation?
The neutrophil/lymphocyte ratio (N/L ratio) of 1.65 can be a reliable inflammatory for subclinical inflammation as it is easily obtained from complete blood count without additional cost and useful as a predictor for the development of amyloidosis.
9. How do off-target antibodies interfere with immunoassay results? Provide two examples of immunoassay test and affect to patient.
Off target antibodies bind-non-specifically to antigen or antibodies in human samples or laboratory reagents causing false positive or elevated results Examples assays for D-dimer, tumour markers, Troponin T and I, hormone levels, and infectious diseases including syphilis. Can cause clinical confusion, patient distress and inappropriate treatment.
10. What is Kode™ technology? And what are kodecytes?
KodeTM is a surface cell attachment technology which enables attachment of synthetic forms of αGal and Fs antigens in the form of function-spacer-lipids (FSLs) constructs. Once FSL attachment has taken place, the red blood cells are called kodecytes and they can specifically bind the corresponding antibodies if present.

The Team at NZIMLS thank you for all your support during 2024!
Our office will be closed from Monday 23 December 2024, reopening Monday 13 January 2025




The Christmas quiz was very successful again last year and the NZIMLS Council has agreed to sponsor another quiz this year. As with last year it is a General Knowledge Quiz and the NZIMLS Council has agreed to award a single prize of $300 towards a laboratory Christmas function for the most correct answers.
While individuals can also complete the quiz, the prize is only to a laboratory team entry to encourage teamwork. This year the question number has been increased from 50 question to 60. There are no CPD points for this quiz and it is not discipline related. The NZIMLS Council have not participated in the question setting or have knowledge of the answers. An answer sheet is available on the NZIMLS website, and the quiz answers can be submitted electronically as for the CPD questions. The answers will be published on the NZIMLS website after the quiz closes and the winner is announced.
The Closing Date for all answers is 8th December 2024 All completed quiz answers with a laboratory contact person should be sent to: mike.legge@nzimls.org.nz and subject ‘Christmas Quiz’. Please indicate if the entry is a team or individual entry No correspondence will be entered into relating to the quiz.
1. In 2013 which mystery thriller was the highest selling novel by an American author?
a. The Da Vinci code
b. Dogs of Riga
c. Lost Souls
d. Fortunes of the dead
2. Which of the following words defines crapulent?
a. Passive
b. Dutiful
c. Enraged
d. Drunk
3. The composer Franz Schubert died from which of the following diseases?
a. Tuberculosis
b. Cholera
c. Syphilis
d. Typhoid
4. George Orwell was a pen name for which person?
a. Wesley Peterson
b. Lawrence Durrell
c. Eric Blair
d. Gerald Wiley
5. Who is believed to be the first person to propose the Earth orbited the Sun?
a. Galileo
b. Aristarchus
c. Johannes Kepler
d. Aristotle
6. Who discovered the inverses square law?
a. Robert Hooke
b. Francis Bacon
c. Isaac newton
d. Robert Boyle
7. In Greek mythology who liberated Prometheus?
a. Apollo
b. Zeus
c. Poseidon
d. Hercules
8. Which painter is strongly associated with landscapes from the West Coast of New Zealand?
a. Bill Hammond
b. Colin McCahon
c. Toss Woolaston
d. Ralph Hotere
9. Who is quoted as saying “he is going around the country stirring apathy”?
a. Geoff Palmer
b. Winston Peters
c. Rob Muldoon
d. David Lange
10. When was the Edmonds Cookbook first published?
a. 1920
b. 1908
c. 1918
d. 1910
11. Which New Zealand sportsman had the nickname, “Paddles”?
a. Richard Hadlee
b. Nathan Cowan
c. Hamish Carter
d. Ian Fergusson
12. Who wrote the 1979 novel “Leaves of the Banyan tree”?
a. Witi Ihimaera
b. Albert Wendt
c. Haruki Murakami
d. Lloyd Jones
13. What is the approximate length of the New Zealand coastline?
a. 8,000km
b. 20,000km
c. 15,000km
d. 11,000km
14. What is Harold Welman considered to be the most influential scientist for?
a. Geothermal research
b. Alpine fault
c. Glaciology
d. Vulcanology
15. In which year is it considered that the Tasman Lake did not exist
a. 1976
b. 1968
c. 1957
d. 1973
16. Who of the following scientists founded the Cavendish Laboratories at Cambridge University in the UK?
a. Joseph John Thomson
b. James Clerk Maxwell
c. William Lawrence Bragg
d. Ernest Rutherford
17. What was the nationality of the first person to demonstrate the properties of electromagnetic waves as radio waves?
a. German
b. Indian
c. English
d. Italian
18. Who originally proposed the “uncertainty principle”?
a. Werner Heisenberg
b. Niels Bohr
c. Albert Einstein
d. Max Planck
19. From which of the following countries did a cartographer produce the first sailing map useful for ocean navigation?
a. Spain
b. Holland
c. England
d. Portugal
20. Where was the opium poppy first domesticated?
a. Pakistan
b. Greece
c. Egypt
d. Italy
21. Who invented the sea-going chronometer?
a. James Cook
b. Nevil Muskelyne
c. John Harrison
d. James Bradley
22. Which European country is believed to be the first to cultivate coffee plants?
a. Italy
b. Holland
c. Germany
d. France
23. Where was the pizza originally created?
a. Florence
b. Rome
c. Naples
d. Pisa
24. Who is considered to have created the first postal system?
a. Cyrus the Great
b. Henry VIII
c. Augustus
d. Herodotus
25. Which of the following battles led to the formation of the Red Cross?
a. Solferino
b. San Martino
c. Magenta
d. Boffalora
26. The “sumptuary laws” in Elizabethan England at the time of Shakespeare were established for what reason?
a. Remove class structure
b. To prevent women being actors
c. To allow boys to take women actor parts
d. Maintain class structure in society
27. What was the nationality of the person believed to have invented the decimal point?
a. English
b. Indian
c. German
d. Italian
28. What does the female monotreme, the duck billed platypus, do with her eggs after laying them?
a. Coat them with mucous to retain moisture
b. Curls her tail around them until hatching
c. Places them in a pouch
d. Burys them in moss to hatch
29. Which ancient civilisation invented the sexagesimal system used in GPS today?
a. Greeks
b. Babylonian
c. Sumerians
d. Egyptians
30. How old was Dame Kiri Te Kanawa when she made her first public broadcast?
a. 8 years old
b. 6 years old
c. 10 years old
d. 12 years old
31. What was the name of the fictitious detective Hercule Poirot’s secretary?
a. Miss Grape
b. Miss Peach
c. Miss Berry
d. Miss Lemon
32. What event initiated the start of the self-operated lifts (elevators)?
a. Reliable lift mechanisms
b. Improved safety mechanisms
c. Lift operators concern for safety
d. Lift operators strike in New York
33. Which of the following authors was considered to have possibly written the first scientific best seller?
a. Mary Sommerville
b. Robert Hooke
c. Laura Bassi
d. Charles Darwin
34. Where would gluons be found?
a. Atomic nucleus
b. Starship Enterprise
c. Dipole-Dipole forces interactions
d. Supernova
35. When were instant noodles first marketed?
a. 1958
b. 1947
c. 1951
d. 1967
36. Uriah Heep is a character in which Charles Dickens novel?
a. David Copperfield
b. Oliver twist
c. A Christmas Carol
d. The Old Curiosity Shop
37. Who was the first person to use the word ’cell’?
a. Albert Von Kolliker
b. Robert Hooke
c. Charles Darwin
d. Anton Von Leuwenhoek
38. Which of the following is the oldest currency still in circulation?
a. US dollar
b. Japanese Yen
c. Russian Ruble
d. British Pound
39. A grimalkin is an old word for which of the following?
a. Cat
b. Orphan
c. Spell book
d. Gnome
40. What nationality was the person who invented the paint tin lid?
a. German
b. New Zealand
c. American
d. Australian
41. Who wrote the “Seven Pillars of Wisdom”?
a. GB Shaw
b. DH Lawrence
c. TE Lawrence
d. Graham Greene
42. In which country was the double entry bookkeeping invented?
a. Italy
b. Holland
c. Germany
d. France
43. Halley’s Comet passed in 1996. When is it next expected to appear?
a. 2056
b. 2061
c. 2042
d. 2082
44. Which chemical element is named after the Greek for “light bearing”?
a. Magnesium
b. Phosphorus
c. Sodium
d. Sulphur
45. Who might wear a “ghillie suit”?
a. Diver
b. Astronaut
c. Sniper
d. Fire fighter
46. Which European city was the first to establish a scientific academy?
a. Berlin
b. Florence
c. Paris
d. London
47. What would the Bruun rule be used for?
a. Retreat of seashore
b. Predicting tides
c. Coastal navigation
d. Predicting ocean currents
48. Where was the Silk Road explorer Marco Polo born?
a. Florence
b. Venice
c. Pisa
d. Rome
49. Harry Potter manuscripts by JK Rowling were rejected how many times by publishers?
a. One
b. Four
c. Twelve
d. Six
50. The word rorulent means which of the following?
a. Opulent
b. Verbose
c. Covered in dew
d. Argumentative
51. Where is the Arafura Sea?
a. South of Japan
b. North of Russia
c. South of Antarctica
d. North of Australia
52. In which year was the ‘pop-up’ toaster patented?
a. 1915
b. 1919
c. 1931
d. 1938
53. What was the name of the cat that featured in the original Alien film?
a. Hermes
b. Moriarty
c. Jonesy
d. Sylvester
54. How many quarks make a baryon?
a. One
b. Two
c. Three
d. Four
55. Which of the Beatles is believed to remortgage his house to fund the making of “Life of Brian”?
a. George Harrison
b. John Lennon
c. Ringo Starr
d. Paul McCartney
56. What is the number of the address of Sherlock Holmes?
a. 221C
b. 221B
c. 221A
d. 221
57. What is the frequency of a male tortoiseshell cat occurring?
a. 1 in 3,000
b. 1 in 1,000
c. 1 in 10,000
d. 1 in 500
58. What was the name of the pub in England where Francis Crick allegedly announced they had discovered the “secret of life”?
a. The Lamb
b. The Fox and Hounds
c. The Eagle
d. The Rabbit and Ferret
59. When was the computing word “bit” invented?
a. 1940s
b. 1950s
c. 1960s
d. 1970s
60. The mountain shown in the Paramount film introduction is styled on which of the following mountains?
a. Ben Nevis
b. K2
c. Annapurna
d. Ben Lomond






Contact - Ajesh Joseph (SIG Convenor)
Email:Ajesh.Joseph@waikatodhb.health.nz






