
2023/24


2023/24
A year of scientific creativity

Meet the interdisciplinary teams taking on cancer’s toughest challenges


$125m for five new teams to take on four challenges

A growing global community of 1,200+ investigators and collaborators
Collaboration and community are essential elements of transformative research. From designing novel T-cell therapies for paediatric tumours to developing new CRISPR-based technologies to study genetic alterations in cancer, over the past year, our Cancer Grand Challenges community has continued to demonstrate the power of global collaboration in making progress against cancer.



For the first time, in this year’s Discover we can begin to reflect on the impact of the work of our first four funded teams – a culmination of seven years of research – as they approach the end of their funding periods. Their work has changed the way we study and understand cancer, and is now revealing potential new avenues for therapy and prevention.

Dr David Scott Director, Cancer Grand Challenges, Cancer Research UK
In tackling our Unusual Mutations Patterns challenge, Team Mutographs has shifted the debate around carcinogenesis by reviving the theory that not all carcinogens cause cancer by directly mutating DNA, but some instead cause cancer through non-mutagenic mechanisms (eg, promotion). For the Lethal versus Non-Lethal Cancers challenge, Team PRECISION has made important strides in unravelling the biology underpinning how ductal carcinoma in situ is initiated and progresses, which are now shedding new light on how ductal carcinoma in situ should be managed clinically. Teams IMAXT and Rosetta, addressing the 3D Tumour Mapping challenge, have been at the vanguard of the spatial biology revolution. Both teams have developed multiple novel technologies to interrogate tumours at the molecular and cellular levels, at a resolution not possible before,
and as a result are opening new avenues for diagnosing and treating cancers. You can read about the science behind some of their discoveries on pages 6, 34 and 40. The progress that our teams are making would not be possible without the creativity of hundreds of PhD students and postdoctoral researchers. Over the past year, we have continued to work with this cohort to develop our Future Leaders Programme, and you can read about the impact that being part of Cancer Grand Challenges has had on the careers of some members of this cohort on page 10. Back in March 2023, we launched our most recent set of challenges, and we were delighted by the response from the community; we received 176 Expressions of Interest, more than ever before. After a rigorous review process, we’re delighted to announce our newest Cancer Grand Challenges teams and the challenges they will be tackling on page 24. It is fantastic to witness the Cancer Grand Challenges community continuing to grow, and we can’t wait to see the collaborations that are sparked across our community as these new teams come on board. We look forward to continuing to work with the global research community to shape the future of our initiative and to drive the research advancements that will ultimately change the lives of people affected by cancer.
We hope you enjoy reading the 2024
Cancer Grand Challenges supports a global community of world-class, interdisciplinary research teams to come together, think differently and take on some of cancer’s toughest challenges, with the ultimate aim of transforming outcomes for people affected by cancer.
Founded by two of the largest funders of cancer research in the world – Cancer Research UK and the National Cancer Institute in the US – Cancer Grand Challenges has the power to create change. The initiative is also supported by Cancer Research UK’s global network of partners and philanthropists who share our ambitions.
By galvanising the international scientific community and giving our researchers the freedom to innovate, we are accelerating the discoveries that are needed to make substantial progress against cancer.



1,200+ investigators and collaborators* $400m invested* 16 interdisciplinary teams* 13 challenges being tackled* 16 countries* 92 research institutions*



*Cumulative figures.

Cancer Grand Challenges leads a global debate with the research community and people affected by cancer to identify cancer’s most complex challenges, and then dares global, interdisciplinary research teams to apply to take them on. Following the announcement of five new teams in March 2024, the Cancer Grand Challenges community has now taken on 13 cancer challenges to date. Read more about the new teams and the challenges they're tackling on page 24.
Cancer inequities
Understand the mechanisms through which genetics, biology and social determinants affect cancer risk and outcomes in diverse populations, to motivate interventions to reduce cancer inequities
Normal phenotypes
Understand how cells and tissues maintain 'normal' phenotypes whilst harbouring oncogenic mutations and how they transition to become a tumour
3D tumour mapping
Find a way of mapping tumours at the molecular and cellular level
Early-onset cancers
Determine why the incidence of early-onset cancers in adults is rising globally
Solid tumours in children (2020)
Develop novel therapies to target unique features in solid tumours in children (2023)
Develop therapeutics to target oncogenic drivers of solid tumours in children
Cachexia
Understand and reverse cachexia and declining performance status in cancer patients
Extrachromosomal DNA
Understand the biology of ecDNA generation and action, and develop approaches to target these mechanisms in cancer
T-cell receptors
Decipher the T-cell receptor cancerrecognition code
Cancer causes
Determine the mechanisms that cause cancer without known mutagenesis, such as obesity, in order to devise novel interventions
Lethal vs non-lethal cancers
Distinguish between lethal cancers that need treating and non-lethal cancers that don’t
Tissue specificity
Devise approaches to prevent or treat cancer based on mechanisms that determine tissue specificity of some cancer genes
Microbiota
Improve treatment responses by manipulating the composition and status of the microbiota
Unusual mutation patterns
Discover how unusual patterns of mutation are induced by different cancer-causing events
In 2015, Cancer Grand Challenges set the Unusual Mutations Patterns challenge, to discover how cancer-causing events induce unusual patterns of mutation. In tackling this challenge over the past seven years, and through uniting the fields of epidemiology and cancer genomics, the Mutographs team has transformed how the field thinks about mutations and the causes of cancer.
Featured challenge: Unusual Mutation Patterns
Featured team: Mutographs

bring this promotion hypothesis to the fore again, by showing that normal, healthy tissues indeed carry many cell clones with cancer driver mutations.
Dangerous levels of exposure to a carcinogen can elicit specific patterns of DNA damage called mutational signatures. If a tumour eventually develops, these mutational signatures can be used to ‘work backwards’ to identify exposure to both known and previously unknown carcinogenic agents. Understanding the mechanisms that lead from an initial exposure to mutation might hold promise in uncovering new preventive targets to stop, delay or weaken carcinogenic effects.
This was the vision for the Unusual Mutations Patterns challenge set by Cancer Grand Challenges in 2015 and, in 2017, the Mutographs team, led by Mike Stratton of the Wellcome Sanger Institute, UK, was awarded funding to take on this challenge. Over its funding period, the team has shifted the consensus around how mutations cause cancer and revolutionised the field of cancer epidemiology. They have discovered multiple widespread human mutagenic exposures, with both known and unknown causes, and highlighted the roles of non-mutagenic carcinogens in human cancer.
Two known types of carcinogens exist: mutagenic and non-mutagenic. Mutagenic carcinogens, such as tobacco smoke and ultraviolet light, increase the chances of cancer ‘driver’ mutations occurring. Non-mutagenic, or ‘promotional’, carcinogens increase the chance that normal cells with pre-existing driver mutations will become cancer cells. This concept is supported by a long history of experimental rodent research, and recent studies, including those by the Mutographs team, have helped
Mutographs has been tackling the challenge of unravelling these phenomena, by uniting global, interdisciplinary teams conducting work at the intersection of epidemiology and genomics. By studying the mutational signatures in the genomes of cancer cells versus normal cells in countries with high and low incidence of specific cancer types, the team has been working to catalogue the mutational processes that cause cancer in humans, understand their causes and apply this knowledge to cancer prevention.
“From an epidemiological perspective, the team represents a significant advancement in our understanding of the environmental and lifestyle causes of cancer,” says Elisabete Weiderpass, director of the International Agency for Research on Cancer (IARC), France. An alternative view of carcinogenesis
One arm of Mutographs has used animal models to identify, characterise and understand the biological processes underlying mutational signatures. Findings from a 2020 Nature Genetics study, led by Mutographs co-investigator Allan Balmain of the University of California San Francisco, US, and Mutographs collaborator David Adams of the Wellcome Sanger Institute, suggested an alternative view challenging the long-held notion that carcinogens directly cause mutations.
In the study, the team conducted the first survey of a broad range of known or suspected environmental carcinogens in mouse models accurately representing exposure. Many of these potential carcinogens were expected mutagens, including trichloropropane (TCP), which is often present in the drinking water near toxic-waste sites. The team’s

aim was to classify these chemicals either as mutagens or as nonmutagenic tumour promoters.
To the researchers’ surprise, only three of the 20 tested compounds, including TCP, showed clear evidence of mutagenic activity. Allan says that mutations can be found in mice due to exposure to mutagens such as TCP, or because of simple mistakes that are made in cells replicating their DNA. The presence of these mutations ensures that cancer has the potential to develop during the entire lifetimes of exposed animals. In some cases, the mutations are essential but not sufficient for tumour development. The role of the non-mutagenic promoters is to awaken these dormant mutated cells so that they can grow and become cancerous.
“Our study indicated that the vast majority of known or suspected human carcinogens are not in fact mutagens,” says Allan. “That was pretty dramatic, because many people suspected there were all these chemicals that in some way cause mutations that contribute to cancer. Our study showed, however, that many chemicals do not seem to do that.”
Mutographs has shown that this research is relevant in human cancers. For example, the team
looked at oesophageal squamous cell carcinoma (ESCC) cases from eight different countries with widely differing incidence rates, including Iran, Kenya and China. Surprisingly, they found no differences in mutational loads or mutational signatures in these tumours – nothing that indicated that a chemical or some other factor caused mutations that led to ESCC.
This landmark study, published in Nature Genetics as well as the seminal discoveries in mice, shifted thinking to reposition promotion as a key mechanism in cancer initiation.
As in all Cancer Grand Challenges teams, patient advocacy is hugely important. For Mutographs, the patient advocates were given the task of helping to spread any public health messages that come from the research of the team. To do this effectively meant that they had to immerse themselves in the research unique to each country where the team had sourced patient samples.
Mutographs patient advocate Mimi McCord participated in the study, travelling to Kenya and other countries. “My involvement helped me to have a perspective of how cancer is perceived and the stigma that it has, particularly in Kenya,” says Mimi. “The patient advocates put a human element into a very science-based project, which may ultimately benefit patients in the future.”
These findings, which really have changed the consensus on the relationship between mutagenic and non-mutagenic carcinogens, as well as insights from normal tissues containing cell clones with cancer-driving mutations, prompted Cancer Grand Challenges to set the Normal Phenotypes challenge in 2020. The challenge called for teams to look beyond the accumulation of mutations and understand in molecular detail how a normal cell harbouring a cancer mutation is restrained from oncogenic progression and then identify the critical steps for progression at the earliest steps of tumour development.
Building on findings from Mutographs, the PROMINENT team, led by Allan alongside Paul Brennan of IARC (who is also a co-investigator in Mutographs) and Núria López-Bigas of the Institute for Research in Biomedicine Barcelona, Spain, is tackling the Normal Phenotypes challenge by investigating the promoter hypothesis as an alternative theory about the very early stages of cancer development.
Cancer promotion, and its potential importance in public health, was highlighted in a study published in Nature in 2023, led by Charles Swanton of The Francis Crick Institute, UK, and member of the Cancer Grand Challenges Scientific Committee.


The OPTIMISTICC team is tackling our Microbiota challenge, which aims to improve treatment responses by manipulating the composition and status of the microbiota.
OPTIMISTICC is studying the gut microbiota to understand how these microbes drive bowel cancer and treatment response.
In a collaboration between Matthew Meyerson, OPTIMISTICC co-team lead of Harvard Medical School, US, and Mutographs members Mike Stratton and Paul Brennan, OPTIMISTICC extended its studies to ESCC to determine whether oral bacteria are linked to the varying incidence rates seen in this cancer type.
The researchers analysed DNA and/or RNA sequencing data of ESCC tumours from patients from high- (Iran, Tanzania, Kenya, Malawi and China) and low-incidence (the US, Ukraine, Vietnam and Russia) regions. Cancer-associated, traditionally oral bacteria were found at high levels in ESCC tumours from patients in the regions of high ESCC incidence.
The researchers found that tiny particles in vehicle exhaust and smoke from fossil fuels are associated with non-small cell lung cancer. These particles promote lung cancer by acting on cells bearing pre-existing mutations in the EGFR and KRAS genes. EGFR mutations are present in approximately half of all people with lung cancer who have never smoked.
“We see differences in mutation loads and mutation patterns in different parts of the world,” explains Mike. “We can’t be absolutely sure what is happening in the whole population, but tens of millions of people could be affected.”
Genome topography drives mutations
in Cancer (COSMIC), the world’s largest, most comprehensive resource for exploring the interaction between mutational signatures and topographic features within and across cancer types.
Paving the way for potential new approaches to cancer prevention

Sanger Institute, UK
The researchers also analysed the microbiomes of matched ESCC saliva and tumour samples from patients in Tanzania. They found a correlation between the genus composition of the saliva microbiome and the ESCC tumour microbiome in these patients.
Featured challenge: Microbiota
Featured team: OPTIMISTICC

Featured team member: Professor Matthew Meyerson OPTIMISTICC co-team lead, Dana-Farber Cancer Institute, US
Featured publication:
Nomburg J et al. Int J Cancer 2022; doi: 10.1002/ijc.34212
OPTIMISTICC is funded by: Cancer Research UK
The findings suggest that environmental triggers awaken normal cells that carry cancer-causing mutations. In combination with the findings from the Mutographs team, this work is revolutionising the field of cancer epidemiology and may lead to the development of new prevention strategies and therapies for lung cancer.
Understanding geographical differences in cancer incidence
Through a powerful alliance between the Wellcome Sanger Institute and IARC, Mutographs has driven a whole new approach to cancer epidemiology.
In emerging research, Mutographs has examined the geographic variation of mutagenic exposures in kidney cancer. Researchers sequenced clear cell renal cell carcinomas, the most common type of kidney cancer, from 11 countries and found that somatic (acquired) mutation profiles differed among the countries. In Romania, Serbia and Thailand, for example, mutational signatures likely to be caused by acids of Aristolochia plants were present in most cases but rare in other countries. In Japan, a distinctive mutational signature was linked to about 70% of kidney cancers, which the researchers did not find elsewhere.
The results indicate the existence of multiple, widespread, geographically variable mutagenic exposures to known and unknown causes, which could contribute to the incidence of kidney cancer.
Underpinning the discoveries from the Mutographs team, Ludmil Alexandrov, Mutographs co-investigator of the University of California San Diego, US, has developed a suite of AI-powered tools, called SigProfiler, to identify and analyse mutational signatures. These computational tools have become widely used by the scientific community.
In a study published in Cell Reports he and Burçak Otlu, a former postdoctoral researcher in Ludmil’s laboratory and now an assistant professor at Middle East Technical University, Turkey, used SigProfiler to show that mutations marked by mutational signatures are affected by the shape, structure and characteristics of the genome. The study integrated mutations from 5,120 wholegenome-sequenced tumours from 40 cancer types with 516 distinct topographical features.
“Our findings suggest that certain mutagenic processes are breaking the rules in protected regions of the genome,” says Ludmil. “Rather than occurring randomly, as many people thought, these mutations accumulate in particular areas based on the architecture of the genome.”
“Studying signatures through a topographical lens will allow us to identify key driver processes and mutations underlying cancer formation,” adds Burçak, “which will ultimately help us to create better diagnostic and prognostic tools.”
The features that Ludmil and his team identified are now available to other researchers through the Catalogue of Somatic Mutations
Mutographs has transformed the field of cancer genomic epidemiology, and has advanced understanding of the lifestyle and environmental causes of cancer. The team has changed the scientific understanding of the mechanisms of carcinogenesis by discovering multiple, widespread human mutagenic exposures of known and unknown causes, and has also changed how cancer biologists think about the mechanisms of carcinogenesis as well as the somatic mutation landscape of normal tissues in health and disease.

Dr Elisabete Weiderpass Director, International Agency for Research on Cancer (IARC), France


Mutographs patient advocate, UK

Dr Paul Brennan PROMINENT co-team lead and Mutographs co-investigator, IARC, France
This research has the potential to improve our ability to reduce cancer risk, develop more effective prevention and treatment strategies and inform government policies to address specific cancer risks.
Elisabete Weiderpass




Professor Núria López-Bigas PROMINENT coteam lead, Institute for Research in Biomedicine, Spain
Professor Charles Swanton Cancer Grand Challenges Scientific Committee member
Professor Ludmil Alexandrov Mutographs coinvestigator, University of California San Diego, US
Dr Burçak Otlu Former Mutographs team member, Middle East Technical University, Turkey
Featured publications
Riva L et al. Nat Genet 2020; doi: 10.1038/s41588-020-0692-4
Moody S et al. Nat Genet 2021; doi: 10.1038/s41588-021-00928-6
Hill W et al. Nature 2023; doi: 10.1038/s41586-023-05874-3
Otlu B et al. Cell Rep 2023; doi: 10.1016/j.celrep.2023.112930
Future leaders are key members of each Cancer Grand Challenges team who are currently in the early stages of their scientific careers. Discover spoke with two future leaders and Cancer Grand Challenges alumni – Mia Petljak, former member of the Mutographs team taking on our Unusual Mutation Patterns challenge, and Peter Kreuzaler, former member of the Rosetta team taking on our 3D Tumour Mapping challenge – who recently established their own laboratories at NYU Langone Health, US, and the University of Cologne, Germany, respectively.


How has Cancer Grand Challenges helped you develop your scientific career?
MP: I joined Cancer Grand Challenges during my transition from a PhD at the University of Cambridge to a postdoctoral position at the Broad Institute of MIT and Harvard, with a brief fellowship at Vertex Pharmaceuticals in between. Joining the Mutographs team enabled me to maintain and expand crucial collaborations, as well as access funding for pursuing outstanding questions following my doctoral studies. Above all, joining Cancer Grand Challenges enabled me to establish an amazing network of colleagues with whom I will certainly collaborate in the future.
PK: Cancer Grand Challenges is unique in that it doesn’t act as only a funding body but instead, through various meetings and outreach activities, strives to build a community beyond the individual Cancer Grand Challenges teams, encompassing an ever-growing and diverse group of scientists. Also, in a scientific environment where it is important to develop a clear and distinct profile, being involved in projects that are easily recognised as being at the forefront of science is exactly what will give you the unique selling point likely to secure your next position.
What did you learn about team science and working on an international, interdisciplinary team?
MP: I’ve gained a deep appreciation for the synergy created by diverse expertise within international, interdisciplinary teams. In science, proficiency transcends borders, making global collaboration essential. I now prioritise working with the best experts, navigating any potential technical challenges linked to international collaboration.
PK: On an interdisciplinary team, we have to give each other time to learn each other’s ‘language’. Our team had biologists, physicists and data scientists all working together, and we all went through a steep learning curve. But, with trust and time, we managed to understand the motivation behind each other’s choices, and that is when things usually started to take off.
Being involved in projects that are easily recognised as being at the forefront of science is exactly what will give you the unique selling point likely to secure your next position. “
Peter Kreuzaler
Joining the Mutographs team enabled me to maintain and expand crucial collaborations, as well as access funding for pursuing outstanding questions following my doctoral studies. “
Mia Petljak
What advice do you have for future leaders engaging in this programme, and how can they make the most out of being part of this community?
MP: Network, network, network. Embrace the power of connecting with both scientists and patient advocates within and beyond your team. Cancer Grand Challenges provides a unique opportunity to forge connections, make friends, blend ideas and foster impactful collaborations.
PK: I would advise any future leader to build their own network of peers. Within the boundaries of the project, engage with those who directly carry out the work – the postdocs and PhD students. Touch base with your PI [principal investigator], but be the driver of your work, and build your team of collaborators as independently as possible.

The Future Leaders Programme offers early career scientists opportunities to network with their peers, exchange ideas and learn from established investigators. It also provides career development opportunities, and the programme is currently expanding in remit and ambitions.
Current opportunities include access to exclusive events, such as:
Future Leaders Conference
Generously supported by Bjorn and Inger Saven. This annual conference comprises scientific sessions, plenary talks, career development and networking, and social programmes.
Cancer Grand Challenges Summit
This annual summit brings together our global research community to celebrate research progress, and enables future leaders to meet and interact with our international team leads, coinvestigators, patient advocates, philanthropists and members of the Cancer Grand Challenges Scientific Committee.
Webinar series
A monthly series in which future leaders showcase their work in progress, expand their knowledge in multiple areas of research and learn from experts in various disciplines.
Team-specific research retreats
These bring together all members of a team to provide progress updates, exchange ideas and brainstorm new research directions.

From using mouse models to interrogate the effects of a ketogenic diet on tumour growth and in cachexia, to identifying a novel mechanism of oncogene amplification in breast cancer following oestrogen-induced DNA breaks, it’s been another exciting year for our teams.
Dr Gemma Balmer-Kemp
Head of Research, Cancer Grand Challenges

The scope of research being undertaken across the Cancer Grand Challenges portfolio is truly impressive. Here are just a few examples of how the teams are continuing to push the boundaries of science to drive progress in their challenge areas.
Dr Andy Kurtz
Programme Director, Center for Strategic Scientific Initiatives,

JAN 2023
Characterising earlyonset colorectal cancer
The incidence of early-onset colorectal cancer (diagnosed before the age of 50 years) has increased. Understanding how the molecular features of these tumours differ by age and location is important for personalised disease management. In an analysis of 3,089 early-onset cases, the OPTIMISTICC team has found that the proportion of abnormal molecular and genetic features in early-onset tumours is generally highest in the transverse and ascending colon, and lowest in the rectum, whereas the prevalence of KRAS mutations in both early-onset and later-onset tumours is highest in the caecum. The findings, published in the American Journal of Gastroenterology, provide a rationale for large-scale studies investigating detailed colorectal cancer subsite data.
Ugai T et al. Am J Gastroenterol 2023; doi: 10.14309/ ajg.0000000000002171

MAY 2023
Sequencing extrachromosomal DNA in single cancer cells
Extrachromosomal circular DNA, small circular DNA particles found in nearly one-third of cancers, enable cells to rapidly change their genomes and drive adaptive evolution in diverse organisms. Questions about the origin, structural dynamics and effects on the intratumour heterogeneity of these molecules remain unanswered. In Nature Genetics, the eDyNAmiC team has described a new method for parallel sequencing of all circular DNA types –regardless of size, content and copy number – and full-length messenger RNA in single cells. The researchers anticipate that this method will provide a resource for future research answering unresolved questions regarding circular DNA.
Chamorro González R et al. Nat Genet 2023; doi: 10.1038/s41588023-01386-y


JUNE 2023
Predicting colorectal cancer survival
The prediction of colorectal cancer survival according to routine tumour node metastasis (TNM) staging is imperfect. The OPTIMISTICC team developed a model, based on a statistical learning technique called Bayesian additive regression trees (BART), that incorporates TNM staging with other clinicopathologic, immune, microbial and genomic factors, to improve mortality risk stratification for advanced-stage patients. Their model, published in npj Precision Oncology, identified seven predictors of stable survival – including positive lymph node count, depth of tumour invasion and extent of extraglandular necrosis – that provide additional patient-specific survival information that can be applied in clinical oncology practice.
Zhao M et al. npj Precis Onc 2023; doi: 10.1038/s41698-023-00406-8


JUNE 2023
Identifying a new mechanism of oncogene amplification in breast cancer
In a study published in Nature the SPECIFICANCER team, taking on the Tissue Specificity challenge, used whole-genome sequencing, RNA sequencing and epigenomic data to uncover the origins of focal oncogene amplifications in breast cancer. The team identified a mechanism they call ‘translocationbridge amplification’ through which oestrogen-induced DNA breaks and their repair by inter-chromosomal translocations initiate a cascade of events that result in oncogene amplification. The findings add to a growing body of work that implicates oestrogen-induced DNA breaks as a potent driver of breast tumour development.
Lee JJ-K et al. Nature 2023; doi: 10.1038/s41586-023-06057-w

JUNE 2023
Understanding the impact of a ketogenic diet on cancer cachexia
Cachexia is a severe wasting syndrome characterised by weakness, and a loss of body weight and muscle mass. A highfat, low-carbohydrate ketogenic diet (KD) has been explored as an intervention in the end stages of cancer associated with cachexia. Utilising two interleukin6-associated mouse models of cancer cachexia, the CANCAN team, taking on our Cachexia challenge, has found that, although a KD slows tumour growth, it shortens overall survival by accelerating cachexia onset. The differing effects were the result of the biochemical interaction of two simultaneously occurring pathways that both depend on the same cofactor molecule –NADPH. The findings, published in Cell Metabolism, may be relevant to researchers investigating nutritional interventions, such as KDs, for patients with cancer.
Ferrer M et al. Cell Metab 2023; doi: 10.1016/j.cmet.2023.05.008

In taking on our Cachexia challenge, CANCAN co-investigator
Norbert Perrimon, in his laboratory at Harvard Medical School, US, is using Drosophila in a cross-species approach to study and understand cancer cachexia.
Featured challenge: Cachexia
Featured team: CANCAN
funded by: Cancer Research UK and the National Cancer Institute
Cachexia is a complex wasting syndrome affecting as many as 80% of people with advanced cancer. The syndrome is characterised by substantial weight loss from both skeletal muscle and fatty tissue, which cannot be reversed by nutritional support. It is estimated to lead to one-third of cancer deaths, often from heart or respiratory failure caused by muscle loss.
“Our lab is studying interorgan communication in Drosophila, in particular in the context of cachectic tumours,” explains Norbert. “This has led us to identify factors derived from tumours that affect the physiology and metabolism of peripheral organs.”
Drosophila is a classical model system for teasing apart the intricate interactions among organs and the roles of hormones in coordinating the state of one organ or tissue with others. These aspects can be investigated under both normal and challenged conditions, including disease and changes in nutrition – both of which are relevant to cachexia.
Norbert and his team are using Drosophila to investigate tumourderived factors and the tissues on which they act to induce wasting. Additionally, in collaboration with CANCAN co-team leads Marcus DaSilva Goncalves of Weill Cornell Medicine, US, and Tobias Janowitz of Cold Spring Harbor Laboratory, US, the team is exploring the relevance of the mechanisms identified in fruit flies to mouse models of cachexia. If their fly discoveries prove relevant in mice, Norbert and his colleagues will investigate whether similar processes and mechanisms occur in patients with cachexia.
“Time is of the essence when we think about how to help patients with cachexia, because they are very sick,” says CANCAN patient advocate Amy Moore. “We are confident that this work in fruit flies will yield new insights that help break open the field in a way that brings meaningful benefit to those impacted by this syndrome.”
“One lesson from cross-species studies, such as the ones underway by the CANCAN team, is the remarkable conservation of mechanisms,” explains Eileen White, CANCAN co-team lead of the Rutgers Cancer Institute, US.
“Over the past several years, many fundamental aspects of cachexia physiology have been found to be conserved between Drosophila and mammals.” New cancer models have also enabled researchers to identify conserved mechanisms underlying the processes through which tumours affect the entire organism.
Modelling yeast to understand extrachromosomal DNA
In the laboratory of eDyNAmiC co-investigator Jef Boeke of New York University, US, PhD student Dominika Wawrzyniak is building a yeast-based model of extrachromosomal DNA (ecDNA) – small circular DNA particles that can drive tumour evolution – to study important genes and molecular processes in cancer. The model, called ecDNA launcher, induces ecDNA biogenesis in a controlled manner from the genome of Saccharomyces cerevisiae commonly known as brewer’s yeast. Genes that regulate DNA rearrangements, which Dominika hypothesises are involved in ecDNA physiology, are highly conserved between S. cerevisiae and humans.
“
Because of the fundamental conservation across species, I have no doubts that what we discover in Drosophila in the context of interorgan communication and cachexia is highly relevant to humans.
Norbert Perrimon
Featured team members





With the yeast ecDNA launcher, Dominika plans to perform high-throughput screening experiments to identify endogenous factors involved in ecDNA physiology. The similarities between S. cerevisiae and humans enable the findings to be extended to human and mammalian analogues, to provide a basis for subsequent studies in human cell models.
“Yeast as a model organism gives us access to a sophisticated molecular toolbox that allows a wide variety of genomic engineering that is too difficult or laborious in mammalian cells,” says Dominika. “Working with yeast is fast and scalable, allowing for chemical screens to identify potential ecDNA-related genes and drugs that interfere with them.”
Featured challenge: Extrachromosomal DNA
Featured team: eDyNAmiC
Featured team members:


eDyNAmiC is funded by: Cancer Research UK and the National Cancer Institute




In 2017, Cancer Grand Challenges funded its first four teams taking on the toughest challenges in cancer. As those teams near the end of their funding, Discover asked the four team leads – Jelle Wesseling of PRECISION, Josephine Bunch of Rosetta, Mike Stratton of Mutographs and Greg Hannon of IMAXT – about their experience in leading these teams and their advice for the future.
Can you share your reflections on the initiative?
JW: You can try to answer scientific questions within the limited space of your own backyard. However, the real puzzles in cancer can only be solved by synergistic team science. This has been very inspiring to us all. Personally, I love to work with different disciplines, with the best experts worldwide, where each discipline delivers a certain piece of the puzzle. Cancer Grand Challenges has empowered this beyond your own comfort zone at an unprecedented scale.
JB: It was an extraordinary opportunity to work with the people I was able to work with. The fact that Cancer Grand Challenges set an unbounded opportunity – where they restricted neither geographical territory, nor sector, nor type of institution, nor career stage – means that they were genuinely interested in the scientific ideas.
MS: Operating at scale has been critical for Mutographs in the alliance between cancer epidemiology and cancer genomics. The funding that each Cancer Grand Challenges team has available to it (up to $25m) makes you think at a different scale from an ordinary grant. It was that difference in mindset that was really critical for us.
GH: We have built relationships and international collaborations that I don’t think would have existed without this platform. We spent $25m to convince everybody that they should be thinking about spatial architecture, and then the whole world moves in that direction –that’s money well invested.
What has Cancer Grand Challenges done well?
JW: Cancer Grand Challenges had the courage to initiate and sustain this initiative. It empowered excellent team science on the big themes in cancer. As such, their vision was to go beyond the ‘my own backyard’ type of research.
MS: The long duration of funding changes your mindset. You get the sense that you are not in Cancer Grand Challenges to put down a couple of deliverables that three years of work will bring. You build a team with a conception about a piece of work that has long enough life to develop itself. Cancer Grand Challenges is also willing to take risks and venture into the unknown. They realise that some of the things that we would do might not give the answer we expected, and their response was very positive.
GH: The different Cancer Grand Challenges teams have different purposes and outcomes. Some are broaching a new field, some are finding new ways to address old questions, and some are very practical, finding a new way to treat cancer.
IMAXT and Rosetta were particularly adventurous from a technical capabilities standpoint. There was an element of ‘Can we do this?’ and an element of ‘If we can do it, does it matter?’
What is your advice to Cancer Grand Challenges and other teams going forward?
GH: To other teams, I would say don’t think you’ve got it right from the beginning. IMAXT only allocated about 60% of the funding at the start. We didn’t know what we were going to do with the rest of it, because we didn’t know what the world would look like in the future. If you lock yourself onto a certain path from the very beginning, that’s probably not the best approach. If you’re truly cutting-edge, you don’t have a clue what two years from now looks like, so flexibility is extremely important.
JW: The real challenge for me in cancer research is how to bridge all of our molecular efforts and nice-to-know and nice-to-have technologies to do something that has clinical impact. It’s the full chain from basic to translational to clinical research. For funding that goes beyond $25m, it’s fair to expect that a team can deliver on building these bridges, because a lot of cancer research is done, and then translation stops.
Cancer Grand Challenges is a prime example that provides the opportunity to build the bridge from A to Z.
MS: There are two things I think work very well, and I would recommend that they continue. Start with a big headline – keep the agenda large for each cancer grand challenge. Under that broad headline, allow a group of people you trust to elaborate on that agenda and develop it according to their own scientific instincts.
JB: Cancer Grand Challenges is supporting cancer research that more traditional funding sources would have been blind to. They’re not funding the usual favourites. That’s how you affect step change.


The accumulation of genetic mutations has long been associated with tumour development, but recent studies suggest that cells often carry many known cancer-causing mutations yet remain phenotypically ‘normal’. Despite having remarkable genetic similarities to cancer cells, these ‘normal’ cells do not form tumours.
Featured challenge: Normal Phenotypes
Featured team: PROMINENT
PROMINENT is funded by: Cancer Research UK, the National Cancer Institute and the Scientific Foundation of the Spanish Association Against Cancer
Featured team members

Dr Mark Taylor
PROMINENT team member, MD student and postdoctoral researcher, University of California San Francisco, US


Professor Allan Balmain
PROMINENT coteam lead, University of California San Francisco, US
Professor Núria López-Bigas
PROMINENT co-team lead, Institute for Research in Biomedicine, Spain
Recent work by the PROMINENT team, taking on the Normal Phenotypes challenge, has broken new ground by investigating dysregulated gene networks that reveal the stem cell transitions that drive carcinogenesis. The study, which emerged following work by the Mutographs team, has identified potential transition points in cells that undergo malignant conversion, and may reveal new targets for tumour prevention and therapy.
Led by Cancer Grand Challenges future leader Mark Taylor, MD student and postdoctoral researcher at the University of California, San Francisco, US, the researchers examined stem cell heterogeneity in bulk geneexpression-network data from genetically heterogeneous mouse skin carcinomas, then applied the findings to single-cell data.
“That let us see how networks are dysregulated repeatedly in cancer,” says Mark.
The data have enabled Mark and his team to create a simplified model of multistage carcinogenesis that identifies distinct stem cell states at different stages of tumour progression. He and his colleagues used long-term mouse breeding to generate populations of mice with genetic diversity mimicking that in humans, then used low levels of external environmental factors to induce tumours. This model essentially simulates how cancer develops in humans.
Most people are exposed to low levels of carcinogens starting early in life, yet they do not suddenly develop cancer when they are
young. Instead, over time, these carcinogens damage normal cells and contribute to tumour development. Consequently, malignant tumours are more likely to develop when people reach their 60s and 70s – the most common ages at which cancers are diagnosed.
“We had all these mice with perfectly normal skin,” says Mark. “They can live their whole lives with these cancer driver mutations, but when we give them a promoter agent, carcinogenesis starts to happen.”
The importance of tumour promotion in the development of cancer is once again becoming a centre piece of research, with an understanding that mutations are necessary but not sufficient for tumour development (read more on page 6). Although people cannot do much to prevent DNA mutations over their lifetime, they may be able to control some of their environmental and lifestyle factors that contribute to tumour promotion.
“This study helped us identify where initiating mutations take place in the skin, where cells acquire these mutations and how these cells change their behaviour in the earliest stages of tumour development,” explains Allan Balmain of the University of California, San Francisco, PROMINENT co-team lead and senior author of the paper.
Mark believes that, by examining the hubs that connect dysregulated gene networks, researchers may find ways to prevent tumour development or even stop the progression of cancer that has already started to form. This may lead to new combinatorial approaches for tumour prevention and therapy in clinical settings.
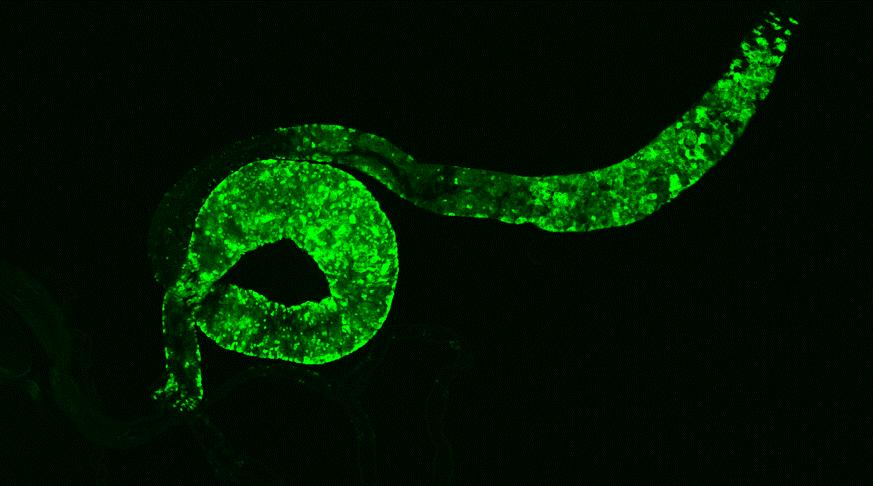
single-cell lineage tracing of Lgr6 cells (left, GFP green) and their progeny (right,
red) in papilloma. Lgr6+ stem cells (green) are localised predominantly in the basal epithelium of the papilloma. These give rise to its progeny (offspring) cells (red) that form a distinct streak rising through the epidermal layers above (suprabasally), representing a clonal expansion of this pre-malignant tumour arising from the Lgr6+ (green) cell-of-origin. Dotted lines represent the epidermal-dermal border, and nuclei were counterstained with DAPI (blue), scale bar = 50um. Credit: Eve Kandyba, University of California San Francisco, US.
Linking chemotherapy to clonal haematopoiesis
There is a very small risk of second cancer with some chemotherapy drugs. In her laboratory at the Institute for Research in Biomedicine Barcelona, Spain, PROMINENT co-team lead Núria López-Bigas is studying how chemotherapy may affect the development of future cancers. Her studies focus on how different types of chemotherapy drugs disrupt the development of healthy haematopoietic stem cells, influence the expansion of pre-existing stem cell clones, and ultimately result in clonal enlargement and the development of malignancies.
After chemotherapy, these enlarged clones can lead to clonal haematopoiesis (CH), a precancerous clonal-expansion state in the blood-forming system that increases the risk
of developing a blood cancer or other conditions, such heart attack, stroke, emphysema or liver disease.
“This is particularly important for childhood cancer survivors, as they have a long life ahead,” says Núria. “We need to better understand the effect of chemotherapy treatment in
healthy tissue as this relates to the increased risk of certain medical conditions.”
Núria and her colleagues are mapping the precise role of chemotherapy in clonal expansion and exploring models of clonal growth before and during treatment.

Patient advocacy is often prominent in clinical research, where people affected by cancer help clinical researchers identify gaps in patient care, among other issues. Cancer Grand Challenges is taking a different tack by embedding patient advocacy in basic discovery science, to ensure that the research underway by funded teams ultimately translates into tangible impact for people affected by cancer worldwide.


Patient advocates are a key pillar of the Cancer Grand Challenges community. In addition to the 12 patient advocates that make up the Cancer Grand Challenges Advocacy Panel, which is instrumental in guiding the initiative’s approach to advocacy, over 40 patient advocates are embedded across the research teams taking on the challenges.
“Patient advocacy is indispensable in basic cancer research and plays a pivotal role in shaping patientcentric studies,” says Yelak Biru, an Advocacy Panel member, and president and CEO of the International Myeloma Foundation, US, who was diagnosed with multiple myeloma at the age of 25.
Each member of the Advocacy Panel brings their own unique perspectives, skills and lived experiences to the initiative. These broad perspectives are important when it comes to helping to shape the challenges and working closely with funded teams.
the team’s research with wider audiences and the public, and ensuring that people affected by cancer have a voice that is heard and reflected in the teams’ scientific strategies.
“It’s so important to build trust between researchers and patient advocates,” says Gita. “It’s vital for collaboration, fostering engagement and bridging gaps between researchers and the people their work is ultimately for.”
Among the many reasons this is beneficial to the scientific community, one has particular resonance for Advocacy Panel member Ivana Cattaneo, of Italy, cancer policy advocate. “Patients are true experts in their disease,” she says. “Therefore, their life experiences are a fundamental source of insights that enable an in-depth understanding of the full impact of a disease.”
She believes that involving patient advocates in discovery science pays dividends in terms of efficiency, improved adherence and efficacy, and better returns for society.
Working with the teams that apply to take on cancer’s toughest challenges
For the most recent funding cycle, the Cancer Grand Challenges Advocacy Panel worked closely with the global research teams that were shortlisted to take on the newest round of challenges. Through online meetings and email exchanges, the panel provided feedback and guidance to each shortlisted team on its proposed approach to involving advocates in its research programme. This is one way the panel has been working to increase communication between its members and the patient advocates that are part of the research teams, from as early in the funding cycle as possible.
Meet our new teams on page 24.
“ The conversations with the shortlisted teams will now continue with the winning teams who have received funding, as a support and encouragement to both their patient advocates and researchers.


Gita Patel, from the UK, whose son passed away after three years with Ewings sarcoma, brings a carer’s perspective to patient advocacy and the panel. “Everyone's journey as a patient or a carer is different,” explains Gita.



“Being part of the Advocacy Panel is about giving your opinion based on your experiences, but being open to suggestions, listening and asking the simple questions that you think might be too silly to ask, so that we can come to the best decision together as a panel.”
Patient advocates embedded in the research teams play active roles through involvement and engagement, helping to share information about –and knowledge gained from –
Patient advocacy is a cornerstone of patient-centred, meaningful and ethical advancements in basic cancer research, contributing to better outcomes and progress in the fight against cancer.


With technological innovation, previously impossible feats are now shaping the nature of cancer research. Advances in technologies – such as cryo-electron microscopy, which generates high-resolution images of how molecules behave, and CRISPR gene-editing technology, which allows scientists to easily change the genetic code of living cells –have opened avenues of cancer research that were once unimaginable.
Featured challenge: Tissue Specificity
Featured team: SPECIFICANCER
is funded by: Cancer Research UK and The Mark Foundation for Cancer Research
Featured team member

SPECIFICANCER co-investigator, NYU Langone Health, US Featured publication
Technology development is a critical part of Cancer Grand Challenges’ mission of supporting a global cancer research community and accelerating transformative research. In one example, in early 2023, members of the team taking on our Tissue Specificity challenge unveiled an innovative tool to create and study chromosome mis-segregation and aneuploidy in cancer.
“We have developed a new tool called KaryoCreate [Karyotype CRISPR Engineered Aneuploidy Technology] that allows us to engineer virtually any type of chromosomal gain or loss in human cells,” explains Teresa Davoli of NYU Langone Health, US, co-investigator in the SPECIFICANCER team who led the research. “The main innovation here is the ability to use any type of cell from any type of tissue and model the desired event.”
Chromosome gains or losses, known as aneuploidy, are a hallmark of cancer. Unlike most cells in the body, more than 90% of cancer cells – particularly in solid tumours – are aneuploid. Aneuploidy is challenging to study, however, because of a lack of straightforward methods to generate models with specific chromosomes added or removed.
To address this lack of methods, SPECIFICANCER’s new CRISPRbased technology enables the creation of cells with chromosomespecific aneuploidies and then allows for these specific aneuploidies to be interrogated in the context of cancer. The method targets human centromeres, the dense and constricted regions of chromosomes that are essential for proper chromosome segregation during cell division.
Illustration of chromosomes. Credit: Science Photo Library.

In a study published in Cell, Teresa and colleagues demonstrated an immediate application of KaryoCreate by inducing chromosome 7 gain and chromosome 18 loss – two of the most frequent events in colorectal cancer – in colon epithelial cells. They found that chromosome 18 loss leads to deregulation of the critical TGF-β signalling pathway in colon cancer and the silencing of several tumour-suppressor genes on the chromosome.
“Being able to model aneuploidies associated with cancer could help us to identify not only the biological consequences of these events,” says Teresa, “but also vulnerabilities and potential drug targets that could hamper the proliferation or survival of the cells with these aneuploidies.”
Although Teresa’s study focused on cancer, KaryoCreate could be applied more generally in the aneuploidy field. For example, the tool could be used to study trisomy 21 (also called Down syndrome) or Klinefelter syndrome (in which a male is born with an extra X chromosome, which can lead to myriad medical problems later in life).
Featured challenge: Extrachromosomal DNA
According to Teresa, the next step is to use KaryoCreate to help elucidate the function of centromeres, which she calls a “mysterious entity in our genome”. By doing so, this tool would enable researchers to better understand genome instability and karyotype evolution in tumours.
The main innovation here is the ability to use any type of cell from any type of tissue and model the desired event. “
Teresa Davoli
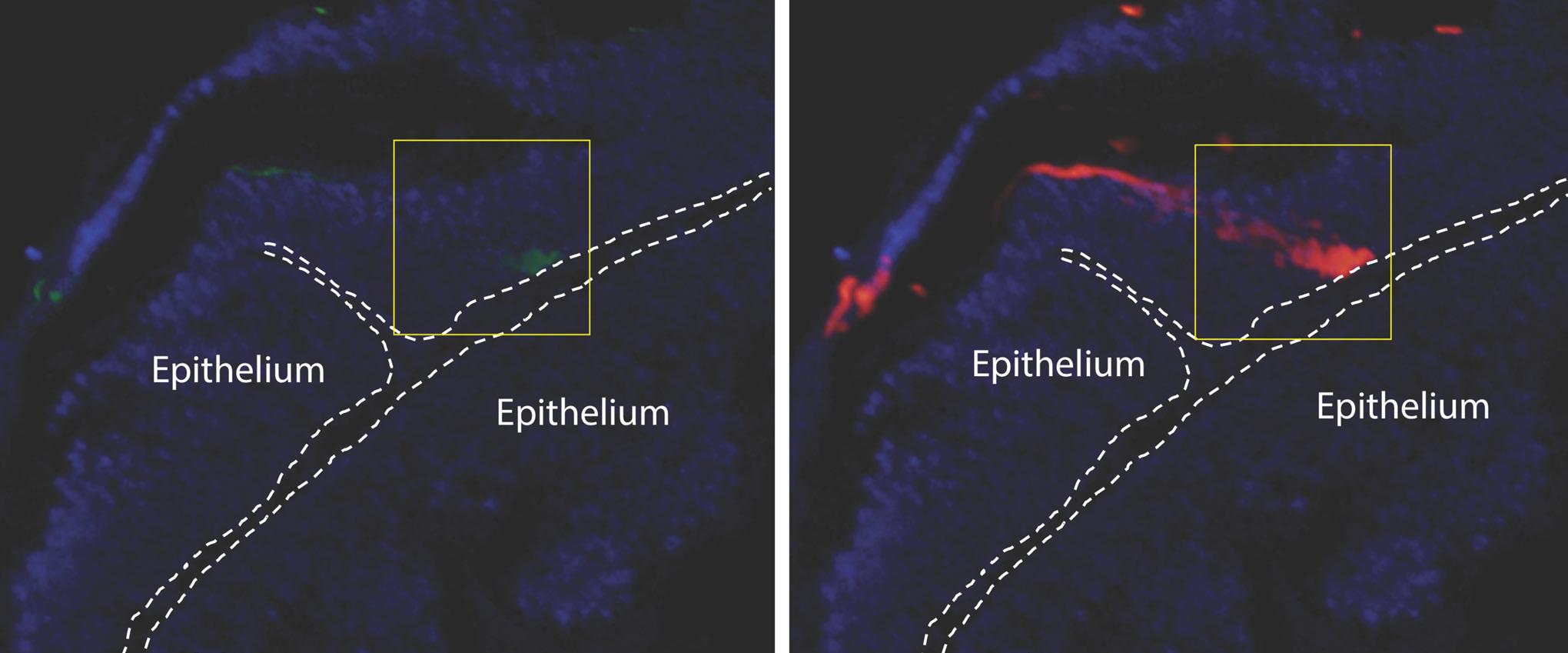
In his laboratory at Stanford University, US, eDyNAmiC coinvestigator Howard Chang invents new techniques for single-molecule analyses to study extrachromosomal DNA (ecDNA). Howard and colleagues have adapted a method called CRISPRCATCH to directly and deeply analyse ecDNA. In a study published in Nature Genetics in 2022, Howard and the eDyNAmiC team showed that CRISPR-CATCH profiling provides insight into the structure, diversity, origin and epigenomic landscape of ecDNA, and can help clarify how ecDNA oncogene amplifications are regulated in cancer cells.
Featured team: eDyNAmiC
team members:

Featured publication Hung KL et al. Nat Genet 2022; doi: 10.1038/s41588022-01190-0

CRISPR-CATCH allows the team to isolate ecDNA (shown in red) from cancer cells and patient tissue. Credit: Xiaowei Yan, Stanford University, US.

This year sees five new teams selected to take on four of the toughest challenges in cancer research. At $125m ($25m awarded to each team), this latest round of funding represents Cancer Grand Challenges’ largest investment to date.
In March 2023, Cancer Grand Challenges announced nine new challenges to the global cancer research community. The challenges were identified through a rigorous selection process, including workshops, consultation and debate, that solicited and distilled ideas from the global research community and people affected by cancer. The Cancer Grand Challenges Scientific Committee then translated these ideas into nine tangible challenges.
After receiving applications from 176 world-class global teams that proposed bold ideas to tackle these challenges, and followed by a competitive process of shortlisting, full applications and interviews, we are pleased to be funding five new teams to take on four challenges.

Understand the mechanisms through which genetics, biology and social determinants affect cancer risk and outcomes in diverse populations, to motivate interventions to reduce cancer inequities.
Team:
SAMBAI, led by Professor Melissa Davis, Morehouse School of Medicine, US
Read more about team SAMBAI on page 25.
Determine why the incidence of earlyonset cancers in adults is rising globally.
Team:
PROSPECT, co-led by Professor Andrew Chan, Massachusetts General Hospital, US, and Dr Yin Cao, Washington University in St. Louis, US
Read more about team PROSPECT on page 26.
Develop therapeutics to target oncogenic drivers of solid tumours in children.
Teams:
KOODAC, co-led by Professor Martin Eilers, University of Würzburg, Germany, and Professor Yaël Mossé, Children’s Hospital of Philadelphia, US
PROTECT, led by Professor Stefan Pfister, Hopp Children's Cancer Center
Heidelberg (KiTZ), Germany
Read more about teams KOODAC and PROTECT on pages 27 and 28.
Decipher the T-cell receptor cancerrecognition code.
Team:
MATCHMAKERS, led by Dr Michael Birnbaum, Massachusetts Institute of Technology, US
Read more about team MATCHMAKERS on page 29.


Societal, ancestry, molecular and biological analyses of inequalities
SAMBAI is funded by: Cancer Research UK and the National Cancer Institute
Inequities in cancer prevention, screening and treatment lead to disparities in cancer incidence and mortality, and are a major public health concern. Led by Melissa Davis of Morehouse School of Medicine, US, SAMBAI will generate a comprehensive database with measurements of social, environmental, genetic

and immunological factors that cause and influence disparate outcomes in diverse populations of African descent.
“Our work will be a catalyst for exponential change," says Melissa.
"In partnership with patients, the resources we will create can galvanise the work of many other groups. We will reposition important research questions in a better scope, with comprehensive data, that represents a global context.”
The team, made up of researchers from 15 institutions in Ghana, South Africa, the US and UK, will focus on breast, prostate and pancreatic cancer. As part of its approach, SAMBAI will characterise germline genomes for the entire SAMBAI study cohort and map genetic variations alongside social and environmental data to identify distinct factors contributing to cancer inequities.

Team members
Team lead:
Professor Melissa Davis Morehouse School of Medicine, US
Co-investigators:
Dr Yaw Bediako Yemaachi Biotech, Ghana
Dr Tiffany Carson Moffitt Cancer Center, US
Dr Isidro Cortes Ciriano European Molecular Biology Laboratory, UK
Professor Zodwa Dlamini University of Pretoria, South Africa
Professor Olivier Elemento Cornell University, US Ricki Fairley TOUCH, The Black Breast Cancer Alliance, US
Dr Fieke Froeling University of Glasgow, UK
Dr Marcin Imieliński New York University, US
Dr Sheeba Irshad King's College London, UK
Dr Lauren McCullough Emory University, US
Professor Gary Miller Columbia University, US
Professor Nigel Mongan University of Nottingham, UK
Dr Nicolas Robine New York Genome Center, US
Professor Clayton Yates Johns Hopkins University, US

Pathways, risk factors and molecules to prevent earlyonset colorectal tumours
Challenge: Early-onset Cancers
PROSPECT is funded by: Cancer Research UK, the National Cancer Institute and Institut National Du Cancer (INCa)
Since the 1950s, the incidence of early-onset cancers – cancers diagnosed in adults under 50 years of age – has risen worldwide. PROSPECT, led by Andrew Chan of Massachusetts General Hospital, US, and Yin Cao of Washington University in St. Louis, US, aims to address the global rise in incidence of earlyonset colorectal cancer (EOCRC) by understanding the pathways, risk factors and molecules involved in its development.

A cross-team focus on earlyonset colorectal cancer
The OPTIMISTICC team, taking on the Microbiota challenge, also has a focus on early-onset cancers. OPTIMISTICC’s goal is to pinpoint how the gut microbiome influences the initiation and development of colorectal cancer, including in early-onset cases.
OPTIMISTICC co-investigator
Shuji Ogino of Brigham and Women’s Hospital, US, led a review, published in Gut Microbes that explores the microbiome and the rise of early-onset cancers, with a focus on digestive system cancers including EOCRC.
“Our vision is to unravel and ultimately intervene upon the intricate network of causal factors that disrupt biological homeostasis to promote colorectal cancer among younger individuals throughout the life course,” says Andrew.
Comprising researchers from nine institutions in France, India, Italy, the US and UK, PROSPECT will leverage data from human cohorts and novel animal models, and conduct exposome studies to identify causal mechanisms underlying risk factors for this cancer type. The team proposes to implement innovative prevention trials in clinical and community settings in the US, UK and India to achieve its aims.

Team members Team leads:
Professor Andrew Chan Massachusetts General Hospital, US
Dr Yin Cao Washington University in St. Louis, US
Co-investigators:
Professor Emily Balskus Harvard University, US
Professor Yasmine Belkaid Institut Pasteur, France
Dr Jason Buenrostro Harvard University, US
Professor Curtis Huttenhower Broad Institute, US
Professor Gary Patti Washington University in St. Louis, US
Professor Nicola Segata University of Trento, Italy
Dr Bhawna Sirohi BALCO Medical Centre, India
Professor Tim Spector King’s College London, UK and Zoe Ltd, UK
Dr Ömer Yilmaz Massachusetts Institute of Technology, US


Developing a suite of oncoprotein degraders for childhood solid cancers
Challenge: Solid Tumours in Children (2023)
Cancer remains a leading cause of death by disease in children globally, and progress in the treatment of and outcomes for children with solid tumours, including brain tumours, has largely stalled.
Shuji and his colleagues summarise current evidence and discuss emerging research opportunities to address the alarming rise in the incidence of these cancer types worldwide.
PROSPECT will address some of the knowledge gaps and research opportunities identified in Shuji’s review, including diet, lifestyle and environmental factors from early life to adulthood, that may contribute to the development of early-onset cancers.
Featured publication
Mima K et al. Gut Microbes 2023; doi: 10.1080/19490976.2023.2269623
Featured challenge: Microbiota
Featured team: OPTIMISTICC
OPTIMISTICC is funded by: Cancer Research UK
Featured team member:

Professor Shuji Ogino
OPTIMISTICC co-investigator, Brigham and Women’s Hospital, US
KOODAC is funded by: Cancer Research UK, Institut National Du Cancer (INCa) and KiKa (Children Cancer Free Foundation)

Team KOODAC, led by Yaël Mossé of Children’s Hospital of Philadelphia, US, and Martin Eilers of the University of Würzburg, Germany, aims to develop a suite of oncoprotein degraders for childhood solid cancers, including orally bioavailable PROTACs and molecular glue degraders, to improve cure rates for children with high-risk tumours.
“Our vision is to develop orally bioavailable protein degradation drugs, an exciting new approach that has the potential to revolutionise cancer therapy by directly targeting five essential oncoproteins unique to high-risk childhood solid tumours,” says Yaël.
The team, from 10 institutions in France, Austria, Germany, the UK and US, is focusing on oncoproteins that are essential to the aetiology and/or metastatic state of neuroblastoma, fusionpositive rhabdomyosarcoma, medulloblastoma, Ewings sarcoma and fibrolamellar hepatocellular carcinomas.
Team members
Team leads:
Professor Martin Eilers
University of Würzburg, Germany
Professor Yaël Mossé
Children’s Hospital of Philadelphia, US
Co-investigators:
Professor Alessio Ciulli
University of Dundee, UK
Dr Olivier Delattre Institut Curie, France
Dr Gwenn Hansen Nurix Therapeutics, US
Dr Charles Keller
Children’s Cancer Therapy Development Institute, US
Professor John Maris Children’s Hospital of Philadelphia, US
Professor Sanford Simon The Rockefeller University, US
Dr Seychelle Vos Massachusetts Institute of Technology, US
Professor William Weiss University of California San Francisco, US
Dr Georg Winter Austrian Academy of Sciences, Austria
Yaël Mossé and colleagues in the laboratory.

Harnessing protein degradation for advanced childhood tumours
Challenge: Solid Tumours in Children (2023)
PROTECT is funded by: Cancer Research UK, the National Cancer Institute, the Scientific Foundation of the Spanish Association Against Cancer and KiKa (Children Cancer Free Foundation)
PROTECT, led by Stefan Pfister of Hopp Children's Cancer Center Heidelberg (KiTZ), Germany, aims to develop protein degraders and molecular glue degraders, to target undruggable selective targets in paediatric cancers. They will also apply targeted protein degradation to both CAR T-cell therapies and small molecules to build on and improve current therapies. Their targets will be relevant oncogenic drivers in Ewings sarcoma, neuroblastoma, synovial cell carcinoma, high-grade glioma, ependymoma and gastrointestinal stromal tumours, which are all paediatric cancers with a high unmet need.
“ This academic team approach to drug discovery for paediatricspecific therapies will be essential to increase cure rates for some of the most challenging cancers in children and decrease lifelong sequelae.
Stefan Pfister

With researchers from 10 institutions in Germany, the Netherlands, Spain, the US and UK, the team’s overarching aim is to create a pipeline of projects from screening through to optimisation of lead compounds that will be actively managed and reprioritised based on the emerging results from the team.

Team members Team lead:
Professor Stefan Pfister
Hopp Children's Cancer Center
Heidelberg (KiTZ), Germany
Co-investigators:
Professor Nathanael Gray Stanford University, US
Professor Kimberly Stegmaier
Dana-Farber Cancer Institute, US
Professor John Anderson University College London, UK
Dr Scott Armstrong Dana-Farber Cancer Institute, US
Dr Ana Banito
German Cancer Research Center (DKFZ), Germany
Professor Louis Chesler
The Institute of Cancer Research, UK
Dr Florencia Cidre-Aranaz
German Cancer Research Center (DKFZ), Germany
Professor Benjamin Ebert
Dana-Farber Cancer Institute, US
Dr Eric Fischer
Dana-Farber Cancer Institute, US
Dr Volker Germaschewski LifeArc, UK
Professor Swen Hoelder
The Institute of Cancer Research, UK
Dr Max Jan Massachusetts General Hospital, US
Dr Cigall Kadoch Dana-Farber Cancer Institute, US
Dr Cristina Mayor-Ruiz Institute for Research in BiomedicineIRB Barcelona, Spain
Professor Frank von Delft University of Oxford, UK
Dr Frank Westermann
German Cancer Research Center (DKFZ), Germany
Dr Judith Wienke
Princess Máxima Center, the Netherlands
Stefan Pfister in the laboratory with a colleague.
Michael Birnbaum with members of his team.


Solving TCR recognition and design via integrated high-throughput screening, structural, functional and computational approaches
MATCHMAKERS is funded by: Cancer Research UK, the National Cancer Institute and The Mark Foundation for Cancer Research
Many cancers harbour tumourinfiltrating T cells that are potentially reactive to (neo) antigens. While it’s possible to sequence the T-cell receptors (TCRs) present on these immune cells, it’s not possible to use the information to comprehensively and at scale infer the antigen that is recognised by the receptor.
Led by Michael Birnbaum of Massachusetts Institute of Technology, US, MATCHMAKERS aims to predict what T cells recognise in a patient’s tumour by using simple laboratory tests and computational prediction.
Uniting researchers from 10 institutions across Germany, the Netherlands, Norway, the UK and the US, the team will develop novel methods and new algorithms, and create large, integrated datasets of TCRs and peptide-major histocompatibility complexes (pMHCs). MATCHMAKERS’ approach will involve structural and molecular studies to transform our understanding of TCR-pMHC interactions.
“We’ve assembled a team with the expertise in immunology, technology development and computational biology needed to create generalised T cell prediction and design,” says Michael. “It’s our goal to establish patient-specific TCR immunotherapies as a routine treatment for patients with cancer.”
Team members
Team lead:
Dr Michael Birnbaum
Massachusetts Institute of Technology, US Co-investigators:
Professor David Baker University of Washington, US
Professor Regina Barzilay Massachusetts Institute of Technology, US
Dr Peter Bruno University of California San Francisco, US
Professor Dirk Busch Technische Universität München, Germany
Dr Brandon DeKosky Massachusetts Institute of Technology, US
Professor Stephen Elledge Brigham and Women’s Hospital, US
Professor Christopher Garcia Stanford University, US
Professor Johanna Olweus University of Oslo, Norway
Professor Sergio Quezada University College London, UK
Professor Ton Schumacher
Netherlands Cancer Institute, the Netherlands
Dr Nikolaos Sgourakis Children’s Hospital of Philadelphia, US


With five new teams on board, the Cancer Grand Challenges research community now spans 16 countries.

























































Cancer
is building a global network of funders and partners who share our ambitions to make radical progress against cancer and accelerate discoveries that will improve the lives of people affected by cancer.


Discover spoke with Isabel Orbe, general director of the Spanish Association Against Cancer, a partner with Cancer Grand Challenges since 2022, and Bruno Quesnel, director of the Research and Innovation Department of the Institute National du Cancer (INCa), which supports our newest round of challenges (page 24), about the power of international partnerships and uniting across borders to drive progress.
Why did your organisation decide to partner with Cancer Research UK to support the Cancer Grand Challenges initiative?
IO: The most important reason is because it’s a worldwide call that puts a focus on basic research. With Cancer Grand Challenges, it was not just about the collaborations but the excellent basic research that is being conducted. It was also a platform for us to gauge the quality of the Spanish scientific network.
BQ: Our research strategy at INCa was to move more onto an international scale. We were looking for an international research partner and, through the Cancer Grand Challenges initiative, Cancer Research UK looked like the right organisation for that. After many meetings, we made the formal decision that we wanted to be part of the process.
What impact do you hope your partnership will have?
IO: This partnership allows Spanish researchers to participate in an excellent research network. It gives them tools to do things
better, accelerate results and benefit Spanish patients in a global way. For Spanish scientists to participate in major cancer research projects is a major opportunity that will not only impact the scientific ecosystem of the country but also aid in reducing inequities in the treatment of our cancer patients.
BQ: Our partnership with Cancer Research UK as part of Cancer Grand Challenges will support the French research community to be part of big international collaborations. We want to make French research highly competitive, and certainly the best way to do that is to join this effort. It’s not a partnership to support a French team, it’s a partnership to reach the global goals defined by Cancer Grand Challenges. We think this is for the common good of patients, regardless of whether they are in France, the UK, the US or elsewhere.
Why is it important to collaborate across geographical borders to make progress against cancer?
IO: Cancer knows no boundaries, and neither should we. Working together [across borders] can reduce the impact of cancer and increase patient survival rates. If we have the same focus, the same aims, why not do it together?
BQ: If you really want to answer the most challenging questions, you need the best people from everywhere. You’re not going to find all the expertise in one country or even in Europe. You need a coordinated effort between top experts across the globe.
An expanding focus on
Perspectives from the Cancer Grand Challenges Scientific Committee
Two of the five new teams that have been awarded funding will focus on populationbased cancer challenges – Cancer Inequities and Early-onset Cancers. Here, Timothy Rebbeck and David Hunter, two expert epidemiologists and members of the Cancer Grand Challenges Scientific Committee, discuss the importance of the addition of these challenges to the initiative’s research portfolio.


“It’s clear that making an impact on cancer incidence and prevention has to be done at population scale,” says Timothy Rebbeck of the Dana-Farber Cancer Institute and the Harvard T.H. Chan School of Public Health, US. “The Cancer Inequities and Early-onset Cancers challenges are oriented toward prevention, early detection and understanding the distribution and determinants of disease in populations. These are critical for addressing things that are going to have an impact population wide.”
Population science examines factors that can influence health outcomes, including individual health behaviours, a person’s environment and social and economic circumstances, and policies. To learn about health and disease among specific populations, these studies often include large numbers of people over long periods of time, to enable researchers to learn how to improve cancer prevention, screening and care.
Population-based studies, however, do not exist in a vacuum. They often involve team science on a global scale. The teams taking on the Cancer Inequities and Early-onset Cancers challenges are both interdisciplinary, uniting basic scientists, clinicians, epidemiologists, population scientists, social scientists and other research professionals to solve these intractable problems.
The SAMBAI team is addressing the Cancer Inequities challenge.
Find out more about the SAMBAI and PROSPECT teams on pages 25 and 26.
It’s clear that making an impact on cancer incidence and prevention has to be done at population scale. “
Timothy Rebbeck
“This challenge is trying to understand a very complex problem, which certainly has some biological basis along with other explanations, such as lack of awareness, lack of access, and maybe including structural racism in the way that people of African ancestry are treated by the health care system,” says David Hunter from the University of Oxford, UK. The PROSPECT team is tackling the Early-onset Cancers challenge, focusing on early-onset colorectal cancer. “By taking a holistic, large-scale approach,” explains Timothy, “they are going to learn things they would never learn in a more constrained environment that looks at just a few risk factors or a few genes.”
With these two challenges, Cancer Grand Challenges is increasing its focus on cancer risk, risk mitigation, early detection and prevention. “A cancer that is prevented is a cancer that doesn’t have to be treated,” says David. “Therefore, a focus on prevention needs to be implemented alongside the very important aspects of what we do when someone is diagnosed with cancer.”

There is a new revolution in cancer research – a spatial biology revolution that is destined to change how scientists capture information about tumours, and may lead to new ways to diagnose and treat cancer. The Cancer Grand Challenges IMAXT and Rosetta teams, tackling the 3D Tumour Mapping challenge, have exploited new and existing technologies to propel spatial biology research even further to the forefront of cancer science.
Featured challenge: 3D Tumour Mapping
Featured teams:
IMAXT is funded by: Cancer Research UK
Rosetta is funded by: Cancer Research UK
In 2015, Cancer Grand Challenges set the 3D Tumour Mapping challenge, challenging the scientific community to develop methods to map the complex interactions among heterogeneous tumour cells, immune cells, tumour cell clones and components of the tumour microenvironment.
At the time the challenge was set, single-cell RNA and DNA sequencing had just taken the field of biology by storm. It was unclear, however, whether single-cell data alone would actually lead to better prognosis and diagnosis of cancer, or provide actionable clinical data.
“If you look at where the field is going,” says IMAXT team lead Greg Hannon of the Cancer Research UK Cambridge Institute, “it’s spatial multi-omics. It will be making multiple types of measurements in a spatial manner – things like clonal distribution of cancer cells, the transcriptome and the proteome – at the same time. That will all be integrated into a larger picture.”
IMAXT and Rosetta, led by Josephine Bunch of the National Physical Laboratory, UK, took the next step of developing techniques to reveal 3D cellular and molecular maps of the mutational and structural heterogeneity in DNA, patterns of conventional and nonconventional RNA transcription and protein expression, and the biochemical signatures of cells and cell types in a tumour, while simultaneously capturing the complexity of the intra-tumour architecture.
“From a technical perspective, we have developed or matured some of the most promising spatial measurement techniques in the field,” explains Greg. “These will have lasting impact.”
Early on, Greg and the IMAXT team realised that no single technology would ever achieve the depth of profiling required to take on some of the toughest questions in cancer biology. The team emphasised the development of tools to integrate data from multiple technologies, enabling single-cell and spatial profiling of tumours with sufficient throughput to allow for largescale studies. This resulted in the establishment of a worldclass data processing centre in Cambridge, built out of a successful collaboration with the Institute of Astronomy. Another critical aspect of the project was finding ways to visualize the enormous amount of data collected for each tumour, and make it accessible to scientists everywhere in the world. A core component of the IMAXT programme is Project Theia, the world’s first virtual reality cancer research laboratory. Theia lets users step inside intricate 3D tumour maps and explore the tumour landscape in unparalleled detail.
By combining existing technologies with new approaches, IMAXT has developed entirely new ways to study cancer, that are changing how cancers are classified, treated and managed, and giving more people a better chance of surviving their disease.
“I have been lucky enough to witness the potential benefits of these new technologies, which enable greater examination of a cancer and its individual cell structure in either two or three dimensions,” says Elaine Chapman, IMAXT patient advocate, UK, who has been living with stage 4 cancer for 17 years. “This opens up a field in which researchers and clinical teams can jointly gain a far greater understanding of cancer, which, in time, will lead to earlier diagnosis and a wave of more targeted treatments.”
Some of the spatial biology technologies developed, or optimised for use in tumour samples, by IMAXT include:
MERFISH (Mutiplexed Error-Robust Fluorescence In-Situ Hybridisation)
An imaging method capable of simultaneously measuring the copy number and spatial distribution of hundreds to thousands of RNA species in single cells. “With MERFISH,” says IMAXT co-investigator Xiaowei Zhuang of Harvard University, US, “we can directly image, in situ, the gene expression profile of individual cells for cell type identification and discovery. A significant advantage of MERFISH is its ability to recover functionally critical spatial information.” In Cell, Xiaowei and her colleagues described an additional MERFISH modality, epigenomic MERFISH, that enables spatially resolved single-cell epigenomic profiling. Using epigenomic MERFISH, Xiaowei generated a high-resolution atlas of hundreds of active promoters and enhancers in embryonic and adult mouse brains.
Multiplexed immunofluorescence (mpIF)
An imaging technique that enables researchers to identify the types of immune cells in a tumour and its microenvironment. Using mpIF combined with whole-genome and single-cell sequencing, IMAXT co-investigator Sohrab Shah of Memorial Sloan Kettering Cancer Center, US, and Ignacio VázquezGarcia, a postdoctoral fellow in Sohrab’s laboratory, discovered a mechanism of immune evasion in distinct mutational subtypes of high-grade serous ovarian cancer tumours. In their work they show that compositional, topographic and functional differences exist among mutational subtypes. They also discovered site-specific properties of the primary tumours and distal peritoneal metastases. The findings, published in Nature, highlight the urgent need for early detection and personalised treatment approaches for ovarian tumours.
“These technologies provide an extraordinary view of the spatial dissemination and interactions of cancer cells in the context of the surrounding tissue microenvironment,” says Ignacio, lead author of the study. The Joyce group also developed a novel multiplexed immunofluorescence technique named HiFI (Hyperplexed Immunofluorescence), which enable the detection of even more markers, which is currently pending publication.
3D imaging mass cytometry (3D IMC)
Used for multiplexed 3D tissue analysis at single-cell resolution. Using 3D IMC, IMAXT coinvestigator Bernd Bodenmiller of ETH Zurich, Switzerland, revealed cellular and microenvironmental heterogeneity and cell-level tissue organisation not detectable in 2D. The study, published in Nature Cancer suggests that 3D IMC may be a powerful tool for studying phenomena, such as tumour cell invasion, that occur in 3D space. The detailed models generated with 3D IMC facilitate comprehensive, single-cellresolution analysis of cellular microenvironments and tissue architecture.
WILD-seq
(Wholistic Interrogation of Lineage Dynamics by sequencing)
A novel strategy for lineage tracing (a way of following the evolutionary history of cell clones in tumours) coupled with single-cell transcriptomics to enable insights into tumour heterogeneity and response to treatment. In a study published in eLife the IMAXT team used WILD-seq to identify a major mechanism of chemotherapy resistance in mouse models of triple-negative breast cancer. Their studies revealed asparagine bioavailability as a druggable vulnerability of taxane-resistant clonal lineages.

IMAXT’s efforts in developing these technologies and the expertise to study the complexity of tumours at the 3D level have enabled the team to begin to address important and fundamental questions in cancer biology that otherwise would not have been possible.
the Cancer Grand Challenges 2023
Mass spectrometry imaging existed before the Cancer Grand Challenges programme, but no project had ever really allowed it to be deployed at such scale, bringing as many different modalities together.
“ Josephine Bunch
Mapping metabolites in the microenvironment
Rosetta adds a metabolic view to spatial biology. The team is using mass spectrometry imaging (MSI) to map the spatial distribution of cellular metabolites in the tumour microenvironment in exquisite detail. The information uncovered through MSI provides a deeper understanding of tumour behaviour, progression and response to therapy by mapping metabolite distribution in relation to individual cells’ genetics and the overarching tumour architecture.
“Mass spectrometry imaging existed before the Cancer Grand Challenges programme,” says Josephine, “but no project had ever really allowed it to be deployed at such scale, bringing as many different modalities together. Throughout the programme, improvements in sample handling, introduction of new modalities and novel data handling and protocols to integrate these measurements have culminated in a very powerful pipeline.”
Rosetta’s pipeline has begun to demonstrate the clinical application of metabolic profiling
and how this technique could be used for patient stratification, monitoring responses to therapy and drug discovery.
Utilising MSI technology, Rosetta co-investigator Mariia Yuneva of The Francis Crick Institute, UK, and former team member Peter Kreuzaler of the University of Cologne, Germany, identified pantothenic acid (vitamin B5) as a metabolite biomarker associated with high expression of MYC in breast cancer. Use of the entire Rosetta MSI pipeline, from low to high spatial resolution, enabled them to see where atoms from specific molecules, such as pantothenic acid, are localised within tumour tissue. In their recent article published in Nature Metabolism they showed that MYC increased levels of vitamin B5 in tumours by upregulating the expression of the multivitamin transporter SLC5A6.
When the researchers put mice on a pantothenic acid-deficient diet, they found that both mouse tumours and patient-derivedxenograft models of breast cancer grew more slowly. This finding suggests that decreasing vitamin B5 availability to the tumour may be advantageous. “All the growth advantage that cells gain by upregulating MYC was lost by taking away one single vitamin,” says Peter. This finding could open up new therapeutic avenues for MYC-driven cancers.
In another study, Rosetta researchers showed that metabolic profiling using a variety of mass spectrometry techniques can be used to stratify tissues according to their underlying mutations in colorectal cancer, and that such techniques can identify new potential targets for cancer treatment.
Johan Vande Voorde, former team member of Rosetta co-investigator Owen Sansom of the Cancer Research UK Scotland Institute (formerly the Beatson Institute), identified an enzyme, called
adenosylhomocysteinase (AHCY), that is upregulated in colorectal cancer compared to with normal intestine, and may be a potential treatment target, according to preclinical findings.
“Our results provide preclinical and clinical evidence that we can use the metabolic profile of intestine samples to understand what the drivers are within a tissue,” says Owen.
The researchers plan to validate their findings in a larger patient study, and aim to test whether targeting AHCY in particular subtypes of colorectal cancer might provide patient benefit.
Johan notes that their studies will explore AHCY as a potential cancer target in more complex models and examine other processes, such as metastasis, to determine how broadly this intervention could be applied as a new target.
The power of the Rosetta pipeline and its potential for clinical impact has been demonstrated through its application to AstraZeneca’s drug development pipeline.
“The Rosetta platform is an integral part of the AstraZeneca spatial-omics workflow and suite of capabilities, which help us to answer important scientific questions,” says Simon Barry, Rosetta co-investigator and Executive Director at AstraZeneca, UK. “We apply the technology across a range of studies – from preclinical to biomarker and mechanistic studies – to gain insight and predict clinical impact.”
“Rosetta has exemplified the value of collaboration in combining a broad set of technologies and approaches,” adds Richard Goodwin, Senior Director at AstraZeneca and Rosetta co-investigator. “The results of these efforts are truly changing the field and have made it easier to integrate the transformational technologies into clinical decision making, with potential for patient benefit.”
An unprecedented view of tumour complexity
As the spatial biology revolution in cancer evolves, IMAXT and Rosetta are continuing to demonstrate the power of spatial profiling for cancer research.
“Together, these teams have given us a ‘Google Earth'-like tool to study the anatomy of cancers at an extraordinary level of detail,” says René Bernards of the Netherlands Cancer Institute and member of the Cancer Grand Challenges Scientific Committee.
“These technologies hold great promise to better understand the interactions between tumours and their microenvironments.”
Since the 3D Tumour Mapping challenge was set, a new era of cancer biology has emerged, characterised by the widespread adoption of singlecell technologies and the gradual emergence and affirmation of spatial-omics technologies. The platforms developed by IMAXT and Rosetta have provided an unprecedented view of tumour complexity, allowing for the identification of new targets for drug development, advancing understanding of why some tumours become resistant to treatment and driving insights that could improve outcomes for patients worldwide.







Professor








Professor Owen Sansom Rosetta co-investigator, Cancer Research UK Scotland Institute, UK
Featured publications
Wild SA et al. eLife 2023; doi: 10.7554/eLife.80981
Vázquez-Garcia I et al. Nature 2022; doi: 10.1038/s41586-02205496-1
Lu T et al. Cell 2022; doi: 10.1016/j. cell.2022.09.035
Kuett L et al. Nat Cancer 2022; doi: 10.1038/s43018-021-00301-w
Vande Voorde J et al. Nat Metab 2023; doi: 10.1038/s42255-02300857-0
Kreuzaler P et al. Nat Metab 2023; doi: 10.1038/s42255-023-00915-7

As part of the NexTGen team tackling the Solid Tumours in Children challenge, coinvestigator Nikolaos Sgourakis of Children’s Hospital of Philadelphia (CHOP), US, is applying his expertise in structural biology to pioneer a novel approach to designing chimeric antigen receptors (CARs) for solid tumours.
Featured challenge: Solid Tumours in Children (2020)
Featured team: NexTGen
NexTGen is funded by: Cancer Research UK, the National Cancer Institute and The Mark Foundation for Cancer Research
CAR T-cell therapy has worked remarkably well for haematologic cancers such as lymphoma, some forms of leukaemia and multiple myeloma. However, CAR T-cell therapy for solid tumours remains a challenge for the scientific community.
Through a combination of structural, biochemical and functional approaches, Nik has been working with NexTGen coinvestigator John Maris of CHOP and their colleagues to develop a peptide-centric CAR, which they hope will be effective in patients with neuroblastoma, a common but difficult-to-treat extracranial childhood cancer.
“Nik is identifying new targets and engineering novel CAR T products which are so key to us realising our goal to deliver next-generation CAR T for the treatment of paediatric patients with solid tumours,” says NexTGen co-team lead Catherine Bollard of the Children’s National Hospital, US.
Traditionally, CAR T-cell-based immunotherapies target extracellular antigens such as CD19, which are expressed on the surfaces of cancer cells. Many proteins, because they are expressed intracellularly, are largely undruggable; however, these inaccessible targets are degraded into small protein fragments called peptides by the body’s natural protein disposal system. These peptides are then presented on the surfaces of cells by major histocompatibility protein class I molecules, which alert the immune system to virus-infected or abnormal cells, such as cancer cells.
The CAR T cells developed by Nik and the CHOP team target one such peptide – derived from the PHOX2B oncoprotein – known to be particularly enriched in neuroblastoma. PHOX2B mutations contribute to neuroblastoma by increasing the proliferation of neural crest cells and making them more likely to become cancerous.
But there was a particular challenge, says Nik, in how to understand which of the potentially hundreds of histocompatibility proteins in patients with neuroblastoma that can display PHOX2B peptide would be recognisable by a CAR T cell.
“Our combined structural and biochemical analysis of the molecular interactions allows

us to address this question,” Nik explains, speaking of the team’s recent study published in Science Immunology, on which he is senior author. “And now CAR T therapy can be administered only to those patients that would benefit from it, improving our success rates and mitigating toxicity risks.”
Peptide-centric CARs overcome several barriers encountered by traditional CAR T cells used against solid tumours, including a phenomenon known as cross-reactivity, in which one T-cell receptor targets multiple different antigens.
“The last thing you want when you build a CAR,” adds Nik, “is this promiscuous set of interactions: you think you’re targeting one antigen, but you have to deal with cross-reactivity against potentially many other antigens.”
In two related studies, both published in Nature Communications, the Sgourakis laboratory released a computational platform, HLA3DB (https://hla3db. research.chop.edu), for predicting the 3D structures of tumour antigens, and solved the first solution structure of the neuroblastoma RAS Q61 antigen by nuclear magnetic resonance spectroscopy. This information can be used to predict potential cross-reactive peptide-centric CARs and mitigate risks in the preclinical development phase.
“It is time to change the standard of care for these diseases by developing new therapies that will give children and families a kinder and more effective treatment option,” says NexTGen patient advocate Abbe Pannucci, who herself experienced stage 4 childhood rhabdomyosarcoma.
“NexTGen’s work gives me hope that we can lessen the severity of treatment for children with cancer diagnoses in the future.”
It is time to change the standard of care for these diseases. “
Abbe Pannucci
With the newest round of funded teams, Cancer Grand Challenges is tripling its spending on research into childhood solid cancers. With two additional teams coming on board to tackle the challenge of solid tumours in children, research underway by the Cancer Grand Challenges community will expand to investigate more potential treatment targets across more childhood cancer types and unite experts in the field from more countries across the globe. These efforts will accelerate the development of effective targeted therapeutics for paediatric solid tumours, to ultimately improve survival and diminish the lifelong toxicities experienced by survivors of these diseases.
Find out more about teams KOODAC and PROTECT, taking on the Solid Tumours in Children challenge (2023), on pages 27 and 28.




Featured publications
Sun et al. Sci Immunol 2023; doi: 10.1126/sciimmunol.adj5792
Gupta S et al. Nat Commun 2023; doi: 10.1038/s41467-02342163-z
McShan et al. Nat Commun 2023; doi: 10.1038/s41467-02343654-9

In 2015, Cancer Grand Challenges set the Lethal vs. Non-Lethal Cancers challenge, with the vision to develop a thorough understanding of features that can distinguish a nonlethal growth from a potentially lethal malignant growth, so that methods could be developed to specifically detect cancers that require intervention. The PRECISION team has been tackling this challenge in ductal carcinoma in situ (DCIS), a potential precursor of invasive breast cancer. By taking a deep dive into DCIS biology, the team has transformed the DCIS field, bringing us closer to distinguishing harmless from hazardous DCIS.
Advances in screening methods mean that we can now detect some cancers, such as breast and prostate cancer, at an early stage. Although this capability is highly beneficial in the cancers that would develop into life-threatening malignancies, the sensitivity of screening methods has led to overdiagnosis and subsequent overtreatment in cases that wouldn’t have become dangerous.
This over-treatment is particularly true for DCIS, in which some cells in the milk ducts of the breast become abnormal but have not started to spread to surrounding breast tissue. Although DCIS is not life-threatening, and its chances of progressing to cancer are low, each year, thousands of patients with DCIS worldwide are treated as if they have invasive breast cancer, and undergo surgery, hormonal and radiotherapy.
“Over the years, it has become clear that up to 4 out of 5 DCIS lesions will never progress to invasive breast cancer if left untreated,” says PRECISION team lead Jelle Wesseling of the Netherlands Cancer Institute (NKI).
“That implies that the majority of women with a DCIS diagnosis undergo invasive treatment without any benefit.”
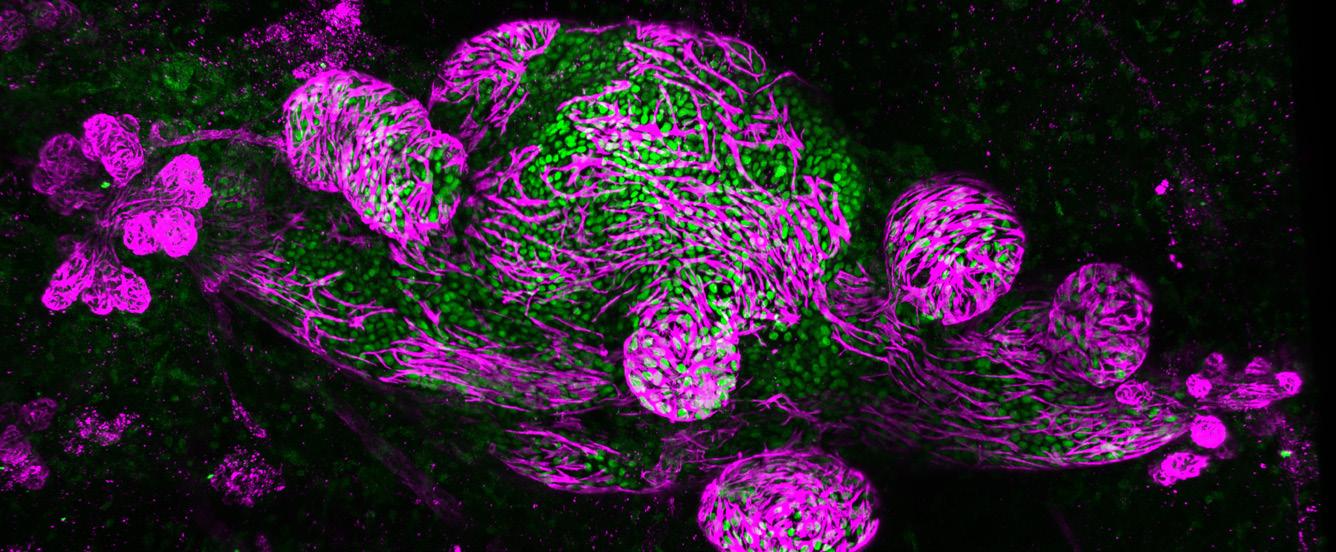
Therefore, the PRECISION team has taken a comprehensive approach to develop the knowledge and tools that could help determine which DCIS cases need treatment and prevent the consequences of over-treatment.
The need for accurate prognostic markers
In the clinic, DCIS size and margin status are two factors often used to stratify the risk of DCIS lesions and determine the course of treatment. However, in a recent study published in the British Medical Journal PRECISION researchers found only a weak association between these clinical factors and the risk of subsequent ipsilateral invasive breast cancer. The team combined data from more than 47,000 women with DCIS from the Netherlands, UK and US, who had received either breastconserving surgery or mastectomy, often followed by radiotherapy or hormone treatment, or both.
The goal of PRECISION is to deescalate DCIS treatment, however, the associations reported in our study are not large enough to drive clinical decisions regarding which patients should be treated and which patients could safely not be, explains Marjanka Schmidt of the NKI, PRECISION co-investigator and senior author of the study.
Part of PRECISION’s goal has been to shift perceptions of a diagnosis of DCIS away from early breast cancer. In the same study, which was the largest of its kind to explore the value of prognostic risk factors after DCIS, the team reported that the 10-year cumulative incidence of ipsilateral invasive breast cancer was just 3.2%.
“Women need much more information about their individual risks from DCIS before they make treatment decisions,” says PRECISION patient advocate Hilary Stobart, UK. “PRECISION is working hard to resolve this dilemma by working together to find a combination of biomarkers,
making very important steps towards achieving this goal so that each woman will soon be able to know her own individual risk.”
Modelling DCIS
To understand the biochemical and genetic pathways that drive cancer, researchers rely on accurate disease models. However, no sufficient models of DCIS had been available to study disease progression until recently.
The Mouse INtraDuctal (MIND) model is the first in vivo model for studying the natural evolution of DCIS. It was developed by PRECISION co-investigator Jos Jonkers and his team at NKI in collaboration with other PRECISION co-investigators, including Fariba Behbod of the University of Kansas Medical Center, US. In this model, patient-derived DCIS epithelial cells are injected into mice to mimic patient histology and biomarker expression.
The PRECISION team has developed a living biobank of 115 patient-derived MIND models and used them to identify risk factors associated with the progression of DCIS to invasive breast cancer. Using multiple omics methods, the researchers compared two groups to characterise the differences between DCIS cases that progressed to invasive cancer and those that did not.
“We saw two distinct growth patterns – replacement growth, which didn’t change the architecture of the mammary gland when the lesion grew, and expansive growth, where the lesion grew perpendicular to the ductal system and put a lot of pressure on the ducts,” explains Jos, senior author of the study. “The latter growth pattern may eventually lead to the breakage of the duct and correlate with invasive progression.”
The study, published in Cancer Cell provides critical information about which markers might potentially predict invasive progression in women with DCIS. Importantly, notes Jos, the biobank includes 19 distributable DCIS
MIND models, accessible through CancerTools.org which have been made available to researchers worldwide to further investigate DCIS molecular subtypes.
“As we and other research groups have shown, most DCIS cases will not become dangerous,” says PRECISION’s Stefan Hutten, postdoctoral researcher at NKI and first author of the Cancer Cell article. “I hope, in the future, we will be able to better assess prognosis in DCIS patients and divide them into three groups: a low-risk group, which receives active surveillance; a high-risk group, which should receive the current standard of care; and a medium-risk group, where the physician makes an informed decision together with the patient about the course of action.”
A prognostic classifier to help guide treatment decision-making
In another effort to avoid unnecessary treatment of DCIS, Elinor Sawyer of King’s College London, UK, and Jelle, together with colleagues from PRECISION have been working to develop a prognostic classifier that can identify patients with DCIS who are unlikely to develop invasive breast cancer. Using RNA sequencing, they have identified more than 100 genes whose expression varies depending on the risk of breast cancer recurrence after DCIS.
According to the expression of these genes, the predictor divides patients into three categories: low risk, intermediate risk and high risk of disease progression. “Rather than having an operation to remove their DCIS,” says Elinor, “women in the low-risk group could follow a watch-and-wait policy with annual mammograms because they are very unlikely to develop invasive breast cancer in the future.”
The predictor may also help to identify patients at low or intermediate risk of DCIS progression who do opt for surgery but would derive little benefit from additional radiotherapy.
Featured challenges: Lethal versus Non-lethal Cancers
3D Tumour Mapping Extrachromosomal DNA
Featured team: PRECISION
IMAXT eDyNAmiC
PRECISION is funded by: Cancer Research UK and the Dutch Cancer Society
IMAXT is funded by: Cancer Research UK
eDyNAmiC is funded by: Cancer Research UK and the National Cancer Institute
Featured publications
Schmitz RSJM et al. BMJ 2023; doi: 10.1136/bmj-2023-076022
Hutten SJ et al. Cancer Cell 2023; doi: 10.1016/j. ccell.2023.04.002
Strand SH et al. Cancer Cell 2022; doi: 10.1016/j. ccell.2022.10.021
Rebbeck CA et al. Nat Commun 2022; doi: 10.1038/s41467-02230573-4
The PRECISION predictor is being validated using samples from COMET (Comparison of Operative versus Monitoring and Endocrine Therapy), a US-based randomised clinical trial for people with low-risk DCIS. “It’s only when we get these results back that we will be able to say whether it’s safe for women in the low-risk group not to have surgery, but to undergo regular surveillance,” adds Elinor.
PRECISION’s approach is complementary to a tool being developed by one of its coinvestigators, Shelley Hwang of Duke University, US, as part of her involvement in the National Cancer Institute’s Human Tumor Atlas Network (HTAN). In work related to HTAN’s Breast Pre-Cancer Atlas, Shelley and colleagues identified 812 genes associated with ipsilateral DCIS recurrence within five years of treatment, as well as stromal expression patterns and immune cell compositions. These markers successfully predicted the risk of both overall recurrence and invasive progression.
“The really exciting part of all of this is that we feel the markers may have promise in terms of predicting which patients may not need to have surgery at all, because their risk of developing cancer is so low,” says Shelley. “These types of biomarkers would address an important clinical need, allowing us to reserve surgery only for those patients who would benefit substantially from it.”
A team-science approach to DCIS
The Cancer Grand Challenges’ team-science approach is helping PRECISION meet the challenge of distinguishing DCIS lesions that require treatment from those that don’t. For example, with Serena Nik-Zainal of Cambridge University, UK, PRECISION is working with the eDyNAmiC team to determine whether extrachromosomal DNAs (ecDNAs) are present in samples from patients with DCIS that ultimately progresses to invasive breast cancer.
Jelle Wesseling and Serena Nik-Zainal at the Cancer Grand Challenges 2023 summit.

With Serena Nik-Zainal of Cambridge University, UK, PRECISION is working with the eDyNAmiC team to determine whether extrachromosomal DNAs (ecDNAs) are present in samples from patients with DCIS that ultimately progresses to invasive breast cancer. Serena, who is a co-investigator in both teams, has applied tools developed by the eDyNAmiC team to show that a high proportion of the DCIS samples PRECISION has studied – about 50% – contain ecDNA. She is now validating the ecDNAs that PRECISION has identified and applying eDyNAmiC’s newest algorithms to understand the structure, origin and function of ecDNA in DCIS.
“This cross-team collaboration is critical,” says Serena. “From our first meeting, it was clear that the expertise from the two teams was critical for us to understand our observations about ecDNA going forwards.”
“DCIS overtreatment is such a big problem that others within the Cancer Grand Challenges community are tackling it, too,” says Jelle. “One team doesn’t have all the answers, but collaborations within the Cancer Grand Challenges community and beyond are leading to important progress and further research in this area, and sparing thousands of patients with DCIS from needless treatment.”
“
One team doesn’t have all the answers, but collaborations within the Cancer Grand Challenges community and beyond are leading to important progress and further research in this area.
Jelle Wesseling
PRECISION’s comprehensive approach to tackling the Lethal versus Non-Lethal Cancers challenge has transformed the DCIS field. The knowledge, models and resources developed by the team are paving the way for the future of the field, helping to reduce the burden of DCIS overtreatment and improve quality of life for thousands of people across the globe.
Featured team members










Professor Jelle Wesseling PRECISION team lead, Netherlands Cancer Institute, the Netherlands
Dr Marjanka Schmidt PRECISION co-investigator, Netherlands Cancer Institute, the Netherlands
Hilary Stobart PRECISION patient advocate, UK
Professor Jos Jonkers PRECISION co-investigator, Netherlands Cancer Institute, the Netherlands
Professor Fariba Behbod PRECISION co-investigator, University of Kansas Medical Center, US
Dr Stefan Hutten PRECISION team member, postdoctoral researcher, Netherlands Cancer Institute, the Netherlands
Professor Elinor Sawyer PRECISION co-investigator, King’s College London, UK
Professor Shelley Hwang PRECISION co-investigator, Duke University, US
Professor Greg Hannon IMAXT team lead, Cancer Research UK Cambridge Institute, UK
Professor Serena Nik-Zainal PRECISION and eDyNAmiC co-investigator, Cambridge University, UK
ecDNA in the cancerous transformation of Barrett’s oesophagus
The eDyNAmiC team has also delved into the biology of a potential cancer precursor lesion called Barrett’s oesophagus, which can develop into oesophageal adenocarcinoma (EAC) in up to 13% of cases.
eDyNAmiC researchers analysed whole-genome-sequencing data from two cohorts of patients with EAC or Barrett’s oesophagus to determine whether ecDNA formation could be an early event in the transition from dysplasia to cancer. In one cohort, the frequency of ecDNA was 43% higher in late-stage EAC than Barrett’s oesophagus; in the other cohort, 33% of patients with Barrett’s oesophagus who developed EAC had at least one biopsy with ecDNA present.
Published in Nature, the study is one of the first to establish that ecDNA appears early, before the development of full-blown cancer, and that its presence is strongly associated with progression to cancer.
“It also means that if you can find ecDNA and discover ways to target it, you can intervene much earlier,” says eDyNAmiC team lead Paul Mischel of Stanford University, US, who led the study.
Paul says that the Cancer Grand Challenges team-science approach was a “game-changer” for this study. “When you’re trying to tackle a really difficult challenge in cancer, the typical approach – one lab, one team, one group – won’t cut it. These are very difficult problems for which you need to bring together diverse experts across a whole spectrum of domains and be able to respond to the science quickly. We have that obligation to the world.”
Featured team member: Professor Paul Mischel eDyNAmiC team lead, Stanford University, US

Featured publication
Luebeck J et al. Nature 2023; doi: 10.1038/s41586-02305937-5
Below:
Chromosomes (blue rods) and ecDNAs (blue circles).
Credit: Kristen Turner, Mischel lab, Stanford University, US.
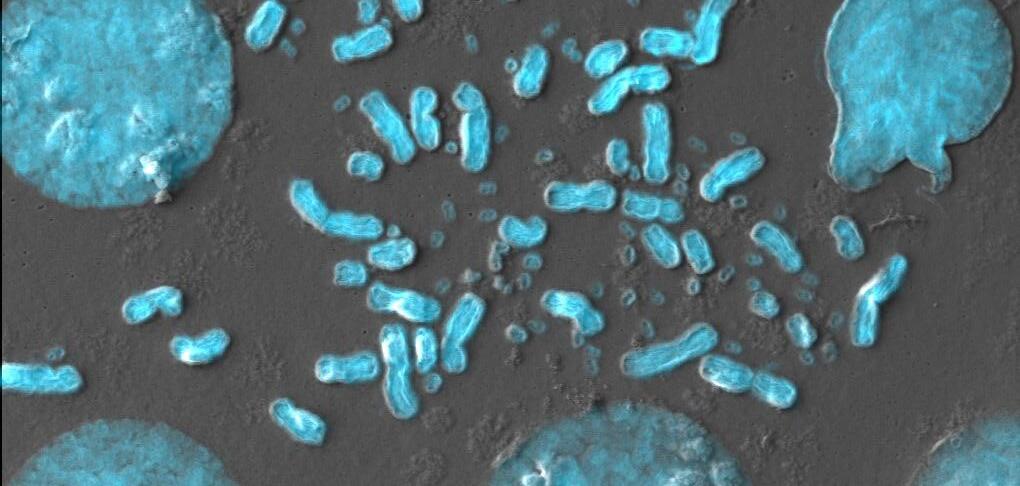
We

The Bowelbabe Fund for Cancer Research UK Nick and Annette Razey Bjorn and Inger Saven Founded by Cancer Research UK’s partners





Be


Professor Sir David Lane
Outgoing chair of the Cancer Grand Challenges Scientific Committee (2021–24)
As the sun sets on my tenure as chair of the Cancer Grand Challenges Scientific Committee, I pause to reflect on the past nine years I have served on this august board and the progress that our very first funded teams have made against some of the toughest challenges in cancer science.
To me, the Mutographs results on mutational fingerprints are pretty amazing. The field got so focused on mutations – and rightly so –that it forgot about all of the early work on promoters. The work of the Mutographs team has caused a reset and opened up exciting new potential for preventing cancer.
Ductal carcinoma in situ (DCIS) is such a major issue, given the enormous numbers of patients who are treated for breast 'cancer'
but certainly don’t benefit from treatment. Through PRECISION’s studies, a clear picture has emerged of this very early stage of disease –not a solution but a better definition of the scale of the problem.
Likewise, Rosetta and IMAXT, in the 3D Tumour Mapping challenge, have made important progress allowing us to look at tumours in ways once unimaginable. This work will revolutionise cancer diagnosis and treatment.
The scale of these and other efforts makes the science coming out of Cancer Grand Challenges so powerful and impactful. These are big projects with long-term funding – as we like to say, science takes time to cook. Cancer Grand Challenges has given scientists the freedom, structure, time and money to conduct research at scale, creating an expectation of excellence that is both important and self-reinforcing.
The future looks very bright for Cancer Grand Challenges. We’ve got terrific new challenges, tackling the issues of cancer inequities and the rising incidence of earlyonset cancers and targeting the oncogenic drivers of solid tumours in children and how T-cell receptors recognise cancer. Some of these areas have been massively under-investigated. Now it’s time for the teams addressing these challenges to shine and to bring the full power of global team science to bear on the work that lies ahead.
I now turn the chair of this committee over to my colleague and friend Charles Swanton, an eminent scientist with great experience of the panel who will help to identify and steward the next grand challenges in cancer research.
What an honour to be taking the position of chair of the Scientific Committee.
I joined the committee in January 2020 as the global conversations got underway to identify the biggest challenges in cancer research for the initiative’s third round of funding. It’s been spectacular to follow the science coming from the funded teams as they address hugely ambitious questions in cancer, and I’m excited about the synergy that’s emerging between teams as they make progress against their challenges. With new teams coming on board, the next chapter for Cancer Grand Challenges promises to be an exciting one as we continue to disentangle the complexity of human cancer and drive the discoveries that will improve outcomes and quality of life for people with cancer.
Professor Charles Swanton
Incoming chair of the Cancer Grand Challenges Scientific Committee (2024–)

Thank you to everyone who contributed to this publication.
Writing, editing and designing
Bethan Warman
Scott Edwards
Phil Prime
Maya Shani
Rebecca Martinho
Contributors
Ludmil Alexandrov
Allan Balmain
Gemma Balmer-Kemp
Simon Barry
René Bernards
Proteeti Bhattacharjee
Yelak Biru
Catherine Bollard
Josephine Bunch
Ivana Cattaneo
Howard Chang
Elaine Chapman
Teresa Davoli
Andrea Delgado-Garcia
Tony Dickherber
Rosalyn Flower
Sharmistha Ghosh-Janjigian
Richard Goodwin
Margaret Grayson
Greg Hannon
Laura Humphreys
David Hunter
Stefan Hutten
Shelley Hwang
Jos Jonkers
Peter Kreuzaler
Andrew Kurtz
Michael Logan
Núria López-Bigas
Mimi McCord
Paul Mischel
Amy Moore
Serena Nik-Zainal
Isabel Orbe
Burçak Otlu
Abbe Pannucci
Gita Patel
Norbert Perrimon
Mia Petljak
Bruno Quesnel
Tim Rebbeck
Lorenzo de la Rica
Owen Sansom
Elinor Sawyer
Marjanka Schmidt
Nikolaos Sgourakis
Sohrab Shah
Anju Singh
Hilary Stobart
Mike Stratton
Charles Swanton
Mark Taylor
Iva Trenevska
Ignacio Vázquez-Garcia
Johan Vande Voorde
Joanna Watson
Dominika Wawrzyniak
Elisabete Weiderpass
Jelle Wesseling
Eileen White
Image credits
Where required, image credits have been provided alongside the relevant images.
