Finding the Function of Mutations that Occur in Caspofungin 9
Mechanism of FKS1 Mutations in Caspofungin-Resistant β-1,3-Glucan Synthase 9
Role of FSK1, GSC2, NDC1, and UTP20 Mutations in Yeast Resistance to Caspofungin 10
Researching FKS1, UTP21 mutations and comparing those results to the mutations in gene GSC2 and FKS1 10
The Effect of Caspofungin on the GSC2 Gene in Saccharomyces cerevisiae Yeast 11
Understanding the Missense Mutations of A08 and A05 through FKS1 11
8JZN’s Purpose and Function within the FKS1 Gene 12
Characterization of AlphaFold2 Protein AF_AQ0PC18F1 14
Characterizing the Functionality of Protein 3L1W 14
Determining the Structure and Function of the 3N2C Protein 15
Discovering the Function of 2O14 15
Functional Characterization of 3CBW 16
Functional Characterization of 4DIU 16
Structural and Functional Characterization of 3DS8 - A Triacylglycerol Lipase from Listeria innocua 17
The Functional Characterization of AlphaFold Proteins Q0P8E0 and Q32HQ2 18
Unlocking 4Q7Q 18
Unraveling the Mysteries of 2QRU 19
3B7F 19
3H04 Enzyme Analysis 19
American Modernism 21
Creative Nonfiction 21
Cross-Comparing the Artistic Styles of Sylvia Plath and Taylor Swift 21
Historical Fiction 22
Impacts of Major Sporting Events: A Historical Lens 22
Linguistics 22
Sustainable Architecture in Cold Climates: Balancing Innovation, Economy, and Environment 23
Sustainable Architecture as a Foundation for Community 23
Women, Witches, and the Wielding of Power 23
The Effects of Communism on the Cuban Healthcare System: An Oral History Examination 24
The Psychology of Color Theory Through The Architecture of Frank Lloyd Wright 24
The Ethics of Election Betting 25
The Federal Reserve’s Interest Rate Mechanism: Decoding Monetary Policy Decisions and Economic Impacts 25
Tracing our Evolutionary Legacy: A Biological Anthropology Exploration 26
Accelerated Molecular Dynamics 28
Coral 28
EmotiMate AI Companion 28
Extended Reality 29
Fluorescent Fish 29
Materials 30
Metagenomics 30
Optimizing the Production of Artificial Collagen for Use as a Biomaterial 31
PETase 31
SmartHeart 32
Structural and Functional Analysis of Shigella Dysenteriae Transcription Factor VirF 32
The Potential for AI in Medicine 33
Keeping Voyager 1 Alive: A Mission of Insane Proportions 34
Alzheimer’s: Investing Hypotheses Regarding Pathophysiology 35
A Promising Study of Ground Squirrels for Cataract Drug Treatment 38
SMART Team - Trapoxin A and Histone deacetylase 39
Robotics 39
Using Differential Equations to Model Gross Domestic Product 40
Wavelet Analysis of GPS Data for Earthquake Prediction in Japan: Investigating Long-Term Patterns and Periodicity 40
The Benefits and Drawbacks of CRISPR/Cas9 Delivery 41
Measuring Spillover Effect in Caribbean Spiny Lobster (Panulirus argus) 43
Bounding the PhISCS-BnB algorithm 43
RWKV-7 “Goose” with Expressive Dynamic State Evolution 44
44
Welcome to the 2025 Research Week edition of the Pingry Community Research (PCR) Journal. We are excited to showcase Pingry’s scientific talent, both in terms of research skills and knowledge of scientific concepts and discoveries.
The PCR Journal provides students the opportunity to publish novel research and share their passion for science with the community. Through a written medium, students demonstrate their in-depth understanding of complex, collegiate-level scientific topics, and their applications of research in and out of Pingry.
This special edition of PCR serves as a written complement to the in-person poster presentations occurring throughout Research Week. Readers can preview abstracts, figures, and summaries reflecting research conducted in advanced courses and extracurriculars, such as AP Biology, Independent Research Teams (IRT), Humanities Independent Research Teams (HIRT), and the Methods in Molecular Biology Research Class, as well as in external collaborations.
We are also excited to announce our collaboration with the Art Fundamentals class for many of the illustrations in this issue! We hope this interdisciplinary collaboration makes the research as visually engaging as it is interesting.
Through the PCR journal, we hope to spark intellectual curiosity and promote scientific inquiry amongst the next generation of Pingry researchers. Dive into the wonders of Pingry Research through this special edition of PCR: Pingry’s foremost journal of scientific research.
Katherine
Jung (V), Editor-in-Chief
Christian Zhou-Zheng (V), Editor-in-Chief
Elbert Ho (VI), Former Editor-in-Chief
Melinda Xu (VI), Former Editor-in-Chief
Editors-in-Chief:
Christian Zhou-Zheng (V), current Katherine Jung (V), current Elbert Ho (VI), former Melinda Xu (VI), former
Head Layout Editor: Julia Ronnen (V)
Head Copy Editor: TingTing Luo (V)
Faculty Advisor: Mr. D. Maxwell
Copy Editors:
Cecilia Caligiuri (V)
Edward Huang (V)
James Draper (V)
Jonah Park (V)
Sarah Yu (IV)
Layout Editors:
Amelia Liu (IV)
Olive Lanao (V)
Samaya Shah (IV)
Sarah Yu (IV)
Shanti Swadia (IV)
by Nyla Goodwin (VI), Harrison Hackett (V), Julia Oussenko (VI), Tanya Patel (V), Mr. D. Maxwell
Saccharomyces cerevisiae is a yeast species that develops genetic mutations when exposed to caspofungin, an antifungal agent with the same active ingredient as Cancidas. In this project, our AP Biology group used lab strains of S. cerevisiae expressing vibrant pigments and gradually exposed them to increasing concentrations of caspofungin (up to 64 µg/mL) to build resistance. The yeast’s resistance stems from genetic mutations when exposed to caspofungin, which impacts cell wall synthesis, lipid metabolism, and other crucial cellular processes. After each exposure, we isolated yeast from the experiment and sequenced their genomes, using online tools and databases to identify their mutations. We hypothesized about how these mutations contribute to the resistance of the evolved strain and how they affect the genes’ protein. We used the different colors of the
yeasts to determine which strain was better at adapting to particular environments. Our results determined a genomic, missense mutation on Chromosome IV, where a tryptophan residue was replaced by arginine in the TSC13 gene. This gene encodes an enoyl reductase enzyme that catalyzes the last step in each long fatty acid elongation cycle through oxidoreductase reactions and helps localize the endoplasmic reticulum while facilitating nuclear-vaculour junctions. These findings can help us better understand evolutionary adaptation and how these specific genetic mutations affect antifungal resistance. Understanding these resistance mechanisms could guide the development of more effective antifungal treatments that can either circumvent or overcome such adaptive responses in the pathogenic fungi.
by Hannah Castiglione (V), Ethan Geppel (V), Stella Reheman (V), Aanvi Trivedi (V), Mr. D. Maxwell
Exposing Saccharomyces cerevisiae to increasing concentrations of caspofungin, an antifungal agent targeting cell wall synthesis, induces targeted missense mutations in the FKS1 gene that suggest a gain of function, while its paralog GSC2 incurs mostly loss of function mutations. The objective of this study was to discover the mechanisms underlying this FKS1-mediated echinocandin resistance. We analyzed structural changes and biophysical parameters—including molecular weight, isoelectric point, and hydrophobicity—resulting from single amino acid substitutions. Three key mutation hotspots were identified: residues 1357–1360, 690-700 and 635–647. All three mutation hotspots sit close to one another on the protein in space. Mutations in the 1357–1360 region introduced larger, more negatively charged side chains that did not significantly alter hydrophobicity but could
disrupt a potential drug-binding site that involves a negatively charged caspofungin and echinocandin lipid tail. In contrast, mutations in the 635–647 and 690-700 hotspots primarily enhanced hydrophobicity without affecting side chain size or charge, potentially implying altered interactions with ordered lipid molecules and modifications in the local membrane microenvironment. These computed biophysical changes support the dual resistance mechanisms proposed by Hu, X. et al. (2023): one mechanism is the direct alteration of the drug-binding interface, while the other entails changes in FKS1’s response to membrane disruptions induced by echinocandin exposure. Collectively, our findings advance the understanding of antifungal resistance and show critical structural features necessary for FKS1 function.
by Keira DeCroix (V), Mena Khan (VI), Mr. D. Maxwell
This research project will explore caspofungin resistance in yeast through missense and nonsense gene mutations of FSK1, GSC2, NDC1, and UTP20. Caspofungin is an antifungal drug used to treat yeast infections in the blood, stomach, lungs and esophagus. Prior to our experiment, we hypothesized that as we continue to transfer the yeast, mutations would arise allowing the yeast to be resistant to caspofungin. We also aimed to determine what specific mutations enable yeast to develop resistance to caspofungin.
We began by inoculating a tube of YPD+G418 growth medium containing 0.0625ug/mL of caspofungin with our yeast culture using a sterile swab. Over several weeks, we transferred the culture, doubling the
caspofungin concentration each time growth was observed. As per our hypothesis, after eleven transfers, we reached a concentration of 64 µg/mL where yeast continued to grow, indicating resistance to caspofungin. Next, we completed sequencing, which revealed the four specific mutations responsible for this resistance: FSK1, GSC2, NDC1, and UTP20. Many of these mutations also arose in our classmates’ sequencing, particularly FSK1, a target for caspofungin.
Our results identify the mutations responsible for caspofungin resistance, guiding further research to understand their role and develop strategies to prevent resistance. This, therefore, can improve caspofungin treatment for yeast and fungal infections in future applications.
by Arjun Kapur (V), Arjun Goel (VI), Dean Zervos (V), Owen Smith (V), Mr. D. Maxwell
Caspofungin resistance in Saccharomyces cerevisiae (S. cerevisiae) develops through the accumulation of mutations that provide a selective advantage in antifungal environments. In our class experiments, we evolved yeast populations by exposing them to increasing concentrations of caspofungin, while monitoring growth dynamics and identifying genetic mutations associated with resistance. We are now comparing two independently evolved yeast strains to identify genetic differences. Our strain acquired a nonsense mutation in GSC2 (W1728*) and a missense mutation in FKS1 (D646Y), while another group’s strain developed missense mutations in both GSC2 and FKS1.
Although both strains exhibit growth at 64 µg/mL caspofungin, the mutations suggest distinct mechanisms of resistance. The nonsense mutation in GSC2 may lead to loss of function, potentially bypassing caspofungin’s inhibitory effect, while missense mutations in FKS1 are known to alter glucan synthase activity, reducing drug binding. By comparing the mechanisms behind these mutations, we aim to gain deeper insights into caspofungin resistance in S. cerevisiae. Specifically, we will evaluate growth rates and fitness across different conditions to determine whether resistance is more effective through partial protein alteration or complete loss of function. Our findings will contribute to understanding the genetic pathways driving antifungal resistance and offer valuable insights for addressing clinical drug resistance.
by Gabby DeLorenzo (V), Kathryn Flanigan (V), Trisha Jetley (V), Julia Ronnen (V), Mr. D. Maxwell
When yeast infections are treated with antibiotics, such as caspofungin, the yeast tends to mutate and develop resistance, prolonging the infection. Thus, understanding these mutations could provide critical insights for developing new antifungal treatments. This research project used experimental evolution to determine the location of mutations in the Saccharomyces cerevisiae genes through 15 rounds of increasing caspofungin concentrations over one and a half months. After sequencing at the University of Washington, the final yeast genome at 64 µL of caspofungin was compared to the ancestral genome to identify genetic mutations. Across 22 experiments, the GSC2 gene on chromosome VII had 18 mutations—the second-highest quantity
among the genes examined. GSC2 codes for enzymes involved in spore wall formation and synthesizes a polysaccharide critical to the structure of the spores. GSC2 mutations prevent yeast cells from becoming spores, ensuring that the cells maintain their vegetative forms. Yeast sporulation interrupts their usual reproduction cycle by entering a dormant state. Vegetative yeast cells become spores to survive in harsh environments, like an antibiotic treatment, by building thick cell walls to protect the yeast’s genetic information. We hypothesize that the mutations on GSC2 promote higher reproduction rates in the yeast, thereby enhancing yeast survival and preventing the efficacy of an antibiotic.
by Sari Berman (V), Sydney Ellsworth (V), Chloe Huang (V), Mr. D. Maxwell
Caspofungin is an antifungal medication that targets and eliminates harmful fungi by inhibiting enzymes crucial for the synthesis of glucans—key components of the fungal cell wall. Since humans lack the enzymes targeted by caspofungin, the medication can effectively target fungi without harming human cells. However, fungi, which are similar to bacteria, can develop resistance to antifungal treatments. In this study, we explored the mechanisms behind fungi’s resistance to caspofungin to develop potential strategies for treating infections caused by caspofungin-resistant fungi.
We investigated caspofungin resistance in yeast strains A05 and A08, which developed reduced susceptibility through direct adaptation. Over several weeks, we gradually exposed the two strains to increasing doses of caspofungin and selected for resistant variants. Once a concentration of 64 µg/mL was reached, the yeast strains underwent a comprehensive analysis, including whole-genome sequencing. In both A05 and A08 strains, we identified four missense mutations linked to resistance. One of the most significant mutations occurred in the FKS1 gene, which encodes a subunit of β-1,3-glucan synthase, the primary target of caspofungin. This study focuses on the functional implications of this mutation and its role in caspofungin resistance.
by Nataly Ruiz (VI), Jack Sherman (V), Ms. B. Farrell
In our AP Biology class, over the course of approximately two months, we conducted an experiment with yeast to observe how it can evolve to continue growing when placed in test tubes with increasing concentrations of caspofungin. Caspofungin acts as an antifungal medication that targets and destroys fungi such as yeast. At the start of the experiment, we placed the yeast in a test tube with a small concentration of caspofungin. This initial concentration did not fully destroy the yeast population. With each class period, we placed the yeast in a new test tube with a concentration of caspofungin that measured twice the amount used in the previous class. Despite the continued increase in caspofungin concentration, the yeast population survived. This survival suggests that the yeast had undergone mutation
and evolved to develop resistance to caspofungin. After the concentration of caspofungin reached a certain threshold, we sent our yeast sample to the lab in order to obtain its DNA sequence. The DNA sequence revealed which genes in the yeast had mutated. This information allowed us to identify specific genes that, when mutated, provide the yeast with resistance to caspofungin.
One of the genes that showed mutation in our yeast sample was FKS1, which we chose to examine in greater detail. Since FKS1 contains a large number of base pairs, we selected a small portion of the gene at random. We focused on the protein 8JZN to understand its purpose, its function, and its contribution to the role of the FKS1 gene.

by Nataly Ruiz (VI), Jack Sherman (V), Ms. B. Farrell

by Sasha Bauhs (VI), Thomas Davis (VI), Ria Govil (VI), Krish Patel (VI), Dr. M. D’Ausilio
Methods in Molecular Biology Research Honors aims to uncover the unknown functions of proteins. While Research 1 follows the BASIL (Biochemistry Authentic Scientific Inquiry) program to predict functions of proteins whose structures have been solved by X-ray crystallography, Research 2 focuses on ten proteins with hypothetical structures predicted by the artificial intelligence algorithm AlphaFold2.
Our in vitro work involves techniques such as bacterial transformation, autoinduction, SDS-PAGE, protein purification, and enzymatic activity assays. Our in silico computational work uses online databases and programs, including Pymol, Foldseek, Brenda, Blast, Dali, SwissDock, Clean, and Chimera to analyze similar proteins with sequential and structural homology. From our initial in silico analysis, we predict AF_AFQ0PC18F1 (AF_A) to be an isochorismatase or a nicotinamidase.
AF_A is a single-chain putative hydrolase with a computed structure model confidence of 97.95% (pLDDT
global). Our in silico work employs tools that assess not only primary structure conservation but also tools that use machine learning to predict enzymatic function from enzyme commission (EC) numbers. Using the SPRITE (now GrAFSS) database to assess RMSD scores of conserved motifs, we identify the active site of AF_A to be GLY62, HIS52, ASP50, and ASP13. Furthermore, we suspect the ligand to either be an isochorismate or a nicotinamide. Using SwissDock–a tool that assesses binding affinities—we have been unable to bind either of these proposed ligands to AF_A’s active site.
Our in vitro work includes expressing our protein in several bacterial cell lines (NEB5α and BL21DE3), SDS-PAGE western blots, cell lysis, and purification to separate AF_A from the biomechanical 6x HIS-TAG motif to be left with exclusively AF_A. Through our in vitro work, we plan to test the function of AF_A and perform enzymatic analysis to confirm if the function matches our hypothesis.
by Ellie Solomon (VI), Ella Tabish (VI), Dr. M. D’Ausilio
Protein function prediction is crucial for understanding enzymatic roles in cellular processes. As a class, we attempted to determine the enzymatic role of various uncharacterized proteins. Our group has focused on the protein 3L1W.
Through phase 1 of our study, we used an E. coli transformation, autoinduction, and SDS-PAGE to confirm proper protein expression. Our SDS-PAGE results confirmed the correct molecular weight of protein 3L1W as 71.6 kDa. In phase 2, we looked to confirm our protein’s function and structure by utilizing computational tools like InterPro, BLAST, DALI, SPRITE,
CLEAN, BRENDA, PyMOL, and Promals3D to analyze its sequence similarity, structural homology, and active site locations. From these methods, we found that protein 3L1W functions as an endonuclease enzyme. An endonuclease is a hydrolase enzyme that proofreads during DNA polymerization in DNA replication to remove mutated DNA structures that arise from problems with DNA replication.
These findings suggest that 3L1W plays a significant role in carbohydrate metabolism, potentially contributing to industrial applications. Future research may focus on substrate specificity to further investigate its functional capabilities.
by Mari Finkelstein (VI), Sam Nkansah (V), Nataly Ruiz (VI), Dr. M. D’Ausilio
We studied the enzyme activity of the protein 3N2C and predicted that it functions as a metal-dependent amidohydrolase that breaks down amide bonds. To test this, we produced and purified 3N2C and confirmed its molecular weight using SDS-PAGE. We then compared its structure and function to known proteins using BLAST, DALI, SPRITE, PyMol, and Promals3D. Our results showed that 3FEQ is the closest match to 3N2C due to its strong structural resemblance with 99.76% sequence identity, 73.1 Z-score, and 0.4 RMSD score.
We then identified six key residues in the active site: His63, His65, Lys188, His229, His249, and Asp321. These amino acids suggest a catalytic process that supports our prediction. Based on this, we proposed that 3N2C breaks down N-acetyl-D-glucosamine 6-phosphate into D-glucosamine 6-phosphate and acetate. To confirm this reaction, we designed an assay to measure acetate production.
Our study helps explain the role of 3N2C in metabolism and highlights its potential use in industrial and medical applications.
by Sari Berman (V), Ms. J. Patel, Dr. M. D’Ausilio
Enzymes play a vital role in biological processes, and understanding their functions is key to uncovering their applications, leading to improved disease treatment and a deeper insight into cellular processes. This study set out to investigate the enzymatic role of 2O14, a protein with a known structure and sequence but an unknown function.
First, 2O14 was successfully expressed using bacterial transformation and autoinduction. SDS-PAGE confirmed the observed molecular weight of 2O14 to be 41.79 kDa. Computational tools such as InterPro, SPRITE, and DALI were then used to compare 2O14 to proteins of known structure and function. This analysis helped identify its conserved domain, active site, and potential enzymatic function. The top homologs of 2O14, determined by full-protein and active site alignment as well as super RMSD scores, were 1J00, 1DEO, and 1PP4. These results revealed that 2O14 may act as an acetyl xylan esterase, hydrolyzing the ester linkages of acetic acid in xylan. By deacetylating xylan, 2O14 enhances xylanase activity, producing simpler sugars such as xylanose and facilitating the degrada-
tion of hemicellulose. Protein purification followed by various enzymatic assays will be used to confirm these results and further characterize the function of 2O14.
Future research may focus on substrate specificity to explore its potential. These findings indicate that 2O14 may play a key role in xylan modification, potentially contributing to industrial applications such as biofuel production.

by Max Ventura (V), Dr. M. D’Ausilio
The characterization of protein function is critical for understanding its mechanism and identifying potential real-world applications. This study focuses on predicting the function of the protein 3CBW using computational tools and lab techniques. We successfully expressed our protein through transformation and autoinduction, as confirmed by gel electrophoresis. To determine the function of 3CBW, we analyzed its structure and amino acid sequence with softwares such as InterPRO, BLAST, DALI, SPRITE, CLEAN, BRENDA, PyMol, and Promals3D. These tools identified 2QHA and 2WHK—both of which are beta-1,4-mannanases—as the most similar proteins in terms of amino acid sequence. 3CBW has a 99% sequence identity and
highly conserved active site residues with these homologs, supporting its classification as a beta-1,4-mannanase (EC 3.2.1.78). The next steps include purifying our protein, predicting substrate binding, and conducting enzymatic activity assays to corroborate our hypothesis that 3CBW is a beta-1,4-mannanase. These findings can also explain 3CBW’s enzymatic role in hydrolyzing β-D-mannosidic linkages, a critical process in the breakdown of cell walls for plant growth. We believe our findings could have broader implications in applications where enzymatic degradation of plant polysaccharides is essential, such as biofuel production, food processing, and drug development.
by Hannah Diao (V), Abigail Neu (V), Ziv Shah (VI), Aarav Sonthalia (VI),
Matan Zelkowicz (V), Dr. M. D’Ausilio, Ms. J. Patel
The function of an enzyme, a type of protein, can be determined through analyzing its molecular structure. The Protein Data Bank (PDB) was created to store and collect these structures, but the function of many entries remains undetermined. One such protein is 4DIU, a synthetically constructed enzyme expressed in Escherichia coli (E. coli). Our study aims to determine the function and enzymatic number of 4DIU by analyzing its molecular structure and similarity to homologs through various computational tools and in vitro work. So far, our group has successfully expressed 4DIU through E. coli transformation and autoinduction and confirmed expression using SDS-PAGE. Analysis of these protocols leads us to
believe that 4DIU is a carboxylesterase of family XIII that breaks down short and medium-chain fatty acids.
Furthermore, 4DIU’s homology to the PDB protein 8HEA suggests that 4DIU could play a role in the degradation process of plastic BHET. Our next steps include purifying our protein sample through nickel chromatography, predicting substrate binding, expressing the purified 4DIU on a large scale, running an activity assay, and introducing mutations to analyze active site amino acids. We hope to confirm 4DIU’s activity in degrading BHET and highlight its potential as a plastic degradation method.
by Edward Huang (V), Siena Kotlewski (VI), Nicolas Rodriguez-Lopez (VI), Dr. M. D’Ausilio
Enzymes play a critical role in metabolism, yet many remain uncharacterized despite their potential biological and industrial significance. This study aims to investigate the structure and function of 3DS8, an uncharacterized enzyme found in the bacteria Listeria innocua, through recombinant expression, computational analysis, and biochemical assays. To accomplish this, a plasmid encoding lin2722, the gene of our target protein, was transformed into E. coli BL21(DE3) cells. Autoinduction was then used to promote protein expression, which we confirmed via SDS-PAGE analysis. Consequently, computational tools such as DALI, CLEAN, and PyMOL were utilized to predict structural features and potential functional similarities by comparing them to known
enzymes. Preliminary research using these bioinformatics tools has predicted that 3DS8 shares structural and functional similarities with triacylglycerol lipases, a type of alpha/beta hydrolase that breaks down lipids by hydrolyzing ester bonds in triacylglycerol proteins. Sequence alignments with known conserved active site residues also support the hypothesis that this protein plays an important role in catalytic lipid metabolism. Future work will involve purifying the protein through cell lysis and nickel column chromatography, followed by a 4-nitrophenyl palmitate enzymatic assay to determine its activity. By characterizing this enzyme, we aim to gain deeper insights into its biological role and broader applications in metabolism and biotechnology.
by Kayla Kerr (VI), Julia Oussenko (VI), Will Pertsemlidis (VI), Dr. M. D’Ausilio
The BASIL (Biochemistry Authentic Scientific Inquiry Laboratory) curriculum allowed students in the Methods in Molecular Biology Research course to conduct novel research on specific proteins, with Q0P8E0 and Q32HQ2 being of interest, to predict structures and unknown functions. In Research 1, students investigate a set of proteins with experimentally determined structures but uncharacterized function. Building upon the same techniques used in Research 1, students in Research 2 are examining ten proteins with predicted enzymatic function and structural models generated by AlphaFold, an artificial intelligence program that predicts protein structures. These two proteins were analyzed to identify their enzymatic functions (UniprotIDs listed). Based on both in vitro and in silico analyses, two proteins—Q0P8E0 (B) and Q32HQ2 (D)—have been identified as strong candidates for future characterization.
A combination of computational and laboratory techniques was employed to investigate these uncharacterized proteins. Initially, plasmids containing the target proteins were transformed into BL21(DE3) E. coli cells
for protein expression. Autoinduction was used to induce protein expression in the bacterial cells, and the results were quantified via gel electrophoresis. To characterize the proteins structurally, three homologs were selected for each AlphaFold-predicted structure. These were analyzed for potential functional insights using Pymol, Sprite, BLAST, and DALI. This information is essential for the class’s ongoing success. Using this data, Protein B was identified as a phosphoglycolate phosphatase with an active site comprising Ile6, Gly11, and Ser16, while Protein D was identified as a pyrimidine utilization protein with an active site of Asp100, His71, and Gly49.
The next steps are to purify proteins B and D from E. coli and to conduct molecular docking studies using SwissDock to predict ligand binding. Enzymatic assays will then be designed and completed to experimentally characterize the enzymatic activity of these proteins. Future directions include mutational analysis of the predicted active sites of both proteins to further support their enzymatic roles as predicted hydrolases.
by Mia Cuiffo (VI), Ms. J. Patel
Enzymes play a crucial role in various biological processes, but there are many unidentified enzymes with undetermined functions and applications in biological research and therapy. This study aims to determine the function of the protein 4Q7Q, a protein with a known structure but unknown function. Initially, E.Coli transformation, autoinduction, and SDS-PAGE, protein gel electrophoresis, were performed to analyze protein expression. SDS-PAGE results confirmed proper protein expression, with a band size of 115kDa, aligning with the predicted size of 4Q7Q and the NusA tag. Subsequently, various databases and computational tools were utilized, including InterPro, SPRITE, DALI, and PyMOL,
to compare the known structure of 4Q7Q to other proteins with known structures and functions. While previous studies suggested that 4Q7Q was a lipase, these databases suggest that 4Q7Q is an acetyl xylan esterase (AcXE), hydrolyzing the ester linkages of acetic acid in xylan. To confirm this hypothesis, purification and various enzyme assays will be conducted to further analyze the function of 4Q7Q. As acetyl xylans are the most abundant hemicellulose in nature, confirming the function of 4Q7Q as an AcXE and further understanding its reaction could have numerous applications.
by Cecilia Caligiuri (V), James Draper (V), Dr. M. D’Ausilio, Ms. J. Patel
Understanding protein function has numerous implications for medical treatments and further research of cellular processes. Our study aims to identify the function of 2QRU, a protein with known structure and sequence but unknown function. We used wet-lab techniques including transformation, autoinduction, purification, and enzyme assays and computational techniques to study the 2QRU’s sequence and structure, including active site configuration, as well as the substrates and products for the reactions it likely catalyzes. In this way, we made an educated prediction of 2QRU’s true function.
First, plasmids were transformed into BL21(DE3 competent) E. coli cells which were then autoinduced. An SDS-PAGE confirmed successful 2QRU expression
since there was a dark band in the experimental lane at the molecular weight of 2QRU, 31.4 kDa. Through computational analysis, 2QRU was predicted to be a dihydrocoumarin hydrolase, an alpha-beta hydrolase that catalyzes the reaction of dihydrocoumarin into melilotate with the addition of water. The top homologs of 2QRU, as determined by full-protein and active site align and super RMSD scores, were 1A7U, 1ZOI, and 3LIP, which are all alpha-beta hydrolases with a Ser-Asp-His catalytic triad to perform esterase, lipase, or perhydrolase activity.
We will use lipase enzymatic assay incorporating spectrophotometric measurement following protein purification. Uncovering the function of 2QRU as it relates to Enterococcus faecalis could lead to discovering highly effective novel E. faecalis treatments.
by Sylvia Ardon (VI), Ms. J. Patel
This study aimed to express 3B7F, a protein with an unknown function, in E. coli BL21(DE3) cells using an autoinduction system and analyze its structural properties through computational tools. BL21(DE3) cells were first transformed with a plasmid containing the 3B7F gene. The culture was then incubated overnight, allowing for protein expression. Using SDS-PAGE, a distinct band corresponding to the expected molecular
weight of 3B7F was observed, confirming successful protein expression. Additionally, computational structural analysis was conducted using PyMOL to visualize the 3-D structure of 3B7F. This analysis provided insights into potential structural features, including secondary structure elements and possible active sites. Comparing these active sites to known proteins may contribute information about the function of 3B7F.
by Jiya Desai (VI), Olive Lanao (V), Sienna Patel (VI), Sofia Wood (VI), Dr. M. D’Ausilio
Scientists may study proteins whose functions are not yet known, where researching the molecular structure of previously uncharacterized enzymes, proteins that catalyze biological processes in all living organisms, may uncover new applications for these enzymes. Our work explores the structure and function of 3H04, the ID representing a previously uncharacterized enzyme. Initially, we performed transformation, autoinduction, and SDS-PAGE, to demonstrate successful protein expression and allow for further functional analysis. We then input the known sequence and structure of protein 3H04 into online databases, such as BLAST, InterPro,
DALI, and PyMol, which compared 3H04 to similar proteins based on amino acid sequence, active site residues, and structural alignment. From this analysis, we hypothesized that 3H04 may be classified as a carboxylesterase that acts on substrate 3-0-acetylpapaveroxine, a compound related to the drug noscapine. This enzyme would break down the substrate into narcotine hemiacetal and acetate, suggesting a role for 3H04 in drug metabolism. Moving forward, we aim to isolate our enzyme through cell lysis and purification, which will allow us to study the protein in its purest form.
by Nataly Ruiz (VI), Jack Sherman (V), Ms. B. Farrell

by
Chloe Huang (V), Durga Menon (V), Lane Purcell (V), Stella Reheman (V), Sriya Tallapragada (VI), Shreeda Sundaram (IV), Mr. B. Burkhart
The essential question of this HIRT is: What does it mean to write in a modernist style? We use William Faulkner’s novel, As I Lay Dying, as a central text for considering modernist ideals. Additionally, this text establishes a framework for thinking about the intersection between literature and philosophy as Faulkner’s prose encourages, and perhaps demands, an experience that mirrors how we experience the world: an experience that includes (re)processing memories, shifting perspectives, and ambiguity. We also pose the question: How does fiction position itself as a form of knowledge? Where does it fail to match world experience and where does it generate its own? What does fiction represent, if anything, and what tensions exist between fiction and the historical and the artistic?
This year, our group deconstructed As I Lay Dying, first through traditional close reading, and then by attempting our own modernist writing. Each of us wrote fictional works where we interpreted and emulated technical and philosophical elements of the text. We completed short fiction writing prompts that each attempted to capture a certain element of Faulkner’s prose, such as point of view, stream of consciousness narration, sensory imagery, or character voice. Our project’s next step will be to analyze our writing and thereby reevaluate our understanding of Modernism. Overall, while we research Modernism as an artistic movement, the ultimate goal of our HIRT is to cultivate original interpretations of the meaning and purpose of modernist literature.
By Olivia Buvanova
(VI), Evan Chen (V), Sheryl Chen (V), Greta Reinhardt (VI), Alex Terpstra (V), Mrs. Johnston, Dr. P. Longo
Creative nonfiction, a genre that adopts techniques of fiction writing to tell “true” stories, by definition blurs boundaries of form. Featuring such literary elements as dialogue, description, and perspective, the genre simultaneously constructs and processes lived experience to generate what leading scholar Lee Gutkind calls the “literature of reality.” The 2024-25 Creative Nonfiction
HIRT reads examples of creative nonfiction that appear in unexpected media, from the Gettysburg Address to JFK’s moonshot speech. The project seeks to understand how and why the “literature of reality” surfaces in texts we do not usually associate with creative writing. In the process, the project interrogates the appeal of nonfiction specifically at moments of political and social disruption.
by Izzy Berger (IV), Hannah Castiglione (V), Catherine Ramesh (III), Ava Goyanes (IV), Tanya Puranik (V), Stella Reheman (V), Fiona Shmuler (IV), Madeline Zhu (III), Ashley Gao (V), Gabby DeLorenzo (V), Avanti Hegde (III), Max Ruffer (V), Aanvi Trivedi (V), Dr. A. Briley, Dr. R. Cottingham
The Lyric and Poetry Analysis HIRT aims to examine the works of important poets and songwriters to investigate their portrayal of themes such as gender and race. We focus on the writings of Sylvia Plath and Taylor Swift, both skilled in confessional poetry—a style that emphasizes deeply personal experiences. We have studied numerous works by both artists, including “Stings” by Sylvia Plath
and “Mad Woman” by Taylor Swift, and conducted an indepth review of scholarly analyses of their work. Through a detailed examination of how both writers convey messages about gender and sexuality, we aim to identify recurring patterns and investigate what makes their expressive styles so compelling to modern audiences.
by Zara Abbasi (V), Lulu Brennan (IV), Sophia Cuiffo (IV), Alexandra DeCillis (V), Keira DeCroix (V), Emily Dicks (VI), Ryan Dicks (III), Briar Hackett (IV), Avery Hoffman (V), Claire Hoffman (IV), Caleb Kalafer (IV), Abigail Neu (V), Rowan Shapiro (IV), Leah Solomon (V), Jake Yang (V)
How does point of view affect historiography and our understanding of history? How does historical fiction shape our perception of the past? We have applied various lenses to our central novel, Trust by Hernan Diaz. This book, with each chapter narrated by a different
character, allows us to analyze the same events from multiple perspectives. Through our analysis using critical lenses, we concluded that point of view significantly influences modern interpretation of history.
By Shaan Barai
(V), Bowen Crosby (V), Matthew Del Vento (V), Moeed Khan (IV), Nate Kosar (III), Lucas Fredella (III), Cooper Goodwin (V), Andrew Hefner (III), Anderson Lee (V), Shea Patel (V), Riya Reddy (III), Aria Saksena (IV), Aryan Saksena (III), Addison Santomassimo (V), Akiv Shah (III), Dylan Shah (III), Jack Sherman (V), Charlie Yang (III)
Almost all of us watch major sporting events, whether it be the Olympics or the FIFA World Cup. We consider how these events affect the economies and communities of host cities through the following questions:
• How does hosting the Olympics affect a country and city’s economy in terms of GDP, unemployment rate, inflation rate, and long-term economic growth?
• What type of economy is optimal for hosting the Olympics?
• How does hosting the Olympics affect the overall morale and quality of life of a community?
Understanding the economic and social impact of the Olympic Games on host cities is crucial. The Games significantly influence a city’s GDP, unemployment, and other economic indicators, as well as the city’s morale and environmental conditions. Our research aims to identify the types of demographics and economies that can most effectively handle and benefit from hosting the Olympics.
Our team is divided into two subgroups: Social Impacts and Economics. The Social Impacts group examines key indicators to assess standard of living and analyzes case studies to understand how the Olympics affects daily life. The Economics group collects and analyzes long-term data on GDP, unemployment, and property prices. To quantify the economic impact of the Olympics, we explore various impact models, including the Synthetic Control Method, time series analysis, and multiple variable regression. These models allow us to compare host cities to similar non-hosting cities, assess trends before and after the Games, and isolate key economic drivers. We will conduct a theoretical analysis to determine what types of economies benefit most from hosting the Olympics. From there, we hope to use our findings to propose new locations and serve as a guide or warning for future host cities.
by Ashley Gao (V), Marc Gautier (V), JingJing Luo (VI), Tingting Luo (V), Fiona Rovito (IV), Dr. P. Longo
The Linguistics HIRT explores how the building blocks of language and the structures that generate meaning–phonemes, morphemes, syntax, semantics and pragmatics–influence communication and interpretation. This year, we are conducting a case study with the novel Lexicon by Max Barry, a thriller that omits traditional expository text and forces us to rely on linguistic structures
and context to construct meaning. We apply speech act theory, a branch of pragmatics that seeks to express language in terms of its applications in performing various speech acts. Our goal is to determine how the text generates or thwarts common ground within the storytelling and between the narrative and the reader.
By Helen Pols (VI), Ms. M. Boone
My research in the Architecture HIRT explores the evolution of sustainable architecture in cold climates, focusing on possible design and economic drawbacks. Through case studies and analysis, I investigate whether sustainability in these extreme environments presents unique economic impacts such as cost efficiencies, long-term energy savings, or ones on local economies. Additionally, I use my findings to draft an original archi-
tectural design: a ski-in, ski-out house that integrates sustainability with functionality. This design incorporates heating, insulation, and innovative building materials to optimize energy efficiency and comfort in cold weather conditions. By examining the intersection of environmental adaptation, economic practicality, and architectural innovation, my project aims to understand sustainable development in extreme climates.
By Jakob Braun (IV), Ms. M. Boone
Architecture is more than just building structures—it influences how communities form and function. This project explores two key ideas: how architecture contributes to community building and how sustainable design impacts both people and the environment. My central questions are: How does architecture foster social connections and a sense of belonging? How do sustainable design choices affect both ecological and human well-being?
To investigate, I am studying real-world examples. One focus is Jennica, an architect and urban planner who is creating a community in Ecuador, demonstrating how physical spaces bring people together. Another focus is an architect specializing in sustainable materials to under-
stand how eco-friendly design choices shape the environment. Our research includes analyzing architectural theory, reviewing project plans, and examining firsthand accounts from architects and community members.
Thus far, I have found that architecture plays a major role in shaping social interactions, whether through intentional design or the organic formation of communities. Meanwhile, sustainable architecture offers solutions for long-term environmental and human well-being. Through this research, I hope to highlight how architecture not only creates spaces but also influences the way people connect and live together.
By Lauren Glasofer (IV), Sylvie Kurtzman (IV), Lily Pereira (IV), Kallie Stern (IV), Ms. L. Cattafi
Throughout history, the accusation of witchcraft has been weaponized to control women who were threats to those in power. Using the gender and feminist critical lenses, the Women, Witches, and the Wielding of Power HIRT examined different case studies of women accused of witchcraft, ranging from the 1600s to the present day. Hundreds of years ago, those accused were often killed, along with the rest of their families. Today, women in various parts of the world accused of partaking in the “dark
arts” are shamed, incarcerated, and often suspected of being involved in mysterious deaths. In the United States, female politicians as recently as 2024 have been accused of using witchcraft to compel the public to cast votes for them. Although the cases change as our world evolves, there is always a common theme: the accusation of witchcraft is used, often by men, to control powerful women who are deemed to be a threat. Our HIRT will create a website and podcast documenting the cases we have studied.
by Matan Zelcowitz (V), Ms. L. Siegel
Oral histories, a method defined by historian Nicholas Mariner as ranging from “anecdotal support to historical inquiry,” are primarily used as a medium to address a research question through a holistic approach. To fully understand the factors influencing the topic of study, a variety of interviews is necessary to grasp the social, political and economic contexts of the time period. For the 2024-2025 Oral Histories HIRT, my central research question was, “How did the Ideology of communism in Cuba affect the Cuban healthcare system?” I conducted a series of interviews to better understand the complexities of the history and current structures of the Cuban healthcare system. In preparation for these interviews, I researched both Cuba’s transition to communism and
the development of its healthcare system. Furthermore, I applied the oral history methods of compiling the individual’s full story through directed discussion during interviews with healthcare workers, medical administrators, historians, and economists. Through my research, I learned about the different components of Cuban healthcare, including the unique role of neighborhood doctors and polyclinics located directly in the community. My interviews revealed that, while the Cuban healthcare system has benefited the country, the quality of medical care has declined due to Cuba’s ongoing economic crisis. In the future, I plan to use this same methodology to analyze the healthcare systems of other Latin American countries, particularly El Salvador and Costa Rica.
By Zoe Snider (IV), Ms. M. Boone
This research delves into the intersection of architecture and psychology, examining the psychological impact of color theory in residential spaces. It focuses on “warm” colors such as reds, yellows, oranges, and greens which evoke feelings of homeliness, intimacy, and warmth. By exploring the design philosophy of Frank Lloyd Wright, particularly through a case study of the Schwartz House, this project analyzes how Wright used color, materials, and spatial arrangement to influence human behavior and emotional well-being. Frank Lloyd Wright’s use of warm, earthy tones in his architecture played a pivotal role in creating an environment that fostered emotional security and connection. In the Schwartz House, for example, Wright integrated rich, natural colors that blended with the surrounding landscape, reinforcing
a sense of harmony between the interior and exterior. The use of deep reds, dark browns, and golden yellows was intentional, designed to evoke a grounded, nurturing atmosphere, while contrasting with the cool blues and fresh greens of the nature outside. Wright also integrated natural materials like wood, stone, and brick, enhancing the tactile experience of warmth and solidity. His signature use of exposed wood beams and textured surfaces not only contributed to the aesthetic beauty of the space, but also provided a psychological sense of intimacy and shelter. This case study of Wright’s architecture explores the deep psychological effects of color and material choice on mood, highlighting how carefully selected hues and materials can promote relaxation, comfort, and social cohesion within works of art.
by Sophie Schachter (IV), Bridget Troy (IV), Dr. B. Ward
The field of ethics and its many frameworks provides us with moral reasoning to help navigate complex situations while focusing on different values. Highlighting ethical frameworks such as deontology, consequentialism, and virtue ethics, the study of ethics attempts to define morality and how it should be implemented in society. The 2024-2025 Ethics HIRT evaluates ethical dilemmas in the modern world by applying a diverse range of moral lenses to various case studies.
For example, our project seeks to explore the ethics behind election betting, primarily focusing on the 2024 United States presidential election. In doing so, our project considers how ethical theories grapple with the tension between personal gain and collective responsibility. Ultimately, we examine whether wagering on democratic outcomes undermines the moral foundations of civic participation, or simply reflects the evolving relationship between politics and finance.
By Suvid
Bordia (III), Henri Brisson (III), William Brisson (V), Ronen Gorbach (V), Arnav Jain (V), Eshaan Jain (VI), Anderson Lee (V), Rishi Mirchandani (III), Jonah Park (V), Catherine Ramesh (III), Gabriel Raykin (V), Rohnik Shah (III), Mr. J. Webber
Understanding how the Federal Reserve sets interest rates is crucial to comprehending the broader economic landscape and making informed financial decisions. Interest rate decisions significantly influence the U.S. and global economies, impacting everything from employment and inflation to currency valuations and international trade. We aim to determine how the Federal Reserve sets interest rates and which economic indicators guide their monetary policy decisions.
To answer these key questions, our team began with a comprehensive deep dive into the history of the Federal Reserve, exploring its role in shaping monetary policy and stabilizing the economy. Primary resources we explored include former Federal Reserve Chair Ben S. Bernanke’s 21st Century Monetary Policy, which provides foundational insights into economic principles and the decision-making process of the Federal Reserve. Supplementary materials include Federal Open Market Com-
mittee (FOMC) meeting minutes and historical data from the publications of the Federal Reserve. Upcoming interviews with industry leaders and simulated meetings will be conducted to investigate applications of our literature review. We will apply our theoretical knowledge to practical scenarios, enhancing our understanding of economic indicators including unemployment, inflation, GDP growth, and international trade balances.
Thus far, our team has analyzed the dual mandate of the Federal Reserve, maximizing employment and maintaining price stability. We have also covered how the Federal Reserve has gained increasing independence from the government, particularly the executive branch. Furthermore, we have examined how the Federal Reserve has been increasingly influenced by real-time data on inflation, employment levels, and global economic conditions. Future predictions of interest rates will incorporate these findings, providing a practical application of our research.
By Cecilia Caligiuri (V), Gisel Calulo (V), Annabelle Elie (IV), Charlotte Hao (III), Risa Patel (III), Dr. P. Haven
What does it mean to be human–socially, culturally, and biologically? How do we compare to other species and our own genus Homo? These are the fundamental questions that drive the Biological Anthropology HIRT, which explores human evolution through scientific inquiry and anthropological literature.
We started the year by distinguishing scientific facts from common beliefs with regard to evolution. Members presented on key evolutionary concepts such as the transmutation hypothesis, natural selection, post-Darwinian theories, scientific racism, and organismal and multilevel evolution. To learn more about the Homo sapiens anatomy, we met with Assistant Professor Hillary Delprete of Biological Anthropology from Monmouth University. She introduced us to linguistic, biological, and forensic anthropology through her study of pelvic morphology analysis over the past 200 years. This helped us deepen our knowledge of human anatomy in comparison to other hominin species for when we analyzed different hominin and hominid skulls with Rutgers anthropology undergraduate students at the end of last year.
We then began constructing our own hominin and hominid family tree, mapping key traits of each of our ancestors. Finally, we analyzed a Neanderthal focused documentary that provided insights to the theories on why and how Homo sapiens led to the downfall of Neanderthals and other Homo species. In the documentary, the researchers used forensic analysis of bone positionings and markings to infer the culture and customs of Neanderthals.
Building on this foundation, we are now preparing to conduct a lab, focusing on the genetic legacy of Neanderthal DNA in Homo sapiens. By analyzing our own DNA, we seek to identify the genetic variance in the DPP4 gene. The percentage of the DPP4 gene we have (humans have 1- 4%) is based on the amount of Neanderthal DNA we possess. The presence of this gene is linked to immune system disorders and varying immune responses including the severity of COVID-19 and other diseases.
This could lead to interesting discussions on what groups of people (ethnically/racially) were hit by the COVID-19 epidemic hardest due to genetics. Through our research, we hope to find a tangible connection between our evolutionary past and its impact on our health today.



In our AP Biology class, over the course of approxi mately two months, we conducted an experiment with yeast to observe how it can evolve to continue growing when placed in test tubes with increasing concentra tions of caspofungin. Caspofungin acts as an antifungal medication that targets and destroys fungi such as yeast. At the start of the experiment, we placed the yeast in a test tube with a small concentration of caspofungin. This initial concentration did not fully destroy the yeast population. With each class period, we placed the yeast in a new test tube with a concentration of caspofungin that measured twice the amount used in the previous class. Despite the continued increase in caspofungin concentration, the yeast population survived. This sur vival suggests that the yeast had undergone mutation and evolved to develop resistance to caspofungin. Af
by Ryan Hao
(IV), Ethan Liu (VI), Katharine Luo (V), Alex Wong (VI), Christian Zhou-Zheng (V), William Zhou-Zheng (IV), Dr. C. Chu
The Accelerated Molecular Dynamics group is focused on developing numerical simulations for modeling motions of interacting particles and harmonic oscillator networks. We optimize our N-Body simulation by varying our propagation and comparing our implementation on CPUs and GPUs. Our simulations are used to study the
responses of materials, modeled as systems of harmonic oscillators, to external perturbations, as well as systems of particles under the influence of given forces. Our research can also be extended to explore more complex systems with different forms of forces, such as crystal lattices and particles interacting under Yukawa potential.
By Sarah Clevenger (IV), Chloe Huang (V), Eshaan Jain (VI), Arjun Kapur (V), Olivia Li (IV), Jack Shea (IV), Aarav Sonthalia (VI), Rohan Yadav (VI), Ms. S. Mygas
Coral polyps have a symbiotic relationship with zooxanthellae, a type of photosynthetic dinoflagellate that provides nutrients and energy vital for coral reef health. However, environmental changes, particularly global warming-induced temperature increases, can trigger corals to expel their zooxanthellae. This expulsion leads to a decline in zooxanthellae populations, causing coral bleaching, heightened stress levels, and potentially death.
Currently, our team is investigating the horizontal transfer of zooxanthellae between neighboring corals. This process involves the movement of zooxanthellae between adjacent corals, which may significantly influence their resilience against environmental stressors. By analyzing the presence and diversity of zooxanthellae among neighboring corals, we aim to uncover patterns of zooxanthellae shifts during horizontal transfer. Understanding these trends could inform strategies to enhance coral longevity and counter stress-induced coral mortality.
Our research includes collecting coral samples from a controlled ecosystem and extracting zooxanthellae for DNA fingerprinting. We will use the DNA fingerprints
to catalog the zooxanthellae in our corals and create a baseline library of zooxanthellae species and their quantities. With this library, we can develop metrics to analyze transfer dynamics within these algal populations. Our group has established a rigorous protocol for tank maintenance to ensure optimal conditions for coral health and growth. We continuously monitor critical tank parameters, such as pH (maintained between 8.2 and 8.6) and salinity (around 35 parts per thousand), making adjustments as necessary by siphoning water or modifying tank conditions. For coral feeding, we administer a specialized coral “dust” directly onto the polyps. We also incubate Artemia (brine shrimp) in a zero-light environment before introducing them into the tank as an alternate food source.
While we have encountered several challenges, including losing our first corals due to stress during a tank transfer, we have refined our DNA extraction protocol for Gymnodinium catenatum and Pyrocystis fusiformis dinoflagellates. This improvement ensures our methods are sound and provide positive controls for future experiments.
By
Connor Francis (VI), Matthew Gilsenan (IV), Bogdan Kuzmanov (VI), Max Li (IV), JingJing Luo (VI), Renny Mirliss (V), Alessio Pasini (V), Victoria Xie (V), Melinda Xu (VI), Dr. M. Jolly
The EmotiMate independent research team investigates how large language models mimic human emotions and facilitate social interactions by creating a chatbot to alleviate loneliness, particularly among the elderly. Our goal is to provide an AI-driven solution to the widespread issue of elderly isolation by developing a chatbot capable of detecting and responding appropriately to user emotions. We used multiple Application Programming Inter-
faces (APIs) to integrate conversational functionality and emotion recognition models. We also experimented with different system prompts with the CAST team members to fine-tune our chatbot’s response style to be empathetic and engaging. Additionally, we implemented a basic avatar that speaks and conveys emotions, experimenting with various tones, inflections, and voices to better resemble human conversation. Through qualitative and quantitative methods,
we integrated findings from cognitive science, human-computer interaction, and artificial intelligence.
We are currently creating a protocol to test our prototype within the Pingry community. Our test results will reveal how our working prototype can more effectively alleviate user loneliness. Ultimately, we seek to contribute to a deeper understanding of AI companionship and its role in human society.
By Ethan Geppel (V), Sia Ghatak (VI), Som Ghatak (IV), Sophia Guild (V), Norah Jacob
The Extended Reality (XR) Independent Research Team explores the potential of different subsets of extended reality to influence real-world behavior and environmental consciousness. Our research investigates the hypothesis that a virtual reality simulation about beach pollution and its impact on biodiversity can influence users’ daily actions concerning single-use plastics. Extended reality is a field with increasing relevance to the future; potential implementations of this powerful tool include assisting psychologists, behavioral studies, and increasing environmental literacy. Moreover, XR has proven to be more effective at influenc-
ing long-term behavior than video or other traditional mediums, which is why we are attempting to determine if XR is an effective tool in increasing climate awareness. To make our simulation, we are using Unity to develop the VR space and interactivity and Blender to create the custom assets. Our simulation will debut within the Pingry community in the near future, as we hope to run a small pilot later in the year. Through this immersive experience, we hope to underscore the significance of individual actions in contributing to a collective environmental impact, promoting more environmentally conscious behavior among the Pingry community and beyond.
By
Sasha Bauhs (VI), Isabelle Chen (VI), James Draper (V), Katherine Jung (V), Stella Reheman (V), Nataly Ruiz (VI), Ziv Shah (VI), Ishaan Sinha (IV), Dr. D. Fried
Understanding the molecular mechanisms behind bioluminescence and fluorescence is important across many areas of biology and medicine, from enhancing our knowledge of ecological interactions to developing new fluorescent tags. Since Roger Tsien was awarded the 2008 Nobel Prize in Chemistry for the discovery and development of green fluorescent protein (GFP), research in fluorescence has increased significantly. In 2013, the Japanese fluorescent freshwater eel (Anguilla japonica) was identified as the first fluorescent vertebrate animal. Since then, other fluorescent marine organisms, such as scorpionfish, algae, and plankton, have also been discovered. Our current project focuses on the expression,
characterization, and mutation of UnaG, the fluorescent protein found in the Japanese fluorescent eel. We have successfully transformed, expressed, and purified this protein using E. coli cells. Since the protein requires a cofactor (bilirubin) for expression, we are preparing to conduct a fluorometric assay with added bilirubin to test the protein’s activity in vitro. Our future goals include testing UnaG active site mutations and exploring different cofactor molecules to determine if we can enhance or alter the color of UnaG fluorescence. These mutated UnaGs could be used to visualize cell structures, detect or track specific molecules, and monitor gene expression.
By Sarah Bonilla (IV), Arjun Goel (VI), Arnav Jain (V), Amelia Liu (IV), Uma Menon (VI), Jonah Park (V), Dean Zervos (V), Dr. J. Keyer, Dr. R. Riman
Concrete, a cornerstone of global infrastructure, contributes 8% of global carbon emissions due to its energy-intensive production. While alternatives like reactive hydrothermal liquid-phase densification (rHLPD), a low-temperature process inspired by biomineralization, reduce cement’s carbon footprint by 70%, scalability challenges persist. This study explores integrating rHLPD with wood composites to develop sustainable, high-strength building materials. Delignified wood, stripped of binding cells, forms a porous cellulose network we use as a scaffold. These pores are then infused with rHLPD-derived composites, which
undergo carbonation to solidify and bond chemically with the wood matrix. The resulting hybrid material has enhanced tensile strength and durability. Initial prototypes have demonstrated compatibility with construction applications such as load-bearing structures. Further optimization is focused on modifying composite viscosity and carbonation factors to maximize CO₂ sequestration and mechanical performance. This approach bridges the gap between bio-based materials and low-carbon cement technologies, offering the potential for emission reductions and structural innovation.
by
Kathryn Flanigan (V), Graham Houghton (VI), Douglas McNaugher (IV), Aiden Suh (IV), Sriya Tallapragada (VI), Ella Wunderlich (VI), Julian Zassenhaus (V), Matan Zelkowicz (V), Ms. B. Farrell, Ms. J. Patel
Metagenomics is the study of genetic material taken from populations of organisms. By performing metagenomic analyses of DNA from the Pingry composter, we aim to understand how the microbial community in the composter responds to changes in environmental conditions. In doing so, we hope to gain practical insights into the biological mechanisms which drive the composting process.
Preliminary data analysis reveals differences between microbial communities at different stages of the decomposition process. Several characteristics in the composter environment, such as temperature, pH, and amount of raw material added, correlate in varying degrees to the abundance of specific bacterial orders. The data provides a natural history of the composter’s functionality, which translates into practical recommendations for improving its future operation. To build upon initial findings, we conduct further ex-
periments to identify the key factors that contribute to the success of the composter. This includes replicating conditions from the main composter in controlled mini-composters to observe how specific environmental variables, such as temperature fluctuations, pH adjustments, and varying input materials, affect microbial composition and decomposition rates. To explore the microbes that play pivotal roles in decomposing organic matter, we perform soil plating experiments that isolate and identify bacterial strains present at different composting stages. These controlled experiments help validate correlations observed in the larger composter and facilitate fine-tuning of operational parameters for maximum efficiency. By integrating results from both metagenomic analysis and practical experimentation, actionable strategies for optimizing compost quality and speed can be developed, which ultimately improves the sustainability and effectiveness of our composting system.
By
Melia Ahn (IV), Rohan Goel (IV), Ria Govil (VI), Daniel Hall (V), Ella Tabish (VI), Max Ventura (V), Julianna Zhang (IV), Carolyn Zhou (VI), Dr. P. Haven
Collagen is the most abundant protein in the human body and it plays an important structural and functional role in many physiological and pathogenic processes. At the molecular level, collagen is made up of three polypeptide chains that form a secondary triple helical structure, which then self-assembles in a lateral staggered association to create fibrils with a unique 67 nm gap-overlap repeat; known as D-period. The unique conformity and behavior of the collagen molecule arise from the repeating Glycine-X-Y pattern in its primary structure. The ultimate goal of our project is to create a collagen mimetic peptide that can model the physical and chemical properties of natural collagen. Our designed collagen peptide, FACT-152 (Fragment of A
Collagen Trimer), didn’t work, so this year we are using a pre-existing plasmid design named V-F877 to start optimizing protein expression in bacteria. Currently, collagen is most commonly purified from animals for use in the medical field. This process is expensive, tedious, and often results in a high degree of variation. Generating collagen mimetic peptides that successfully replicate human collagen could provide a safer and more cost-effective alternative. We plan to provide critical research on these foundational steps to deepen our knowledge of collagen’s structure as well as determine the best methods of creating viable artificial collagen that can be used as a biomaterial.
Sofia Wood (VI), Sloane Mandelbaum (VI), Sari Berman (V), Edward Huang (V), Julia Ronnen (V), Jasmine Zhou (IV), Noah Reichman
Polyethylene terephthalate (PET) is one of the most widely used plastics in the United States of America, however; PET takes up to 450 years to degrade in landfills. As the severity of plastic-related damage to the environment grows, the scientific community searches for an efficient way to recycle PET. Our group aims to optimize PETase, an enzyme with the unique ability to break PET polymers into their monomers, such as TPA and EG, which can be reassembled into strong plastic polymers for consumer use. This nuanced recycling method is termed “enzymatic recycling” and has gained attention for its minimal environmental impacts. However, the mutation of PETase that is used has to be optimal for this process to be most efficient. To determine the
efficiency of the PETase enzyme in our lab, we perform a PCL plate-clearing assay, in which suspended PCL, a plastic nearly identical to PET, is plated in a solution with LB/agar. A colony of BL21-DE3 Escherichia coli expressing the PETase enzyme is then incubated on the plate overnight, and the radius of a “clearing halo” surrounding the originally placed bacteria expressing PETase is observed. By using the diameter of the clearing halo, the enzyme’s efficiency at degrading PCL and similar plastics can be quantified. Using this assay, our group hopes to research and document the effects of different mutations on PETase’s efficacy, further advancing the scientific community’s knowledge of the enzyme and contributing to the global effort to recycle PET plastic efficiently.
By Eric Chen (IV), Elbert Ho (VI), Derek Peng (IV), Vinav Shah (VI), Arjun Subramanian (V), Albert Wu (V), Dr. M. Jolly
Atherosclerosis is a chronic disease of the arteries that affects nearly half of all Americans between the ages 45 and 84. Cholesteryl Ester Transfer Protein (CETP) converts high-density-lipoproteins to low density lipoproteins, which accumulate in the walls of blood vessels. Despite CETP’s critical role in atherosclerosis, all existing drugs targeting CETP’s mechanism have failed to reach the market. We are developing a diffusion-based machine learning model to generate novel molecules and improve existing inhibitors of the CETP protein. In the past, we converted Simplified Molecular Input Line Entry System (SMILES) molecules to a lower-dimensional latent space using an autoencoder. Now, we are using a 3D equivariant graph representation to better capture the chemical and spatial properties of the mol-
ecules. Sampling from a standard Gaussian distribution, our model gradually converts the represented molecule into pure noise, before training a neural network to denoise it. We generate new molecules by denoising a graph of random noise. Once generated, the molecules are screened using molecular docking to measure binding affinity, the Quantitative Estimate of Drug-Likeness (QED), and the Synthetic Accessibility Score (SAS). Our models have generated molecules that perform at least as well as those currently being tested in clinical trials, demonstrating the promise of diffusion methods in drug discovery. We are currently exploring the use of more accurate molecular representations during the diffusion process to either accelerate the process or significantly improve its efficacy through guided diffusion.
by Mia Cuiffo (VI), Hannah Diao (V), Leah Holmes (IV), Tingting Luo (V), Krish Patel (VI), Riya Prabhu (IV), Aanvi Trivedi (V), Keira Troy (VI), Dr. M. D’Ausilio
Shigella, a major cause of bacterial foodborne illness globally, relies on the transcription factor VirF, a member of the AraC superfamily, to regulate its virulence cascade. Despite its significance, the structure of VirF remains unsolved, hindering further understanding of its role in Shigella pathogenicity. Previous studies have shown that other AraC superfamily members, like ToxT from Vibrio cholerae, contain a binding pocket that interacts with a small fatty acid to inhibit DNA binding and affect virulence. We seek to understand whether VirF functions in a comparable manner to ToxT, given its similar structur-
al feature and function, and if its activity can be inhibited by fatty acids. To address this, the VirF independent research team is focused on expressing, purifying, and crystallizing VirF to find out its three-dimensional structure. Additionally, biochemical assays will be employed to test for functional similarities to ToxT. The purification process is progressing well, with ongoing efforts to optimize crystallization conditions. We aim to uncover the molecular mechanisms behind transcriptional regulation in Shigella dysenteriae that could contribute to the development of targeted treatments for Shigella infections.
By Griffin Dragert (III)
Drug discovery can be extraordinarily difficult. In 100 years of modern medicine, “we’ve found treatments for only around 500 of the roughly 7,000 rare diseases,” according to David Pardoe of Evotec in Germany. Incredibly high costs and time required to develop treatments prevent us from attempting to treat the roughly 6,500 diseases that remain uncured. However, to the excitement of many, researchers are now working to implement artificial intelligence into modern medicine and potentially achieve stronger results while addressing both cost and time challenges.
If used properly, AI should be able to apply previous data to construct the three-dimensional structure of a potential drug, then depict the binding of the drug-like molecule to its targeted protein. Another goal is for AI to potentially consider the interactions between the drug and the patient, preventing unwanted interactions as well as unnecessary side effects. Other functions of implementing AI into modern medicine include providing advice on diagnosis, analyzing X-ray scans to detect abnormalities, and generating a personalized treatment plan based on the patient’s age and lifestyle. While implementing AI into modern medicine has many positive effects, researchers have frequently ex-
pressed concerns about potential repercussions and have questioned the complete replacement of physicians. AI lacks essential human qualities, such as empathy and compassion, which are indispensable in patient care. Furthermore, AI’s effectiveness is limited by the insufficient availability of information. Although there is a vast accumulation of research in the field, many studies—whether successful or not—have not been publicized for the use of others and AI. As Miraz Rahman of King’s College London articulates, “What is being published? Always the success story.” The outcome seems simpler when companies decide not to publicize their negative results. If the quality, quantity, and consistency of medical research are improved, AI’s understanding of these topics can be strengthened, preparing it for more effective use in the medical field.
It is almost guaranteed that AI will not completely replace physicians’ jobs. However, collaboration between humans and AI will strengthen the foundation of modern medicine. Integrating AI, though, will take time and facilitation. Once we implement it into everyday medical use, our general medical expertise will be enhanced in the years to come.

by Ryan Hao (IV)
Since their launch in 1977, the Voyager 1 and 2 space probes have witnessed and documented the many mysteries of outer space. Even in 2025, when they are 169 AU away, they continue to escape our solar system at 3.6 AU and 3.3 AU per year, respectively. Though they were intended to last only five years, they still function, sending signals back to Earth about spatial information via NASA’s deep space network. However, as the Voyagers’ nuclear batteries are slowly depleting, NASA is desperately trying to recover the probes.
Thus far, Voyager 1 has made many discoveries about orbits in our solar system and beyond. It discovered a thin ring around Jupiter, and two new Jovian moons, Thebe and Metis. As it passed Saturn in its journey, it found five new moons and a new ring, now named the G-ring. As the first satellite to pass the heliosphere, it now represents humanity’s best connections with forces beyond our solar system.
Because of the explorational value that Voyager 1 holds, NASA is willing to sacrifice some useful functionalities to increase its lifespan, as any second may bring an insightful discovery. Therefore, Voyager 1
was designed to not rely on solar panels, instead using radioisotope thermoelectric generators (RTGs). RTGs convert heat from the natural decay of radioactive material (alpha decay) into usable electricity. With a power output of 470 watts at launch, Voyager 1 only loses 4 watts of power each year from its fuel degradation. However, it has already decreased to 70% of its original power output, and would have been on track to lose all functionality within the next few months. By remotely shutting down power usage for some of the less essential components of the Voyager 1, scientists predict that the space probe may retain functional until the 2030s. To transmit discoveries, it still retains key technologies like cosmic ray sensors and deep space communication devices.
Weatherbed, Jess. “NASA is making sacrifices to keep the Voyager mission alive.” The Verge, 6 Mar. 2025, www. theverge.com/news/625182/nasa-voyager-mission-instruments-shut-down-power. Accessed 6 Mar. 2025. NASA. 9 Jan. 2018, science.nasa. gov/mission/voyager/voyager-1/.

by Daniel Hall (V)
Even before its discovery in 1906, Alzheimer’s disease (AD) has ravaged the brains of billions of individuals, and its exponential trajectory indicates no signs of slowing down. In 2020, an estimated 5.8 million Americans were living with this disease, and every five years, the number of people aged 65 and older living with Alzheimer’s doubles [1]. While prevalent, this disorder is also one of the top 10 primary causes of death globally and is ranked as the sixth leading cause of death among adults in the US [1,11].
Named after Alois Alzheimer, the German psychiatrist who first identified this form of dementia, AD is a fatal neurological disorder characterized by progressive neuron damage. The damage typically begins in the entorhinal cortex and hippocampus—regions of the brain responsible for memory—and later spreads to other areas responsible for language, reasoning, and behavior [5]. This pathway is similar to the progression of symptoms observed in AD patients. Initial signs of memory loss and cognitive decline are sometimes diagnosed as Mild Cognitive Impairment (MCI). The loss of other functions follows slowly over time. However, a diagnosis of MCI does not guarantee that symptoms will progress into AD.
Currently, the amyloid-beta (Aβ) hypothesis is the predominant theory explaining the cause of AD. In short, researchers attribute neuronal degradation in AD to the aggregation of Aβ proteins in the brain. Aβ can assemble into various plaque formations, each having a great effect on the brain. At the core of each configuration is the amyloid-beta monomer, which is derived from a series of cleavages, and in some cases, genetic mutations. In this article, we aim to explore the structures into which Aβ aggregates and their effects on the brain. Additionally, we seek to investigate how the Aβ monomer is formed and what genetic factors contribute to its formation.
and neuritic plaques
Aβ aggregates into three structures: oligomers, diffuse plaques, and neuritic plaques. Oligomers are molecules
composed of multiple repeating monomers. In the case of AD, Aβ oligomers form when two to twelve Aβ monomers accumulate (although little is known about the specific structure these monomers adopt) [7]. These soluble aggregates can further cluster into larger soluble oligomers, known as soluble protofibrils (protofibrils referring to Aβ monomers), and eventually form larger, insoluble fibrils (Fig. 1) [7]. However, it is the smaller, soluble Aβ oligomers that are considered the most neurotoxic. For instance, they have been found to create makeshift calcium ion channels, either by destabilizing or puncturing the lipid bilayer or by triggering ionotropic receptors (i.e., NMDAr or AMPAr - Fig. 2), further enabling calcium ions to flood the cell [2]. This calcium influx disrupts cellular homeostasis, inducing mitochondrial dysfunction and oxidative stress [7]. Furthermore, Aβ oligomers have been shown to inhibit the first step of memory formation—hippocampal long-term potentiation (LTP)— thereby impairing the brain’s ability to store information long-term and limiting learning and memory [7].
When Aβ oligomers aggregate, they play a crucial role in the formation of insoluble protofibrils. Both diffuse plaques (DP) and neuritic plaques (NP) are composed of fibrous Aβ. However, NPs are distinct, consisting of dystrophic neurites and altered glial cells intricately woven into a denser core of insoluble fibrils (Fig. 3) [6,10]. Dystrophic neurites, which are collections of altered
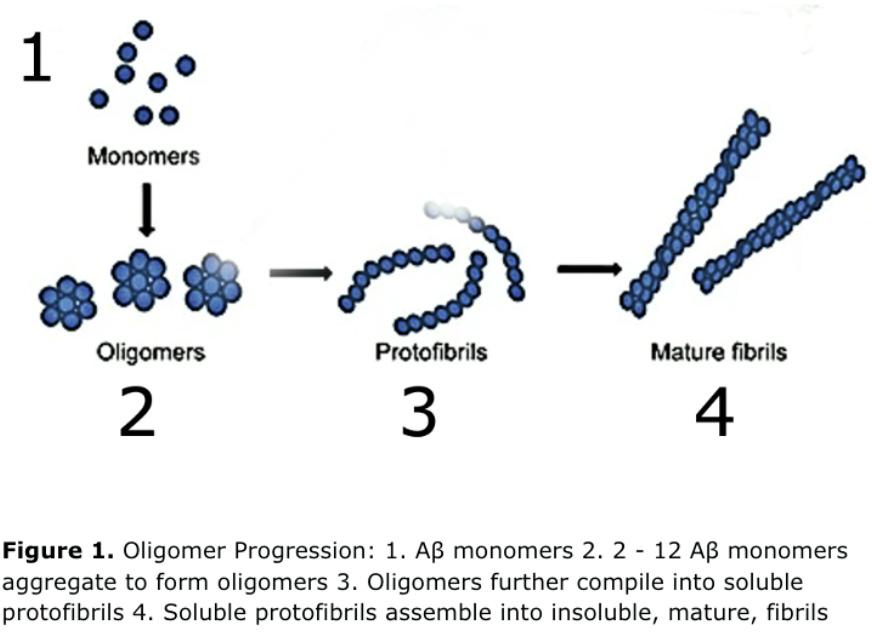
axons and swollen cellular components (e.g. mitochondria) [8], are a clear indication of neuronal degeneration. These abnormal axonal structures are commonly observed in patients with various neurodegenerative diseases, such as traumatic brain injury and cortical dysplasia. In AD, dystrophic neurites are often found bound to Aβ fibrils. Therefore, while not yet fully proven, it is strongly believed that Aβ aggregates serve as the catalyst for neuronal deterioration and the subsequent formation of swollen axonal structures, which ultimately bind to the plaques that initiated their damage [8].
Additionally, a halo of glial cells, often microglia and astrocytes, is frequently observed encircling the core of Aβ fibrils and their swollen neuritic attachments [6]. As the central nervous system’s primary defense system, glial cells are crucial in maintaining brain homeostasis and removing foreign substances and damaged neurons. The presence of neuritic plaques triggers a response from microglial cells and astrocytes, inducing the release of multiple proinflammatory cytokines [8]. This Aβ-induced inflammation is highly detrimental to the brain and is believed to further damage neurons.

At the core of every Aβ composition, from the oligomer to the neurotic plaque, lies the Aβ monomer. As previously mentioned, Aβ stands for amyloid-beta protein. The term “amyloid” refers to protein fragments, and “beta” indicates that these fragments are cleaved from the precursor protein by proteases. This is precisely what the Aβ monomer is—a fragment of protein separated from a larger protein. Specifically, amyloid-beta is the cleavage product of APP, or amyloid precursor protein. APP is a transmembrane protein typically cleaved by the enzyme alpha-secretase (α-secretase) at amino acid position 688 (on an APP isoform 770 amino acids long), yielding sAPP-alpha [4,4]. This protein is believed to play a role in the brain’s memory and learning processes and helps
protect the brain from stress. However, in neurodegenerative conditions, APP is cleaved by β-amyloid cleaving enzyme (BACE) or β-secretase at amino acid position 672, resulting in sAPP-beta and a C99 fragment (Fig. 4) [9,4]. Gamma-secretase (γ-Secretase) then cleaves the C99 fraction at amino acid positions 40 or 42, producing one of two isoforms, Aβ 40 or Aβ42 [4,4]. It is important to note that the two extra amino acids at the C-terminal residue of Aβ42 enhance the monomer’s propensity to aggregate into plaques [9]. Therefore, Aβ plaques contain more Aβ42 than Aβ40, even though the concentration of Aβ40 in the brain is generally greater than that of Aβ42 [9].
Furthermore, specific mutations have been identified that affect the production of Aβ. For example, a rare mutation in the N-terminus region of the β-secretase cleavage site substitutes threonine for alanine at amino acid position 673 (A673T), the second position of the β-secretase cleavage site [9]. This substitution reduces Aβ production and its tendency to aggregate [9]. This mutation is considered a protective variant, and studies in Iceland have revealed that older, non-demented individuals are five times more likely to carry the A673T substitution than individuals with AD [3]. However, when alanine is replaced with valine at this position (A673V), proteolysis of APP is increased, ultimately escalating Aβ production. Additionally, mutations at the γ-secretase cleavage site can alter the ratio of Aβ40 to Aβ42, and mutations in the mid-domain of the Aβ region augments Aβ’s ability to form oligomers and fibrils.
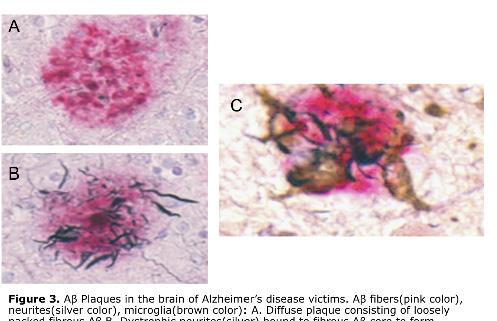
The Aβ monomer plays a critical role in the development of Alzheimer’s disease. Aβ can aggregate into various structures—oligomers, neuritic plaques, diffuse plaques—which disrupt CNS homeostasis, in-
cite inflammation, and catalyze neurodegeneration. Ultimately, Aβ is the product of proteolytic activity, beginning with the proteolysis of APP and ending with γ-secretase’s cleavage of the C99 fragment. Furthermore, researchers have identified certain genetic mutations that affect Aβ production and behavior.
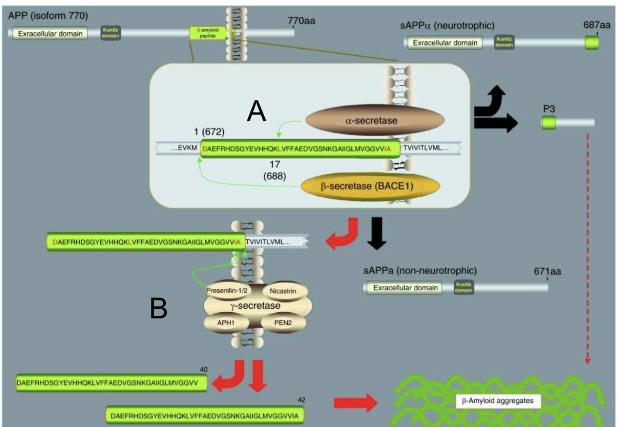
1. Centers for Disease Control and Prevention. (2020, October 26). Alzheimer’s Disease and Related Dementias. CDC; CDC. https:// www.cdc.gov/aging/aginginfo/alzheimers.htm
2. Fani, G., Mannini, B., Vecchi, G., Cascella, R., Cecchi, C., Dobson, C. M., Vendruscolo, M., & Chiti, F. (2021). Aβ Oligomers Dysregulate Calcium Homeostasis by Mechanosensitive Activation of AMPA and NMDA Receptors. ACS Chemical Neuroscience, 12(4), 766–781. https://doi.org/10.1021/acschemneuro.0c00811
3. Hampel, H., Hardy, J., Blennow, K., Chen, C., Perry, G., Kim, S. H., Villemagne, V. L., Aisen, P., Vendruscolo, M., Iwatsubo, T., Masters, C. L., Cho, M., Lannfelt, L., Cummings, J. L., & Vergallo, A. (2021). The Amyloid-β Pathway in Alzheimer’s Disease. Molecular Psychiatry, 26(10). https://doi.org/10.1038/s41380-021-01249-0
4. MacLeod, R., Hillert, E.-K., Cameron, R. T., & Baillie, G. S. (2015). The role and therapeutic targeting of α-, β- and γ-secretase in Alzheimer’s disease. Future Science OA, 1(3). https://doi.org/10.4155/fso.15.9
5. National Institute on Aging. (2021, July 8). What Is Alzheimer’s Disease? National Institute on Aging. https://www.nia.nih.gov/health/alzheimers-and-dementia/what-alzheimers-disease
6. Neuritic Plaques - an overview | ScienceDirect Topics. (n.d.). Www.sciencedirect.com. https:// www.sciencedirect.com/topics/biochemistry-ge -
netics-and-molecular-biology/neuritic-plaques
7. Tolar, M., Hey, J., Power, A., & Abushakra, S. (2021). Neurotoxic Soluble Amyloid Oligomers Drive Alzheimer’s Pathogenesis and Represent a Clinically Validated Target for Slowing Disease Progression. International Journal of Molecular Sciences, 22(12), 6355. https://doi.org/10.3390/ijms22126355
8. Uddin, M. S., & Lim, L. W. (2022). Glial cells in Alzheimer’s disease: From neuropathological changes to therapeutic implications. Ageing Research Reviews, 78, 101622. https://doi.org/10.1016/j.arr.2022.101622. (n.d.).
9. Walker, L. C. (2020). Aβ plaques. Free Neuropathology, 1, 31–31. https://doi. org/10.17879/freeneuropathology-2020-3025
10. Wippold, F. J., Cairns, N., Vo, K., Holtzman, D. M., & Morris, J. C. (2007). Neuropathology for the Neuroradiologist: Plaques and Tangles. American Journal of Neuroradiology, 29(1), 18–22. https://doi.org/10.3174/ajnr.a0781
11. World Health Organization. (2020, December 9). The Top 10 Causes of Death. World Health Organization; WHO. https://www.who.int/news-room/ fact-sheets/detail/the-top-10-causes-of-death
by Brynne Dragert (IV)
The development of cataracts in humans is most commonly the result of aging, but may also be due to family history of cataracts, an injury to the lens of the eye. Cataract formation occurs over time as the proteins and fibers within the lens of the eye begin to break down and clump together, clouding the lens. Continuous cataract growth can and does have permanent, detrimental effects on people’s vision, as perceived light becomes increasingly difficult to intake through the lens, causing projected images to appear blurred or hazy. As of 2025, there is no real treatment or cure for eye cataracts, although a surgical procedure can be done to remove the clouded lens and replace it with a clear one. Nearly 4 million cataract surgeries are performed each year in the United States, with 90% restoring 20/20 vision with glasses. However, those who have an eye condition such as glaucoma may not meet similar success, and all surgeries, in general, come with a chance of potential risk to the patient [1]. Scientific research searching for the ultimate cure for all cataract cases is ongoing and has not yet defined plausible solutions, although one study recently done on ground squirrels may prove to establish an answer.
As published in an article in September of 2024, an NIH (National Institutes of Health) research team conducted a study on 13-lined ground squirrels in hibernation to further advance the research of possible cataract drug creation. What led to the 13-lined ground squirrel becoming of utmost interest to the scientists in finding a cure for human cataracts? During hibernation in the winter, 13-lined ground squirrels develop cataracts due to a significant drop in body temperature, specifically to 4 degrees Celsius. What makes this creature unique is its ability to undo

the damage of the cataracts as the body temperature is rewarmed. The main experiment performed in the study consisted of comparing the man-made cataract formations in squirrel and rat eye lenses. Both species’ lenses were cooled down to a temperature of 4 degrees Celsius in 24 hours, allowing cataracts to form in simulation of those which form during hibernation. After rewarming the lenses, the squirrel’s returned to normal as damaged or misfolded proteins were destroyed and replaced, but the rat’s remained partially clouded. The group further studied the eye tissues and found that the rewarming of the squirrel’s lens sets off a process called ubiquitination [2]. This process involves the assistance of RNF114, a protein-coding gene that enforces protein ligase ubiquitin activity [3]. Through this occurrence, the protein CRYAA, which is connected to the formation of cataracts, is broken down by RNF114 and replaced with healthy proteins to clear the 13-lined ground squirrel’s lens. When RNF114 was added to another rat lens in the same manner of experiment, the cataract was cleared, proving the researchers’ discovery to be factual and quite the breakthrough for a non-surgical cataract cure in humans [2].
[1]Cataracts. (n.d.). Retrieved March 5, 2025, from https://www.mayoclinic.org/diseases-conditions/cataracts/symptoms-causes/syc-20353790
[2]Potential target for cataract drug development. (n.d.). Retrieved March 6, 2025, from https://www.nih. gov/news-events/nih-research-matters/Potential-target-cataract-drug-development
[3]RNF114 ring finger protein 114. (n.d.). Retrieved March 6, 2025, from https://www.ncbi.nlm.nih.gov/ gene/55905

by Hannah Castiglione
(V), Isabel Caldis (V), Michael Cardona (V), D’Ausilio, Morgan, Max Ventura (V), Matan Zelkowicz (V), Shea Patel (V), Trisha Jetley (V), Gabby DeLorenzo (V), Ms. S. Mygas
Histone Deacetylases (HDACs) are enzymes that remove acetyl groups from lysine residues on histones, causing chromatin compaction. This causes decreased gene expression and silencing. Upregulation of HDAC activity, which disrupts transcription of tumor suppressor genes that are critical in preventing unregulated cell proliferation, is closely linked to tumorigenesis. Inhibition of HDACs can induce therapeutic effects by allowing chromatin remodeling and therefore reinitiating transcription. Trapoxin A is a microbial cyclic tetrapeptide exhibiting potent antitumor effects by irreversibly binding to class I HDACs. The crystal structure of Trapoxin A complexed with HDAC8, a metal-dependent class I HDAC, reveals the importance of the Trapoxin A’s α,β-epoxyketone moiety. This moiety mimics the side chain of acetyl-L-lysine, an HDAC substrate. The ketone carbonyl group of the α,β-epoxyketone moiety undergoes nucleophilic attack by water upon binding to HDAC8, forming a gem-diolate, similar to an intermediate form of acetyl-L-lysine. This gem-diolate stabilizes Trapoxin A in HDAC8 by
coordinating with the active site Zn2+ ion and forming three hydrogen bonds with catalytically important amino acids in the active site. Although the epoxide ring of the α,β-epoxyketone moiety does not change in structure or establish hydrogen bonds, it was found to be essential for irreversible inhibition of HDACs. It is believed that the epoxide ring’s energetically favorable conformation confers Trapoxin A the ability to tightly bind to HDAC8. To further investigate the structure of Trapoxin A, The Pingry School SMART (Students Modeling A Research Topic) Team used the 3-D modeling program, Jmol, and printing technology from 3D Molecular Designs to create and print a model of the Trapoxin A-HDAC8 complex. Our model highlights the interactions between the α,β-epoxyketone moiety of Trapoxin A and the active site amino acids of HDAC8 to visualize their binding. This visualization aids in understanding the mechanism of Trapoxin A, allowing for the future use as a therapeutic agent in HDAC-induced cancer.
by Zach Abrahams (III), Neil Amin (III), Joe Cridge (III), Alex DeLorenzo (IV), Matthew Dicks VI), Sia Ghatak (VI), Sophia Guild (V), Ryan Hao (IV), Aashritha Kolli (III), James Kotsen (VI), Rhys Llewellyn-Jones (III), Ryan Mirchandani (III), Aadi Stewart (V), Christian Zhou-Zheng (V), William Zhou-Zheng (IV)
Do you like robots? Of course you do! At the Pingry Robotics research week exhibit, people have the opportunity to drive some of the team’s robots, see the team’s robots that made championships, and ask questions to the brilliant team behind them. Throughout the course of this year, the Pingry robotics team has spent its time building two robots that complete tasks in time-constrained
competitions to win points. We compete in FTC (FIRST Tech Challenge) and FRC (FIRST Robotics Competition), while also mentoring the middle school team. This year’s FTC competition focused on shooting rectangular prisms into buckets at different height levels. This year’s FTC competition involves placing long tubes of pipe onto a scoring element of different angles and heights.
by Amar Janki Bhan (V), Katharine Luo (V), Dr. A. Lasevich
Throughout modern history, economists have desired to understand how capital accumulation and consumption drive the Gross Domestic Product (GDP) growth. Differential equations have provided a powerful window to approximate and predict the overall trajectory of GDP over time. This study aimed to simulate and compare the GDP projections of two distinguished differential equation models, the Solow-Swan and Ramsay-Cass-Koopmans (RCK). First, the Solow-Swan model was analytically solved to get an equation to model capital per worker in the steady state. On the other hand, for the RCK model, which has no analytical solution, a Python program implementing the fourth-order Runge-Kutta method was used to obtain a numerical solution for the capital and con-
sumption per worker. Then, both models were plugged into a modified Cobb-Douglas production function to estimate the overall GDP of America for the next 100 years. The Solow-Swan model predicts America’s GDP will reach $43.2 trillion in 10 years, $54.1 trillion in 20 years, and $133.7 trillion in 50 years, showing steady exponential growth. Similarly, the RCK model has projections of $42.56 trillion, $57 trillion, and $169.48 trillion for the same intervals. Both models show that higher savings and lower depreciation lead to faster capital accumulation, and thereby larger future GDPs. These models can aid government policy decisions, as they depict how changes in savings, depreciation, and population growth affect long-term economic progress.
By David Sirichenko (V), Antonios Marseilles
Earthquakes pose significant risks to human safety and infrastructure, making it crucial to understand and predict their occurrences and impacts. This is especially important in Japan, an extremely densely populated region, which is well known for its frequent high-magnitude earthquakes. Our research aims to determine whether there are long-term patterns in GPS data that can predict future earthquakes.
The methods employed were wavelet analysis on GPS time series data using the R programming language. We applied a 30-day Kolmogorov-Zurbenko (KZ) moving average filter to find repeating pat-
terns and periodicity in the GPS data. Our research aims to use the pattern and periodicity of the data to determine if a deviation from the pattern could be used as an early indicator for an earthquake.
Depending on the direction of the GPS data, there were periods found of approximately 90, 120, 180, or 360 days, which most likely represent seasonal changes. Although this pattern and associated periods were clear, there was no correlation between this pattern and any deviations from it resulting in earthquakes. Therefore, additional methods and data analysis are needed to correlate these patterns to precursors for earthquakes.
by Abdullah Sultan (III), Ashley Pearson
Gene editing has the potential to positively impact many fields such as crop production, biofuel production, and cancer therapy. CRISPR/Cas9 has emerged as a useful and impactful tool to assist with the gene editing process. However, CRISPR/Cas9 requires the delivery of multiple components and has multiple delivery methods, each with its benefits and drawbacks. In this review, we discuss three types of CRISPR/Cas9 delivery methods: transfection, transduction, and encapsulation. We highlight how each delivery method is useful in different contexts and how it functions. CRISPR/Cas9 has the potential to enable precise gene editing, and its delivery methods can control the limits of its precision.
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) create a breakthrough tool in genetic engineering. The two main parts of CRISPR/Cas9 are the Cas9 enzyme, which cuts DNA at specific spots like molecular scissors, and the guide RNA (gRNA), which points Cas9 toward the target DNA sequence. By letting the gRNA bind to a complementary DNA sequence, this method enables Cas9 to precisely induce a double-strand break at that location. CRISPR/Cas9 simplifies gene editing for a wide range of uses because of its ability to make precise changes to the genome. Common applications include gene knockout studies, in which scientists disrupt genes to study their function, and therapies, such as fixing genetic changes that cause disease. Furthermore, CRISPR/Cas9 is utilized in agriculture to create crops with desirable characteristics. Understanding these components and methods is critical for investigating the numerous uses of CRISPR/Cas9 in science and industry (Doudna and Charpentier 2014; Jinek 2012).
The goal of CRISPR/Cas9 is to make precise genome alterations possible, which has enormous research and therapeutic application implications. It is frequently employed to fix genetic abnormalities, introduce certain
mutations, and produce gene knockouts. Biotechnology, where CRISPR/Cas9 is used to engineer microorganisms with industrial applications; medicine, where it is used to create models of human diseases and develop gene therapies; and agriculture, where it is used to develop crops with desirable traits (Tavakoli 2021). Despite its promise, CRISPR/Cas9 cannot be used effectively in clinical settings unless a number of obstacles are addressed, the most important of which is making sure that the CRISPR components enter the target cell.
For the CRISPR/Cas9 technology to be successfully used, target cells must be supplied with the necessary components. These techniques, used to insert the Cas9 enzyme and gRNA into cells, are referred to as delivery. Effective distribution is crucial because it affects the specificity and efficiency of the genome editing process. Delivering the CRISPR components to the target cells without degradation, without any side effects, is substantially difficult. A number of delivery strategies have been created to avoid these obstacles. For in vitro applications, transfection techniques including electroporation and lipid-mediated transfection are frequently used. By encasing the CRISPR components in lipid nanoparticles, which subsequently fuse with the cell membrane to deliver their payload, lipid-mediated transfection is achieved. Through the process of electroporation, brief holes in the cell membrane are made by electrical pulses, which permits the CRISPR components to enter the cells. For in vivo applications, transduction techniques such as lentivirus and adenovirus usage are frequently used. These viral vectors are effective in introducing genetic material into various cell types. To increase the efficacy and specificity of CRISPR administration, other techniques like hydrogels and nanoparticle-based delivery systems are currently being investigated (Lino 2018).
This review paper highlights the benefits and drawbacks of the different CRISPR/Cas9 delivery strategies. The paper highlights how crucial it is to maximize delivery methods to maximize the therapeutic
potential of CRISPR/Cas9. This research highlights the use of hydrogels and nanoparticles, two novel techniques that show promise for enhancing the delivery of CRISPR components in clinical settings. Researchers want to fully utilize CRISPR/Cas9 for treating genetic illnesses and promoting biological research by tackling the delivery-related obstacles (Lino 2018).
For gene editing to be successful, CRISPR/Cas9 components must be delivered to target cells. Three main delivery strategies have been covered in this review: transfection, transduction, and encapsulation. Transfection is the process of changing the genetic makeup of eukaryotic cells by inserting foreign nucleic acids. Lipid nanoparticle-based chemical transfection has demonstrated remarkable efficacy in vitro, but physical transfection techniques such as nanostructure-mediated electroporation provide accuracy with negligible cellular harm. Viral vectors are used in transduction, which introduces genetic material into cells. Lentiviral vectors allow for stable, long-term gene editing in human cells, whereas Adeno-associated virus (AAV) vectors have shown promise for in vivo applications. Innovative CRISPR/Cas9 delivery methods include hydrogels and nanoparticle-based systems. These techniques provide targeted delivery to particular cells or tissues and regulated release of CRISPR components, which may improve the accuracy and effectiveness of gene editing.
Even with the encouraging developments in CRISPR/ Cas9 delivery techniques, a number of drawbacks still exist. The possibility of off-target effects, in which accidentally altered genomic locations have unanticipated repercussions, is a major obstacle. In clinical settings where accuracy is crucial, this problem is especially worrisome. The immune system’s reaction to viral vectors employed in transduction is another drawback. Many people have already experienced a viral vector as a child, which might decrease the efficiency of gene delivery and perhaps cause unfavorable immunological responses. Delivery of the CRISPR/Cas9 system is complicated by its size, particularly when utilizing viral vectors with constrained packaging capacity. Because of this limitation, it is frequently necessary to employ numerous vectors or smaller Cas9 variations, which could reduce editing efficiency. The field of CRISPR/Cas9 delivery is developing quickly, and to overcome present constraints and broaden applications, researchers are concentrating on sever-
al important areas. Researchers aim to improve guide RNA design algorithms and develop high-fidelity Cas9 variants to increase the accuracy of CRISPR/Cas9 editing. To avoid the drawbacks of viral vectors, there is also rising interest in creating safer and more effective non-viral delivery systems, like lipid nanoparticles and cell-penetrating peptides. Additionally, scientists are developing methods to increase the tissue specificity of CRISPR/Cas9 delivery, such as the application of tissue-specific promoters and specially designed nanoparticles that have targeting ligands. Lastly, improving the temporal regulation of CRISPR/Cas9 distribution through the development of hydrogels and other controlled release systems may increase editing efficiency and decrease off-target consequences. As the area develops, different technologies may converge and combine the advantages of different delivery systems to produce safer, more accurate, and efficient gene editing. To effectively apply this formidable gene-editing technology outside of the lab—perhaps transforming the way genetic illnesses are treated and expanding our knowledge of gene function—continued improvement of CRISPR/Cas9 delivery methods will be essential.
1. Lino, C. A., Harper, J. C., Carney, J. P., & Timlin, J. A. (2018). Delivering CRISPR: a review of the challenges and approaches. Drug delivery, 25(1), 1234-1257.
2. Tavakoli, K., Pour-Aboughadareh, A., Kianersi, F., Poczai, P., Etminan, A., & Shooshtari, L. (2021). Applications of CRISPR-Cas9 as an advanced genome editing system in life sciences. BioTech, 10(3), 14.
3. Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., & Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. science, 337(6096), 816-821.
4. Doudna, J., & Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. (2014). Science, 346, 6213.
by Emilio Gomez (IV), Isabel Genier, Nicholas Higgs, Marco Turner
Spiny Lobsters (Panulirus argus) are crucial to the Bahamian fishery, economy, and culture, particularly on the island of Eleuthera. However, further research is necessary to better understand the species. Spiny Lobsters live in the warm waters of Florida and the Caribbean. As juveniles, they live in warm, shallow, grassy-bottomed water, moving to deeper water and reefs as they grow. Marine Stewardship Council (MSC) is the primary Spiny Lobster fishery, certified as sustainable. The Bahamas is the largest producer of Spiny Lobster in all the Americas, producing approximately 2,500 tons and $90 million per year. Our research focused on measuring changes in Spiny Lobsters’ size and location. We conducted freediving surveys at various sites, capturing and tagging lobsters. We discovered that their growth follows a step-
ladder pattern rather than a continuous curve, as they molt or shed periodically until their death. A key aspect of our research was analyzing spillover effects when species migrate from a Marine Protected Area (MPA) to surrounding waters. Our goal was to provide evidence in order to establish a greater MPA in the area surrounding the sand banks of Eleuthera. However, our findings revealed no significant difference in lobster health or size between MPA and nonMPA areas. We concluded that this was due to the MPA’s light restrictions, which still permitted fishing. Our group presented this project at a Cape Eleuthera Institute conference to members of the Bahamian Government and resident researchers. Over three months, we recaptured more than 50 lobsters, contributing valuable data to conservation efforts.
by Arjun Subramanian (V), Juan Luque, Dr. Cenk Sahinalp
In cancer research, it is often important to understand which mutations are associated with cancer and how the accumulation of these mutations can lead to tumor formation. We are investigating ways to accelerate the existing PhISCS-BnB algorithm using an upper and lower bound on the branch-and-bound (BnB) algorithm. More specifically, we are given a Single Cell Sequencing data matrix, where the rows represent sequenced cells and the columns are either 1 or 0, depending on whether the corresponding cell has a particular mutation. The BnB technique switches the 0s to 1s with a certain cost, creating a new node in a branching tree. Having an upper or lower bound on the number of switches needed to reach a viable matrix helps prune out branches that do not need to be explored, where a viable matrix is one such that any given pair of columns has no conflicts. So far, we have tested two techniques. The first maps each
zero as a vertex in a graph and draws edges between them if they conflict with each other, with the lower bound being the minimum vertex cover of this graph. The second technique involves constructing a linear programming (LP) problem, where each cell in the graph is a variable, and we create a graph with no conflicts through minimal switches. This method provides a lower bound for the optimal solution, as determined by integer LP, with a factor of 2. The latter method has an alternative version, where we only create constraints between columns if they are already in conflict. Our empirical findings show that the LP method with all columns and the LP method with just the conflict columns provide the exact same bound, but the latter runs in half the time. The vertex cover lower bound gives 30% smaller values but runs two orders of magnitude faster.
by Christian Zhou-Zheng (V), Bo Peng*, Ruichong Zhang*, Daniel Goldstein*, Eric Alcaide, Haowen Hou, XingJian Liu, Janna Lu, Kaifeng Tan, Guangyu Song, Saiteja Utpala, Johan S. Wind, Nathan Wilce, Tianyi Wu
We present RWKV-7 “Goose”, a new sequence modeling architecture, along with pre-trained language models that establish a new state-of-the-art in downstream performance at the 3 billion parameter scale on multilingual tasks, and match current SoTA English language performance despite being trained on dramatically fewer tokens than other top 3B models. Nevertheless, RWKV-7 models require only constant memory usage and constant inference time per token. RWKV-7 introduces a newly generalized formulation of the delta rule with vector-valued gating and in-context learning rates, as well as a relaxed value replacement rule. We show that RWKV-7 can perform state tracking and recognize all regular languages, while retaining parallelizability of training. This exceeds the capabilities of Transformers under standard complexity
conjectures, which are limited to TC0. To demonstrate RWKV-7’s language modeling capability, we also present an extended open source 3.1 trillion token multilingual corpus, and train four RWKV-7 models ranging from 0.19 billion to 2.9 billion parameters on this dataset.
To foster openness, reproduction, and adoption, we release our models and dataset component listing at https://huggingface.co/RWKV, and our training and inference code at https://github.com/ RWKV/RWKV-LM all under the Apache 2.0 License.
*Equal first authorship. Pingry students listed first, then first authors, then others listed alphabetically. This paper was submitted to COLM 2025 and is published on arXiv at https://arxiv.org/abs/2503.14456
By Christian Zhou-Zheng (V), Jeff Ens, Nathan Fradet, Davide Rizzotti, Paul Triane, Philippe Pasquier
Computer-assisted composition (CAC) explores the partial or complete automation of music composition tasks. The Multi-Track Music Machine (MMM) is a controllable, style-agnostic CAC system for the MIDI symbolic music format that allows a set of bars to be edited, selected, and resampled, affording users a fine degree of control over music generation. Past work on the MMM has leveraged the Transformer architecture to great effect with the MIDI-GPT model, leading to music albums and synthesizer integration, game music composition software, and digital audio workstations. However, Transformers struggle with longer sequence processing and lack real-time opti-
mization for most artists’ workstations. We address this problem by training a MMM model based on the novel RWKV-7 architecture, which exhibits comparable quality to MIDI-GPT while running faster and operating on longer sequences. We release the model under the Open RAIL-M license and a suite of integrations for professional use. We also release new evaluation methods to compare model performance in the symbolic music domain to foster further model development. Future work includes the development of real-time musical agents based on MIDI-RWKV, expanding the set of attribute controls, and continuing the integration of the MMM into real-world products and industry practices.
Email:
Christian Zhou-Zheng (V): czhouzheng2026@pingry.org
Katherine Jung (V): kjung2026@pingry.org
Mr. D. Maxwell: dmaxwell@pingry.org