FINAL REPORT (2008-2010) SEAGRANT PROJECT: R-92-3-08
Molecular Ecology of Integron-encoded Antibiotic Resistance and Prevalence of Fungicide Resistance in Microbial Populations from Critical Coastal Habitats Impacted by Sewage, Animal Waste, and Wastewater Treatment Plant Discharges.
Carlos M. Rodríguez-Minguela, Rafael Montalvo-Rodríguez, and Sandra L. MaldonadoRamírez. University of Puerto Rico Mayagüez Campus, Department of Biology, PO Box 9012, Mayagüez, PR 00681.
ABSTRACT
Sewage discharges are major contamination sources of ecologically and economically important coastal environments. Moreover, fecal bacteria from clinical settings have been identified as key hosts of multiple antibiotic resistance (ABR) genes encoded by integrons. Integrons are genetic elements that mediate the assimilation of DNA molecules encoding a variety of adaptive functions that may promote competence under unfavorable circumstances. Using a PCR-based approach we detected a prevalence pattern of clinical integrons and integron-encoded antibiotic resistance genes (class 1) associated with mangrove, beach and estuarine habitats exposed to high levels of wastewater pollution or anthropogenic impact. The application of these molecular techniques to the analysis of culturable bacteria also revealed the presence of similar resistance determinants among conventional fecal indicators as well as in reference isolates from hospital-related settings. We similarly detected the dominance of fluconazole resistant yeasts in association with human influence. Moreover, phylogenetic analysis of ITS sequences identified various fluconazole-resistant yeast isolates that were highly similar to opportunistic pathogens that are routinely treated with this medicament. Our data indicate that polluted coastal habitats are stable reservoirs of antimicrobialresistant bacterial and fungal populations and that cassette PCR assays targeted at class 1 integrons have the potential to serve as dual indicators of fecal contamination and inconspicuous risks to human health.
INTRODUCTION
The widespread use of antimicrobials in human and veterinary medicine as well as in agricultural production settings have resulted in a large scale release of these chemicals into natural environments. These observations have raised serious concerns within the clinical sector which also have environmental implications of great significance (Kümmerer, 2004). Recently, integrons, a genetic system carried by bacteria, have been implicated in the establishment and dispersal of antibiotic resistance traits which include protection mechanisms against the most important antibiotics used for the treatment of infection in humans as well as in plants and animals of economical importance. This system is based on the activity of a site-specific recombination mechanism catalyzed by an integrase which captures and inserts circular DNAmolecules (gene cassettes) downstream a promoter for subsequent expression (Figure 1).Although
these genetic elements are known to be horizontally transferred and commonly found among intestinal bacteria of clinical significance, little is known about their prevalence and persistence outside the clinical scene. Similarly, the occurrence in the environment of yeast strains resistant to fungicides commonly used to treat infections in humans is an understudied subject. Since integron-encoded antibiotic resistance has been associated with fecal bacteria and the spread of potentially harmful microorganisms is also linked to sewage and nonpoint sources of fecal contamination, we proposed to study the prevalence of clinical integrons (classes 1, 2 and 3), class 1 integron-encoded antibiotic resistance genes, and fungicide (fluconazole) resistant yeasts in coastal settings of ecological, economical and recreational importance. Likewise, in view of the fact that fluconazole is the most widely used fungicide for treating infections in humans, this chemical was used as to evaluate potential risks associated with impacted environments.
Figure 1. Schematic representation of a class 1 integron presenting the region common to all integrons (conserved 5’end) which generally consists of an integrase coding gene (intl) the site-specific recombination sequence attI and two promoters. Expression of the integrase relies on the Pint promoter while Pc drives the expression of inserted gene cassettes. The region designated as the conserved 3’end is characteristic of class 1 integrons detected in clinical strains and includes genes conferring resistance against quaternary ammonium compounds (qacE1) and sulfonamide drugs (sul1).
Based on a previous survey of integron integrases in terrestrial and marine settings (Rodríguez-Minguela, et al., 2009) we hypothesized that the level of fecal or anthropogenic impact in coastal environments correlates with the prevalence of antibiotic and fungicide resistance traits within the microbial community. Hence, to asses relationships between the degradation of coastal ecosystems due to wastewater-related disturbances and potential risks to human health we used molecular and culture-based techniques to study the dominance of clinical integrons and integron–associated antibiotic resistance genes in total DNAand culturable bacteria recovered from disturbed and relatively undisturbed coastal ecosystems. In parallel we optimized isolation procedures and implemented standard methods for the recovery and detection of fluconazole resistant yeasts. The sites analyzed included mangrove ecosystems (water, sediments and rhizosphere) beaches (water and sand) and estuarine environments (water and sand) which were compared with respect to reference samples recovered from wastewater treatment plants as well as from terrestrial and river habitats (Table 1).
Table 1. Description of reference and experimental sampling sites analyzed for the culture independent survey of integrons and integron-associated antibiotic resistance genes.
Sample code
UI*
Level of fecal or
Source and sample type description anthropogenic impact
Liquid phase, influents, urban secondary high wastewater treatment plant.
UO* Outflow, urban secondary high wastewater treatment plant.
RI*
Liquid phase, influents, rural, high secondary wastewater treatment plant.
RO* Outflow, rural secondary high wastewater treatment plant.
PP Wet sand ,Pico de Piedra Beach, high Aguada.
MGW** Water, estuarine habitat at high Guanajibo Beach, Mayagüez.
MGS Wet sand, estuarine habitat at high Guanajibo Beach, Mayagüez.
LBW Water, estuarine habitat at high La Boca, Barceloneta.
US Urban soil, UPRM Campus. high
ISO Pipeline outflow near Sardinera beach, high Isabela.
ISR
PMW
Impacted sea water, pipeline outflow high near Sardinera Beach, Isabela.
Water from mangrove habitat, high La Parguera, Lajas.
PMS Sediment from mangrove habitat, high La Parguera, Lajas.
PMR
Rhizosphere from mangrove habitat, high La Parguera, Lajas.
Table 1. Continued.
Level of fecal or
Sample code Source and sample type description anthropogenic impact
EDW Water from mangrove habitat, moderate Ensenada Dakity, Culebra.
EDS
EDR
Sediment from mangrove habitat, moderate Ensenada Dakity, Culebra.
Rhizosphere from mangrove habitat, moderate Ensenada Dakity, Culebra.
PB Wet sand from the shorline at Peña Blanca moderate Beach, Aguadilla.
FR** Water, Fanduco reef, ~1.25Km minimal off the Cabo Rojo coast.
CRU River water, upsteam region, minimal Barrio Cupeyes, Sabana Grande.
CRD River water, downsteam region, pristine Barrio Cupeyes, Sabana Grande.
FS Forest soil, Miradero, Mayagüez. moderate
GF Soil, secluded area, Guajataca minimal forest, Isabela.
* anonymous sampling sites
** Bonkosky et al., 2008.
MATERIALS AND METHODS
Extraction of Environmental DNA. To collect microbial biomass from water, sand, sediment and mangrove rhizosphere, samples were homogenized to suspend cells and then processed using the membrane filter technique. Water samples were shaken manually (25 times) following the filtration of a volume of 500 ml in 0.22 µm (pore size) membrane filters (Whatman, Germany). Sand samples (300 g) were mixed with 300 ml of dilution buffered water (USEPA, 1997) and homogenized in a shaker (200 rpm) for 30 min at room temperature and finally left to settle for 20 min. Mangrove rhizosphere and sediment samples were processed similarly except that 50 g of the appropriate material was diluted in buffered water (USEPA, 1997). The supernatant (100-300 ml) of the washed sand, sediments and mangrove rhizosphere material were filtered as described above.
Total DNA, from soil and filter-captured biomass was extracted using the FastDNA SPIN Kit for Soil (Qbiogene Inc., Carlsbad, CA) according to the manufacturer’s manual except for the following modifications. Cell lysis by bead beating
was carried out in a vortex using a Mo-Bio adapter (Mo Bio Inc., Carlsbad, CA) for 5 minutes at maximum speed followed by a centrifugation of 10 minutes at 10,000 rpm. Samples were centrifuged at 14,000 rpm for 15 min after the addition of the PPS reagent. The lysate was mixed with the DNA Binding Matrix Suspension for 10 minutes, followed by two washes with the SEW-S solution. Finally the DNA was eluted in 150 µl of DES, electrophoresed in a 1% agarose gel and quantified (optical density at 260 nm) using a nanodrop instrument (ND-1000, Thermo Scientific, Wilmington, DE).
PCR amplification of intI loci of clinical integrons and gene cassettes associated with class 1 elements. Small sub-unit 16S rRNA genes were amplified to test the quality of the extracted DNA using previously described primers (8F, 5’-
AGAGTTTGATCMTGGCTCAG-3’ [Giovannoni, 1991]; 1392R, 5’-ACGGGCGGT
GTGTACA-3’ [Amman et al., 1995]). Amplification reaction mixtures consisted of 12.5 µM dNTPs; 0.025 U/µl of Taq polymerase (Promega, Madison, WI); 0.5X Buffer, 3.5 mM MgCl2, 100 µg of BSA (New England® Biolabs, Ontario, Canada), 6.25 p/moles of each primer (827 FWD and 1392 REV, Table 2) and 22 ng of template DNA, in a final volume of 25 µl. Cycling conditions for the amplification of 16S rRNA genes were, initial denaturation at 95ºC for 3 minutes, followed by 25 cycles of melting at 94ºC for 45 sec, 45 sec of annealing at 57ºC, 1 min 30 sec of extension at 72ºC and a final extension of 72ºC for 7 min. Four replicates of PCR reactions were prepared for each sample and genomic DNA from E. coli (strain R-388) was used as template in positive controls.
Integrase genes (classes 1-3) and the variable region of class 1 integrons were independently amplified using the above mentioned PCR cocktail adjusted for a 12.5 µl reaction volume and ~30 ng of template DNA. The appropriate primers (Table 2) were added at a final concentration of 6.25 pmoles. Cycling conditions for integrase genes were, initial denaturation at 94ºC for 2 minutes, followed by 35 cycles of melting at 94ºC for 30 sec, 1 min of annealing at 59 ºC, 30 sec of extension at 72ºC and a final extension of 72ºC for 7 min. Four replicate PCR reactions were prepared for each sample while genomic DNA from E. coli strains harboring class 1, class 2 and class 3 integrons were used as positive controls (Avila and de la Cruz, 1988; Dillon et al., 2005 and Arakawa et al., 1995).
Gene cassettes carried by class 1 integrons were amplified using previously described primers (Lévesque et al., 1995 and Arduino et al., 2003) (Table 2) and 30 ng of environmental DNA as template, in a final volume of 25 µl. Cycling parameters consisted of initial denaturation at 94ºC for 2 minutes, followed by 30 cycles of melting at 94ºC for 30 sec, 1 min of annealing temperature at 62ºC, 5 min 30 sec at 72ºC of extension and a final extension of 72ºC for 7 min. All PCR reactions were carried out in a Robocycler Gradient 96 instrument (Stratagene, La Jolla, CA).
Cloning, sequencing and phylogenetic analysis. PCR products were electrophoresed in a 1.5% agarose gel to determine the product size and to extract amplicons of interest using the IBI Gel/PCR DNA Fragments Extraction Kit (IBI Scientific, Peosta, IA). The extracted PCR product was ligated into the pCR4-TOPO plasmid vector for subsequent transformation of E. coli cells using the TOPO TA cloning for sequencing kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions, except for the following modifications: the ligation reactions were carried out for 3 hours and the entire transformation mix was spread on LB plates (~75 µl per plate) containing 50 µg/ml of kanamycin. The cloned inserts were sequenced and
identified relative to matching sequences in GeneBank using BLAST. Multiple sequence alignments were constructed with ClustalX in order to conduct phylogenetic analyses using the MEGA 4 software package as previously described (Rodríguez-Minguela et al., 2009).
Table2PrimerpairsusedforPCRamplification
Primer Target Sequence(5’-3’) Position
Primer
Annealing
Product Size(pb)
Temperature (°C)
References
Int1F CAGTGGACATAAGCCTGTTC 2734–2751
IntI1
Int1R CCCGAGGCATAGACTGTA 2874–2891
Int2R GTAGCAAACGAGTGACGAAATG 11524–11545
IntI2
Int2F CACGGATATGCGACAAAAAGGT 12291–12312
Int3R ACGGATCTGCCAAACCTGACT 1697–1717
Int3F GCCTCCGGCAGCGACTTTCAG 738–758
hep35 TGCGGGTYAARGATBTKGATTT 2372–2393
IntI3 IntI
hep36 CARCACATGCGTRTARAT 1903–1920
59 788
59 979
52
59 160 491
Koelemanet al.,2001)
Mazeletal., 2000)
(Mazeletal., 2000)
Variable 62 (Whiteetal. 2000)
5’-CSF IntI GGCATCCAAGCAGCAAG 24-43 (Lévesqueet al.,1995)
3’-CSR ΔqacE AGCCCCATACCTACAAAGCC 2160–2181 (Arduinoet al.,2003)
sul1
Prevalence of antibiotic resistance traits and integrons in culturable bacteria. Samples were serially diluted and plated in R2A and TSA media containing cyclohexamide (100 mg/ml) sulfadiazine (200 ug/ml) and kanamycin (100 ug/ml). Cyclohexamide was added as an inhibitor of fungi while the later two antibiotics were added for the enrichment of bacterial isolates carrying class 1 integrons. To probe for the presence on integrons among the isolated colonies, a master plate containing all the recovered strains was independently prepared for all the processed samples. Total DNA was extracted from the colonies grown on the master plates, which was PCR-screened with primers specific for integron encoded integrases.
Quantification of fecal indicators and prevalence of resistance integrons. To determine the relation between fecal contamination and the presence of resistance integrons (Classes 1-3) molecular analyses were coupled to the culture-based methods for the enumeration fecal coliforms and mEI and mFC agar plates, respectively (USEPA, 1997 and 2003). Four replicates were processed for each sampling location. Representative strains of coliform and enterococci isolates were screened for the presence of clinical integrons and resistance determinants.
Inventory of fluconazole-resistant yeasts from impacted and undisturbed environments. All samples were processed using a standard dilution procedure (Leboffe and Pierce, 2002). The three quadrant method was used to purify the cultures. The recovered yeast isolates were screened for resistance against fluconazole (MIC>64μg/mL, NCLSI M27-A3) and characterized based on their reaction profiles in API 20 C AUX strips (bioMérieux SA, Marcy l'Etoile, France), chlamydospore and
capsule tests using CMA (corn meal agar), and phylogenetic analysis of internal transcribed spacers regions of rRNA genes using universal primers ITS1 and ITS4 (Korabecná et al., 2003).
RESULTS AND DISCUSSION
Prevalence patterns of clinical integrons and resistance genes encoded by class 1 integrons in environmental DNA. A PCR protocol targeted at conserved regions among integron encoded integrases (intl genes, Figure 1) was implemented as an initial screening method to evaluate the presence of integrons in wastewater. Integrase genes were PCR-amplified, cloned and sequenced (N=40) from liquid material retrieved from a sedimentation tank in an urban, secondary treatment wastewater plant. Three intl clones showed predicted amino acid sequences 98-100% identical to that encoded by intI genes of class 1 integrons previously described in antibiotic resistant strains of Klebsiella pneumoniae, Salmonella enterica, Pseudomonas aeuruginosa, and Morganella morganii which have been associated with hospital-acquired infections and outbreaks (Figure 2, GenBank accession numbers: CAJ13499, CAF18331, CAD20932 and AAX46051). Additionally, five clones representative of putative, novel integrons (<75% amino acid identity relative to previously described IntI elements) were uncovered, indicating that the integron pool in wastewater is diverse and that integrons described in human pathogens and associated with fecal waste are detectable within the material already present in a wastewater treatment facility (Figure 2).
IntI1K.pneumoniaeCAJ13499.1
clone9-36
IntI1M.morganiiCAF18331.1
IntI1S.entericaCAD20932.2
P.aeruginosaAAX46051.1
clone3-18
clone3-9
IntI3K.pneumoniaeAAO32355
IntIP.stutzeriAAN16061.1
IntIM.flagellatusYP544521.
clone5-2
clone6-28
clone11-34
clone2-18
IntI8unculturedAAK00304.1
IntIN.mobilisZP01128918.1
MONA-Int-A9ABD62557
MONA-Int-C10ABD62558.1
clone6-42
cloneSLV-Int-A12ABD62682.1
IntIC.aggregansZP01516571.1
IntI2S.sonneiAAT72891
VchIntI499031763
XerDP0A8P8
XerCP0A8P6
Figure 2. Neighbor-joining dendrogram based on partial amino acid sequence analysis (119 residues) of integron-encoded integrases amplified from liquid material recovered from a sedimentation tank from an urban wastewater treatment plant. GenBank accession nos. follow the designation of reference sequences). Representative sequences (XerC and XerD) of the closest relatives of integron encoded integrases are also included as references. Red, black and blue squares depict the positions of sequences related to class 1 integrases, integrases from clinical integrons (classes 1-3) and novel integrases, respectively.
Based in these results, PCR methods using primers specific to class 1 class 2 and class 3 integrases were preformed to detect clinical integrons in the influents and effluents of a rural and an urban, secondary wastewater treatment plant, in different impacted coastal environments as well as in coastal and terrestrial reference sites with moderate or minimal human influence (Table 1). Results from these tests using equal amounts of total community DNA (11ng) from each environmental sample revealed different distribution and prevalence trends among integrases from clinical integrons (Figure 3). Class 1 and class 2 integrases were detected among impacted and relatively undisturbed sites. However their detection signals were stronger in DNA from sites exposed to a high fecal or anthropogenic impact (wastewater and urban soil, Mayagüez) with respect to those from less disturbed environments (forest soil, Miradero, Mayagüez and wet sand from a secluded beach, Peña Blanca, Aguadilla). In contrast, class 3-like integrases 93% identical to an Intl3 described in Serratia marcescens an intestinal bacteria known to be a coral pathogen (Patterson 2002) were detected only in the outflow of the rural, secondary, wastewater treatment plant further supporting previous associations between fecal impact and the deterioration of coastal habitats as well as suggesting a linkage with the intestinal flora of individuals from a particular geographical location (Figure 4).
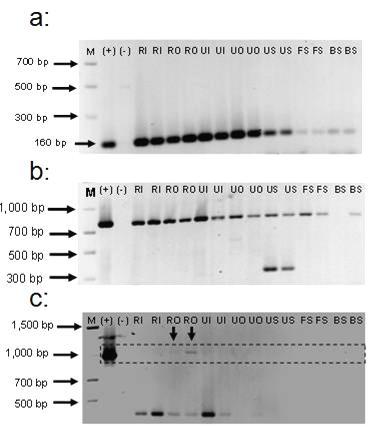
Figure 3 Agarose gel electrophoresis showing representative results of the amplification of integrase genes from class 1 (Panel [a]) class 2 (Panel [b]) and class 3 (Panel [c]) integrons from total community DNA recovered from environments with different levels of fecal impact. M = 24kb Max DNA Ladder (Fisher BioReagents, Pittsburgh, PA); (+) = positive control, template DNA containing the target intI sequence, (-) = negative control, no template DNA added, RI= rural wastewater treatment plant influent, RO = rural treatment plant outflow, UI = urban treatment plant influent, UO = urban treatment plant outflow, US = urban soil, FS = forest soil adjacent to urban soil, Mayagüez, PR, BS = beach sand with moderate human influence (Pena Blanca site, Aguadilla, PR).
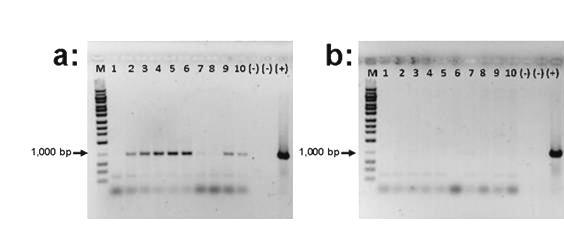
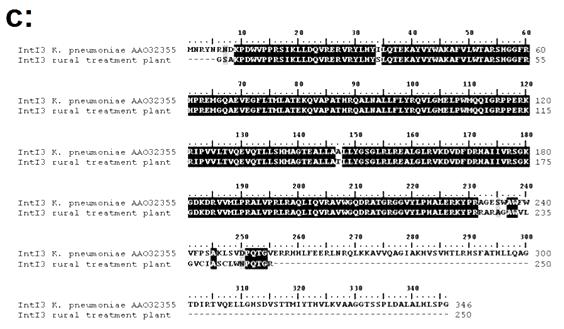
Figure 4. Panel (a): Ten replicate reactions showing the PCR amplification of intl3 genes from total DNA recovered from the outflow of a rural wastewater treatment plant. Panel (b): Results for 10 replicate PCR reactions of the amplification of intl3 genes from total DNA extracted from the outflow of a rural wastewater treatment plant. All experimental and control reactions were run in the same batch. Panel (c): Alignment of the predicted amino acid sequence of the PCR product recovered from the outflow of the of the rural wastewater plant using primers targeted at intl3elements against that of a class 3 integrase described in Klebsiella pneumoniae. The Genbank accession number follows the designation of the reference Intl3 sequence.
IntI2 sequences 98% identical (amino acid identity) to class 2 integrases were found at Peña Blanca suggesting the existence of a natural pool of class 2 integrons or some degree of anthopogenic impact (Figure 5, Panel A). Although class 2 integrases appeared to be widespread in nature as judged by their detection at Peña Blanca, they showed a lower frequency in relatively undistrubed habitats. This was demonstrated through an experiment conducted with soil DNA from an isolated sector of the Guajataca forest. When using this soil DNA, intI2 elements were not clearly detected whereas they were easily amplified when the template was spiked with DNA from a bacterial control strains harboring a class 2 integron (Figure 5, Panel B and C).
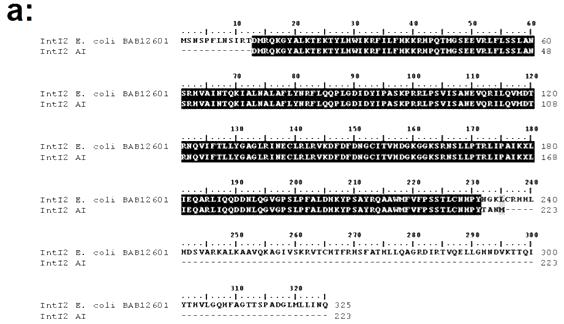

Figure 5. Panel (a): Alignment of the predicted amino acid sequence of the PCR product recovered from Peña Blanca beach (Aguadilla) using primers targeted at intl2 elements against that of a class 2 integrase described in Escherichia coli. The Genbank accession number follows the designation of the reference Intl2 sequence. Panel (b): PCR amplification of intl2 genes from total soil DNA (undisturbed area Guajataca Forest). Panel (c): PCR amplification of intl2 genes from total soil DNA (undisturbed area Guajataca Forest) spiked with increasing amounts of intl2 template DNA. Numbers in white and yellow describe the respective mass (ng) of soil DNA and intl2 template used for each reaction. All experimental and control reactions were run in the same batch.
Since class 1 and class 2 integrons appeared to be widespread and class 3 integrons showed a more restricted distribution, we implemented and additional PCR assay specifically designed for the detection of class 1 integrons loaded with antibiotic resistance gene cassettes. The purpose of this cassette-targeted PCR method was to evaluate any pattern on the prevalence of antibiotic resistance mechanisms and human
influence under the hypothesis of dominance of class 1 integrons carrying resistance mechanisms in impacted habitats. This approach revealed the presence of an amplicon within the 1,000-1500 base pairs range in influents of an urban wastewater plant (Figure 6).

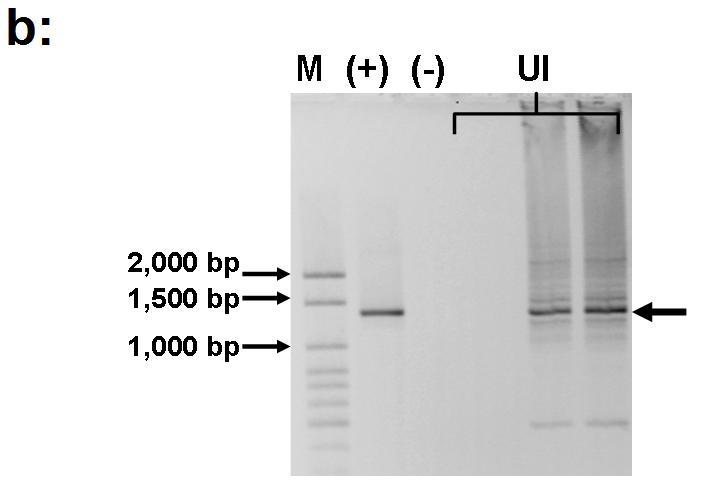
Figure 6. Panel (a): Diagram depicting the structure of a class 1 integron and the relative position of primers targeted at conserved regions flanking the gene cassettes insertion site. Panel (b): Agarose gel electrophoresis showing the amplification (three replicate reactions) of gene cassettes (~1.3 Kb long) associated with class 1 integrons detected in DNA recovered from the influents of an urban wastewater treatment plant. M=100bp ladder DNA marker (Applied Biological Materials Inc., Richmond, BC, Canada); (+) = positive control; (-) = negative control.
Partial sequencing analysis of the aforementioned fragment confirmed the occurrence of genes associated with class 1 integrons encoding a putative dehydrofolate reductase (Dhfr) (Figure 7, clone 1.5-T7-13) that was ~98.7% identical to that previously detected in pathogenic strains of Klebsiella pneumoniae, Acinetobacter baumannii, Enterobacter faecalis, Salmonella enterica and Escherichia coli (GenBank accession numbers: ABC50093, ABJ15828, BAD77819, AAT45231 and ACD02027). Genomic and integron-encoded Dhfr are involved in the synthesis of folic acid a precursor of proteins and nucleic acids. However, integron-encoded Dhfr is insensitive to inhibition by trimethoprim, an antibiotic frequently used for treating urinary tract infections. Furthermore, a gene encoding an enzyme capable of inactivating the antibiotic chloramphenicol (a protein synthesis inhibitor) and a determinant coding for an efflux pump (qacΔE1) which protects against disinfectants widely used in hospitals (quaternary ammonium compounds [qac]) were also recovered (Figures 8 and 9). The former showed an amino acid identity of 97% with respect to a chloramphenicol modifying enzyme
1.5-T7-13_
reported in Klebsiella pneumoniae (GenBank accession number: YP_001965795) while the later was 100% identical to a qac pump detected in Klebsiella pneumoniae (GenBank accession number: ACF06163).
DhfrC.freundiiNP_775043
DhfrK.pneumoniaeABC50093
DhfrA.baumanniiABJ15828
DhfrE.faecalisBAD77819
DhfrS.entericaAAT45231
DhfrE.coliACD02027
MNSESVRIYLVAAMGANRVIGNGPNIPWKIPGGQKIFRRLTEGKVVVMGRKTFESIGKPL 60
MNSESVRIYLVAAMGANRVIGNGPNIPWKIPGEQKIFRRLTEGKVVVMGRKTFESIGKPL 60
MNSESVRIYLVAAMGANRVIGNGPNIPWKIPGEQKIFRRLTEGKVVVMGRKTFESIGKPL 60
MNSESVRIYLVAAMGANRVIGNGPNIPWKIPGEQKIFRRLTEGKVVVMGRKTFESIGKPL 60
MNSESVRIYLVAAMGANRVIGNGPNIPWKIPGEQKIFRRLTEGKVVVMGRKTFESIGKPL 60
MNSESVRIYLVAAMGANRVIGNGPNIPWKIPGEQKTFRRLTEGKVVVMGRKTFESIGKPL 60
MNSESVRIYLIAAMGANRVIGNGPNIPWKIPGEQKIFRRLTEGKVVVMGRKTFESIGKPL 60
DhfrNocardioidessp.ABL81817 MTPGGKRVVLVAAVARNGVIGDGPDIPWQLPGEQRLFKGLTWGHILVMGRATYDSIGRPL 60
hyp.prot.M.caseolyticusYP_ M----I-ISLIAAISSNYVIGKDKDIPWKIPGEQVRFKDLTMGKSVIMGRKTFESIGQPL 55
DhfrG.uraniireducensYP_0012 MV-----ISLIAAMAENRVIGRNNAIPWDIPADRKRFRALTLGHPVIMGRKTFESLAGPL 55

DhfrGeobactersp.YP_00253720 MI-----ISLIAAMSENRVIGDKNTIPWDLPADRKRFRSLTLGHPVIMGRKTFESIGFPL 55
1.5-T7-13_ PNRHTLVISRQAN
DhfrC.freundiiNP_775043 PNRHTLVISRQAN
DhfrK.pneumoniaeABC50093 PNRHTLVISSQAN
DhfrA.baumanniiABJ15828 PNRHTLVISRQAN
DhfrE.faecalisBAD77819 PNRHTLVISRQAN
DhfrS.entericaAAT45231 PNRHTLVISRQAN
DhfrE.coliACD02027 PNRHTLVISRQAN
DhfrNocardioidessp.ABL81817 PGRTTIVLTRSPD
hyp.prot.M.caseolyticusYP_ PNRKTIIISKSKD
DhfrG.uraniireducensYP_0012 PGRKNIIITRQMD
SEVHQTFEGDAFF

1.5-T7-13_
DhfrC.freundiiNP_775043 SEVHQTFEGDAFF
DhfrK.pneumoniaeABC50093 SEVHQTFEGDAFF
DhfrA.baumanniiABJ15828 SEVHQTFEGDAFF
DhfrE.faecalisBAD77819 SEVHQTFEGDAFF
DhfrS.entericaAAT45231 SEVHQTFEGDAFF
DhfrE.coliACD02027 SEVHQTFEGDAFF
DhfrNocardioidessp.ABL81817 SEVDLEPQGDAFY
hyp.prot.M.caseolyticusYP_ TIIEKEYEGNIFF
DhfrG.uraniireducensYP_0012 TIIHSRFDGDTRF
DhfrGeobactersp.YP_00253720 TVIHQVYEGDTYF

1.5-T7-13
DhfrC. freundii NP 775043
DhfrK. pneumoniaeABC50093
DhfrA. baumannii ABJ15828
DhfrE. faecalis BAD77819
DhfrE. coli ACD02027
DhfrS. entericaAAT45231
hyp. prot. M. caseolyticus YP 002559382
DhfrG. uraniireducens YP 001229711
DhfrGeobactersp. YP 002537209
DhfrNocardioides sp. ABL81817
Figure 7. Multiple sequence alignment (Panel [a]) and Neighbor Joining tree (Panel [b]) depicting the relationships between database matches and the partial amino acid sequence of a class 1 integron gene cassette encoding a dehydrofolate reductase (clone 1.5-T7-13) detected in the influents of an urban wastewater treatment plant. The GenBank accession number follows the designation of reference sequences.
a:
1.5-T7-3
CatB2K.pneumoniaeYP_0019657
MTNYFESPFK
MTNYFESPFK
CatMarinomonassp.YP_0013417 MNNYFESPFK
CatB10P.aeruginosaCAI47810
MTNYFESPFK
CatP.aeruginosaYP_002085096 MGNYFESPFR
CatMoritellasp.ZP_01896524 MNNYFESPFV
CatS.stellataZP_01745082 MENAFESPFR

LLPDRDDVDQLIIG 60
LLPDRDDVDQLIIG 60
LLPDRDDVDQLIIG 60
LLPDRDDIDQLIVG 60
LMPDRDDVDKLVIG 60
LLPDRSDVDKLIIG 60
LPPD-EGADRLLIG 59
CatPseudovibriosp.EEA93073 MPNYFESPFKGIPLQEQVTNPNIIVGKHSYYSGFYHAHSFDECARYLSAEEDEQDKLIIG 60
1.5-T7-3 SFCSIGSGAA
CatB2K.pneumoniaeYP_0019657 SFCSIGSGAA
CatMarinomonassp.YP_0013417 SFCSIGSGAS
CatB10P.aeruginosaCAI47810 SFCSIGTGAS
CatP.aeruginosaYP_002085096 SFCSIGSGAA
CatMoritellasp.ZP_01896524 SYCSIGSGAV
CatS.stellataZP_01745082 SFCSIGSGAA
CatPseudovibriosp.EEA93073 SYCSIGSGAV Q
1.5-T7-3
SEAMIMPGIK
CatB2K.pneumoniaeYP_0019657 SEAMIMPGIK
CatMarinomonassp.YP_0013417 SEAMIMPGIK
CatB10P.aeruginosaCAI47810 SEAMIMPGIN
CatP.aeruginosaYP_002085096 TEAMFMPGVR
CatMoritellasp.ZP_01896524 TEAMIMSGVK
CatS.stellataZP_01745082 SEAIVMPGIR

RAGDTVIGSDVWIG 120
RAGDTVIGSDVWIG 120
SAGDTVIGSDVWIG 120
KAGDTVIGSDVWIG 120
PAGDTLIGHDVWIG 120
RAGDTVIGNDVWIG 119
PAGDTIVGNDVWIG 119
PSGSTCIGNDVWIG 119

CatPseudovibriosp.EEA93073 SEAMIMPGVQIGNGALIGSRAVVTKDVPAYAVVAGNPAKVIR 161
b:
CatB2 K. pneumoniae YP 001965795.1
Cat Marinomonas sp. YP 001341796
CatB10 P. aeruginosa CAI47810
Cat P. aeruginosa YP 002085096
Cat S. stellata ZP 01745082
Cat Moritella sp. ZP 01896524
Cat Pseudovibrio sp. EEA93073
Figure 8. Multiple sequence alignment (Panel [a]) and Neighbor Joining tree (Panel [b]) depicting the relationships between database matches and the partial amino acid sequence of a class 1 integron gene cassette encoding a chloramphenicol adenyltransferase (clone 1.5-T7-3) detected in the influents of an urban wastewater treatment plant. The GenBank accession number follows the designation of reference sequences.
1.5-T3-13
MKGWLFLVIAIVGEVIATSALKSSEGFTKLAPSAVVIIGYGIAFYFLSLV 50 Orf3/QacEdelta1K.pneumoniae
MKGWLFLVIAIVGEVIATSALKSSEGFTKLAPSAVVIIGYGIAFYFLSLV 50 QacP.aeruginosaABW25065
MDEprot.S.entericaYP_00204
QacA.baumanniiAAK72475
QacGC.japonicusYP_001983646
MDRprot.O.anthropiYP_00137
MDRprot.G.sulfurreducensNP
1.5-T3-13
Orf3/QacEdelta1K.pneumoniae
QacP.aeruginosaABW25065
MDEprot.S.entericaYP_00204
MKNWIFLAVAIFGEVIATSALKSSHGFTRLVPSVVVVAGYGLAFYFLSLA 50
MKGWLFLVIAIVGEVIATSALKSSEGFTKLAPSAVVIIGYGIAFYFLSLV 50
MKGWLFLVIAIVGEVIATSALKSSEGFTKLAPSAVVIIGYGIAFYFLSLV 50
MNPWIFLSIAIIAEVIATSALKASDGFSNTLPATIVIVGYGAAFYFLSLT 50
MPVYAILAIAIVSEVIGTLSLKASEGFTRLGPSLIVVVAYGLAFYFLSMT 50
MHKWLYLLVAIISEVAGTTALKSAEGFTRLWPSCVVVAGYASAFYFLSLT 50
LKSIPVGVAYAVWSGLGVVIITAIAWLLHGQKLDAWGFVGMG 92
LKSIPVGVAYAVWSGLGVVIITAIAWLLHGQKLDAWGFVGMG 92
LKSIPVGIAYAVWAGLGIVLVAAIAWIFHGQKLDFWAFIGIG 92
LKSIPVGVAYAVWSGLGVVIITAIAWLLHGQKLDAWGFVGMG 92
QacA.baumanniiAAK72475 LKSIPVGVAYAVWSGLGVVIITAIAWLLHGQKLDAWGFVGMG 92
QacGC.japonicusYP_001983646
MDRprot.O.anthropiYP_00137
MDRprot.G.sulfurreducensNP
LKAIPVGIAYAVWSGAGIVLVTAIAWILYGQKLDVWAFVGIG 92
LKSIPVGIAYAVWSGIGVTLVALIGWLVFGQKLDLAAVLGMG 92
LKAIPVGIAYAIWSGLGTALVAVVAWVFMGQRLDLPAVFGIL 92
1.5-T3-13
MDE prot. S. enterica YP 002045451
Orf3/QacEdelta1 K. pneumoniae ACF06163
Qac A. baumannii AAK72475
QacG C. japonicus YP 001983646
Qac P. aeruginosa ABW25065
MDRprot. O. anthropi YP 001370804
MDRprot. G. sulfurreducens NP 951765
Figure 9. Multiple sequence alignment (Panel [a]) and Neighbor Joining tree (Panel [b]) depicting the relationships between database matches and the deduced, partial amino acid sequence of the region corresponding to the qac gene associated with the conserved 3’ end of class 1 integrons. (clone 1.513). The GenBank accession number follows the designation of the reference sequences.


DNA samples from other impacted and relatively undisturbed environments (Table 1) were submitted to the cassette PCR assay. These sites were sampled during the dry and rainy seasons. Again, an amplicon of similar size (~1000-1500 bp) to that detected at the wastewater treatment plant and to that from an E. coli strain containing a class 1 integron was consistently detected in disturbed sites. Nevertheless, this PCR product was undetected in environments with limited human impact such as the Fanduco Reef, the upstream region of the Cupeyes River and samples from undisturbed forest reserves, indicating that the targeted integron-encoded genes were associated with wastewater pollution or fecal impact. (Figures 10-16).
Results from mangrove habitats with minimal impact (Figure 12, Ensenada Dakity, Culebra) suggest that natural factors may influence the prevalence of integronencoded genes as these were undetected during the dry season but were found during the rainy season. During this sampling a significant activity of waterbirds was observed at the site. Wild and domesticated avian populations have been reported to be implicated on the dispersal of fecal indicator organisms presenting antibiotic resistant traits (Middleton and Ambrose, 2005; Lanthier et la., 2010; Literak et al., 2010 ). Nevertheless, ABR gene cassettes were detected only in samples from sediments and rhizosphere material indicating that mangrove habitats may serve as transient reservoirs of ABR genes when the impact is moderate. In contrast, sampling of the disturbed mangrove site (La Parguera, Lajas) indicates that a sustained impact of wastewater pollution may turn the mangrove ecosystem into a stable reservoir of resistance determinants (Figure 12). Additionally, human traffic to secluded beaches seem to contribute to increased levels of class 1 integrons as these elements were detected in sand samples during the dry season and not during the rainy season when beaches are less frequented (Figure 14).
Representative sequences of the variable region of class 1 integrons were submitted to comparative alignment analysis. Functional and structural features of class 1 integrons were indentified in all of them, confirming class 1 elements as the source of the recovered resistance determinants (Figures 17 and 18). The amino acid sequences were deduced for each of the detected genes and a phylogenetic tree was constructed which included their most closely related matches as reveled by searches in public databases. A total of eighth different antibiotic resistance genes were detected across the tested sites. These included three variants of aminoglycosyde modifying enzymes, a dehydrofolate reductase, an extended spectrum beta-lactamase, a chloramphenicol modifying enzyme and 2 variants of quaternary ammonium compounds efflux pumps (Figure 19). All of them were closely related 98-100% amino acid identity) to resistance determinants described in known human pathogens harboring class 1 integrons (Cloeckaert et al., 2006; Chen et al., 2007; Chen et al., 2009; Nawaz et al., 2009; Bogaerts et al., 2010; Shi and Yamasaki, unpublished, GenBank accession no. BAE66663).
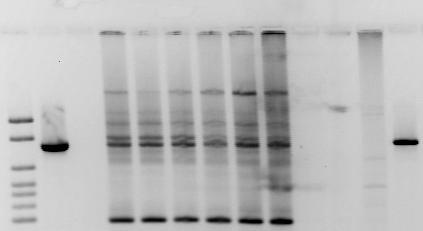
Figure 10. Panel (a): Agarose gel electrophoresis showing representative results for the amplification of gene cassettes associated with class 1 integrons. The red line highlights the detection of a ~1,300 bp amplicon encoding antibiotic resistance genes in the Isabela samples (wastewater pipeline outfall (ISO) and receiving sea water (ISR) near Sardinera Beach). The amplicon was undetected in water samples from the Fanduco Reef which has minimal anthropogenic impact (Bonkosky et al., 2008).
2,000bp 1,500bp 1,000bp
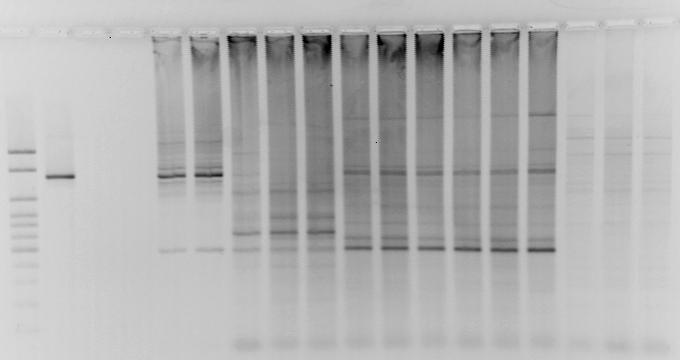
Figure 11. Agarose gel electrophoresis showing the amplification of gene cassettes associated with class 1 integrons detected in DNA recovered from: the outflow of a urban wastewater treatment plant (UO), water form the Guanajibo beach at Mayaguez (MGW), a coastal pipeline outfall (ISO) and receiving sea water (ISR) near Sardinera beach, Isabela and undisturbed forest soil, Guajataca forest (GF). The dashed lines depict the detection of a ~1,300 bp amplicon previously associated with antibiotic resistance genes in wastewater plants influents (UI). The amplicon was not detected in water samples from the Fanduco reef which has minimal anthropogenic impact. M=100bp ladder DNA marker (Applied Biological Materials Inc., Richmond, BC, Canada); (+) = positive control; (-) = negative control.
disturbed

minimal impact

Ensenada Dakity, Culebra, PR

Season Site Coliforms Enterococci cassettes Sample dry Parguera + + + water + + + sediment + + + rhizosphere
Dakity - - - water - - - sediment - - - rhizosphere rainy Parguera + + + water + + + sediment + + + rhizosphere
Figure 12. Summarized results for samples retrieved from impacted (La Parguera) and relatively undisturbed (Ensenada Dakity) mangrove ecosystems. The table includes results for the presence of coliforms and eterococci above permissible levels (200 cfu [USEPA Method 9222] and 35 cfu [USEPA Method 1600] per 100 ml respectively) as well as for the detection of integron-encoded antibiotic resistance


Dakity - - - water - - + sediment - - + rhizosphere


ABR
Season Site
Coliforms Enterococci cassettes Sample dry Isabela + + + outfall + + + water

PicodePiedraBeach,Aguada(disturbed)

Pico de + + + water
Piedra + + + sand rainy Isabela + + + outfall + + + water
. Summarized results for samples retrieved from impacted beaches (open wastewater discharge, Isabela and Pico de Piedra Beach, Aguada). The table includes results for the presence of coliforms and eterococci above permissible levels (200 cfu [USEPA Method 9222] and 35 cfu [USEPA respectively) as well as for the detection of integron-encoded antibiotic resistance
Pico de + + + water
Piedra + + + sand
PeñaBlancaBeach(Aguadilla) (minimalimpact)


. Summarized results for samples retrieved from Peña Blanca Beach, Aguadilla. This site was representative of a beach habitat exposed to moderate anthropogenic influence and was used as a reference with respect to disturbed beaches (open wastewater discharge, Isabela and Pico de Piedra Beach, Aguada). The table includes results for the presence of coliforms and eterococci above permissible levels (200 cfu [USEPA Method 9222] and 35 cfu [USEPA Method 1600] per 100 ml respectively) as well as for the detection of integron-encoded antibiotic resistance (ABR) gene cassettes.
ABR
Site Season Coliforms Enterococci cassettes
Sample
Peña dry - - + water
Blanca - - + sand rainy - - - water - - - sand
ABR
Figure 15. Summarized results for samples retrieved from disturbed estuarine habitats (La Boca, Barceloneta and Guanajibo, Mayagüez). The table includes results for the presence of coliforms and eterococci above permissible levels (200 cfu [USEPA Method 9222] and 35 cfu [USEPA Method 1600] per 100 ml respectively) as well as for the detection of integron-encoded antibiotic resistance (ABR) gene cassettes.

Figure 16. Summarized results for samples retrieved from the downstream and upstream regions of the Cupeyes River (Barrio cupeyes, Sabana Grande). These sites were representative of moderately impacted (downstream) and pristine (upstream) river habitats and were used as references with respect to disturbed estuarine sites (La Boca, Barceloneta and Guanajibo, Mayagüez). The table includes results for the presence of coliforms and eterococci above permissible levels (200 cfu [USEPA Method 9222] and 35 cfu [USEPA Method 1600] per 100 ml respectively) as well as for the detection of integron-encoded antibiotic resistance (ABR) gene cassettes.
ABR Season Site Coliforms Enterococci cassettes Sample dry Cupeyes - - + water downstream
Cupeyes - - - water upstream
a:
5'primer-Lavesqueetal.,1995 -----------------------------ggcatccaagcagcaag V.choleraeintI1GQ214169.1
GACTGTTTTTTTGTACAGTCTATGCCTCGggcatccaagcagcaagCGCGTTACGCCGTG
PS12CRaad -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
TI12CDaad -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
AAS12CRaad -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
MZH1.5Caad -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
MZS1.2Caad -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
RI3.4Caad -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
PR33CRaad -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
PS11CRaac -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
TI3.3Caac -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
MZH3.3Caac -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
RI3C4aac -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
MZH3.1CRESBLGES-14 -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
TI17CDESBLGES-14 -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
PR12CRESBLGES-14 -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
PS23CRESBLGES-14 -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
IUWP1.5-T7-13dhfr -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
IUWP1.5-T7-3cat -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
TI2.5CqacG -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
RI2C4Orf3/Qacdelta -----------------------------ggcatccaagcagcaagCGCGTTACGCCGTG
5'primer-Lavesqueetal.,1995 V.choleraeintI1GQ214169.1
GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGCAGCAGGGCAGTCGCCCTAAA PS12CRaad GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGCAGCAGGGCAGTCGCCCTAAA TI12CDaad GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGCAGCAGGGCAGTCGCCCTAAA
AAS12CRaad
GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGTAGCAGGGCAGTCGCCCTAAA MZH1.5Caad
GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGTAGCAGGGCAGTCGCCCTAAA MZS1.2Caad
RI3.4Caad
GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGTAGCAGGGCAGTCGCCCTAAA
GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGTAGCAGGGCAGTCGCCCTAAA PR33CRaad
GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGCAGCAGGGCAGTCGCCCTAAA PS11CRaac
TI3.3Caac
MZH3.3Caac
GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGCAGCAGGGCAGTCGCCCTAAA
GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGCAGCAGGGCAGTCGCCCTAAA
GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGCAGCAGGGCAGTCGCCCTAAA RI3C4aac
GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGTAGCAGGGCAGTCGCCCTAAA MZH3.1CRESBLGES-14
GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGCAGCAGGGCAGTCGCCCTAAA TI17CDESBLGES-14
GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGCAGCAGGGCAGTCGCCCTAAA PR12CRESBLGES-14
GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGCAGCAGGGCAGTCGCCCTAAA PS23CRESBLGES-14
GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGCAGCAGGGCAGTCGCCCTAAA IUWP1.5-T7-13dhfr
GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGCAGCAGGGCAGTCGCCCTAAA IUWP1.5-T7-3cat
GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGCAGCAGGGCAGTCGCCCTAAA TI2.5CqacG
GGTCGATGTTTGATGTTATGGAGCAGCAACGATGTTACGCAGCAGGGCAGTCGCCCTAAA RI2C4Orf3/Qacdelta
5'primer-Lavesqueetal.,1995 V.choleraeintI1GQ214169.1
GGTCAATGTTTGATGTTATGGAGCAGCAACGATGTTACGCAGCAGGGCAGTCGCCCTAAA
ACAAAGTTAAACATCATGAGGGAAGCGGTGATCGCCGAAGTATCGACTCAACTATCAGAG PS12CRaad ACAAAGTTAGGCATCATGAGGGTAGCGGTGACCATCGAAATTTCGAACCAACTATCAGAG TI12CDaad ACAAAGTTAGACATCATGAGGGTAGCGGTGACCATCGAAATTTCGAACCAACTATCAGAG AAS12CRaad ACAAAGTTAGACATCATGAGTGAAAAAGTGCCCGCCGAGATTTCGGTGCAACTATCACAA MZH1.5Caad ACAAAGTTAGACATCATGAGTGAAAAAGTGCCCGCCGAGATTTCGGTGCAACTATCACAA MZS1.2Caad ACAAAGTTAGACATCATGAGTGAAAAAGTGCCCGCCGAGATTTCGGTGCAACTATCACAA RI3.4Caad ACAAAGTTAGACATCATGAGTGAAAAAGTGCCCGCCGAGATTTCGGTGCAACTATCACAA PR33CRaad ACAAAGTCAGACATCATGAGTGAAAAAGTGCCCGCCGAGATTTCGGTGCAACTATCACAA PS11CRaac
ACAAAGTTAGGCCCGCACGGAATCAACATCTCATGTCCGCCAACAATGCCGCAATAGCTC TI3.3Caac
ACAAAGTTAGGCCCGCACGGAATCAACATCTCATGTCCGCCAACAATGCCGCAATAGCTC MZH3.3Caac
ACAAAGTTAGGGATGTAGAGTGCAAAGAAAAATTGTTCTTCTGCTAGTGCTGTCCTCTAT RI3C4aac ACAAAGTTAGACATCATGAGTGAAAAAGTGCCCGCCGAGATTTCGGTGCAACTATCACAA MZH3.1CRESBLGES-14
ACAAAGTTAGACGGGCGTACAAAGATAATTTCCATCTCAAGGGATCACCATGCGCTTCAT TI17CDESBLGES-14
ACAAAGTTAGACGGGCGTACAAAGATAATTTCCATCTCAAGGGATCACCATGCGCTTCAT PR12CRESBLGES-14
ACAAAGTTAGACGGGCGTACAAAGATAATTTCCATCTCAAGGGATCACCATGCGCTTCAT PS23CRESBLGES-14
ACAAAGTTAGACGGGCGTACAAAGATAATTTCCATCTCAAGGGATCACCATGCGCTTCAT IUWP1.5-T7-13dhfr
ACAAAGTTAGCCATATGAACTCGGAATCAGTACGCATTTATCTCGTTGCTGCGATGGGAG IUWP1.5-T7-3cat
ACAAAGTTAGGCGACGCGTGGAGTCGCTCTAGAATTTTCGGGTACAAATTTTATGACGAA TI2.5CqacG
ACAAAGTTAGATGCTTTGCTGTGCGCACAAATTTCGGCCTGCAACAAGACTGTTTTTTTT RI2C4Orf3/Qacdelta
ACAAAGTTAGATGCACTAAGCACATAATTGCTCACAGCCAAACTATCAGGTCAAGTCTGC
b:
Figure 17. Panel (a): Partial alignment of representative, PCR-amplified nucleotide sequences corresponding to the conserved 5’end of class 1 integrons (Panel (b) red doted lines) detected in environmental samples. The blue and green arrow depict the positions of the -10 and -35 signals of the intl1 promoter, respectively, The orange bar shows the position of the attI1 site. Numbering is based on the V. cholerae intI1 (GenBank accession no. GQ214169 ).
3'primercomplArduino2003 ------------------------------------------------------------
K.pneumoniaepGDKA1EU722351.2
RI2C4Orf3/Qacdelta
MZO1.2COrf3/Qacdelta
MZH12CROrf3/Qacdelta AAAGGCTGGCTTTTTCTTGTTATCGCAATAGTTGGCGAAGTATCGCAACATCCGCATTAA
3'primerArduino2003(3'-5') ------------------------------------------------------------
3'primercomplArduino2003 ------------------------------------------------------------
K.pneumoniaepGDKA1EU722351.2 GGCAATCACCCAACTACTGGGAGCACCAGGCATCTTACTCATAGGCTTGGCACTATGGCA
RI2C4Orf3/Qacdelta
MZO1.2COrf3/Qacdelta AATCTAGCGAGGGCTTTACTAAGCTTGCCCCTTCCGCCGTTGTCATAATCGGTTATGGCA
MZH12CROrf3/Qacdelta AATCTAGCGAGGGCTTTACTAAGCTTGCCCCTTCCGCCGTTGTCATAATCGGTTATGGCA
3'primerArduino2003(3'-5') ------------------------------------------------------------
3'primercomplArduino2003 ------------------------------------------------------------
K.pneumoniaepGDKA1EU722351.2
RI2C4Orf3/Qacdelta
3'primercomplArduino2003 ------------------------------------------------------------
RI2C4Orf3/Qacdelta
MZH12CROrf3/Qacdelta
3'primerArduino2003(3'-5') --------------ccgaaacatccataccccga 3'primercomplArduino2003 --------------ggctttgtaggtatggggct K.pneumoniaepGDKA1EU722351.2 AGCTTGATGCGTGGggctttgtaggtatggggct RI2C4Orf3/Qacdelta AGCTTGATGCGTGGggctttgtaggtatggggct MZO1.2COrf3/Qacdelta AGCTTGATGCGTGGggctttgtaggtatgggcgt MZH12CROrf3/Qacdelta AGCTTGATGCGTGGggctttgtaggtatgggcgt
Figure 18. Panel (a). Partial alignment of representative, PCR-amplified nucleotide sequences corresponding to the conserved 3’end of class 1 integrons (Pabel [b]) detected in environmental samples. The orange bar shows the position of the qacEΔ1 gene. Numbering is based on the K. pneumoniae intI1 (pGDKA1) (GenBank accession no. EU722351).
E.coli aadA7AAF44098
RI3C4aad
AAS12CRaad
PR33CRaad
RI3.4Caad
MZH1.5Caad
MZS1.2Caad
V.choleraeaadA2BAE66663.1
PS12CRaad
TI12CDaad
E.cloacaeAac(6)YP002791508.1
PS11CRaac
TI3.3Caac
E.coli dhfrACD02027.1
IUWP1.5-T7-13dhfr
A.baumannii ESBLGES-14ADC91899.1
MZH3.1CRESBLGES-14
TI17CDESBLGES-14
PR12CRESBLGES-14
PS23CRESBLGES-14
K.pneumoniaecatB2YP001965795.1
IUWP1.5-T7-3cat
Uncultbacterium QacGFJ172386.1
TI2.5CQacG
S.entericaQacE ABG36699.1
IUWP1.5-T3-13
aminoglycoside nucleotidyltransferases
aminoglycoside acetyltransferases
dehidrofolate reductase
extended-spectrum beta-lactamases
chloramphenicol adenyltransferase
QACeffluxproteins
Figure 19. Unrooted Neighbor-joining tree illustrating the relationship of representative intI1associated gene cassettes sequences recovered from a variety of impacted coastal habitats. Values represent the percentage of 2,000 bootstrap replications that supported the branching order. GenBank accession numbers follow the designation of the reference sequences.
Prevalence of antibiotic resistance traits and integrons in culturable bacteria.The culture-dependent component of this project consisted of the isolation and characterization of bacteria carrying integron-based antibiotic resistance. For this part of the study, exploratory work was conducted by sampling sites of high and minimal fecal impact.. The results indicated the presence of culturable strains harboring integrons in samples with high fecal impact (Figure.20)
Since the isolation of culturable integron hosts was feasible, our efforts were focused on the recovery of fecal indicator organisms (coliforms and enterococci ) that could serve as carriers of class 1 integrons loaded with antibiotic resistance genes. To this end, 66 axenic cultures isolated and presumptively characterized as coliforms using the USEPA 9200 method were submitted to our cassette-targeted PCR assay. Similarly, 66 isolates presumptively identified as enterocci according to the USEPA 1600 method were also analyzed. These isolates were representative of the Guanajibo and Isabela sites (Table 1). From this pool of cultures, six coliform isolates and six enterococci strains were positive for the presence of ABR genes encoded by class 1 integrons. Subsequent sequencing and comparative alignment analyses revealed the presence of gene cassettes 99-100% identical to determinants conferring resistance against beta-lactam antibiotics, trimethoprim, and quaternary ammonium compounds (Figures 21-23). Furthermore we applied our casseete PCR assay to a collection of 15 clinical strains of Klebsiella pneumoniae from the Vertans Affairs Hospital to evaluate the degree of similarity between integron-encoded determinants detected across impacted coastal habitats and those associated with the clinical scene. As observed in our environmental samples, we identified partial sequences of a dehydrofolate reductase, an aminoglycoside adenyltransferase and an efflux pump which are implicated in conferring resistance against trimethoprim, amino glycoside antibiotics and quaternary ammonium compounds respectively (Figures 24-26). These were 97-100% similar to variants described in clinical other clinical isolates. This findings demonstrate that integron encoded antibiotic resistance is present in local Hospitals and in impacted coastal settings in association with wastewater pollution and bacteria from intestinal niches.
Figure 16. Detection of integron integrases in master plates grids of bacterial isolates recovered from influents from the urban wastewater treatment plant (500 bp fragments lanes1-4). M= 100 bp ladder DNA marker (New England Biolabs, Ipswich, MA) .
ADC91899beta-lactamaseGES-14
enterococci90Mayaguez
ADC91899beta-lactamaseGES-14
AMCSTFKFPLAALVF-ERIDSGTERGDRKLSYGPDMIVEWSPATERFLASGHMTVLEAAQ enterococci90Mayaguez AMCSTFKFPLAALV-LERIDSGTERGDRKLSYGPDMIVEWSPATERFLASGHMTVLEAAQ
ADC91899beta-lactamaseGES-14
AAVQLSDNGATNLLLREIGGPAAMTQYFRKIGDSVSRLDRKEPEMSDNTPGDLRDTTTPI enterococci90Mayaguez AAVQLSDNGATNLLLREIGGPAAMTQYFRKIGDSVSRLDRKEPEMSDNTPGDLRDTTTPI
ADC91899beta-lactamaseGES-14
enterococci90Mayaguez
Figure 21. Alignment of the deduced amino acid sequence for a gene cassette detected in an enterococci strain isolated from the Guanajibo (Mayagüez) site with respect to that of an extended beta lacatamase (GenBank accession no. ADC91899). The sequences share a 99% identity level.
Figure 22. Alignment of the deduced amino acid sequence for a gene cassette detected in a coliform strain isolated from the Guanajibo (Mayagüez) site with respect to that of a dihydrofolate reductase (GenBank accession no. YP_209335). The sequences share a 100% identity level.
Figure 23. Alignment of the deduced amino acid sequence for a gene cassette detected in an enterococci strain isolated from the Isabela site with respect to that of a quaternary ammonium efflux pump (GenBank accession no. ACF06163). The sequences share a 99% identity level.
Figure 24. Neighbor-Joining tree depicting the phylogenetic relationships of a dehydrofolate reductase (red triangle) encoded by a class 1 integron detected in a clinical strain of K. pneumoniae (isolate 542, Veterans Affairs Hospital, Río Piedras, PR) with respect to reference sequences retrieved from GenBank. GenBank accession nos. of reference sequences precede the designation of the source organisms.
YP 002045451 S enterica
ACF06163 K pneumoniae
542 K pneumoniae
NP 044260 Eaerogenes
ABI85513 R anatipestifer
AAM03346 P mirabilis
YP 209355 S enterica
ABX52005 S enterica
NP 539962 B melitensis
YP 342650 N oceani
YP 001635386 C aurantiacus
Figure 25. Neighbor-Joining tree depicting the phylogenetic relationships of an efflux pump of quaternary amonium compounds (red diamond) encoded by a class 1 integron detected in K. pneumoniae (isolate 542, Veterans Affairs Hospital, Río Piedras, PR) with respect to reference sequences retrieved from GenBank. GenBank accession nos. of reference sequences precede the designation of the source organisms.
1.8-T7-II
aad K. pneumonia YP 001338814
aad S. typhimurium ABA56506
aad K. pneumoniae ABN11922
aad A. xylosoxidans CAD38272
aad C. freundii AAL59386
aad A. baumannii AAN85830
aad Acinetobacter NP 049451
aad P. aeruginosa AAX93309
aad P. aeruginosa CAD38272
aad A. adenylyltransferase NP 490175
Figure 26. Neighbor-Joining tree depicting the phylogenetic relationships of an aminoglycoside adenyltransferase (green diamond) encoded by a class 1 integron detected in K. pneumoniae (isolate 502, Veterans Affairs Hospital, Río Piedras, PR) with respect to reference sequences retrieved from GenBank. GenBank accession nos. of reference sequences follow the designation of the source organisms.
Inventory of fluconazole-resistant yeasts from impacted and undisturbed environments. The distribution of fluconazole-resistant yeasts was evaluated across a variety of impacted and relatively undisturbed sites (Figure 27, Table 1). A total of 41 fluconazole resistant strains (MIC>64μg/mL, NCLSI M27-A3) were recovered. Resistant isolates were ultimately identified by phylogenetic analysis of ITS seqeuences associated with rRNA genes. Nearly 63% of the resistant strains recovered were highly related (99100% identical) to recognized, opportunistic human pathogens. The dominant genotypes detected were related to Aureobasidium pullulans (n=8) and Candida tropicalis (n=7). These were recovered from wastewater treatment plants effluents and impacted beaches. Candida tropicalis ranks among the most common pathogenic yeast species (Leinberger et al., 2005) while Aureobasidium pullulans has been associated with a case of a disseminated nosocomial infection in a severely traumatized patient, which eventually recovered after fluconazole therapy (Bolignano et al., 2003). Additionally, phylotypes (n=2) related to a common human pathogen, Hortaea werneckii, the causative agent of tinea nigra (a superficial infection of the outermost layer of the epidermis) and several opportunistic strains implicated in hospital-acquired outbreaks were detected. These included: Exophiala jeanseimei (n=1), Acremonioum strictum (n=2), Pseudozyma sp. (n=4) and Pichia anomala (n=2) (Nucci et al 2002; Abliz et al., 2003; Novicki et al., 2003; Pasqualotto et al., 2005; Hwang et al., 2010). Since fluconazole therapy is routinely used for the successful treatment of infections caused by opportunistic strains genetically related to several representatives from our collection of fluconazole-resistant yeasts, it is recommended to further study potential risks posed by the prevalence of resistance traits to this medicament in association with wastewater pollution of coastal habitats (Chakrabarti et al., 2001; Yalaz et al., 2003; Bolignano et al., 2003; da Matta et al., 2007; Rossetto et al., 2010)
Figure 27. Unrooted Neighbor-joining tree illustrating the relationship of environmental, fluconazole-resistant yeasts (MIC >64μg/mL) based on comparative analysis of ITS regions amplified with universal primers (ITS1 / ITS4). Red branches illustrate the clustering of isolates related to species reported as opportunistic or common human pathogens whereas green and blue branches indicate the position of isolates that grouped with species described in marine or plant–related niches. Values represent the percentage of 2,000 bootstrap replications that supported the branching order. GenBank accession numbers precede the designation of the reference sequences.
CONCLUSIONS
Our results indicate that sustained impact from direct and non point sources of wastewater pollution has influenced the establishment of stable reservoirs of multiple ABR genes encoded by class1 integrons across critical coastal environments. The application of culture dependent and DNA-based methods demonstrated that cassette PCR assays targeted at class 1 integrons have the potential to be further developed as a dual indicator of fecal contamination and inconspicuous risks to human health. Integronencoded resistance was also detected in representative strains of fecal indicators recovered from impacted coastal settings as well as in pathogenic bacterial isolates from clinical sources, indicating potential health risks associated with exposure to contaminated habitats and resistance determinants commonly assumed to be exclusive of secluded facilities such as hospitals or animal husbandry operations. Moreover, impacted coastal habitats and wastewater discharges were shown to comprise an environmental reservoir of fluconazole resistant yeast highly related to opportunistic pathogens. Hence, the discharge of wastewater containing resistant microorganisms that have been selected and enriched after the use of antimicrobial compounds associated with demographic development seems to be a key factor in the aggravation of resistance problem and further deterioration of impacted ecosystems. Additional epidemiological studies complemented with quantitative molecular methods targeted at specific antibiotic resistance determinants and analytical detection methods focused on commonly used antibiotics and their respective degradation products are needed to gain a more comprehensive assessment of risks posed by wastewater pollution in coastal settings.
ACKNOWLEDGMENTS
Special thanks to Dr. Harold Stokes of Macquarie University, Sydney, Australia and Dr. Sonia Saavedra of the Veterans Affairs Caribbean Healthcare System (Río Piedras, PR) for reference strains used in integron-related PCR assays. We are also grateful to Daniel Irizarry for facilitating sampling at the Fanduco Reef.
REFERENCES
Abliz P, Fukushima K, Takizawa K, Miyaji M, Nishimura K. Specific oligonucleotide primers for identification of Hortaea werneckii, a causative agent of tinea nigra. Diagn Microbiol Infect Dis. 2003 Jun;46(2):89-93.
Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995 Mar;59(1):143-69.
Arduino SM, Catalano M, Orman BE, Roy PH, Centrón D. Molecular epidemiology of orf513-bearing class 1 integrons in multiresistant clinical isolates from Argentinean hospitals. Antimicrob Agents Chemother. 2003 Dec;47(12):3945-9.
Arakawa Y, Murakami M, Suzuki K, Ito H, Wacharotayankun R, Ohsuka S, Kato N,
Ohta M. A novel integron-like element carrying the metallo-beta-lactamase gene blaIMP. Antimicrob Agents Chemother. 1995 Jul;39(7):1612-5.
Avila P, de la Cruz F. Physical and genetic map of the IncW plasmid R388. Plasmid. 1988 Sep;20(2):155-7.
Bogaerts P, Naas T, El Garch F, Cuzon G, Deplano A, Delaire T, Huang TD, Lissoir B, Nordmann P, Glupczynski Y. GES extended-spectrum β-lactamases in Acinetobacter baumannii isolates in Belgium. Antimicrob Agents Chemother. 2010 Nov;54(11):4872-8.
Bolignano G, Criseo G. Disseminated nosocomial fungal infection by Aureobasidium pullulans var. melanigenum: a case report. J Clin Microbiol. 2003 Sep;41(9):4483-5.
Bonkosky M, Hernández-Delgado EA, Sandoz B, Robledo IE, Norat-Ramírez J, Mattei H. Detection of spatial fluctuations of non-point source fecal pollution in coral reef surrounding waters in southwestern Puerto Rico using PCR-based assays. Mar Pollut Bull. 2009 Jan;58(1):45-54.
Chakrabarti A, Singh K, Narang A, Singhi S, Batra R, Rao KL, Ray P, Gopalan S, Das S, Gupta V, Gupta AK, Bose SM, McNeil MM. Outbreak of Pichia anomala infection in the pediatric service of a tertiary-care center in Northern India. J Clin Microbiol. 2001 May;39(5):1702-6.
Chen YT, Lauderdale TL, Liao TL, Shiau YR, Shu HY, Wu KM, Yan JJ, Su IJ, Tsai SF. Sequencing and comparative genomic analysis of pK29, a 269-kilobase conjugative plasmid encoding CMY-8 and CTX-M-3 beta-lactamases in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2007 Aug;51(8):3004-7.
Chen YT, Liao TL, Liu YM, Lauderdale TL, Yan JJ, Tsai SF. Mobilization of qnrB2 and ISCR1 in plasmids. Antimicrob Agents Chemother. 2009 Mar;53(3):1235-7.
Cloeckaert A, Praud K, Doublet B, Demartin M, Weill FX. Variant Salmonella genomic island 1-L antibiotic resistance gene cluster in Salmonella enterica serovar Newport. Antimicrob Agents Chemother. 2006 Nov;50(11):3944-6.
da Matta VL, de Souza Carvalho Melhem M, Colombo AL, Moretti ML, Rodero L, Duboc de Almeida GM, dos Anjos Martins M, Costa SF, Souza Dias MB, Nucci M, Levin AS. Antifungal drug susceptibility profile of Pichia anomala isolates from patients presenting with nosocomial fungemia. Antimicrob Agents Chemother. 2007 Apr;51(4):1573-6. Epub 2007 Jan 29.
Dillon B, Thomas L, Mohmand G, Zelynski A, Iredell J. Multiplex PCR for screening of integrons in bacterial lysates. J Microbiol Methods. 2005 Aug;62(2):221-32.
Giovannoni, S. J. 1991. The polymerase chain reaction, p. 177–201. In E. Stackenbrandt (ed.), Nucleic acids techniques in bacterial systematics. John Wiley & Sons, New York, NY.
Hwang S, Kim J, Yoon S, Cha Y, Kim M, Yong D, Chang JH, Jeong SH, Uh Y, Lee K. First report of brain abscess associated with Pseudozyma species in a patient with astrocytoma. Korean J Lab Med. 2010 Jun;30(3):284-8.
Koeleman JG, Stoof J, Van Der Bijl MW, Vandenbroucke-Grauls CM, Savelkoul PH. Identification of epidemic strains of Acinetobacter baumannii by integrase gene PCR. J Clin Microbiol. 2001 Jan;39(1):8-13.
Korabecná M, Liska V, Fajfrlík K. Primers ITS1, ITS2 and ITS4 detect the intraspecies variability in the internal transcribed spacers and 5.8S rRNA gene region in clinical isolates of fungi. Folia Microbiol (Praha). 2003;48(2):233-8.
Kümmerer K. Resistance in the environment. J Antimicrob Chemother. 2004 Aug;54(2):311-20.
Lanthier M, Scott A, Lapen DR, Zhang Y, Topp E. Frequency of virulence genes and antibiotic resistances in Enterococcus spp. isolates from wastewater and feces of domesticated mammals and birds, and wildlife. Can J Microbiol. 2010 Sep;56(9):715-29.
Leboffe, M.J. and B.E. Pierce, 2002. Microbiology Laboratory Theory and Application. 1st Edn., Morton Publishing, USA.
Leinberger DM, Schumacher U, Autenrieth IB, Bachmann TT. Development of a DNA microarray for detection and identification of fungal pathogens involved in invasive mycoses. J Clin Microbiol. 2005 Oct;43(10):4943-53.
Lévesque C, Piché L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995 Jan;39(1):185-91.
Literak I, Dolejska M, Janoszowska D, Hrusakova J, Meissner W, Rzyska H, Bzoma S, Cizek A. Antibiotic-resistant Escherichia coli, Including Strains with the Extended-spectrum Beta-lactamase and qnrS Genes, in Waterbirds on the Baltic Sea Coast of Poland. Appl Environ Microbiol. 2010 Oct 15.
Mazel D, Dychinco B, Webb VA, Davies J. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob Agents Chemother. 2000 Jun;44(6):1568-74.
Middleton JH, Ambrose A. Enumeration and antibiotic resistance patterns of fecal indicator organisms isolated from migratory Canada geese (Branta canadensis). J Wildl Dis. 2005 Apr;41(2):334-41.
National Committee for Clinical Laboratory Standards, Reference method for broth dilution antifungal susceptibility testing of yeasts: approved standard (3rd ed), M27-A3 National Committee for Clinical Laboratory Standards, Wayne, PA (2008).
Nawaz M, Khan AA, Khan S, Sung K, Kerdahi K, Steele R. Molecular characterization of tetracycline-resistant genes and integrons from avirulent strains of Escherichia coli isolated from catfish. Foodborne Pathog Dis. 2009 Jun;6(5):553-9.
Novicki TJ, LaFe K, Bui L, Bui U, Geise R, Marr K, Cookson BT. Genetic diversity among clinical isolates of Acremonium strictum determined during an investigation of a fatal mycosis. J Clin Microbiol. 2003 Jun;41(6):2623-8.
Nucci M, Akiti T, Barreiros G, Silveira F, Revankar SG, Wickes BL, Sutton DA, Patterson TF. Nosocomial outbreak of Exophiala jeanselmei fungemia associated with contamination of hospital water. Clin Infect Dis. 2002 Jun 1;34(11):1475-80.
Pasqualotto AC, Sukiennik TC, Severo LC, de Amorim CS, Colombo AL. An outbreak of Pichia anomala fungemia in a Brazilian pediatric intensive care unit. Infect Control Hosp Epidemiol. 2005 Jun;26(6):553-8.
Patterson KL, Porter JW, Ritchie KB, Polson SW, Mueller E, Peters EC, Santavy DL, Smith GW. The etiology of white pox, a lethal disease of the Caribbean elkhorn coral, Acropora palmata. Proc Natl Acad Sci U S A. 2002 Jun 25;99(13):8725-30.
Rodríguez-Minguela CM, Apajalahti JH, Chai B, Cole JR, Tiedje JM. Worldwide prevalence of class 2 integrases outside the clinical setting is associated with human impact. Appl Environ Microbiol. 2009 Aug;75(15):5100-10.
Rossetto AL, Dellatorre G, Pérsio RA, Romeiro JC, Cruz RC. Subcutaneous phaeohyphomycosis on the scrotum caused by Exophiala jeanselmei: case report. An Bras Dermatol. 2010 Aug;85(4):517-20.
Yalaz M, Hilmioglu S, Metin D, Akisu M, Nart D, Cetin H, Ozturk C, Isik E, Kultursay N. Fatal disseminated Acremonium strictum infection in a preterm newborn: a very rare cause of neonatal septicaemia. J Med Microbiol. 2003 Sep;52(Pt 9):835-7.
White PA, McIver CJ, Deng Y, Rawlinson WD. Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol Lett. 2000 Jan 15;182(2):265-9.
IMPACT AND DISSEMINATION OF RESULTS
Educational and Societal Impact. We have acquired basic equipment to be used for the proposed experiments and recruited three graduate students (Odalys Álvarez, and Glendalys Vargas [PI Rodríguez-Minguela], Lourdes Galarza [Co-PI MontalvoRodríguez] and Roberto Román [Co-PI Maldonado-Ramírez]) which have been trained in bacteriological, mycological and molecular techniques and conducted the research under the supervision of the investigators. Further details on each graduate student involved in the project are summarized in Table 2. The following undergraduate students have also participated in the project: Paloma Monroig (PI Rodríguez-Minguela), Alitza Santiago, Isabelita Martínez, and Harriel Acosta (Co-PI Maldonado-Ramírez). Additionally, graduate and undergraduate students from Co-PI Maldonado’s laboratory participated in a workshop offered by PI Rodríguez-Minguela in which they received training in molecular techniques to be used in the identification of fungicide resistant yeasts (Figure 28).

Figure 28. Results obtained during the workshop on DNA extraction and PCR ITS genes from yeast isolates. Panel ( ): Agarose gel electrophoresis of Genomic DNA (1.5%) (lanes1 Panel (b): Agarose gel electrophoresis of PCR amplified ITS genes (1.5%) (lanes 1 DNA marker.


Table 3. Graduate students impacted in this initiative.
Expected
Student's Starting graduation Thesis name date date status
Odalys Álvarez* Jan-08 Mar-11 In progress
Glendalys Vargas Aug-09 ND In progress
Roberto Román* Aug-08 Mar-11 In progress
Lourdes Galarza* Aug-08 ND In progress
*Students received a stipend of $917 per month ($1,834 per student/summer) for two summers. This monthly salary is equivalent to that of a full (6cr) teaching assistantship at the Biology Department. ND = not determined.
`This study sets the frame work for the development of measures in support of the protection of coastal environments and improvements for public safety. Our results have uncovered potential risks to human health commonly assumed to be inherent of hospital settings which have not been previously evaluated within an environmental context in Puerto Rico. We have identified a linkage between wastewater pollution and the prevalence of traits among microorganisms from impacted habitats that could compromise the effectiveness of treatment options for bacterial and fungal infections. Our integron-targeted PCR assays seem to have the potential to be refined as a quantitative, molecular method for the development of an alternative indicator of water quality and human influence. These may become important tools for epidemiological and risk assessment studies that may provide useful information to the general public, urban planners, health professionals, resource users and managers.
Dissemination of Results. The activities in which results from this investigation were presented include:
1. Sigma Xi Poster Day (UPRM, April 2008).
2. Sea Grant 3rd Annual Symposium for Coastal and Marine Applied Research (UPRM, October 2008).
3. Annual Biomedical Research Conference for Minority Students (Orlando, FL, November 2008).
4. 29th Puerto Rico Interdisciplinary Scientific Meeting (UPR-RP, March 2009).
5. Biominds Annual Poster Day (Bioprocess Development and Training Complex ,Mayagüez, PR, March 2009)
6. Signa Xi Poster day (UPRM, April 8th, 2010).
7. General Meeting of the American Society for Microbiology (San Diego, CA, May 2010).
8. Sea Grant Research Symposium (UPRM, October 29th, 2010).
9. Special lecture for the undergraduate level course Microbial Ecology, BIOL 4365 (Biology Department, UPRM, November, 2010)
Future presentations will be scheduled with the Department of Natural Resources and Fish and Wild Life. Additionally, we plan on publishing our results in peer reviewed journals.
