Epidemiological investigation of outbreaks including WGS of isolates from The Netherlands
1996-2022
J. Wiegel, R. Buter, Royal GDAim

Epidemiological investigation of unrelated outbreaks of septicaemia in broilers with Ornithobacterium rhinotracheale (O.r.) as causative agent. Investigation of the genetic differences of O.r. isolates from different outbreaks, recurrent outbreaks on a farm and cases with septicaemia versus respiratory cases.
Introduction
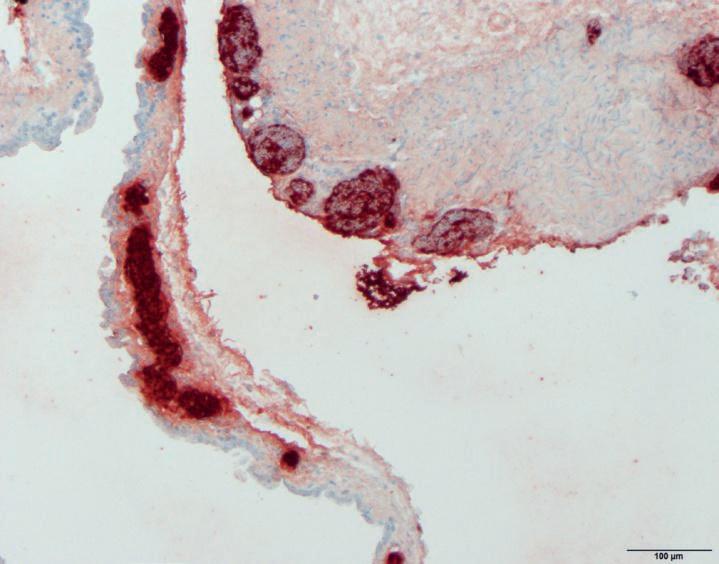
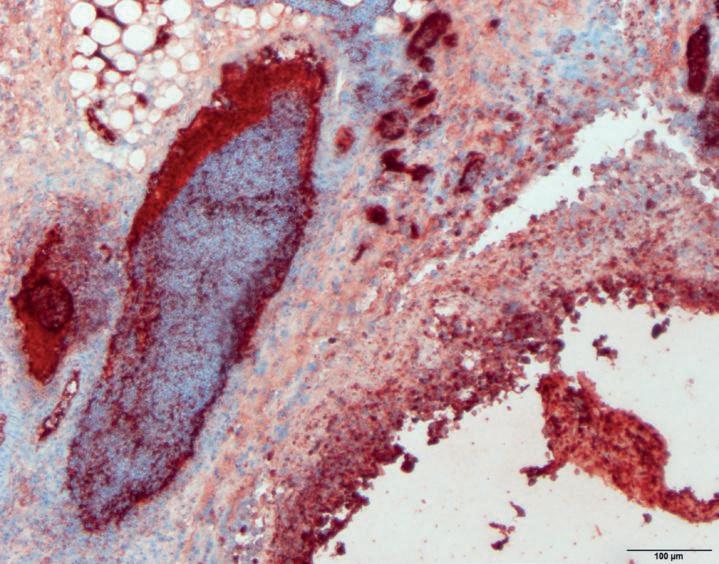
Two separate and unrelated cases occurred, on multi age broiler farms in 2020, continuing in 2021 and 2022. Acute mortality due to septicaemia started at 23 days of age, responded to antimicrobial treatment and resulted in increased condemnations at slaughter. On post mortem, no classical lesions of O.r. infection were seen. Instead it was indicative of septicaemia with hepatomegaly and splenomegaly. Septicaemia and the absence of air sac lesions due to O.r. was confirmed by immunohistochemistry (IHC) on HE samples of air sacs (Pictures 1 and 2). O.r. was cultured from bone marrow, spleen and liver, also indicative of septicaemia. No epidemiological link between the two farms was found. A third broiler farm was also confronted with cases of septicaemia due to O.r., earlier in 2018 and also in 2022. Isolates from the three outbreaks were supplemented with isolates and O.r. from respiratory cases from the years 1996 to 2022 from private collections added to the investigation.

Materials & Methods
• 49 isolates of O.r. from 27 locations: 27 from broilers, 16 from meat turkeys, 3 from rearing breeders, 2 from breeders and 1 from a backyard chicken.
• Whole genome sequencing (WGS) using Oxford Nanopore Technology (ONT).
• Data analysis using CLC Genomic Workbench 22: Single Nucleotide Polymorphism (SNP) analysis and in silico Multi Locus Sequence Typing (MLST).
Results
• Based on MLST three sequence types (ST) 1, 9 and 11 were found showing high correlation with three main branches in the SNP tree (Figure 1):
- Isolates of ST1 originated from meat type chickens (broilers and rearing parent stock) and meat turkeys;
- Isolates of ST9 originated from meat turkeys with respiratory lesions and from chickens with both respiratory lesions (only parent stock) and septicaemia (broilers and parent stock) between 2015 and 2022;
- Isolates of ST11 originated from meat turkeys and from a backyard chicken with respiratory lesions between 2017 to 2021.
• WGS showed a higher discriminating power compared to MLST.

• Using a cut-off of 100 SNPs difference, 7 different WGS clusters were assigned (clusters A to F) visualized by different coloured nodes.
• Multiple isolates from one outbreak showed high genetic similarity with a variation of max. 19 SNPs.
• Isolates from farms with multiple outbreaks on were divided over different genotypes, except for one farm (farms are visualized by different coloured blocks on the right in the SNP tree):
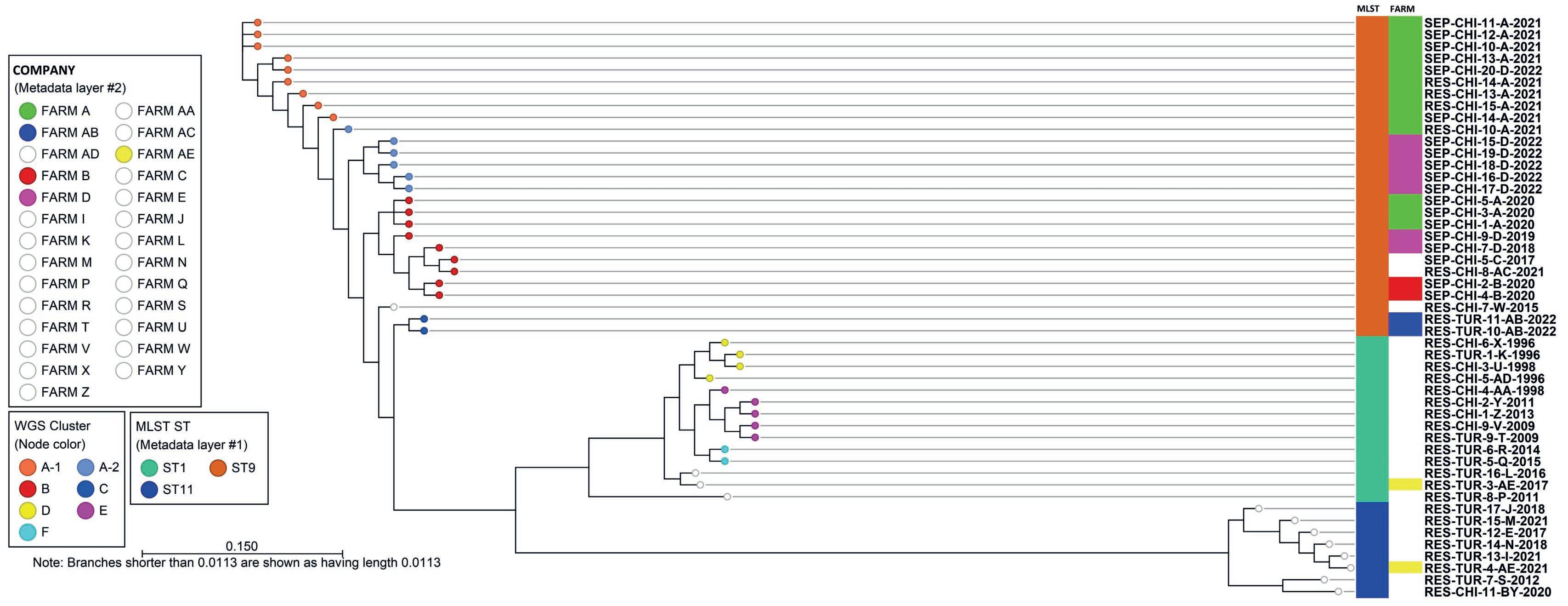
- Isolates from Farm A showed a difference in time; isolates from 2020 belonged to cluster B, all isolates from 2021 and 2022 belonged to cluster A;
- Isolates from Farm B showed high genetic similarity (3 SNPs). Outbreaks were in the same year, 2.5 month in between;

- Isolates from Farm D showed a difference in time; isolates from 2018 and 2019 belong to cluster B, the isolates from one outbreak in 2022 belong to cluster A;
- Isolates from Farm AE from outbreaks in 2017 and 2021 showed great genetic difference (cluster were not defined due to low numbers of isolates).
• Isolates from chickens with septicaemia belonged to clusters A and B with a variation of max. 129 SNPs.
Picture 1 & 2. Microscopic pictures of air sacs of a broiler with septicaemia due to O.r. The O.r. bacteria are visible as red dots (IHC) and the pictures show that they are present in large numbers on the surface of the air sacs and in the blood vessels.
Conclusions & Discussion
Although there is a gap in time in the isolation of genotyped O.r., it can be tentatively concluded that ST1 has been largely replaced by two other ST’s, being ST11 in turkeys and a backyard chicken and ST9 in chickens. Isolates from one outbreak showed high genetic similarity. Multiple genotypes were involved in different outbreaks on farms, except for one farm with two outbreaks with only 2.5 month in between. It is not yet clear whether isolates from birds with O.r. septicaemia are more pathogenic or that this manifestation of disease is related to other factors, such as complicating infections.
We thank drs. André Steentjes (Veterinair Centrum Someren), drs. Suzanne Bosman (Poultry Vets), drs. Bart Machielsen (Avecur) for their contribution. This research was supported by AVINED.
j.wiegel@gdanimalhealth.com

