Sharing our achievements
Wellcome Centre for Anti-Infectives Research
Six Year Report

Image on the Front Cover shows a cross-section of a mouse small intestine infected with Cryptosporidium parasites
The image was taken by Dr Ross Bacchetti, a previous PhD student in Dr Mattie Pawlowic’s lab Dr Bacchetti worked to investigate proteins that are involved in parasite transmission The surface of intestinal cells are stained in white and their DNA is stained in cyan, and the Cryptosporidium parasites appear magenta. Cryptosporidium causes diarrhea which can be deadly for young, malnourished children At WCAIR we are working on technology to learn more about this important parasite and to create new medicines to treat the disease
The WCAIR mural on Craigie Street in Stobswell was created through an open competition organised by WCAIR, the Stobswell Forum and OpenClose Dundee The selected artist was Glasgow-based artist Lewi Quinn, aka Boiiing, who painted the mural with his assistant Finnula in July 2022 The images in the mural were inspired by conversations between Lewi and WCAIR scientists, particularly x-ray crystallographers Alice and Paul

Foreword page 4
About WCAIR
Awards to WCAIR members
Setting our sights on infectious diseases
Research at WCAIR
Focus on anti-infective oxaboroles
Technology development – genetic approaches for the trypanosomatids
The Mode of Action group
page 6
page 8
page 11
page 12
page 14
page 15
page 16
page 17 and transfer of revolutionary screening platforms to industry
Case Study: Delivery of new and repurposed drugs for visceral leishmaniasis
Cryptosporidiosis
Overcoming the challenges for Chagas disease drug discovery
Innovation in Chemistry
Enhancing target-based screening
Innovation within Drug Metabolism and Pharmacokinetics
page 18
page 19
page 21
page 22
page 23
page 24 35 years of the Ferguson Lab in Dundee (1988 – 2023).
A journey in parasite glycobiology and scientific infrastructure:
Case Study: Adoption of a new rapid diagnostic test for
page 27 Human African Trypanosomiasis
6 years of the WCAIR Training Programme
WCAIR Public Engagement at 6
Closing Remarks
WCAIR Management Board
Selected Publications
page 28
page 31
page 35
page 37
page 38
WCAIR Timeline page 42
WCAIR Global Partners page 44
WCAIR Advisory Board
c o n t e n t s
page 46
Wellcome Centre for Anti-Infectives Research Six Year Report I page 3
foreword
As we pass six years of operations, it’s a real pleasure to introduce our achievements documented in this report and I want to extend a huge thank-you to all our truly outstanding Wellcome Centre for Anti-Infectives Research (WCAIR) teams. We have made excellent progress across a range of activities, improving our understanding of pathogen biology, and delivering new technologies, new anti-infective drugs, a trained workforce, and a more engaged public.

WCAIR is a world-leading hub for neglected tropical disease drug discovery and technological innovation, with a focus on research, training, and public engagement. With over £18 million in funding from Wellcome since the Centre’s inception, we are developing and exploiting new methods, technologies and processes that help us understand and combat infectious diseases, changing the drug discovery paradigm, and making drug discovery faster and smarter. We are passing on our skills and knowledge to researchers in countries most affected by NTDs; Ghana and Brazil, in particular We also aim to inform and inspire others in relation to how science can combat disease and poverty.
Technological advance can fast-forward the pace of scientific discovery. Similarly, technology-enabled, and data-driven drug discovery can help to deliver high-quality, life-changing, and durable drugs to the right patients at the right time. We are connecting basic research and drug discovery to address those gaps that currently hamper
successful drug discovery. To do so, WCAIR supports a broad range of short- and longer-term projects seeking to develop new approaches. Several of these projects have been identified through calls for proposals that are open to any member of the Centre.
To accelerate the development of new drugs, we need a greater understanding of the pathogens, and our parasitology experts work with multiple global partners to achieve this. Goals include a better understanding of pathogen biology and prioritization of potential drug targets. Our world-leading parasitology teams, and Mode-of-Action group, develop new and versatile technologies, including new genetic and proteomic methods. Identification of the targets of compound series then helps to bring together phenotypic- and target-based approaches across our drug discovery portfolio.
to his drug discovery efforts
Wellcome Centre for Anti-Infectives Research Six Year Report I page 4
It was not luck, it was bloody hard work.
Sir James Black (Chancellor, University of Dundee 1992 – 2006) referring
Progress in science depends on new techniques, new discoveries and new ideas, probably in that order.
Sydney Brenner (‘opened’ Sir James Black Centre, University of Dundee in 2006)
We are developing improved in vitro and in vivo imaging to better predict clinical efficacy and to allow us to understand why compounds work or fail. This includes an enhanced understanding of pathogen distribution before and after drug treatment, and the ability of compounds to kill parasites in clinically relevant states
We are radically reforming our drug discovery design-make-test cycle to accelerate projects to key milestones, or project closure. WCAIR is integrating computational chemistry design with high throughput approaches to chemistry optimization and synthesis. This includes robotics to enable chemistry and assays, testing unpurified compounds in both parasiteand target-based assays to reduce timescales and costs, and integration of improved disease models.
Other avenues we are exploring to increase productivity include improved efficiency of protein production for X-ray crystallography and cryo-electron microscopy studies that enable structure-based drug discovery. New compound sets are being assembled to expand available chemical space. New equipment purchases also help to ensure that all areas of our work are underpinned by state-of-the-art technology.
I have been proud to serve as Interim WCAIR Director for just over a year and want to extend a special thanks to Paul Wyatt, as WCAIR Director from inception to February 2022, for his outstanding leadership and for building WCAIR to an integrated and highly effective
Centre. I also extend thanks to all members of our superb advisory board; we are truly grateful for the excellent support, advice, and guidance you provide. Collaboration is at the heart of what we do Therefore, thanks to the many partners we have both in Dundee and across the globe. Thank you especially to Wellcome for funding the Centre and associated projects. We also work in partnership with many other funding agencies and acknowledge them for their help and support. Our relationships with them are very much partnerships These include the Bill & Melinda Gates Foundation, Medicines for Malaria Venture (MMV), Drugs for Neglected Diseases Initiative (DNDi), the UKRI Medical Research Council, and GHIT (Global Health Innovation Technology Fund). Our staff and teams’ other achievements, including several promotions and professional awards, are described in more detail later in these pages.
Professor David Horn Interim Director Wellcome Centre for Anti-Infectives Research (April 2022– April 2023) Head of Division of Biological Chemistry and Drug Discovery, University of Dundee

Wellcome Centre for Anti-Infectives Research Six Year Report I page 5
about WCAIR

The vision of the Wellcome Centre for Anti-Infectives Research (WCAIR) is to support the 2012 London Declaration to eliminate Neglected Tropical Diseases (NTDs). We have created a world-leading hub for NTD drug discovery and are a collaborator of choice for academics, BioPharma and Product Development Partnerships (PDPs) in the translation of discovery science into drug candidates.
WCAIR is part of the Division of Biological Chemistry and Drug Discovery in the School of Life Sciences, University of Dundee It encapsulates parasitology discovery biology, Mode of Action group and the Dundee Drug Discovery Unit. Within WCAIR these groups work together to deliver innovative methods and technologies, training to scientists from disease endemic countries and engagement with local and global public communities.
Our Research goal is to position us and our partners at the cutting edge of drug discovery for major infectious diseases of low- and middle-income countries.
Together, we are developing new tools and methods to radically increase the rate of delivery and success of drug discovery projects Creating a fundamentally redesigned and rigorous approach to drug discovery for these diseases

Our Training goal is to create a skilled and knowledgeable cohort of drug discovery scientists within disease endemic countries. Empowering them to deliver medicines for their communities.
Our Public Engagement goal is to inform and inspire our publics about the many ways science fights poverty and disease. We want to reach a common understanding of each other’s needs so that we can co-create mutually beneficial outcomes.

Wellcome Centre for Anti-Infectives Research Six Year Report I page 6
Over the next decade R & D at the Centre will generate a legacy of new therapeutics, new drug discovery approaches, novel targets and tool molecules that will impact and enrich the wider scientific, medical infectious diseases communities and patients.
Parasite Discovery Biology
Mode of Action
Accelerating and innovating drug discovery research
Training and Capacity Building
Public Engagement
WCAIR was created in April 2017 after an underpinning 5 year grant of £13 6 million In 2020 the Centre funding was extended by £5.6 million for a further 2 years. Over this period Wellcome have provided an additional £12.3 million to continue support of the kinetoplastid drug discovery programme for visceral leishmaniasis and Chagas disease. This programme is carried out with our partners GSK and DNDi. Wellcome have also provided £7.3 million to the Mode of Action group The major funders of the Centre’s TB, Malaria, cryptosporidiosis and COVID-19/coronavirus drug discovery programmes are the Bill and Melinda Gates Foundation, Medicines for Malaria Venture, GHIT, UKRI Medical Research Council and the COVID-19 Therapeutics Accelerator (initiated by the Bill & Melinda Gates Foundation, Wellcome and Mastercard), respectively. In 2021 we have initiated a Wellcome funded drug discovery programme for Schistosomiasis with Prof. Karl Hoffman at the University of Aberystwyth.
WCAIR in numbers
172 current members
38 WCAIR trainees from 15 countries
201 papers since April 2017
£12.3 million
£1.5 million internal funding for 30 projects
£2.25 million internal funding for equipment
Visceral Leishmaniasis
2 clinical candidates (Ph1)
41 compound series molecular target/ MoA identified
> 65,000 engagements with public
1 International conference
160 attendees
Total number of people under the Centre funded through multiple sources, March 2023. 29 staff directly funded by Centre grant
Number of trainees completed a training period in Dundee as of March 2023
Total number of papers published by all WCAIR groups since April 2017
Average yearly income from all funders. Wellcome funding is 62% of total
WCAIR funding allocated to internal innovation projects (consumables and postdoc salaries)
WCAIR funding allocated to support equipment purchases to expand capabilities
Clinical development candidates delivered by the DDU/GSK Portfolio Award, working through different novel mechanisms of action
Compound series sourced globally, many identified targets are novel
Includes activities as part of Science Festivals and Careers Hive, visitors to LifeSpace exhibitions, visitors to NMS exhibition, activities hosted at WCAIR e.g. Medicine Maker badge and Dundee Rep.
A conference to share experiences and expertise around drug discovery across multiple diseases from bedside to bench
Wellcome Centre for Anti-Infectives Research Six Year Report I page 7
Awards to WCAIR members
Knighthood for Professor Mike Ferguson

Professor Mike Ferguson, Regius Professor of Life Sciences at the University of Dundee received a knighthood in the 2019 New Year’s Honours list On receiving his award Prof Ferguson said “I am thrilled to receive this great honour, but minded immediately that it recognises the efforts of many at the University of Dundee – my home for 30 years and an institution where collaboration and cooperation are truly valued, and where advancing knowledge into solutions is highly-prized Together, we have managed to build in Dundee a truly world-class environment for science, and I feel very privileged to have been able to contribute to that. I am particularly grateful to my life- and science-partner, Dr Lucia Güther, and my family for support of every kind ”
Dr Mattie Christine Pawlowic recognised as a leading Early Career Researcher
Dr Pawlowic has had a winning start to her career at WCAIR At the end of 2018 Mattie won a prestigious Sir Henry Dale Fellowship from Wellcome and the Royal Society In April 2019 Dr Pawlowic won the inaugural British Society of Parasitology’s President’s medal This is awarded annually to an outstanding early-career researcher who has produced international-quality research and demonstrates the potential to become a world leader in parasitology Dr Pawlowic’s continued impact as a research leader and mentor has been recognised by two awards. Mattie is the 2023 recipient of the Royal Society of Edinburgh’s (RSE) Dame Anne McLaren Medal in Life Sciences, which will be awarded in November. Mattie is also the recipient of a Biochemical Society Early Career Researcher Award for 2024.
Dr Pawlowic’s research focuses on how the parasite Cryptosporidium protects itself from destruction, particularly a protective outer shell that makes it resistant to low-cost water treatment The models developed by the Pawlowic group and their research into the life cycle of Cryptosporidium are a key component of the drug discovery programme for cryptosporidiosis led by the DDU and carried out in collaboration with the Pawlowic lab
William Trager award for Professor David Horn
WCAIR Deputy Director (and Interim Director 2022– 23) Prof David Horn received the William Trager award at the 2019 Annual Meeting of the American Society of Tropical Medicine and Hygiene

The award recognised the breakthroughs Prof. Horn has made in understanding the basic biology of parasites which has unlocked new areas of research The award committee hailed Professor Horn’s “transformative work, significant for both its fundamental and translational impact” The world leading research of David Horn and his group was also recognised through the receipt of a £2.1 million Wellcome Investigator award in 2019
C.A. Wright Memorial Medal for Dr Susan Wyllie


In 2023 Dr Susan Wyllie was awarded the prestigious C A Wright Memorial Medal from the British Society for Parasitology in recognition of her outstanding contribution to the discipline Dr Wyllie is a Principal Investigator and head of the Mode of Action Group in the Wellcome Centre for Anti-Infectives Research in the School of Life Sciences, with more than twenty years of experience studying kinetoplastid biology. Her studies have predominantly focused on deciphering drug mechanism(s) of action and mechanisms of drug resistance – in particular in Leishmania, the causative agent of visceral leishmaniasis
David Horn said “Congratulations to Susan on this well-deserved award, and particularly for the truly transformative target identification work she and her team have delivered Target identification yields important chemical biology insights, and Susan’s work has had a huge impact on how anti-infective drug discovery projects are progressed, and also on key decisions to register compounds for clinical trials ”
In 2018, Susan’s team was awarded GlaxoSmithKline’s Annual Scientific Termination of Projects (STOP) Award The team showed that a compound series killed the parasites by a mechanism that had potential to damage human cells Dr Susan Wyllie explained “Although it sounds negative, it’s a huge positive because identifying compound series that are not likely to be successful in the drug discovery process means we can divert much-needed resources into the development of more promising compounds.”
Wellcome Centre for Anti-Infectives Research Six Year Report I page 8
DDU Malaria team win MMV Project of the Year 2018
The Medicines for Malaria Venture (MMV) prize is awarded annually to the scientific partners involved in that year’s most exciting project from the MMV portfolio The 2018 prize was awarded to the DDU Malaria team led by Prof Ian Gilbert, Prof Kevin Read and Dr Beatriz Baragaña The team are developing compounds to inhibit an enzyme involved in protein synthesis

“The Dundee team was awarded MMV Project of the Year 2018 as their work with MMV really represents a step forward from business-as-usual drug discovery,” said Dr Timothy Wells, MMV’s Chief Scientific Officer “Having a greater understanding of the structure of the drug target has been like shining a bright light on the work to optimise the drug series – it means we can be really precise as we improve the selectivity and potency of the compounds.”
Professor David Gray honoured in HM Queen’s birthday honours 2021
David Gray, Head of Biology and Professor of Translational Biology at the University’s Drug Discovery Unit, has been given a British Empire Medal (BEM) for services to the Delivery of Testing during the Covid-19 pandemic

Prof Gray helped establish Scotland’s central Covid-19 testing facility, Glasgow Lighthouse Laboratory, in support of the national response to the coronavirus pandemic He had an integral role in the design and implementation of the Scottish testing centre, and also advised on operating procedures.
Prof Gray said, “This honour belongs to the entire amazing Glasgow Lighthouse team It has been a privilege for me to work with them in delivering high quality and high throughput Covid diagnostic testing. I am grateful to them and for the support that my colleagues at Dundee have given me. However, the biggest thank you must go to my wife, Nicola She kept our family fed, educated and mostly smiling, giving me time to be able to contribute to the testing centre She is the one that deserves a medal ”
Glasgow’s Lighthouse Laboratory also won the Knowledge Exchange/Transfer Initiative of the Year trophy at the seventeenth annual Times Higher Education (THE) Awards in 2021
Professor Ian Gilbert elected as fellow of prestigious scientific Societies
WCAIR’s new Director, Prof Ian Gilbert, was elected a Fellow of the Royal Society of Edinburgh in 2020 and a Fellow of the Academy of Medical Sciences in 2021. At the time Prof. Gilbert was Head of Chemistry in the DDU which he helped establish in 2006 As a medicinal chemist, Ian’s research interests are primarily in the design and synthesis of potential drugs with a particular focus on infectious diseases which affect Low- and Middle-Income Countries such as malaria and visceral leishmaniasis. He is also interested in novel approaches to drug discovery and studies to understand the mode of action of biologically active compounds In October 2022 Prof Gilbert become Head of the DDU

Dr Joana Faria
Dr Joana Faria was a postdoctoral research fellow in the lab of Professor David Horn. Joana’s research focuses on understanding how african trypanosomes (parasites responsible for African Sleeping Sickness) try and avoid detection by the human immune system Dr Faria left WCAIR in 2021 to set up her own laboratory at the University of York after she successfully obtained a Sir Henry Dale Fellowship
While at WCAIR Dr Faria’s research on the control of monogenic expression of variant surface glycoproteins on the surface of Trypanosoma brucei was recognised by Scottish Universities Life Sciences Alliance (SULSA) Early Career Researcher award 2020 in the Development and Regulation category. Also, in 2020 Joana won an award for her research talk at the Genetics Society of America’s Molecular Parasitology Meeting XXI. As part of her research Joana captured beautiful microscope images of the trypanosomes. The image she entitled ‘The VEX’ed Trypanaosome’ earnt her a finalist place in the Infectious Diseases Hub photograph competition in 2019.
Joana’s promise as a future leader was recognised by the award of the Wellcome-Beit prize. The prize is awarded by Wellcome to their most promising fellows who are starting to lead their own independent research programmes Joana was considered while interviewing for her Sir Henry Dale Fellowship Joana also won the British Society for Parasitology’s President’s medal in 2022

Wellcome Centre for Anti-Infectives Research Six Year Report I page 9
a w a r d s
Biochemical Society Awards 2024

Two WCAIR individuals and a WCAIR team have won awards from the Biochemical Society Dr Mattie Pawlowic has won an Early Career Researcher award and Prof Sir Mike Ferguson has been invited to deliver the Morton Lecture for outstanding contribution to lipid biochemistry The DDU-GSK Kinetoplastid Drug Discovery Team led by Dr Manu De Rycker and Dr Tim Miles have won the Industry & Academic Collaboration Award
Dr Manu De Rycker said, “It is great recognition of a truly integrated industry-academia collaboration that has delivered multiple clinical candidates for neglected tropical diseases The success of our collaboration is down to a brilliant group of like-minded scientists working together towards a single aim.”
Public Engagement Awards
Julia Haddow (née Wcislo) was named runner-up in the Medical Research Council’s Max Perutz Science Writing Award 2020. Julia, an MRC funded PhD student in Kevin Read’s lab in the Drug Discovery Unit, received a runner-up prize of £750 for her article, ‘The game of hide-and-seek‘ Julia’s article focussed on her research to develop methods to determine the localisation of the parasite which causes Chagas disease within the body Julia said “I tried to portray the struggles and achievements I have encountered so far in an engaging, simple and humorous style The article is aimed at a non-scientific audience so that people from all backgrounds can understand what my research is about and why it matters ”

A number of the public engagement initiatives supported by WCAIR have received recognition at the School of Life Sciences annual awards The 2018 prize for Engaged Researcher was jointly shared by Lesley-Anne Pearson and Lauren Webster The Project of the Year was ‘Kirsty’s project: Search for a new medicine’ a children’s book conceived by Suzanne Duce and Tracey Baylis and illustrated by Daisy
 MacGowan
MacGowan
In 2019 the Project of the Year was awarded to the WCAIR team who created the Medicine Maker badge for Girl Guides. The team won again in 2021 for their ongoing collaboration with Girlguiding including participation in Wander the World (2020) and hosting the Virtual Sleepover 2021

Dr Irene Hallyburton was highly commended in the engaged researcher category in 2021 for her long lasting and sustained commitment to engaging members of the public in the work of the School of Life Sciences. In 2022 Irene’s idea the WeeCAIR Medicinal Garden was highly commended in Project of the Year.

The School of Life Sciences, of which WCAIR is a part, was awarded a Gold Engage Watermark by the National Co-ordinating Centre for Public Engagement in 2017 WCAIR activities also contributed to the University of Dundee Institutional Gold Engage Watermark in 2020

Wellcome Centre for Anti-Infectives Research Six Year Report I page 10
The success of our collaboration is down to a brilliant group of like-minded scientists working together towards a single aim.
Dr Manu De Rycker
Setting our sights on infectious diseases
In May 2019 WCAIR hosted its first international conference, ‘Setting our sights on infectious diseases’, SOSID. The conference aimed to harness cross-disciplinary and cross-disease learning to accelerate drug discovery and reduce drug candidate attrition rates for infectious diseases affecting low- and middle-income countries (LMICs). This included discussion on malaria, tuberculosis, HIV, kinetoplastid infections, bacterial infections and helminth infections such as schistosomiasis There were a surprising number of lessons learned between the different disease areas.


Conference sessions spanned the whole pipeline from the initial start of a drug discovery programme through to clinical trials We started with an introductory session on the current global disease situation and then worked backwards from clinical development, combination therapy, pharmacokinetic/pharmacodynamic (PK/PD) studies, drug discovery pathways to new start points and targets. Overall, the conference aimed to learn lessons from across different disease areas and between the preclinical and clinical phases, with the aim of exploring how we can improve and speed up the drug discovery, translational and clinical development processes.
We were keen to ensure that viewpoints of researchers from disease endemic countries were captured during conference discussions We therefore raised sponsorship to support 19 travel bursaries for early career researchers from LMICs. Each bursary

paid for registration, travel and accommodation
The WCAIR management thank the generosity of Medicines for Malaria Venture (MMV), the Bill and Melinda Gates Foundation, Novartis Institutes for Biomedical Research and Drugs for Neglected Diseases initiative (DNDi) for their support

The 160 people attending the conference included participants from Africa, South America, Cuba, Europe, North America, Malaysia, India and Australia. Evaluation of the conference showed very positive feedback with 98% saying they would attend another WCAIR conference; 94% Agreed or strongly agreed they now have a better understanding of drug discovery for infectious diseases in LMICs; 92% Agree or Strongly Agree they now have new ideas to solve challenges they face in their own work; 90% Agree or Strongly Agree SOSID has allowed them to make new connections.
Ian Gilbert has coordinated a paper with contributions from all the conference speakers to capture the topics, learnings and discussions which took place You can read the viewpoint paper in ACS Infectious Diseases, https://doi.org/10.1021/acsinfecdis.9b00371

As well as many scientific discussions and learnings, the meeting helped to cement existing collaborations and to initiate new interactions This is a valuable output from a resource-poor scientific area
We thank all our speakers and the Division of Biological Chemistry and Drug Discovery admin team for their contribution to a successful conference

Wellcome Centre for Anti-Infectives Research Six Year Report I page 11
Research at WCAIR
 This image shows fluorescent labelled trypanosomes taken by Mark Field’s group
This image shows fluorescent labelled trypanosomes taken by Mark Field’s group
Research at WCAIR is tackling a variety of gaps and issues to create impact across the whole of the anti-infective drug discovery continuum These include:

> Gaining a deeper understanding of parasite biology and mode of action of successful therapeutics.
> Defining critical paths and assay cascades predictive of human efficacy.
> Applying and integrating state-of-the-art approaches to compound design and synthesis.
Strategic research and technology implementation are designed, planned, coordinated and adjusted by the Director and WCAIR management team This is augmented by supporting and developing internally peer-reviewed “bottom-up” ideas from WCAIR staff to capture innovation relevant to our mission and to encourage forward thinking in the team
Strategic developments to date include:
> Improving our ability to manipulate pathogens at a genome-wide scale (e.g. genome-wide under- and over-expression) or at a precision level (e g gene editing)
> Applying these genetic and also proteomic methods, via the cutting-edge Mode-of-Action group, who continue to develop platforms for drug target deconvolution and exploitation This allows rational decisions on the progression of the drug-lead portfolio, ensuring individual targets are not over-represented and the transition of phenotypic projects into structure-enabled projects
> Developing advanced quantitative chemical proteomics for drug target deconvolution
> Expanding our range of assay systems, including replacing in vitro reporter assays with a RapidFire® mass spectrometry-based high throughput compound screening system
> Enabling our structure-based drug discovery projects by introducing high throughput construct design and expression system testing, and by collaborating with Diamond, Oxford to establish and optimise X-ray fragment screening of thousands of crystal soaks.
> Radically redesigning the compound design-make-test cycle to accelerate drug discovery projects to key milestones or project closure For example, enabling better and quicker decision making for compound series, by integrating improved computational compound design with high throughput chemistry synthesis (e g chemistry based on 384 micro-titre plate formats). This is enabled by robotics and testing unpurified compounds in biochemical and parasite assays
> Developing better models of disease that predict animal model and clinical efficacy, allowing us to understand why compounds work and why they don’t For example, for Chagas disease this includes developing an understanding of parasite distribution before and after drug treatment and the ability of compounds to kill parasites in clinically relevant states The latter includes dormant/persister parasites that are more difficult to kill than replicating forms These insights enable us to better focus our compound optimisation projects.
> Introducing cryptosporidiosis into our repertoire of biology and disease expertise WCAIR co-funded the set-up costs to recruit Dr Mattie Pawlowic (Sir Henry Dale Fellow)
The Research highlights that follow build on those reported in the WCAIR 3 year report, which can be read here, https://wcair.dundee.ac.uk/wcair-3-year-report/
Wellcome Centre for Anti-Infectives Research Six Year Report I page 13
>
Focus on anti-infective oxaboroles
Small molecules that contain boron have yielded several anti-infective drugs Indeed, a recent phase IIb/III clinical trial with acoziborole, carried out by DNDi, demonstrated excellent efficacy against sleeping sickness In a New Scientist article, David Horn, Interim WCAIR Director said, “The findings show acoziborole to be a safe, effective, oral therapy for the treatment of human African trypanosomiasis. The target set by the World Health Organisation is to interrupt disease transmission by 2030 and the challenges here must not be under-estimated, but the improvements that acoziborole offers over current alternative therapies could prove pivotal in helping to reach this goal ”
Oxaboroles are now in veterinary trials to treat the cattle disease, nagana, and in phase I trials for the treatment of Chagas disease and leishmaniasis. These compounds also display antiviral, antibacterial, antifungal, and anti-inflammatory activity and are under development for the treatment of malaria and cryptosporidiosis
WCAIR teams have been specifically interested in identifying the target of acoziborole, and in understanding how other oxaboroles are metabolised by specific trypanosome enzymes
Thanks to work carried out by WCAIR teams, including the Horn lab, the Field, lab, the Wyllie Mode-of-Action team, and Drug Discovery Unit teams, we now know how these drugs work, why they are selective for trypanosomatid parasites, and how resistance may emerge in the field
A key collaboration with colleagues at Anacor pharmaceuticals facilitated access to a diverse set of oxaboroles, as well as expertise in boron-based chemistry Initially, these compounds were profiled using genome-scale RNA interference screens, and further assessed using chemical biology, and structural biology approaches, revealing a specific class of pro-drugs that are activated in two steps, involving first host and then parasite enzymes 1 The resulting novel drug activation mechanism, catalysed by a parasite aldehyde dehydrogenase, may now be further exploited to improve therapeutic index

References
To identify the target of acoziborole, we developed a trypanosome genome-scale over-expression library. Screens carried out using this library showed that acoziborole targets an RNA-processing enzyme called CPSF3 2 Further structural modelling, and gene editing studies, revealed key differences between human and parasite CPSF3, at the site where the drug binds, helping to explain why the drug is safe and non-toxic to humans. Notably, we also found CPSF3 to be the target of the related oxaborole currently in veterinary trials against animal trypanosomiasis.
Working closely with DNDi, the Wyllie lab established that the oxaborole DNDI-6148, a pre-clinical candidate in development for visceral leishmaniasis, also targets CPSF3 3 The team demonstrated that overexpression of CPSF3 in L. donovani, as well as specific mutation of this enzyme, was sufficient to modulate the potency of DNDI-6148. WCAIR computational chemists were able to confirm the precise binding site of this oxaborole at its molecular target and identify new vectors for exploration, should a back-up series for visceral leishmaniasis be required
Further studies on the veterinary oxaborole, carried out in collaboration with our colleagues at the Wellcome Centre for Integrative Parasitology at the University of Glasgow, revealed another distinct and novel pro-drug activation mechanism, involving a peptidase-like enzyme 4 In this case, the drug activation mechanism was characterised in Trypanosoma brucei and in Trypanosoma congolense, a major causative agent of nagana Mutations that prevent drug activation also dramatically reduce drug accumulation in parasites.
We’ve now identified several drug transporters that allow drugs to enter parasites and now also several enzymes that modify drugs, converting them to their active form, often also a form that remains ’trapped’ and concentrated inside the parasite cell The oxaboroles are having major impacts on clinical and veterinary practice against the trypanosomatids and we now have a clearer view as to how these drugs work. WCAIR teams will continue to use cutting-edge technologies to further characterise these drugs and to develop understanding in this area
1 Host-parasite co-metabolic activation of antitrypanosomal aminomethyl-benzoxaboroles Zhang N, Zoltner M, Leung KF, , Horn D, Field MC PLoS Pathog 2018 Feb 9;14(2):e1006850
2 Clinical and veterinary trypanocidal benzoxaboroles target CPSF3 Wall RJ, Rico E, Lukac I, Zuccotto F, Elg S, Gilbert IH, Freund Y, Alley MRK, Field MC, Wyllie S, Horn D Proc Natl Acad Sci U S A 2018 Sep 18;115(38):9616-9621
3 DNDI-6148: A novel benzoxaborole preclinical candidate for the treatment of visceral leishmaniasis Mowbray, C, Braillard, S, Glossop, PA, and Wyllie, S J Med Chem 2021 Nov 11; 64(21):16159-16176
4 Veterinary trypanocidal benzoxaboroles are peptidase-activated prodrugs Giordani F, Paape D, Vincent IM, Pountain AW, Fernandez-Cortes F, Rico E, Zhang N, Morrison LJ, Freund Y, Witty MJ, Peter R, Edwards DY, Wilkes J, Van der Hooft J, Regnault C, Read KD, Horn D, Field MC, Barrett MP PLoS Pathogens 2020 Nov 3;16(11):e1008932
Wellcome Centre for Anti-Infectives Research Six Year Report I page 14
Technology development – genetic approaches for the trypanosomatids
Genetic tools and technologies, applied to the trypanosomatids, have had substantial impacts on efforts to characterize protein function, and to identify drug resistance mechanisms and drug targets. To increase throughput, ‘loss-of-function’, ‘gain-of-function’ and tagging approaches have all been scaled up, typically to achieve genome-scale coverage Indeed, high-throughput approaches have been parallelized, whereby millions of parasites, each with a specific single protein depleted or overexpressed, can be screened in one experiment WCAIR teams have pioneered the development and application of these approaches to drug mode-of-action and drug resistance studies. A current focus is on developing gene editing technologies
We have used RNA interference loss-of-function screening to facilitate drug mechanism-of-action studies for over a decade, as reviewed in 2022 1 , whereas overexpression gain-of-function screening has emerged more recently and more readily yields direct drug-target identification
CRISPR–Cas9-based approaches have revolutionized biotechnology by enabling RNA-programmed targeting of specific chromosomal loci, and Cas9-based gene editing is increasingly impacting drug discovery efforts against parasites
Precision editing of drug targets, for example, facilitates the generation of drug-resistant strains and provides insight into structure–activity relationships Quantitative measures of drug resistance can also be important in determining whether specific mutations have had, or are likely to have, a detrimental impact in a clinical setting
We developed a tightly regulated inducible Cas9-based editing system for T brucei 2 The approach has been used for one-step double-allele knockout, or for precision gene
tagging and subcellular localisation studies; we also established a transient delivery format, and the reagents have been shared with the wider research community (https://www.addgene.org/David Horn/). Precision templated base-editing has been particularly powerful for investigating drug-resistance mechanisms


Another new parasite gene editing technology is oligo targeting, which is Cas9-independent, and has now been used to edit priority drug targets in T brucei, T cruzi and Leishmania 3; the WCAIR team won the School of Life Sciences best innovation prize 2022 for this work We are currently scaling up oligo-targeting to increase throughput. The approach can now be used to generate ‘libraries’ of >1000 mutants for parallelized drug-resistance screening against priority drug targets, and to profile all possible mutations around each drug-binding pocket. Oligo-targeting subsequently has the potential to be the method of choice to address a variety of important questions, and to greatly facilitate studies on drug resistance and more.
Thus, WCAIR have developed and implemented several novel genetic approaches, innovative technologies that broadly facilitate research on the parasites we study
These developments meet a key goal for WCAIR, which is to develop new technologies that make drug discovery for neglected tropical diseases faster and smarter, and we look forward to delivering further technological advances in this area
Wellcome Centre for Anti-Infectives Research Six Year Report I page 15
References 1 Genome-scale RNAi screens in African trypanosomes. Horn D. Trends Parasitol. 2022 Feb;38(2):160-173. 2 Inducible high-efficiency CRISPR-Cas9-targeted gene editing and
in African trypanosomes Rico E, Jeacock L, Kovářová J, Horn D Sci Rep 2018 May 21;8(1):7960 3 Oligo targeting for profiling drug resistance mutations in the parasitic trypanosomatids Altmann S, Rico E, Carvalho S, Ridgway M, Trenaman A, Donnelly H, Tinti M, Wyllie S, Horn D Nucleic Acids Res 2022 Aug 12;50(14):e79
precision base editing
Dr Simone Altmann and Dr Melanie Ridgway are developing the oligo-targeting tools described below
The Mode of Action group
The development of new drugs to treat kinetoplastid and other infectious diseases has been hampered by a severe lack of robustly validated drug targets This has left drug discovery programs heavily reliant upon phenotypic screening in which large compound libraries are screened directly against parasites to identify suitable chemical start points, ‘hits’ This provides direct evidence of compounds that are known to kill or hinder growth of parasites However, the downstream development and optimisation of these phenotypically active compounds are often hindered by lack of information regarding mechanism(s) of action and/or molecular target(s) Specifically, knowledge of molecular targets is often crucial in developing strategies to overcome issues such as poor pharmacokinetics and toxicity
When the targets of active compounds are identified this information can then be used to support target-based and structure-enabled drug discovery programmes. Thus, mode of action studies can effectively integrate these two major, often disconnected, approaches to drug discovery
The Mode of Action (MoA) group, led by Dr Susan Wyllie, was established in 2015 specifically to focus on determining the MoA and/or molecular targets of compounds that are phenotypically active against kinetoplastid parasites

Historically, MoA studies have been of secondary consideration for drugs being developed for NTD If these studies were carried out at all, they were initiated after the development of pre-clinical or clinical candidates A significant point of difference for the MoA group, compared to others working in this area, is their ability to provide MoA input and information in real-time for on-going drug discovery programmes This enables the data they provide to guide and indeed drive the evolution of the best possible drug candidates. The team now effectively underpins the drug discovery programmes within WCAIR and works with multiple academic groups and key stakeholders in NTD drug discovery from around the world. Indeed, through the work of the MoA group, the molecular targets of the majority of assets in preclinical and clinical development within the DNDi kinetoplastid portfolio have now been established 2 . Ultimately, their studies play an important role in accelerating the delivery of better, safer compounds to clinic
This multidisciplinary approach to drug target deconvolution has proven extremely effective, with the molecular targets of over 40 compound series (including 4 pre-clinical candidates) identified to date 1
References
Capitalising on their experience in kinetoplastids and supported by a pump-priming grant from WCAIR, the team is now applying its multidisciplinary approach to support drug target deconvolution in other pathogens, in particular Plasmodium 3 , the parasite responsible for malaria The team are now key members of the prestigious the Bill & Melinda Gates Foundation funded “Malaria Drug Accelerator” consortium (MalDA)
1 Preclinical candidate for the treatment of visceral leishmaniasis that acts through proteasome inhibition Wyllie S, Brand S, Thomas M, De Rycker M, et al Proc Natl Acad Sci U S A 2019 May 7;116(19):9318-9323
2 DNDI-6148: A novel benzoxaborole preclinical candidate for the treatment of visceral leishmaniasis Mowbray, C, Braillard, S, Glossop, PA, and Wyllie, S. J. Med. Chem 2021 Nov 11; 64(21):16159-16176.
3 Potent acyl-CoA synthetase 10 inhibitors kill Plasmodium falciparum by disrupting triglyceride formation Bopp S, Pasaje CFA, Summers RL, Magistrado-Coxen P, et al Nat Commun 2023 Mar 16;14(1):1455
Wellcome Centre for Anti-Infectives Research Six Year Report I page 16
Dr Susan Wyllie featured in the exhibition ‘Parasites: Battle for Survival’ at the National Museum of Scotland.
The group has developed a unique approach to MoA studies, employing an array of approaches in the fields of high throughput genetics, cell biology and chemical proteomics.
Case Study Delivery of new and repurposed drugs for visceral leishmaniasis and transfer of revolutionary screening platforms to industry
There are estimated to be 600 million people at risk of visceral leishmaniasis (VL) across the globe with 50,000 to 90,000 new cases every year, mainly among the poorest of the poor The disease is caused by Leishmania parasites and is spread through the bite of infected sandflies Limited therapeutic options make the treatment of this neglected disease very challenging Each drug currently in use has serious drawbacks, such as difficulty in administration, length of treatment, toxicity, cost, and emerging drug resistance For example, miltefosine is the only oral drug, but is contraindicated in women of childbearing age as it can cause birth defects
In 2010, the global not-for-profit Drugs for Neglected Diseases initiative (DNDi) reported ‘rediscovery’ of the abandoned drug fexinidazole for the treatment of Trypanosoma brucei (causing African sleeping sickness), a parasite related to Leishmania

The drug was thought to require activation by an enzyme in the parasite Despite the prevailing consensus at DNDi that fexinidazole was inactive against Leishmania, the Dundee team identified that Leishmania had a gene for a similar activating enzyme In 2011, Prof Alan Fairlamb and Dr Susan Wyllie in collaboration with Prof Kevin Read and the University of Dundee Drug Discovery Unit (DDU) undertook a project to examine the efficacy of fexinidazole against Leishmania donovani This culminated in 2012 with the demonstration that fexinidazole displayed excellent parasite killing activity in vivo1 with activity dependent on oral administration and fexinidazole transformation in the liver into its active metabolites The work was reported pre-publication to DNDi and in 2013 they instigated a Phase II proof-of-concept clinical trial for the treatment of adults with VL in East Africa. In parallel, given the paucity of front-line treatments for VL, the DDU in 2011 initiated a major research programme to uncover new drugs for this disease, including developing novel assay platforms to revolutionise leishmaniasis drug discovery 2, 3 A ‘Mode of Action’ research team was also established to discover the molecular targets of drug candidates that were at an advanced stage of development.4, 6 Success of the programme is evidenced by the discovery and development of two clinical drug candidates for VL, each with different mechanisms of action The first candidate arose from a compound series previously discovered in
References
Dundee that inhibited growth of the related Trypanosoma brucei. Through clever iterative changes to the chemical core scaffold, the Dundee team were able to produce a compound active against the Leishmania parasite The work, published in 2018, also identified the target of the compound as cyclin-dependent related kinase 12 (CRK12), providing Pharma with a completely novel validated drug target for Leishmania 4 Discovery of the second clinical drug candidate also began with compounds discovered in Dundee to be active against a different related parasite, Trypanosoma cruzi, which causes Chagas disease Repurposing and optimisation in Dundee 5, 6 produced a compound with potent activity against Leishmania that was shown to act through inhibition of the proteasome.6
An important impact of Dundee’s research has been adoption by industry of a suite of screening and assay platforms developed by the DDU for VL drug discovery These were transferred to GSK (following training of GSK staff in Dundee) and are now widely used by industry and Product Development Partnerships In total, DNDi and GSK have screened >500,000 and 1 8 million compounds respectively using DDU assays, a feat previously impossible with earlier, laborious assays. The Mode of Action group screening cascades also provided DNDi, GlaxoSmithKline and others with knowledge that saved resources by enabling decisions not to proceed with compounds that might have negative outcomes. Overall, the DDU’s drug discovery expertise has enhanced the productivity of the global VL drug discovery portfolio DNDi comment on DDU’s gold standard screening assays thus:
“…continued development of high capacity, physiologically relevant assays using Leishmania by the Dundee DDU has been transformational for visceral leishmaniasis (VL) drug discovery. The high throughput [parasite-killing] assay and high content imaging platform for intracellular assays have revolutionized the capacity and efficiency of screening, enabling many millions of compounds to be screened for new chemical starting points… The Dundee L.donvani screening platform allowed rapid identification of hit series of molecules for both Dundee and other groups around the world, including major Pharma companies ”
Dr Charles Mowbary, Discovery Director, DNDi.
1 The Anti-Trypanosome Drug Fexinidazole Shows Potential for Treating Visceral Leishmaniasis Wyllie, S, Patterson, S, Stojanovski, L, Simeons, FRC, Norval, S, Kime, R, Read, KD & Fairlamb, AH (2012), Science Translational Medicine, vol 4, no 119, pp119re1
2 Comparison of a high- throughput high-content intracellular Leishmania donovani assay with an axenic amastigote assay De Rycker, M, Hallyburton, I, Thomas, J, Campbell, L, Wyllie, S, Joshi, D, Cameron, S, Gilbert, IH, Wyatt, PG, Frearson, JA, Fairlamb, AH & Gray, DW (2013), Antimicrobial Agents and Chemotherapy, vol 57, no 7, pp 2913-2922
3 Development and validation of a novel Leishmania donovani screening cascade for high-throughput screening using a novel axenic assay with high predictivity of Leishmanicidal intracellular activity. Nühs, A, De Rycker, M, Manthri, S, Comer, E, Scherer, CA, Schreiber, SL, Ioset, J-R & Gray, DW (2015), PLoS Neglected Tropical Diseases, vol 9, no 9, e0004094
4 Cyclin-dependent kinase 12 is a drug target for visceral leishmaniasis Wyllie, S, Thomas, M, Patterson, S, et al (2018), Nature, vol 560, no 7717, pp 192-197
5 'Identification of GSK3186899/DDD853651 as a Preclinical Development Candidate for the Treatment of Visceral Leishmaniasis Thomas, MG, De Rycker, M, Ajakane, M, et al (2019), Journal of Medicinal Chemistry, vol 62, no 3, pp 1180-1202
6 Preclinical candidate for the treatment of visceral leishmaniasis that acts through proteasome inhibition Wyllie, S, Brand, S, Thomas, M, De Rycker, M et al (2019), Proceedings of the National Academy of Sciences, vol 116, no 19, 201820175, pp 9318-9323
Wellcome Centre for Anti-Infectives Research Six Year Report I page 17
Cryptosporidiosis

Cryptosporidiosis remains a threat to the lives of young, malnourished children and immunocompromised people around the world There are currently no vaccines or effective treatments for these patients for cryptosporidiosis Using genetics, proteomics-based techniques, and new culture methods, we are uncovering more about the biology of this important parasite at WCAIR.
Cryptosporidium is a fecal-oral pathogen, spread through contaminated food and water. Its robust shell, called the oocyst wall, protects the parasite from environmental factors including common water treatments like chlorination. Recently, the Pawlowic Lab described the proteome of the oocyst wall Excitingly, this list included for the first time all 9 members of a family of proteins called the Cryptosporidium Oocyst Wall Proteins (COWPs) COWPs are unusual in that every 10–12 amino acid residues is a cysteine. This is fascinating because cysteines can form very strong chemical bonds We think that these bonds could be the critical ingredient that strengthens the parasite’s shell We are using CRISPR/Cas9 to determine which COWPs are required for parasite transmission, and if it is the COWPs cysteines that make the shells so strong.


The Pawlowic lab also works closely with the Mode of Action group, led by Dr Susan Wyllie, to adapt chemical proteomics approaches to identify the target of anti-cryptosporidial compounds We have successfully performed thermal proteome profiling in Cryptosporidium for the first time The data from our first experiments provides direct evidence that the Drug Discovery Unit’s lead compound is on-target. Now we are working to apply this approach to compounds whose target is unknown This is really exciting as identifying the target of a drug is very important to understanding how the drug works
We are also developing new tools for studying this parasite We have partnered with Dr Curtis Thorne’s lab the University of Arizona to culture fluorescent Cryptosporidium with human “enteroids”, aka miniature intestines Using microscopy we can create movies that will allow us to investigate the timings of the life cycle in more depth This will enhance our understanding of the basic biology of the parasite, drug mode of action, and improve our understanding of what happens to the intestinal tissue during infection
This year the Pawlowic Lab saw some milestones: 3 PhD students successfully completed their viva, the lead optimization project with the Drug Discovery Unit finished, and we submitted our first manuscript for publication
Our Drug Discovery Unit Apicomplexan Portfolio team, led by Dr Beatriz Baragaña, in collaboration with the Pawlowic lab, Prof. Chris Huston at University of Vermont and Eisai have developed two potent late lead molecules capable of clearing Cryptosporidium infections in mouse and calf models. The drug discovery project started with hit molecules with activity against a protein that is essential for the parasite survival. We optimized the potency and drug-like properties of these compounds using a structure-driven target-based approach
Wellcome Centre for Anti-Infectives Research Six Year Report I page 18
The left hand image is a mouse intestine (cell surfaces stained white) infected with Cryptosporidium parasites (appear cyan) (Dr Emma Sands); Middle image shows Cryptosporidium oocysts labelled to show the proteins of the oocyst wall in magenta (Dr Ross Bacchetti); right hand picture shows a cross-section of a mouse small intestine infected with Cryptosporidium parasites (Dr Ross Bacchetti). The surface of the intestinal cells are stained in white and their DNA is stained in cyan, and the Cryptosporidium parasites appear magenta.
We are excited about our progress on this project because our advanced lead molecules can potentially be effective treatments for cryptosporidiosis in young children and immunocompromised patients.
Overcoming the challenges for Chagas disease drug discovery

Wellcome Centre for Anti-Infectives Research Six Year Report I page 19
>
Chagas disease is caused by the parasite Trypanosoma cruzi (T. cruzi ) and affects millions of people. The drug discovery pipeline for Chagas disease remains very sparse due to the challenges associated with developing new drugs for this disease As Chagas disease is a chronic disease caused by a very low parasite burden we are aiming to develop compounds that can eradicate all parasites from an infected person Identifying such compounds is a major challenge To improve our chances of success in this area we are developing multiple new approaches and technologies in WCAIR.
1 Hit-discovery screening cascade
Our cascade is designed to quickly identify the most promising compounds for entry into lead-optimisation The process starts with a high-throughput screen to test hundreds of thousands of compounds for their ability to kill intracellular parasites, without affecting their host cells. Hits that act through an enzyme called CYP51 are then rapidly removed, as this mode of action has been invalidated through clinical studies. The remaining hits are profiled in further mode of action assays to identify compounds that act through known targets The rate-of- kill assay developed in WCAIR is then used to determine how fast compounds can kill the parasites This helps to prioritise compounds and design dosing regimens for animal models of disease Finally, the most promising compounds go through a two-month long washout assay to check they can kill every single T cruzi parasite1 We treat cells and parasites with compounds for up to 16 days. Then compounds are removed, and we observe the cells for up to two months to assure that no viable parasites emerge
To date this assay provides the best in vitro predictor of the ability for a compound series to achieve sterile cure in an animal efficacy model As such this assay is now a key gatekeeper before we move compounds into animal model studies
2 Combination treatment
We have built a combination screening platform and are testing combinations of compounds with different modes of action to assess if they can eradicate all parasites in vitro and in vivo Our findings are encouraging and indicate that some combinations are indeed able to clear all parasites rapidly, even when each compound alone cannot achieve this. This opens the door for the development of short course treatments that would be transformative for patients

Interestingly we are also finding combinations that are less good at clearing the parasites compared to the individual compounds Why we see this apparent antagonism with some combinations remains unknown
References
3 Expression of multi-subunit targets for structure-guided drug discovery
Purification of large protein complexes from parasites for drug screening efforts (e.g. the proteasome) is a major challenge, due to long timelines and bio-safety concerns To overcome these issues, we have developed the technology to express and purify parasite proteasomes, allowing us to carry out high-throughput screening campaigns and identify new inhibitors Additionally, we have also generated various mutant proteasomes that provide detailed information on the mode of inhibition of new compounds. Furthermore, we are implementing cryo-EM technology at the University of Dundee to facilitate structure-guided drug discovery programmes on large protein complexes
4 More physiologically relevant host cell models
T cruzi parasites live inside a wide range of mammalian host cells, and little is known about the impact of host cell type on drug susceptibility, in particular in the context of physiologically relevant cell models In collaboration with the University of Dundee Stem Cell Facility we have generated human host cells relevant to T. cruzi infection and are characterizing the behaviour of the parasites in these host cells Our main aim is to explore any host-cell dependent differences in compound susceptibility and the underlying reasons for this
5 New modalities
Targeted protein degradation approaches are currently being explored extensively for the treatment of non-communicable diseases such as cancer and neurodegeneration We are seeking to apply these approaches, using modalities such as PROTACs and molecular glues, to protozoan parasites
Based on work carried out in Prof. Mark Field’s group, we have identified candidate E3 ligases and are assessing their tractability for targeted protein degradation approaches in kinetoplastid parasites
1 Development of Trypanosoma cruzi in vitro assays to identify compounds suitable for progression in Chagas’ disease drug discovery MacLean LM, Thomas J, Lewis MD, Cotillo I, Gray DW, De Rycker M (2018) PLoS Negl Trop Dis 12(7): e0006612
Wellcome Centre for Anti-Infectives Research Six Year Report I page 20
Innovation in Chemistry
Chemistry is fundamental to the drug discovery process. In WCAIR we have been looking at ways in which we can increase the efficiencies of our chemistry processes and the integration of chemistry with other disciplines to improve the design-make-test-analyse cycle.
The Synthetic Methodologies team has the remit of developing and delivering new chemical methods and technologies to the in-house medicinal chemistry teams The team has several project areas of current interest: increasing the rate of compound synthesis; increasing the number of compounds that can be synthesised at one time; and expanding the range of chemical space that is readily available to project teams The technologies developed and validated by the team has allowed chemistry to be delivered to project teams that would otherwise have been difficult or impossible The approaches developed have increased the range of molecules available to synthetic chemists and allowed faster delivery of compounds to speed drug discovery efforts
Plate Based Chemistry
To speed up chemistry synthesis and screening we have invested heavily in the development and introduction of automated plate-based synthesis methodology This approach can be carried out in 96- or 384-well-plates, allowing large numbers of analogues to be prepared in a format that allows transfer directly into screening assays It makes extensive use of computational chemistry in the design process, to generate many virtual compounds which can be filtered and selected by desired properties and then synthesis can begin We make extensive use of robotics to automate the processes as much as possible Another advantage is that it can be carried out on a very small scale, typically of the order of 0.1 mg per compound which is two hundred times less than traditional methods used within the DDU The DDU Compound Management team have been heavily involved in delivery of the project using their knowledge of reagent, compound and plate tracking and the use of automated liquid dispensing which is pivotal to the success of the technique
Late Stage Functionalisation and Diversification
We have been identifying and validating a dedicated set of reactions which allow functionalisation of elaborated molecules without extensive synthesis. This is useful to allow fast optimisation of late-stage molecules reducing the timelines to optimise compounds of interest
Flow chemistry
We have developed a fully capable flow chemistry set up that has allowed the synthesis and use of unstable intermediates and the safe generation and use of carbon monoxide. The system has also been upgraded and used for electrochemistry and light driven chemistry, increasing access to chemical space using these reagents and techniques
Biotransformations
We are investigating the use of biotransformations of molecules using enzymes. Some of this work is being carried out in collaboration with a small company called Hypha Discovery Ltd We are investigating which approaches give us access to molecules which are not easily made by conventional organic chemistry.
Route Development and Scale Up
The route development and scale up section of the team has two main responsibility areas. The first is to work with project teams and take chemistry off the critical path where a particular route or transformation is slowing the synthesis of final compounds. The second role is in developing and executing scale-up routes to allow the safe and efficient delivery of intermediates and final compound to project teams The team are regularly engaged in solving issues faced by project teams, synthesising useful intermediates and delivering final material at extremely high purity for in vivo testing.
Covalent Inhibitors
We have started to look at covalent inhibitors as a potential modality We have developed some mass spectrometry assays to identify residues which may be suitable for covalent modification and set up an NMR assay to determine the relative reactivity of different warheads.

New Chemical Space: screening Natural Product Libraries
Current phenotypic screening against the parasites often finds new series which inhibit the same targets as other chemical series. It is relatively rare that a novel drug target is revealed by a phenotypic screen, with the current small molecule synthetic libraries With a view to discovering potential new drug targets, we have started screening collections of natural products, accessed through collaborators. Hits are being fed back to our Mode of Action group to carry out target deconvolution
Wellcome Centre for Anti-Infectives Research Six Year Report I page 21
Enhancing target-based screening High throughput protein production and X-ray crystallography
A target-based approach to drug discovery requires the ability to gain structural information by ‘visualising’ fragments and compounds bound to the target of interest
A high throughput methodology to produce the structural data allows this information to be gathered in just a few hours The WCAIR Molecular Interactions Team have worked with scientists at the Diamond Light Source, Harwell to adapt their XChem technology to the needs of the WCAIR screening programmes and establish it in Dundee

The drivers for XChem are to work on a very small scale, carry out as many tasks as possible in parallel, use computers to automatically do all the note-taking, and use robotics to do most (but not quite all) of the intricate work Crystals are grown in droplets of liquid of just a few hundred nanolitres, into which a specialist acoustic dispenser then deposits a few nanolitres of the chemical fragment being tested They are then left for a several hours to allow the fragment to soak into the crystal If, and only if, the fragment binds specifically to a site on the protein will we be able to see it in the final structures via X-ray crystallography
Getting crystals ready for an X-ray diffraction experiment is the one part of the procedure that robotics cannot perform, and we have to rely on the skills of our protein crystallographers to do this. However, we have in place the ‘shifter’, a piece of equipment that makes this easier and significantly faster than standard manual harvesting. Various software tools are then used to process the diffraction data collected, easing some of the load on the crystallographers as they search through the numerous datasets for the successful fragments Decoding the X-ray patterns to show the fragment bound to the target requires the specialist knowledge of the WCAIR team of computational chemists
XChem relies on being able to make and crystallise the target protein in a form that replicates the natural shape of the protein in the cell. In parallel with setting up the XChem screen, WCAIR has developed high throughput protein production methods. The methods and technologies under development allow us to test different ways of making the protein in parallel with the aim of finding a suitable construct as efficiently as possible. The best method is then scaled up to produce enough protein for the XChem screen and potentially other biophysical screening methods
The high throughput protein production methods implemented have been crucial in a number of DDU projects allowing production of the target protein when conventional methods had failed.
For example, by producing 12 chimeric proteins for a malaria target the team went from gene to high resolution crystal structure in 9 weeks, after previous failures by another group to produce a protein which would crystalise It also allows rapid and easy manipulation of a target protein to explore crucial residues in the binding site of the compound and explore potential resistance. This has been applied to study the trypanosome proteasome as a drug target for visceral leishmaniasis and Chagas disease
The results of XChem are just the starting point The structures guide the chemists, both computational and medicinal, in how to build up the fragments into larger molecules with more interactions with the target protein The detailed molecular interactions data generated by crystallography and other biophysical techniques empower the computational chemists to design more efficient drugs and new machine learning based software tools to improve their modelling. Combined with the direct to biology high throughput chemistry approaches (described in previous section) the drug discovery project team are able to move around the design-make-test cycle more rapidly and efficiently as they progress from hits to leads to candidate drugs.
Wellcome Centre for Anti-Infectives Research Six Year Report I page 22
Innovation within Drug Metabolism and Pharmacokinetics
8HUM – A
new mouse model to improve compound selection in drug discovery
Demonstration of in vivo efficacy in a mouse model of disease is a key milestone in anti-infective early drug discovery programmes Although an effective criterion by which to select the most promising preclinical candidates, differences in pathways of drug metabolism between mouse and human frequently result in medicinal chemistry resource being given to ensuring that mouse-specific metabolic liabilities are eliminated, despite these having no bearing on compound progress towards the clinic
In 2019, University of Dundee researchers introduced the “8HUM” mouse model, a line in which 33 murine cytochrome P450 (CYP) enzymes from the major xenobiotic metabolising subfamilies were replaced with 6 human enzymes, CYP1A1, CYP1A2, CYP2C9, CYP2D6, CYP3A4 and CYP3A7, and in which the transcription factors Pregnane X Receptor (Pxr) and Constitutive Androstane Receptor (Car), key regulators of CYP induction in response to xenobiotic challenge, were replaced with their human orthologues 1 This extensive genetic humanisation was shown to remove many species differences in metabolism, with the added benefit that drug exposure was typically greatly increased. Since 2020, we have been evaluating 8HUM within our anti-infective drug discovery workflows as a platform technology for the bypass of mouse-specific issues of compound metabolism. In vitro, in incubations with hepatic microsomes or primary hepatocytes from 8HUM, we have found that species differences in intrinsic metabolic clearance between mouse and human are removed for the vast majority of new chemical entities. Further, the course of infection of 8HUM with T. cruzi and L donovani matches that of the wild-type mouse strains used routinely in Dundee and, as shown by collaborators at GSK Tres Cantos, this is also the case in an acute model of M. tuberculosis infection. Combining these observations in an exemplification of the value of 8HUM within kinetoplastid drug discovery we have shown that mouse-specific metabolic liability is no longer a barrier to compound progression
(manuscript in preparation). Furthermore, in a project funded by the Tres Cantos Open Lab Foundation we have shown that, in the 8HUM mouse line, the pharmacokinetics, metabolite profiles, and magnitude of drug-drug interactions for a test set of 30 approved medicines are in much better alignment with clinical observations than equivalent data generated in wild-type mice Collectively, our results with the 8HUM line have demonstrated that the elimination of species differences in drug metabolism through the application of 8HUM, in place of wild-type mice within a drug discovery programme, has the potential to significantly increase the proportion of compounds that can be considered for progression during lead optimisation, increase speed of workflow, as well improve translational relevance of the data generated We are currently working with collaborators to validate 8HUM in additional models of infectious disease
Metabolomics

Measurement of changes in the abundance of endogenous small molecules within a biological sample in response to chemical challenge can indicate dysregulation of specific metabolic pathways. As has been demonstrated for the Medicines for Malaria Venture Malaria Box compound set 2 , hits from a screening campaign can be profiled for their effect on the endogenous metabolome and thereby classified into groups which are likely to be acting through common Modes of Action (MoA) In collaboration with the MoA team in WCAIR we are developing a medium-throughout LCMS-based metabolomics capability at the DDU which will allow us to define the metabolic fingerprint of 20–50 hit compounds in 24 hours, and thereby expedite prioritisation of those compounds with novel or preferential MoA for hit expansion To date, we have added chromatographic retention time and fragmentation spectra for approximately 400 authentic reference standards of endogenous metabolites to an in-house database This database will be used to give high confidence in metabolite annotations during sample analysis. Initially our validation work will focus on hits from screens against T cruzi but, if successful, the platform will be rolled out to other projects
1 An Extensively Humanized Mouse Model to Predict Pathways of Drug Disposition and Drug/Drug Interactions, and to Facilitate Design of Clinical Trials Henderson CJ,
Wellcome Centre for Anti-Infectives Research Six Year Report I page 23
References
J,
LA,
Drug
Dispos
Jun;47(6):601-615
EL, Painter HJ, Samra J, Carrasquilla M, Llinás M Antimicrob.
Kapelyukh Y, Scheer N, Rode A, McLaren AW, MacLeod AK, Lin D, Wright
Stanley
Wolf CR
Metab
2019
2 Metabolomic Profiling of the Malaria Box Reveals Antimalarial Target Pathways Allman
Agents Chemother 2016 Oct 21;60(11):6635-6649
A journey in parasite glycobiology and scientific infrastructure: 35 years of the Ferguson Lab in Dundee (1988–2023)

Mike is not retiring, but he is closing his research laboratory this summer. Here he reflects on what his lab has achieved, and some of the contributions he has made to the scientific infrastructure that supports WCAIR and Life Sciences in general at the University of Dundee.
The science
Recruited by Philip Cohen, Mike Ferguson arrived in Dundee from Oxford, along with Steve Homans, in 1988 Steve, a superb NMR expert, Mike and many collaborators from around the world spent several years “structure busting” – determining the primary and 3-dimensional structures of the carbohydrate components of the glycoproteins and glycolipids that dominate parasite surfaces – such as the variant surface glycoproteins (VSGs) and procyclins of T brucei, surface mucins of T cruzi and the lipophosphoglycans (LPGs) and proteophosphoglycans (PPGs) of leishmania Mike’s lab has continued this function up to the present day, developing along the way many analytic methodologies. Arguably, their heyday for structure-busting was the late 80s and 90s when they defined the surface molecular architectures of various parasite life cycle stages This work has provided a framework with which to understand parasite survival, lifestyles and tropisms in the host and vector and “images” of these parasite cell surfaces are now taken for granted.

This period was also marked by determining the structures of glycosylphosphatidylinositol (GPI) anchors from across eukaryotic evolution – defining the conserved and species/tissue/life cycle stage-specific structural features of the GPI family of molecules.
Structures are a prerequisite for studying biosynthesis, and understanding biosynthesis is a prerequisite for exploiting parasite biology for therapeutic gain Mike’s lab contributed to the detail of GPI anchor biosynthesis in African trypanosomes, leishmania and fungal pathogens and, using radiolabelling of whole cells and cell-free systems, demonstrated exploitable differences between parasite and human GPI pathways and provided genetic and chemical validation of their drug target potential
Wellcome Centre for Anti-Infectives Research Six Year Report I page 24
P h o t o s b y S o p h i e G e r r a r d
Probing the substrate and inhibitor specificities of the enzymes of the GPI pathway required access to GPI sub-structures and analogues thereof One of the challenges of working on a newly discovered pathway is that there are no reagents – let alone kits. In collaboration with Prof John Brimacombe, a superb synthetic carbohydrate chemist in our then Chemistry Department, Mike and colleagues set about synthesising unique GPI molecules and, with Andrei Nikolaev, expanded this into leishmania LPG sub-structures This era of chemical biology (much of it performed before that term was coined) was extremely productive and set this interdisciplinary team apart in the field.
Eventually, Mike’s team turned to protein N-glycosylation in trypanosomes Initial studies on the N-glycans of T brucei VSGs had suggested these were not so different to host N-glycans. However, they became interested in the “gunk” that fills the flagellar pocket of bloodstream form T brucei and found that it is made predominantly of unique giant poly-N-acetyllactosamine (PNAL) containing N-glycans With interest piqued, his team investigated N-glycan biosynthesis in T brucei and discovered that, uniquely, it has two N-glycosylation systems running in parallel – one to add complex N-glycans to glycosylation sites in acidic environments on the glycoprotein surface and another to add oligomannose structures to the remaining sites Using machine learning, they built an algorithm to predict the kinds of N-glycan present on every putative glycosylation site in the organism The unusual oligosaccharyltranferases of trypanosomes are currently being exploited in biotechnology to increase the beneficial glycosylation of recombinant glycoproteins
As with all research, one thing leads to another and one of the perplexing issues implicit in the GPI and N-glycan structures Mike’s team had determined in trypanosomes was which glycosyltransferases (GTs) were doing what?


The parasite genome is devoid of several canonical GT families and replete with members of a trypanosomatidunique one: family GT67 They therefore undertook a painstaking mapping of GT gene to GT function by constructing GT mutants and determining the biochemical lesions in GPI and/or N-glycan structures in bloodstream and procyclic form parasites. This required the development of sophisticated analytical strategies The results were worth it, and we now understand the GT67 family and its sub-families and what they do, and in the process provided the first evidence for convergent evolution in GT enzymes
One recent foray into a non-GT67 GT in T brucei (and in collaboration with Steve Beverley in L major) was to study a GT11 fucosyltransferase (FUT1) This turns out to be located in, and essential for, the parasite mitochondrion This is a radical discovery and much remains to be uncovered about mitochondrial glycosylation in general and its therapeutic potential for kinetoplastid parasites. All protein and lipid glycosylation requires nucleotide sugars as the direct or indirect donors of glycosylation reactions, making them key essential metabolites Mike’s team originally took an analytical approach by developing an LC-MS/MS method to detect and quantify nucleotide sugars (an approach later modified for mRNA cap structures) This showed GDP-fucose in all trypanosomatids, which led them to discover the trypanosomatid mitochondrial FUT1s, and stimulated them to define the biotransformations in T. brucei that make UDP-glucose, UDP-galactose, UDP-N-acetylglucosamine, GDP-mannose and GDP-fucose. Over several years, they cloned, expressed, characterised, crystallised and localised most of the thirteen enzymes involved and, crucially, discovered that all of them are localised to the glycosomes in bloodstream and procyclic form parasites This led them to discover and characterise an entirely new class of nucleotide sugar transporter, necessary to move UDP-sugars made inside the glycosomes into the cytoplasm
Wellcome Centre for Anti-Infectives Research Six Year Report I page 25
>
Throughout, Mike’s group have been heavy users of mass spectrometry, and this led Mike to set up and develop, with other colleagues like Angus Lamond and Doreen Cantrell, the School of Life Sciences proteomics facility. Open to all, it is heavily and productively used by many groups – not least Susan Wyllie’s superb mode-of-action team who have also added equipment to it. Mike’s team have contributed several fundamental proteomic data sets to the parasitology community including the first bloodstream/procyclic form proteome and phosphoproteome comparisons and comprehensive proteincomplex, cell-cycle, protein turnover and glycoproteome data.
award from Wellcome, and by making key initial appointments of Julie Frearson and Ian Gilbert and then subsequently Paul Wyatt. Kevin Read was recruited shortly thereafter and David Gray succeeded Julie Frearson in 2010 The DDU team has been enormously successful over the years and is now 130-strong with a turnover in excess of £12m pa, five drugs in clinical trials and nine assets licenced to BioPharma companies The translational capabilities of the DDU were also central to the establishment of the Wellcome Centre for Anti-Infectives Research in 2017, one of only fifteen Wellcome Centres
The burgeoning success of the DDU meant that it was outgrowing its laboratories and, in 2009, Mike initiated the fund raising for, and the design of, the Discovery Centre for Translational and Interdisciplinary Research. This £25m development opened in 2014 and allowed the DDU to double in size It also provided purpose-built accommodation for our proteomics facility (one of the largest in the world).
His longest collaborator has been Lucia Güther, also his life-partner, who has played such a key role in performing original research and organising and coordinating the lab No amount of thanks are enough! Other key contributors, collaborators and fellows in the lab over the years include Angela Mehlert, Malcom McConville, Wayne Masterton, Jane Thomas-Oates, Sylvain Cottaz, Igor Almeida, Alvaro Acosta-Serrano, Terry Smith, Art Crossman, Abdel Atrih, Francoise Routier, Luis Izquierdo, Karina Marino, Mick Urbaniak, Sabine Kuettel, Manuela and Seb Damerow, Sam Duncan, Rupa Nagar and Michele Tinti. Huge thanks to all, and to Dougie Lamont and his team for managing the proteomics facility so brilliantly
Scientific infrastructure and recruitment
Mike was the first molecular parasitologist to arrive in Dundee. He was able to convince Alan Fairlamb to move here in 1995 from the London School of Hygiene and Tropical Medicine, as well as David Horn from the same place and Mark Field from Cambridge in 2013 and Mattie Pawlowic from the U S in 2017 Alan’s move, as well as that that of Angus Lamond, was instrumental in Philip Cohen convincing the Wellcome Trust to invest £10 million into the Wellcome Trust Building which opened in 1997. Alan also attracted Bill Hunter to bring X-ray crystallography to Dundee from Manchester, and Alan, Mike and Bill shared a vision for advancing drug discovery for neglected tropical diseases


The next phase required more building – Mike and Philip Cohen jointly led the campaign to fund and build the Sir James Black Centre, which opened in 2006 This included the first Drug Discovery Unit (DDU) laboratories and Mike and Alan initiated drug discovery the same year with an extraordinary strategic
Since 2017, Mike and Morag Martin have been involved with the Tay Cities Deal (TCD), shaping and justifying the £25m TCD investment in Growing the Tay Cities BioMedical Cluster, raising additional funds from the Garfield Western and Wolfson Foundations and making the case for University of Dundee strategic investment. Construction of the Life Sciences Innovation Hub began on the Dundee Technopole in March 2023 and will complete in 2024 It will provide laboratory and office space for spin-out companies, reversing the current trend for the University’s highly-invested companies to move elsewhere – thus retaining jobs and opportunities in this economically depressed region Together with Alessio Ciulli’s Centre for Targeted Protein Degradation, this is the first step towards developing a wider Life Sciences Innovation District for the prosperity of the region

Summing up
In summarising his career so far, Mike said: “It has been a privilege to work with so many talented scientists and to collectively add something to the canon of molecular parasitology and eukaryotic biology. It has also been a pleasure, if somewhat time consuming, to help provide facilities and infrastructure for the next generation of scientists to thrive. Dundee has been a fantastic home and work environment for me, and I hope it is and will be for many others for many years to come.”
Wellcome Centre for Anti-Infectives Research Six Year Report I page 26
Altogether, Mike’s lab in Dundee has published 233 peer-reviewed articles, graduated 28 PhD students and trained over 50 post-doctoral scientists.
Photos by UoD creative Services
From L to R: Lucia Güther, Dougie Lamont, Michele Tinti
Focus on anti-infective oxaboroles Case Study Adoption of a new rapid diagnostic test for Human African Trypanosomiasis
Human African Trypanosomiasis (HAT), or sleeping sickness, is caused by subspecies of the parasite Trypanosoma brucei transmitted by infected tsetse flies. It is generally fatal if untreated. Although the prevalence of HAT decreased dramatically over the past 20 years due to strict control measures, history has shown that it can quickly re-establish to epidemic proportions if surveillance is relaxed It is endemic in 36 sub-Saharan African countries with 57 million people at risk
The WHO has targeted HAT for elimination of community transmission by 2030. Accurate testing to identify infected individuals is critical to enable timely treatment and for active surveillance to identify re-emergence in areas of previous disease elimination When Ferguson’s team began the research, HAT screening used the relatively primitive CATT (Card Agglutination Test for Trypanosomiasis) test where trypanosomes fixed to a card are used to test patient’s serum for the presence of antibodies. The CATT needs a refrigerated ‘cold-chain’ during distribution, and trained personnel with electrical lab equipment for patient testing It has severe limitations for large-scale use for community screening in remote locations
Prof Sir Mike Ferguson recognised that new approaches were needed In research published in 2013 1 , he identified improved diagnostic antigens and a more suitable platform for inexpensive field-based testing. Taking an unbiased approach, he used antibodies from the blood of HAT patients to probe a mixture of trypanosome proteins and see which proteins they bound to This identified a trypanosome surface protein, the Invariant Surface Glycoprotein ISG65, as the best diagnostic antigen Mike obtained experimental tools from Prof Mark Carrington at the University of Cambridge and subsequently produced ISG65 in an engineered bacterium (recombinant ISG65) ensuring unlimited future supply without the need to grow parasites To progress this research towards a usable diagnostic, Mike and colleagues developed a prototype lateral flow Rapid Diagnostic Test (RDT) in collaboration with Dundee-based immunoassay development and manufacturing company BBI Solutions. The performance of this recombinant antigen prototype RDT 1 , and a subsequent version incorporating a second native antigen also discovered in the Ferguson lab2 , fulfilled the desired characteristics of a priority
References
diagnostic test for FIND, the Foundation for Innovative New Diagnostics, a global not-for-profit product development partnership accelerating the development of diagnostic tests for poverty-related diseases
Prior to Mike’s involvement, the first-generation RTD commissioned by FIND for HAT used native antigens purified from live parasites This has several disadvantages; few laboratories have the specialized facilities and expertise needed to grow the parasites and purify the antigens, and batch-to-batch quality issues can interrupt production of tests WHO and FIND therefore required second-generation RTDs to be developed using recombinant antigens 1 At Mike’s suggestion, FIND commissioned an independent (blinded) three-way side-by- side comparison of the existing dual native antigen test (SD BIOLINE HAT), a dual recombinant prototype of the same antigens, and the Dundee dual antigen prototype. Statistical analysis of the data showed that one of the two best recombinant diagnostic antigens was Dundee’s ISG65 3 All of Dundee’s knowhow and reagents were transferred free of charge to FIND who contracted Abbott (Standard Diagnostics Inc, South Korea, hereafter SD Inc) to manufacture a secondgeneration RDT (BIOLINE HAT 2.0) for front-line use in Africa with recombinant ISG65 as one of its two diagnostic antigens The new kit is cheap, can be stored at room temperature, is easy to use even in remote resource-poor settings and will give rapid results in 15 minutes from a pin-prick of blood

1 Proteomic selection of immunodiagnostic antigens for human African trypanosomiasis and generation of a prototype lateral flow immunodiagnostic device
Sullivan, L, Wall, SJ, Carrington, M & Ferguson, MAJ (2013), PLoS Neglected Tropical Diseases, vol 7, no 2, e2087
2 Identification of sVSG117 as an immunodiagnostic antigen and evaluation of a dual-antigen lateral flow test for the diagnosis of human african trypanosomiasis
Sullivan, L, Fleming, J, Sastry, L, Mehlert, A, Wall, SJ & Ferguson, MAJ (2014), PLoS Neglected Tropical Diseases, vol 8, no 7, pp e2976
3 Evaluation of the diagnostic accuracy of prototype rapid tests for human African trypanosomiasis Sternberg, JM, Gierlinski, M, Biéler, S, Ferguson, MAJ & Ndung u, JM (2014), PLoS Neglected Tropical Diseases, vol 8, no 12, pp e3373
Wellcome Centre for Anti-Infectives Research Six Year Report I page 27
“FIND is grateful for Professor Ferguson’s continued engagement and assistance in technology transfer to Standard Diagnostics and Abbott… we acknowledge the University of Dundee's significant research contributions to achieving FIND’s goals in supporting the elimination of HAT.” FIND Senior Project Manager.
r a i n i n g
The aim of the WCAIR training programme is to support capacity building for drug discovery in the low- and middle-income countries most affected by the diseases we research. Training has been split into three main categories – training in Dundee, short courses and online content, and partnerships.
WCAIR Trainees in Dundee
Since January 2018, we have delivered 279 months of training in Chemistry, Biology, DMPK and Parasitology This training has been delivered to 38 trainees, from 15 different countries

Each programme is designed to suit the individual trainee, based on their pre-existing skillset and their research. While the trainees have access to our labs and associated equipment, we are mindful that these are unlikely to be available at their home institute Great care is taken to train them with techniques that will be applicable in their own institution and skills that they can pass onto colleagues
TWe ensure that they have as rounded an experience as possible with the opportunity to learn about all aspects of drug discovery research This is achieved through group seminars, access to our undergraduate modules in drug discovery and access to the seminars and lectures available to all WCAIR staff

To maintain links and help the trainees implement their new skills when they return to their home institutions, they can apply for small project grants (up to £30K) to continue collaborating with WCAIR

Short Courses
In collaboration with the Wellcome Connecting Science team, we have run the ‘Practical Aspects in Small Molecule Drug Discovery’ for over a decade. In 2019 we took the course to University of Cape Town where it was open to scientists from the African continent only Funded places removed financial barriers to attendance, allowing 26 participants from 19 institutes from 11 different countries to attend. After a long delay due to travel restrictions, we ran the course again in South America in November 2022 with support from the Institut Pasteur de Montevideo, Uruguay 24 participants from 9 different countries attended that course from South and Central America



Wellcome Centre for Anti-Infectives Research Six Year Report I page 28
The WCAIR Training Team: Suze Farrell (Training manager), Lauren Webster (Scientific and Pedagogy Lead), Cedric Graebin (Medical Chemistry), Nicole Mutter (DMPK)
WCAIR Training team and trainees July 2019
Uruguay 2022
The WCAIR training team have designed a number of new short courses. Making use of the improved online webinar and collaborative tools developed through the COVID pandemic, these have been created to be delivered online, thus reducing financial barriers for researchers from disease endemic countries to participate The courses cater to a variety of needs; some provide an overview of drug discovery, while others focus on a specific topic. For each course, we set up a Slack group so that participants can continue to discuss content and ask us questions, also allowing them to collaborate with each other Online programmes are all recorded so that individuals can rewatch the content The full list of courses can be found at https://wcair.dundee.ac.uk/training/short-courses/ They will all be running in 2023 Examples include:
Target to Candidate: An Introduction to Drug Discovery is designed to give scientists a flavour of how a drug project moves from idea to pre-clinical candidacy It highlights how to start and progress a drug discovery campaign with real life examples from experts working in this area of science, both academia and industry. This programme has run twice with collaborators in South Africa and Brazil, both online due to travel restrictions

Medicinal Chemistry 101 and DMPK 101 are online programmes, made up of 12 sessions, once a week for 1.5 hours. Designed for chemists and pharmacologists respectively, they each cover key concepts those working in these disciplines within drug discovery need to understand to be successful

Drug Discovery Mission has been delivered 4 times, both in person and online. It has been designed as an interactive tool to support the understanding of the processes involved in drug discovery It exposes participants to all aspects of the pre-clinical drug discovery process Regardless of discipline, scientists will be asked (as a team) to make critical analytical decisions and conclusions based on the data presented to them. The programme has been designed to support learning scientific content, but also to promote team working and highlight the multi-disciplinary nature of drug discovery



Online Content




We are aware it is not possible for everyone to attend training in Dundee, or even have the opportunity to attend a short course To cater for this group, we have developed some free resources. Content ranges from short written pieces on drug discovery in general, but also includes some short talks on topics such as medicinal chemistry and DMPK. There are helpful hints in the form of Padlets (a way of grouping useful free-to-access resources) and workshops on data management From a more practical perspective, we have included some ‘how to’ sections, showing people how to do specific assays and a series of videos showing chemistry experimental set ups. We have also developed short animations to help people understand the concepts of absorption and metabolism All online content is available for free at https://wcair.dundee.ac.uk/training/training-resources/

Wellcome Centre for Anti-Infectives Research Six Year Report I page 29
Partnerships
Dialogue with our trainees has helped us understand where our support can make most of a difference in their home countries With this in mind we are focusing our resources at a few strategic partnerships.

Ghana. We are focusing our efforts on Ghana as there are several institutes working in drug discovery, and they have some infrastructure in place already Following visits in 2019, we are acting as a coordinator to bring interested institutes together to form a drug discovery hub. See ‘Focus on Ghana’.
Brazil. We have had scientific collaborations in Brazil for several years, so it is a natural choice within South and Central America Much of the support that Brazil needs is in terms of training rather than infrastructure A Newton Fund (MRC-FAPESP) award allowed us to work with 3–4 partner institutes in São Paulo state and deliver a training course to their staff (Target to Candidate: An Introduction to Drug Discovery)
Focus on Ghana

Building on visits in 2019, WCAIR partnered with several academics in Ghana to start developing a joint research portfolio These were small pots of money from the University of Dundee Global Challenges Research Fund (GCRF) There were two projects, one on anti-malarial chemistry, and the other a project on Cryptospordium parvum, both of which were with the University of Ghana Building on these awards, the scientists we worked with wanted to bring everyone working in drug discovery together Funding from the Academy of Medical Sciences allowed a network to be developed. We now support several working groups who each have their own agendas to take forward e g the analytical chemistry group would like to develop training for technical staff as well as establish structural elucidation support. As well as individual scientific goals, the groups want to be able to further the understanding of the entire drug discovery process within the academic institutes of Ghana
The next step was to help support the relevant parties to develop a working drug discovery network in country There were groups working on parts of drug discovery but no coordinated work We supported the University of Ghana and KNUST (Kwame Nkrumah University of Science and Technology) with an application to the Bill & Melinda Gates Foundation This bid was successful, and the award began in February 2022
We have recently delivered a Drug Discovery Mission programme to a group of 10 chemists with support from University of São Paulo, Ribeirão Preto campus Malawi. There are strong links between Scotland and Malawi at a governmental level coordinated by the Scotland-Malawi Partnership. Wellcome also invest in research through the Malawi-Liverpool-Wellcome Trust Clinical Research Programme and the Wellcome Centre for Integrative Parasitology, University of Glasgow Our partnership with Kamuzu University of Health Sciences, Blantyre compliments these existing projects We have delivered 2 weeks of intense practical training at the College in 2018 and 2019 A technician from the College worked with us for 6 months in Dundee, and he helped develop and deliver a training programme to his colleagues on plant extraction techniques This series of workshops was designed to be delivered to MSc students and we hope to develop more sets of workshops around the drug discovery theme.
Once travel restrictions allowed, we held a meeting in Ghana, which served as the final meeting of the Academy of Medical Sciences networking award, and the launch of the Ghana Drug Discovery Hub This meeting allowed all the partners of the Gates Foundation award to meet in Ghana and for some this was the first time they had been in Ghana to see the facilities
In the first year of the award, the team in Ghana have been working incredibly hard. They have synthesised over 100 new compounds, and these are now able to be tested in a malaria parasite assay in Ghana rather than being sent overseas. The next major milestone will be the establishment of a Drug Metabolism and Pharmacokinetics (DMPK) facility in KNUST They have one assay running but are waiting on delivery of a new LC-MS-MS unit for the remaining assays. Once this is in place, it will be the first time that a compound made in Ghana can be tested for both pharmacological parameters and in parasitic assays in country
A visit from the University of Dundee Principal in February 2023 is helping to further develop the research links between the University of Dundee and institutions in Ghana, embedding our work in a wider University strategy. The overall aim of our support to the groups in Ghana, is not only to further science within Ghana, but also within the region as they have strong research links with other academic groups in West Africa

Wellcome Centre for Anti-Infectives Research Six Year Report I page 30
H
From Left to right: Launching the Gates Foundation Award in Ghana, 2022; Celebrating the end of a NMR short course, Malawi 2019; Drug Discovery Mission, Brazil 2023
WCAIR Public Engagement at 6 >


4 key concepts:
> Community > Connectivity > Culture > Creativity
Community
We have worked in a close partnership with the Stobswell Forum since 2018 The relationship has been incredibly beneficial for both sides We were able to use our digital skills to keep the Forum functioning throughout COVID The Forum was, ultimately, the only one of Dundee’s community councils to have a weekly Zoom meeting.
As we became able to work in person again, we were able to tackle a new project that both the Forum and WCAIR were interested in We worked with Open Close Dundee, who pioneered our local street art trail, which Stobswell wished to add to. Following an open call, and extensive consultation with locals, scientists, and other colleagues across the University, we created a huge new piece with artist Lewi Quinn, transforming a blank façade on a local care home
The work has had a fantastic response throughout the city – indeed, it is visible from across parts of the University campus and even across the River Tay. The piece has cemented our place locally, and made a lasting mark on Dundee, and is featured in Dundee City Council’s next 10-year plan

Connectivity

A partnership with Girlguiding was originally established through the co-creation of the Medicine Maker Badge in 2019 In early 2020 we were a key stop on the Scotland-wide game ‘Wander the World’, which saw 700 Girlguides from across the nation join us in the School of Life Sciences

This connection continued throughout COVID In March of 2021 we ran an incredibly successful ‘virtual sleepover’, which saw audiences from the length of the UK and beyond. Over 1,000 people took part, and our resources saw over 6,000 downloads. The Facebook group was a marvellous way of engaging with audiences, with many hundreds of photos shared, and thousands of comments

Wellcome Centre for Anti-Infectives Research Six Year Report I page 32
The period from 2020–23 has seen huge changes in how we live and work. It has presented challenges, opportunities, and a chance to look at what we do afresh. While it has had difficult moments, refocusing on what really matters to us as a Centre has led us on to our biggest projects yet. We found that our core mission and refreshed strategy could be distilled down into
WCAIR Public Engagement Team: Ali Floyd (PE Manager) and Ailsa Mackintosh (PE officer)
Photo Ali Floyd
In the 2019 WCAIR symposium, one of our researchers proposed the idea of a physic garden on campus as a public engagement project The idea was very well received and the Centre board approved funding, to begin development in 2020, as a partnership with the University of Dundee Botanic Garden

Of course, this became a protracted process, but by mid-2021 the garden was indeed built and planted up It had evolved from a physic garden to a medicinal garden, and the long gestation had allowed a great deal of maturation in our ideas In particular, it allowed us to build new partnerships with organisations also interested in science and gardening This has so far included:
> A sensory garden at Dundee Science Centre

> A physic garden at Hospitalfield House in Arbroath
> Dundee Museum of Transport’s new garden area
We are also investigating other places, including the new ‘pocket parks’ in Stobswell, which are within sight of our new mural We are also working with HMS Unicorn, one of Dundee’s most iconic tourist attractions The garden has provided inspiration for a whole new range of activities to make science approachable to a wide variety of ages It has also allowed us to begin an Instagram, thanks to the visual nature of the project, again reaching new audiences.
Culture
The Wellcome Centre is part of both the School of Life Sciences and the wider University of Dundee community. As such, we are in a position to create a great deal of good as part of our own internal culture.
One key area where we have excelled is in training We have long been a key part of the School of Life Sciences internal training schedule, but in the last 2 years that has expanded The University’s Organisational and Professional Development department asked us to share the training we developed with the entire community. This has now reached around 100 people, who have taken inspiration in areas such as developing new activities, making short stories for YouTube, and even developing games to bring research to life
At the same time, we are also an active part of the University’s wider culture of equality, diversity, and inclusion. The Centre PE team’s skills have been essential to running LGBT+ History Month at the University of Dundee, supporting diverse voices and stories.
e n g a g e m e n t
Wellcome Centre for Anti-Infectives Research Six Year Report I page 33
e n g a g e m e n t
Creativity
Our mural has not been our only creative work In early 2020, we began to work with artist Emily Fong, with the hope of her becoming our artist in residence As we all left the building shortly after, this was impossible – but we came up with an alternative plan.

Emily spent much of 2020 meeting with our senior scientists on Microsoft Teams They talked about science, art, and life. From these conversations, Emily created a beautiful series of portraits and blogs, which grew on our website Emily formed a particularly strong bond with Professor Alan Fairlamb, and the two have met online or in person weekly since that time, Alan leading Emily in science, and Emily inspiring Alan’s growth as an artist
As we came out of lockdowns and returned to in-person working in late 2021 and early 2022, we were ready with the next phase of this experiment Laboratory Art Binge was a series of workshops, where Emily invited Centre staff to be creative Working in small groups, they transformed a set of materials, including ex-crab-pots salvaged from a Fife beach, into something useful. Some groups created sculptures which brought their research to life in a new way, while others created more abstract work. In each session, people discussed, candidly, their work, science, and how they do and don’t feel creative The sessions were a great chance for our staff to make new connections and look at their work afresh We filmed them at work, and have a vast repository of photography and footage of them explaining complex concepts in beautiful, simple language.
Our next step brought the project full circle, back into LifeSpace Instead of using the gallery simply for an exhibition, Emily and Alan decided to occupy the space, living there for a full week This piece of performance art activated conversation in the School of Life Sciences in a totally new way Their concept was to live as parasites in the building, adapting it to their needs, without harming their host This has ultimately led on to a superb new exhibition, Bonding: electrons are FREE.
Information about all WCAIR’s Public engagement activities can be found on our website, where you can also play the online versions of a number of games we have created and download the Medicine Maker badge pack https://wcair.dundee.ac.uk/public-engagement/

Wellcome Centre for Anti-Infectives Research Six Year Report I page 34
closingremarks

Wellcome Centre for Anti-Infectives Research Six Year Report I page 35
We would like to thank Wellcome for their support for the Wellcome Centre for Anti-Infective Research. Drug discovery needs constant innovation, to improve the rate at which new candidates are produced and their quality. For the most part, for the neglected infectious diseases that we tackle, the disease biology and drug discovery pathways are poorly understood. The funding in WCAIR is allowing us to develop tools to understand disease biology and relevant assays to develop the drug discovery pathways and to gain further understanding of critical issues such as pharmacokinetic/ pharmacodynamic relationships. Further WCAIR funding has been important in improving how we produce and assay proteins and chemically synthesise compounds more efficiently.
We have opportunities to explore new modalities for drug discovery. Computational chemistry is moving at a fast pace; through our WCAIR funding, we are developing new computational tools to guide the selection of compounds for synthesis, with the aim of speeding up the drug discovery process. The phenomenal success of the Mode of Action group has transformed our drug discovery. Through identification of the molecular targets of phenotypic hits, we have been able to prioritise promising drug targets, de-prioritise targets that do not look promising and to identify and rank compound series hitting the same targets. Genetic tools have become increasingly important; we have been able to develop new tools which help us to identify drug targets. These new tools will also be important in understanding potential for drug resistance It is really exciting to see the progress that we have made over the last 6 years. It is important that we continue to innovate over multiple areas as we go forwards.
It is key that we can explain what we are doing and why it is important to the general public. COVID has shown us how important this is. We are fortunate to have people involved in public engagement.
The WCAIR training team has been working with scientists in Low- and Middle-Income Countries, through a variety of different means. Recently we have seen the development of the concept of “hubs” in Ghana and Brazil, with the aim of building critical mass and focusing our efforts. It is great to see these develop.
None of this work would be possible without the great team of people within WCAIR; many thanks to all of you.
Prof. Ian Gilbert Director of WCAIR and Head of the DDU

Wellcome Centre for Anti-Infectives Research Six Year Report I page 36
Drug discovery needs constant innovation, to improve the rate at which new candidates are produced and their quality.
WCAIR Management Board



Congratulations to Dr Manu De Rycker and Dr Beatriz Baragaña who have recently been promoted to Principal Investigators in the School of Life Sciences




Welcome to Prof Marcus Lee who has joined WCAIR in 2023 from the Wellcome Sanger Institute Marcus is interested in the molecular basis of drug resistance in the human malaria parasite Plasmodium falciparum, and in developing molecular genetics approaches to interrogate gene function. One of his longstanding research interests has been to understand the mechanisms available to the parasite to develop resistance, which often comes at a cost in terms of fitness in the absence of drug pressure. He hopes the labs research will guide the development and prioritisation of future therapeutic targets and gain fundamental biological insights into critical parasite pathways.







Wellcome Centre for Anti-Infectives Research Six Year Report I page 37
Prof. Ian Gilbert, FRSE FMedSci FRSC, Director and Head of the DDU
Prof. David Horn FRSE, Deputy Director, Professor of Parasite Molecular Biology
Dr Catharine Goddard, Centre Manager
Dr Charlotte Green, Business Development Manager
Prof. David Gray BEM, Professor Translational Biology, DDU Head of Biology, Academic lead for PE
Prof. Alan Fairlamb CBE FLS FRSE FMedSci FRSB, Emeritus Professor of Biochemistry and Wellcome Principal Research Fellow
Prof. Sir Mike Ferguson, CBE FRS FRSE FMedSci FRSB, Regius Professor Life Sciences
Prof. Mark Field, FRSB, Professor Cell Biology and Parasitology
Prof. Marcus Lee, Professor of Parasite Molecular Genetics
Dr Mattie Pawlowic, Sir Henry Dale Fellow
Dr Manu De Rycker, Head of Translational Parasitology and DDU Portfolio Leader for Kinetoplastid Drug Discovery
Dr Beatriz Baragaña, DDU Portfolio leader Apicomplexans and Schistosomiasis
Prof Kevin Read, Professor of Quantitative Pharmacology, DDU Head of DMPK, Academic lead for Training
Dr Susan Wyllie, Head of Mode of Action
Selected Publications
Researchers at WCAIR have contributed to over 200 publications since April 2017. A full list of publications can be found on our website, wcair.dundee.ac.uk .
2017
Bayliss T, Robinson DA, Smith VC, Brand S, McElroy SP, Torrie LS, et al Design and Synthesis of Brain Penetrant Trypanocidal N-Myristoyltransferase Inhibitors J Med Chem. 2017;60(23):9790–806
Crozier TWM, Tinti M, Larance M, Lamond AI, Ferguson MAJ Prediction of Protein Complexes in Trypanosoma brucei by Protein Correlation Profiling Mass Spectrometry and Machine Learning Mol Cell Proteomics 2017;16(12):2254–67
Field MC, Horn D, Fairlamb AH, Ferguson MAJ, Gray DW, Read KD, et al Anti-trypanosomatid drug discovery: An ongoing challenge and a continuing need Nat Rev Microbiol 2017;15(4):217–31
Hallyburton I, Grimaldi R, Woodland A, Baragaña B, Luksch T, Spinks D, et al Screening a protein kinase inhibitor library against Plasmodium falciparum Malar J 2017; 16(1):446 Heikamp K, Zuccotto F, Kiczun M, Ray P, Gilbert IH Exhaustive sampling of the fragment space associated to a molecule leading to the generation of conserved fragments Chem Biol Drug Des 2017;91(3):655–67
Jinnelov A, Ali L, Tinti M, Güther MLS, Ferguson MAJ Single-subunit oligosaccharyltransferases of Trypanosoma brucei display different and predictable peptide acceptor specificities J Biol Chem 2017;292(49):20328–41
Ray PC, Kiczun M, Huggett M, Lim A, Prati F, Gilbert IH, et al Fragment library design, synthesis and expansion: nurturing a synthesis and training platform Drug Discov Today 2017;22(1):43–56
Torrie LS, Brand S, Robinson DA, Ko EJ, Stojanovski L, Simeons FRC, et al. Chemical Validation of Methionyl-tRNA Synthetase as a Druggable Target in Leishmania donovani ACS Infect Dis 2017;3(10)
2018
Cleghorn LAT, Ray PC, Odingo J, Kumar A, Wescott H, Korkegian A, et al Identification of Morpholino Thiophenes as Novel Mycobacterium tuberculosis Inhibitors, Targeting QcrB J Med Chem 2018;61(15):6592–608
De Rycker M, Baragaña B, Duce SL, Gilbert IH Challenges and recent progress in drug discovery for tropical diseases Nature 2018;559 498-506
Fairlamb AH, Horn D Melarsoprol Resistance in African Trypanosomiasis Trends Parasitol 2018 34(6) 481-492
Jeacock L, Faria J, Horn D Codon usage bias controls mRNA and protein abundance in trypanosomatids elife 2018;7:e32496
Kovářová J, Nagar R, Faria J, Ferguson MAJ, Barrett MP, Horn D Gluconeogenesis using glycerol as a substrate in bloodstream-form Trypanosoma brucei PLOS Pathog 2018;14(12):e1007475
Luise N, Wyatt PG Diversity-oriented synthesis of bicyclic fragments containing privileged azines Bioorg Med Chem Lett 2018;29(2):248–51
MacLean LM, Thomas J, Lewis MD, Cotillo I, Gray DW, De Rycker M Development of Trypanosoma cruzi in vitro assays to identify compounds suitable for progression in Chagas’ disease drug discovery PLoS Negl Trop Dis 2018;12(7):e0006612
Murugesan D, Ray PC, Bayliss T, Prosser GA, Harrison JR, Green K, et al 2-Mercapto-quinazolinones as inhibitors of NDH-2 and Mycobacterium tuberculosis: Structure-activity relationships, mechanism of action and ADME characterization. ACS Infect Dis. 2018; 4(6) 954-969.
Pinger J, Nesic D, Ali L, Aresta-Branco F, Lilic M, Chowdhury S, Kim HS, Verdi J, Raper J, Ferguson MAJ, Papavasiliou FN, Stebbins CE African trypanosomes evade immune clearance by O-glycosylation of the VSG surface coat Nature Microbiol 2018;3: 932-938
Prati F, Zuccotto F, Fletcher D, Convery MA, Fernandez-Menendez R, Bates R, et al Screening of a Novel Fragment Library with Functional Complexity against Mycobacterium tuberculosis InhA ChemMedChem 2018;13(7):672–7
Quintana JF, Canavate R, Pino D, Yamada K, Zhang N, Field MC Adaptation and Therapeutic Exploitation of the Plasma Membrane of African Trypanosomes Genes 2018; 9(7) 368
Rico E, Jeacock L, Kovářová J, Horn D Inducible high-efficiency CRISPR-Cas9-targeted gene editing and precision base editing in African trypanosomes Sci Rep 2018;8(1):7960
Thomas J, Baker N, Hutchinson S, Dominicus C, Trenaman A, Glover L, Alsford S, Horn D Insights into antitrypanosomal drug mode-of- action from cytology-based profiling PLoS Negl Trop Dis 2018; 12(11):e0006980
Thomas MG, De Rycker M, Cotillo Torrejon I, Thomas J, Riley J, Spinks D, et al 2,4-Diamino-6-methylpyrimidines for the potential treatment of Chagas’ disease Bioorganic Med Chem Lett 2018; 28(18):3025–30
Thomas MG, De Rycker M, Ajakane M, Albrecht S, Álvarez-Pedraglio AI, Boesche M, et al Identification of GSK3186899/DDD853651 as a Preclinical Development Candidate for the Treatment of Visceral Leishmaniasis J Med Chem 2018; 62(3) 1180-1202
Venkatesh D, Zhang N, Zoltner M, del Pino RC, Field MC Evolution of protein trafficking in kinetoplastid parasites: Complexity and pathogenesis Traffic 2018; 19(11) 803–12
Wall RJ, Rico E, Lukac I, Zuccotto F, Elg S, Gilbert IH, et al Clinical and veterinary trypanocidal benzoxaboroles target CPSF3 Proc Natl Acad Sci 2018; 62(8) 1-15
Wall RJ, Moniz S, Thomas MG, Norval S, Ko E-J, Marco M, et al Antitrypanosomal 8-Hydroxy-Naphthyridines Are Chelators of Divalent Transition Metals Antimicrob Agents Chemother 2018; 62(8):1–15
Wyllie S, Thomas M, Patterson S, Crouch S, De Rycker M, Lowe R, et al Cyclin-dependent kinase 12 is a drug target for visceral leishmaniasis Nature 2018; 560(7717):192–7
Zhang N, Zoltner M, Leung K-F, Scullion P, Hutchinson S, del Pino RC, et al Host-parasite co-metabolic activation of antitrypanosomal aminomethyl-benzoxaboroles PLOS Pathog 2018; 14(2):e1006850
Zoltner M, Krienitz N, Field MC, Kramer S Comparative proteomics of the two T brucei PABPs suggests that PABP2 controls bulk mRNA PLoS Negl Trop Dis 2018;12(7) e0006679
Wellcome Centre for Anti-Infectives Research Six Year Report I page 38
2019
Baragaña B, Forte B, Choi R, Nakazawa Hewitt S, Bueren-Calabuig JA, Pisco JP, et al Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis Proc Natl Acad Sci 2019 Mar 20;201814685
Boehm C, Field MC Evolution of late steps in exocytosis: conservation and specialization of the exocyst complex [version 2; peer review: 3 approved] Wellcome Open Res 2019;4(112) Corpas-Lopez V, Moniz S, Thomas M, Wall RJ, Torrie LS, Zander-Dinse D, et al Pharmacological Validation of N-Myristoyltransferase as a Drug Target in Leishmania donovani ACS Infect Dis 2019 Jan 11;5(1):111–22
Fairlamb, Alan H and Patterson S Current and Future Prospects of Nitro-compounds as Drugs for Trypanosomiasis and Leishmaniasis Current Medicinal Chemistry 2019 26(23) 4454-4475
Faria J, Glover L, Hutchinson S, Boehm C, Field MC, Horn D Monoallelic expression and epigenetic inheritance sustained by a Trypanosoma brucei variant surface glycoprotein exclusion complex. Nat Commun. 2019;10(1):3023.
Glover L, Marques CA, Suska O, Horn D Persistent DNA Damage Foci and DNA Replication with a Broken Chromosome in the African Trypanosome Burleigh B, editor MBio 2019; 10(4):e01252-19
Homeyer N, van Deursen R, Ochoa-Montaño B, Heikamp K, Ray P, Zuccotto F, et al A platform for target prediction of phenotypic screening hit molecules J Mol Graph Model 2019;107485
Luise N, Wyatt EW, Tarver GJ, Wyatt PG A Continuous Flow Strategy for the Facile Synthesis and Elaboration of Semi-Saturated Heterobicyclic Fragments European J Org Chem 2019;1–10
Martin JS, MacKenzie CJ, Fletcher D, Gilbert IH Characterising covalent warhead reactivity Bioorg Med Chem 2019;27(10):2066–74
Norcross NR, Wilson C, Baragaña B, Hallyburton I, Osuna-Cabello M, Norval S, et al Substituted aminoacetamides as novel leads for malaria ChemMedChem 2019;14(14) 1329-1335
Pawlowic MC, Somepalli M, Sateriale A, Herbert GT, Gibson AR, Cuny GD, et al Genetic ablation of purine salvage in Cryptosporidium parvum reveals nucleotide uptake from the host cell Proc Natl Acad Sci 2019; 116(42):21160–21165
Tinti M, Güther MLS, Crozier TWM, Lamond AI, Ferguson MAJ Proteome turnover in the bloodstream and procyclic forms of Trypanosoma brucei measured by quantitative proteomics Wellcome Open Res 2019; 4(152)
Torrie LS, Zuccotto F, Robinson DA, Gray DW, Gilbert IH, De Rycker M Identification of inhibitors of an unconventional Trypanosoma brucei kinetochore kinase PLoS One 2019;14(5):1–16
Trenaman A, Glover L, Hutchinson S, Horn D A post-transcriptional respiratome regulon in trypanosomes Nucleic Acids Res 2019; 47(13):7063–77
Wyllie S, Brand S, Thomas M, De Rycker M, Chung C, Pena I, et al Preclinical candidate for the treatment of visceral leishmaniasis that acts through proteasome inhibition Proc Natl Acad Sci 2019; 201820175
Zmuda F, Sastry L, Shepherd SM, Jones D, Scott A, Craggs PD, et al Identification of novel Trypanosoma cruzi proteasome inhibitors using a luminescence-based high-throughput screening assay Antimicrob Agents Chemother 2019; AAC 00309-19
Zmuda F, Shepherd SM, Ferguson MAJ, Gray DW, Torrie LS, De Rycker M Trypanosoma cruzi phosphomannomutase and guanosine diphosphate-mannose pyrophosphorylase ligandability assessment Antimicrob Agents Chemother 2019 Aug 12;AAC 01082-19
2020
De Rycker M, Horn D, Aldridge B, Amewu RK, Barry CE, Buckner FS, et al Setting Our Sights on Infectious Diseases ACS Infect Dis 2020, 6(1) 3-13
Floyd A, Yeoman H, Southern J, Dillon R, Cook S, Fairlamb A, et al Para-site-seeing: Departure Lounge: LifeSpace science art research gallery, University of Dundee [Internet] 2020 Available from: https://discovery dundee ac uk/en/publications/para-site-seeing-exhibition-catalogue
Giordani F, Paape D, Vincent IM, Pountain AW, Fernández-Cortés F, Rico E, et al Veterinary trypanocidal benzoxaboroles are peptidase-activated prodrugs PLOS Pathog [Internet] 2020;16(11):1–20
Harrison JR, Sarkar S, Hampton S, Riley J, Stojanovski L, Sahlberg C, et al Discovery and Optimization of a Compound Series Active against Trypanosoma cruzi, the Causative Agent of Chagas Disease J Med Chem 2020;63(6):3066–89
Innes EA, Chalmers RM, Wells B, Pawlowic MC A One Health Approach to Tackle Cryptosporidiosis Trends Parasitol 2020; 36(3):290–303
Izquierdo M, Lin D, O’Neill S, Zoltner M, Webster L, Hope A, et al Development of a High-Throughput Screening Assay to Identify Inhibitors of the Major M17-Leucyl Aminopeptidase from Trypanosoma cruzi Using RapidFire Mass Spectrometry SLAS Discov Adv Sci Drug Discov 2020; 25(9) 1064-1071
Quintana JF, Bueren-Calabuig J, Zuccotto F, de Koning HP, Horn D, Field MC. Instability of aquaglyceroporin (AQP) 2 contributes to drug resistance in Trypanosoma brucei. PLoS Negl Trop Dis. 2020;14(7):1–26.
Sateriale A, Pawlowic M, Vinayak S, Brooks C, Striepen B Genetic Manipulation of Cryptosporidium parvum with CRISPR/Cas9 BT – Cryptosporidium: Methods and Protocols In: Mead JR, Arrowood MJ, editors New York, NY: Springer New York; 2020 p 219–28
Thomas MG, De Rycker M, Ajakane M, Crouch SD, Campbell L, Daugan A, et al Identification of 6-amino-1H-pyrazolo[3,4-d]pyrimidines with in vivo efficacy against visceral leishmaniasis RSC Med Chem 2020; 11(10) 1168-1177
Thomas MG, De Rycker M, Wall RJ, Spinks D, Epemolu O, Manthri S, et al Identification and Optimization of a Series of 8-Hydroxy Naphthyridines with Potent In Vitro Antileishmanial Activity: Initial SAR and Assessment of In Vivo Activity J Med Chem 2020; 63(17) 9523-9539
Torrie LS, Robinson DA, Thomas M, Hobrath J V, Shepherd SM, Post JM, et al Discovery of an Allosteric Binding Site in Kinetoplastid Methionyl-tRNA Synthetase ACS Infect Dis 2020;6(5):1044–57
Wall RJ, Carvalho S, Milne R, Bueren-Calabuig JA, Moniz S, Cantizani-Perez J, et al The Q(i) site of cytochrome b is a promiscuous drug target in Trypanosoma cruzi and Leishmania donovani ACS Infect Dis 2020;6(3):515-528
Wang Q, Boshoff HIM, Harrison JR, Ray PC, Green SR, Wyatt PG, et al PE/PPE proteins mediate nutrient transport across the outer membrane of Mycobacterium tuberculosis Science 2020; 367(6482):1147–51
Zoltner M, Campagnaro GD, Taleva G, Burrell A, Cerone M, Leung K-F, et al Suramin exposure alters cellular metabolism and mitochondrial energy production in African trypanosomes J Biol Chem 2020; 295(24):8331–47
Zoltner M, del Pino RC, Field MC Sorting the Muck from the Brass: Analysis of Protein Complexes and Cell Lysates BT – Trypanosomatids: Methods and Protocols
In: Michels PAM, Ginger ML, Zilberstein D, editors New York, NY: Springer US; 2020 p 645–53
Wellcome Centre for Anti-Infectives Research Six Year Report I page 39
Aguado ME, González-Matos M, Izquierdo M, Quintana J, Field M C, González-Bacerio J Expression in Escherichia coli, purification and kinetic characterization of LAPLm, a Leishmania major M17-aminopeptidase Protein Expression and Purification 2021; 183, 105877
Arendse LB, Wyllie S, Chibale K, Gilbert IH Plasmodium Kinases as Potential Drug Targets for Malaria: Challenges and Opportunities ACS Infectious Diseases 2021; 7(3), 518–534
Bandini G, Damerow S, Sempaio Güther ML, Guo H, Mehlert A, Paredes Franco JC, Beverley S, Ferguson MAJ An essential, kinetoplastid-specific GDP-Fuc: β-D-Gal α-1,2-fucosyltransferase is located in the mitochondrion of Trypanosoma brucei ELife 2021; 10, e70272
Butterfield ER, Abbott JC, Field MC Automated Phylogenetic Analysis Using Best Reciprocal BLAST Methods in Molecular Biology 2021; 2369, 41–63
Corpas-Lopez V, & Wyllie S Utilizing thermal proteome profiling to identify the molecular targets of anti-leishmanial compounds STAR Protocols 2021; 2(3), 100704
Duncan SM, Nagar R, Damerow M, Yashunsky DV, Buzzi B, Nikolaev AV, Ferguson MAJ A Trypanosoma brucei β3 glycosyltransferase superfamily gene encodes a β1-6 GlcNAc-transferase mediating N-glycan and GPI anchor modification Journal of Biological Chemistry 2021; 297(4) 101153
Faria J, LuzakV, Müller LSM, Brink BG, Hutchinson S, Glover L, HornD, SiegelTN Spatial integration of transcription and splicing in a dedicated compartment sustains monogenic antigen expression in African trypanosomes Nature Microbiology 2021; 6, 289-300
Ferguson MAJ, Tinti M, Kelner-Mirôn A, Marriott LJ (2021) Polysomal mRNA Association and Gene Expression in Trypanosoma brucei Wellcome Open Research, 6 https://doi org/10 12688/wellcomeopenres 16430 1
Gissot M, Pawlowic, MC Modrzynska KK, & Francia ME (2021) Editorial: Molecular Basis of Stage Conversion in Apicomplexan Parasites Frontiers in Cellular and Infection Microbiology, 11 https://doi org/10 3389/fcimb 2021 680184
Jenni A, Knüsel S, Nagar R, Benninger M, Häner R, Ferguson MAJ, Roditi I, Menon AK, Bütikofer P (2021) Elimination of GPI2 suppresses glycosylphosphatidylinositol GlcNAc transferase activity and alters GPI glycan modification in Trypanosoma brucei Journal of Biological Chemistry, 297(2), 100977
Ji Z, Tinti M, & Ferguson MAJ Proteomic identification of the UDP-GlcNAc: PI α1–6 GlcNAc-transferase subunits of the glycosylphosphatidylinositol biosynthetic pathway of Trypanosoma brucei PLoS ONE 2021; 16(3) e0244699
Lukac I, Wyatt PG, Gilbert IH, Zuccotto F. Ligand binding: evaluating the contribution of the water molecules network using the Fragment Molecular Orbital method. Journal of Computer-Aided Molecular Design 2021; 35, 1025-1036
Martin JS, Mackenzie CJ, Gilbert IH Synthesis of a Series of Diaminoindoles The Journal of Organic Chemistry 20-21; 86(17), 11333–11340
Michalska K, Jedrzejczak R, Wower J, Chang C, Baragaña B, Gilbert IH, Forte B, Joachimiak A Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation Nucleic Acids Research 2021; 49(9), 5351–5368
Mowbray CE, Braillard S, Glossop PA, Whitlock GA, et al DNDI-6148: A Novel Benzoxaborole Preclinical Candidate for the Treatment of Visceral Leishmaniasis
Journal of Medicinal Chemistry 2021; 64(210 16159-16176
Pacheco J da S, Costa D de S, Cunha-Júnior EF, Andrade-Neto VV, et al Monocyclic Nitro-heteroaryl Nitrones with Dual Mechanism of Activation: Synthesis and Antileishmanial Activity ACS Medicinal Chemistry Letters 2021; 12(9), 1405-1412
Padilla-Mejia NE, Makarov AA, Barlow LD, Butterfield ER, Field MC Evolution and diversification of the nuclear envelope Nucleus 2021; 12(1), 21–41
Paradela LS, Wall RJ, Carvalho S, Chemi G, Corpas-Lopez V, Moynihan E, Bello D, Patterson S, Güther MLS, Fairlamb AH, Ferguson MAJ, Zuccotto F, Martin J, Gilbert IH, Wyllie S. Multiple unbiased approaches identify oxidosqualene cyclase as the molecular target of a promising anti-leishmanial. Cell Chemical Biology. 2021; 28(5), 711-721 e8
Pearson L -A, Green CJ, Lin D, Petit A-P, Gray DW, Cowling VH, Fordyce EAF Development of a High-Throughput Screening Assay to Identify Inhibitors of the SARS-CoV-2 Guanine-N7-Methyltransferase Using RapidFire Mass Spectrometry SLAS Discovery 2021; 26(6) 749-756
Quintana JF, Field MC Evolution, function and roles in drug sensitivity of trypanosome aquaglyceroporins Parasitology 2021; 148(10) 1137-1142
Ray PC, Huggett M, Turner P A, Taylor M, Cleghorn LAT, et al T Spirocycle MmpL3 Inhibitors with Improved hERG and Cytotoxicity Profiles as Inhibitors of Mycobacterium tuberculosis Growth ACS Omega 2021; 6(3), 2284–2311
Sampaio Guther ML, Prescott AR, Kuettel S, Tinti M, Ferguson MAJ Nucleotide sugar biosynthesis occurs in the glycosomes of procyclic and bloodstream form Trypanosoma brucei PLOS Neglected Tropical Diseases 2021; 15(2), e0009132
Svensen N, Wyllie S, Gray DW, De Rycker M Live-imaging rate-of-kill compound profiling for Chagas disease drug discovery with a new automated high-content assay PLOS Neglected Tropical Diseases 2011; 15(10), e0009870
Thomas M, Brand S, De Rycker M, Zuccotto F, Lukac I et al Scaffold-Hopping Strategy on a Series of Proteasome Inhibitors Led to a Preclinical Candidate for the Treatment of Visceral Leishmaniasis Journal of Medicinal Chemistry 2021; 64(9), 5905–5930
Van den KerkhofM, Leprohon P, Mabille D, Hendrickx S, Tulloch LB, Wall RJ, Wyllie S, et al. Identification of Resistance Determinants for a Promising Antileishmanial Oxaborole Series Microorganisms 2021; 9(7) 1408
Yang T, Ottilie S, Istvan ES, Godinez-Macias KP, Lukens AK, Baragaña B, et al (2021) MalDA, Accelerating Malaria Drug Discovery Trends in Parasitology 2021; 37(6) 493-507
Wellcome Centre for Anti-Infectives Research Six Year Report I page 40
2021
2022
Altmann S, Rico E, Carvalho S, Ridgway M, Trenaman A, Donnelly H, TintiM, Wyllie S, Horn D. Oligo targeting for profiling drug resistance mutations in the parasitic trypanosomatids Nucleic Acids Research, 2022; 50(4) e79
Amewu RK, Amoateng P, Arthur PK, Asare P, et al Drug discovery research in Ghana, challenges, current efforts, and the way forward PLOS Neglected Tropical Diseases 2022; 16(9), e0010645
Benns HJ, Storch M, Falco JA, Fisher FR, Tamaki F, et al CRISPR-based oligo recombineering prioritizes apicomplexan cysteines for drug discovery
Nature Microbiology 2022; 7 1891-1905
Bravo Ruiz G, Tinti M, Ridgway M, Horn D Control of Variant Surface Glycoprotein Expression by CFB2 in Trypanosoma brucei and Quantitative Proteomic Connections to Translation and Cytokinesis MSphere 2022; 7(2), e00069-22
Czarna A, Plewka J, Kresik L, Matsuda A, et al Refolding of lid subdomain of SARS-CoV-2 nsp14 upon nsp10 interaction releases exonuclease activity Structure 2022; 30(8), 1050-1054 e2
Duncan SM, Ferguson MAJ Common and unique features of glycosylation and glycosyltransferases in African trypanosomes Biochemical Journal 2022; 479(17), 1743–1758 Field MC, Rout MP Coatomer in the universe of cellular complexity Molecular Biology of the Cell 2022;33(14), pe8
Green SR, Davis SH, Damerow S, Engelhart CA, Mathieson M, et al T Lysyl-tRNA synthetase, a target for urgently needed M tuberculosis drugs Nature Communications 2022; 13(1), 5992
Horn D Genome-Scale RNAi screens in African trypanosomes Trends in Parasitology 2022; 38(2), 160–173
Horn D A profile of research on the parasitic trypanosomatids and the diseases they cause PLOS Neglected Tropical Diseases 2022; 16(1), e0010040
Inoue AH, Domingues PF, Serpeloni M, Hiraiwa PM, Vidal NM, Butterfield ER, del Pino R C, Ludwig A, Boehm C, Field MC, Ávila, AR Proteomics Uncovers Novel Components of an Interactive Protein Network Supporting RNA Export in Trypanosomes Molecular & Cellular Proteomics 2022; 21(3) 100208
Kovářová, J, Novotná M, Faria J, Rico E, Wallace C, Zoltner M, Field MC, Horn D CRISPR/Cas9-based precision tagging of essential genes in bloodstream form African trypanosomes Molecular and Biochemical Parasitology 2022; 249, 111476
Lima, ML, Tulloch LB, Corpas-Lopez V, Carvalho S, et al Identification of a Proteasome-Targeting Arylsulfonamide with Potential for the Treatment of Chagas’ Disease Antimicrobial Agents and Chemotherapy 2022; 66(1), e01535-21
Lukeš J, Kachale A, Votýpka J, Butenko A, Field MC African trypanosome strategies for conquering new hosts and territories: the end of monophyly? Trends in Parasitology 2022; 38(9), 724–736
Mak K-K, Epemolu O, Pichika MR The role of DMPK science in improving pharmaceutical research and development efficiency Drug Discovery Today 2022; 27(3), 705–729
Marques, CA, Ridgway M Tinti M, Cassidy A, Horn D Genome-scale RNA interference profiling of Trypanosoma brucei cell cycle progression defects Nature Communications 2022; 13(1), 5326
McGonagle K, Tarver GJ, Cantizani J, Cotillo I, Dodd, PG, et al Identification and development of a series of disubstituted piperazines for the treatment of Chagas disease European Journal of Medicinal Chemistry 2022; 238, 114421
Milne R, Wiedemar N, Corpas-Lopez V, Moynihan E, Wall RJ, et al Toolkit of Approaches To Support Target-Focused Drug Discovery for Plasmodium falciparum Lysyl tRNA Synthetase ACS Infectious Diseases 2022; 8(9) 1962-1974
Obado SO, Rout MP, Field MC Sending the message: specialized RNA export mechanisms in trypanosomes Trends in Parasitology 2022; 38(10), 854–867
Roberts A, Nagar R, Brandt C, Harcourt K, Clare S, Ferguson MAJ, Wright GJ The Leishmania donovani Ortholog of the Glycosylphosphatidylinositol Anchor Biosynthesis Cofactor PBN1 Is Essential for Host Infection MBio 2022; 13(3), e00433-22
Smith A, Wall RJ, Patterson S, Rowan T, Rico Vidal E, et al Repositioning of a Diaminothiazole Series Confirmed to Target the Cyclin-Dependent Kinase CRK12 for Use in the Treatment of African Animal Trypanosomiasis Journal of Medicinal Chemistry 2022;, 65(7), 5606–5624
Summers RL, Pasaje, CFA, Pisco JP, et al Chemogenomics identifies acetyl-coenzyme A synthetase as a target for malaria treatment and prevention Cell Chemical Biology 2022; 29(2), 191-201 e8
Tamaki F, Fisher F, Milne R, Terán Fernando S-R, Wiedemar N, Wrobel K, Edwards D, Baumann H, Gilbert IH, Baragana B, Baum J, Wyllie S High-Throughput Screening Platform to Identify Inhibitors of Protein Synthesis with Potential for the Treatment of Malaria Antimicrobial Agents and Chemotherapy 2022; 66(6), e00237-22
Tinti M, Ferguson MAJ Visualisation of experimentally determined and predicted protein N-glycosylation and predicted glycosylphosphatidylinositol anchor addition in Trypanosoma brucei Wellcome Open Research 2022; 7, 33 https://doi org/10 12688/wellcomeopenres 17640 1
Tinti M, Ferguson, MAJ Visualisation of proteome-wide ordered protein abundances in Trypanosoma brucei Wellcome Open Research 2022; 7, 34 https://doi org/10 12688/wellcomeopenres 17607 1
Toviwek B, Riley J, Mutter N, Anderson M Webster L, Hallyburton I, Gleeson D, Read KD, Gleeson MP Preparation, biological evaluation and QSAR analysis of urea substituted 2,4-diamino-pyrimidine anti-malarials RSC Medicinal Chemistry 2022; 13(12), 1587–1604
Wilson C, Ray P, Zuccotto F, et al Optimization of TAM16, a Benzofuran That Inhibits the Thioesterase Activity of Pks13; Evaluation toward a Preclinical Candidate for a Novel Antituberculosis Clinical Target Journal of Medicinal Chemistry 2022; 65(1), 409–423
2023
Basu S, Pawlowic M, Hsu F-F, Thomas G, Zhang K Ethanolaminephosphate cytidyltransferase is essential for survival, lipid homeostasis and stress tolerance in Leishmania major; BioRxiv. 2023; 2023.01.10.523530. https://doi.org/10.1101/2023.01.10.523530
Bopp S, Pasaje CFA, Summers RL, et al (2023) Potent acyl-CoA synthetase 10 inhibitors kill Plasmodium falciparum by disrupting triglyceride formation Nature Communications, 14(1), 1455
De Rycker M, Wyllie S, Horn D, Read KD, Gilbert IH (2022) Anti-trypanosomatid drug discovery: progress and challenges
Nature Reviews Microbiology 2023; 21 35-50
Makarov A, Began J, Mautone IC, Pinto E, Ferguson L, Zoltner M, Zoll S, Field MC The role of invariant surface glycoprotein 75 in xenobiotic acquisition by African trypanosomes Microbial Cell 2023; 10(2), 18–35
Moreira CM do N, Kelemen C D, Obado SO, Zahedifard F, Zhang N, Holetz FB, Gauglitz L, Dallagiovanna B, Field MC, Kramer S, Zoltner M Impact of inherent biases built into proteomic techniques: Proximity labeling and affinity capture compared Journal of Biological Chemistry 2022; 299(1), 102726
Smith RJ, Milne R, Lopez VC, Wiedemar N, Dey G, Syed AJ, Patterson S, Wyllie S Chemical pulldown combined with mass spectrometry to identify the molecular targets of antimalarials in cell-free lysates STAR Protocols 2023 4(1), 102002
Wellcome Centre for Anti-Infectives Research Six Year Report I page 41
WCAIR Timeline
July 2018: DDU, GSK and Wellcome announce new pre-clinical candidate for visceral leishmaniasis
January and
February 2018: WCAIR’s first Trainees arrive from Brazil
Dr Mattie Pawlowic joins WCAIR
November 2017: WCAIR runs Street Food for the first time in Dundee Science Festival
April 2017: WCAIR starts with £13.6 million funding from Wellcome
2018 2017
October 2018: Mode of Action group win GSK ‘STOP’ award
Mattie Pawlowic awarded Sir Henry Dale fellowship
WCAIR help WCS run 1st overseas Practical Aspects of Drug Discovery in South Africa
April 2019: Dr Mattie Pawlowic wins inaugural BSP President’s Medal
February 2019: WCAIR and Girlguiding Dundee launch Medicine Maker badge
September 2018: WCAIR researchers use Mode of Action studies to identify new target for sleeping sickness
May 2019: WCAIR hosts its 1st International conference on NTDs ‘SOSID’
WCAIR and GSK declare 2nd pre-clinical candidate for visceral leishmaniasis
WCAIR opens its 1st LifeSpace exhibition
Para-Site-Seeing: Departure Lounge created by Jen Southern and Rod Dillon
July 2019: DDU Malaria Team win MMV Project of the year 2018
Susan Wyllie promoted to Principal Investigator in School of Life Sciences
November 2019: David Horn wins William Trager Award
December 2019:
Parasites: Battle for Survival exhibition opens at National Museum of Scotland
February 2020: WCAIR 2nd Life Space Show ‘Translations’ opens A collaboration with Dundee Print Collective
March 2020: Ian Gilbert elected Fellow of the Royal Society of Edinburgh
August 2020: WCAIR joins CARE (Corona Accelerated R&D in Europe)
April 2020: Emily Fong becomes WCAIR’s first artist in virtual residence
2019 2020
DDU receives $5 3 million from COVID-19 Therapeutics Accelerator (initiated by the Bill & Melinda Gates Foundation, Wellcome and Mastercard) to work on anti-coronavirus agents
Wellcome Centre for Anti-Infectives Research Six Year Report I page 42
October 2020: Julia Haddow (née Wcislo) is runner up in the UKRI MRC Max Perutz Science Writing Award
In conversation with webinars launched
September 2020: Dr Joana Faria wins SULSA Early Career Researcher prize
Mode of Action group receive £4 9 million funding from Wellcome
March 2021: WCAIR and Girlguiding host a virtual sleepover as COVID means the girl guides can still not meet in person
May 2021: Prof Ian Gilbert is elected Fellow of Academy of Medical Sciences
WeeCAIR Medicinal Garden is built
June 2021: Prof David Gray is awarded the British Empire Medal in Queen’s Birthday honours for services to delivery of testing during COVID-19 pandemic
October 2021: Dr Joana Faria, who was awarded a Sir Henry Dale Fellow in June, wins the Wellcome-Beit prize
July 2022: WCAIR inspired mural is painted on 7 storey wall in Stobswell by artist Lewi Quinn
June 2022: Launch of Drug Discovery Hub in Ghana with funding from the Gates Foundation and support from MMV, H3D and LGenia
April 2022: Wellcome extend funding for WCAIR and Chagas diseases drug discovery
Goodbye to Prof Paul Wyatt, WCAIR’s 1st Director
March 2022: Prof. Sir Mike Ferguson awarded Honorary membership British Society Parasitology
Wellcome award £2 5 M to continue drug discovery for Schistosomiasis in Dundee
January 2022:
DDU take on leadership of SDDC with $5M award from from the Gates Foundation
January 2023: Prof. Marcus Lee joins WCAIR
February 2023: Emily Fong and Alan Fairlamb’s exhibition ‘Bonding: electrons are FREE’ opens in LifeSpace
August 2022: MSD Richard T Clark Fellows for Global Health start 3 month placement with Ghana Hub
October 2022: Prof. Ian Gilbert announced as new Head of DDU
November 2022: Training team finally get to Uruguay to deliver Practical Aspects Drug Discovery with WCS
March 2023: WCAIR’s interactive short course Drug Discovery Mission is delivered at USP, Brazil
April 2023: Awards to Dr Mattie Pawlowic (Early Career Research awards from RSE and Biochemical Soc ); Dr Susan Wyllie (CA Wright Medal, BSP); Prof. Sir Mike Ferguson (Morton lecture, Biochemical Soc ); DDU-GSK Kinetoplastid Drug Discovery Team (Industry & Academic Collaboration Award, Biochemical Soc )
Prof Ian Gilbert becomes Director of WCAIR
19
2021
2022
2023
Wellcome Centre for Anti-Infectives Research Six Year Report I page 43
WCAIR Global Partners
University of Glasgow
University of Edinburgh
Moredun Institute
Wellcome Sanger Institute
Imperial College, London
Spain & Portugal
GSK, Tres Cantos
Universidad Pablo de Olavide-CSIC, Seville
Canada
University of Alberta
USA
University of Washington
Seattle Children’s Research Institute
Washington University St Louis
AbbVie
Novartis NITD
Emeryville
Takeda USA
Hackensack
Meridian Health
Eisai USA
Eli Lilly
CALIBR
MIT
Broad Inst.
Harvard
Colorado State
Columbia University
Cornell University
UCSD
University of Vermont
University of Georgia
NIAID
Thomas Jefferson University
John Hopkins
University of Arizona
Merck & Co
TB Alliance
Gates Foundation
MALDA
Univ. Chicago/ Argonne Labs
University of Porto
Ghana
Noguchi Memorial Institute for Medical Research, Universityof Ghana
Kwame Nkrumah University of Science and Technology
University of Cape Coast
University of Health and Allied Sciences
Cameroon
University of Yaoundé 1
Brazil
Federal U Sāo Paolo
Oswaldo Cruz Foundation
Adolfo Lutz Foundation
Sāo Paolo
Uruguay
Insititut Pasteur de Montevideo
University of Campinas
UK
LSHTM
University of Aberystwyth
University of East London
University of Cambridge
University of Cardiff
University of Oxford
University of York
GSK, Stevenage
Wellcome Centre for Anti-Infectives Research is funded by the Wellcome Trust awards
203134/Z/16/Z and 203134/A/16/Z
We also thank our other funders and partners who support research within WCAIR: Academy of Medical Sciences, Bill and Melinda Gates Foundation, Drugs for Neglected Diseases initiative (DNDi), GlaxoSmithKline plc (GSK), Medicines for Malaria Venture (MMV), Medical Research Council (MRC), National Institute of Health (NIH)
WCAIR is a member of the Tuberculosis Drug Accelerator (TBDA), Structure-guided Drug Discovery Coalition (SDDC), Malaria Drug Accelerator (MALDA), IMI European Regimen Accelerator for Tuberculosis (ERA4TB) and IMI Corona Accelerated R&D in Europe (CARE)
Central Europe
Institut Pasteur
DNDi
MMV
Swiss TPH
ABScience
Enamine
Evotec
University of Kiel
Helmholtz Institute
Czech Academy of Sciences, Inst. Parasitology
Charles University, Prague
Ludwig-Maximilians-Universität
University of Berne
India
TCGLS
Asia
WuXi AppTec, China
GHIT, Japan
Malawi
Kamuzu University of Health Sciences
Malawi Uni Science & Technology
South Africa
H3D, University of Cape Town
Uni of Pretoria
Australia
Bio21 Melbourne Science & Biotech Institute, University of Melbourne
The Australian National University
Monash University
Eskitis Institute, Griffith University
Epichem
Wellcome Centre for Anti-Infectives Research Six Year Report I page 45
International Scientific Advisory Board
Current members
Dr Graeme Bilbe Chair Senior Advisory and previous Research and Development Director, DNDi
Professor Mike Blackman Group leader, Division of Parasitology, Francis Crick Institute and Professor of Molecular Microbiology, London School of Hygiene & Tropical Medicine
Professor Paul Herrling Professor of Drug Discovery Science, University of Basel (emeritus) (Wellcome representative)
Professor John Kelly Professor of Molecular Biology, Department of Infection, London School of Hygiene and Tropical Medicine
Dr Paul Leeson FRSC Independent Medicinal Chemistry Consultant, Paul Leeson Consulting Ltd Honorary Professor of Medicinal Chemistry, University of Nottingham
Professor Julian Rayner Director Cambridge Institute for Medical Research, University of Cambridge, Director Wellcome Genome Campus Connecting Science
Steve Rees OBE Vice President Discovery Biology, AstraZeneca, Former Chair and SAB member of European Laboratory Research & Innovation Group (ELRIG)
Dr Isabela Ribeiro Viral Diseases Cluster Director, DNDi
Past members
Professor Simon Croft FRSB Professor of Parasitology, Faculty of Infectious and Tropical Diseases, London School of Hygiene and Tropical Medicine
Professor Meg Philips The Sam G Winstead and F Andrew Bell Distinguished Chair in Biochemistry and Chair of the Department of Biochemistry
UT Southwestern
Professor Jason Swedlow OBE FRSE Professor of Quantitative Cell Biology, School of Life Sciences, University of Dundee
Chemistry Scientific Advisory Board
Professor Varinder Aggarwal Professor in Synthetic Chemistry, University of Bristol
Professor Tim Cernak Assistant Professor of Medicinal Chemistry and Chemistry, University of Michigan
Dr David Lathbury Astute Chemical Development Consulting Ltd
Dr Paul Leeson Director at Paul Leeson Consulting Ltd
Professor Adam Nelson Professor of Chemical Biology, University of Leeds
Dr Steve Taylor Founding Director at Celbius Ltd
Public Engagement Advisory Board
Dr Hazel Lambert Public Engagement with Research Manager, University of Edinburgh
Dr Emily Scott-Dearing Museum and Science Communication consultant
Lynne Short Councillor, Dundee City Council
Past member Pamela MacLean Teacher, Grove Academy, Dundee
Training Review Board 2020
Dr Rachel Berkson Education Manager, Wellcome Connecting Science
Dr Jeremy Burrows Vice President, Head of Drug Discovery at MMV
Dr Ben Perry former Senior Discovery leader, DNDi; now at Medicxi
Professor Jean Ker former National lead for Clinical Skills, NHS Education Scotland
Professor Julian Rayner Director Cambridge Institute for Medical Research, University of Cambridge, Director Wellcome Genome Campus Connecting Science
Alastair Strickland former GCRF Development Manager, Research and Innovation, University of Dundee
Acknowledgements
A big thank you to BCDD Admin and Lab management teams, the SLS Stores team, the SLS postgraduate team and all other SLS core teams without whom all the activities of the Centre would not run so smoothly
Peachsnaps photograpghy for photographs at Setting our Sights on infectious Diseases Erika Stevenson for photographs of Para-Site-seeing: Departure Lounge
John Post for photographs of WCAIR members in the lab and Bonding: Electrons are FREE Exhibition
Other photographs from members of WCAIR Design and Layout: Emma Quinn Design
Wellcome Centre for Anti-Infectives Research Six Year Report I page 46




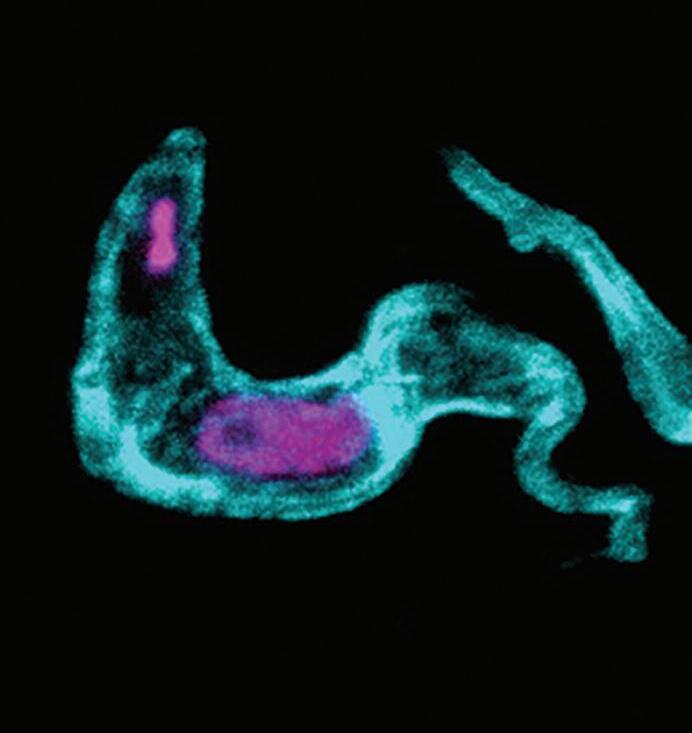







Wellcome Centre for Anti-Infectives Research
School of Life Sciences
University of Dundee
Dow Street
Dundee DD1 5EH
WCAIR@dundee.ac.uk
www.wcair.dundee.ac.uk
@WCAIRDundee


















 MacGowan
MacGowan









 This image shows fluorescent labelled trypanosomes taken by Mark Field’s group
This image shows fluorescent labelled trypanosomes taken by Mark Field’s group










































































