3 minute read
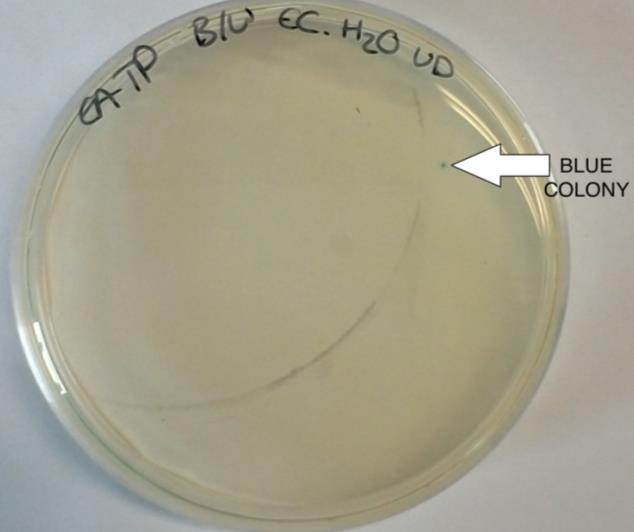
Figure 5. Blue/white colonies selection of E.coli
from silk
Petri dishes and duplicates containing SGG medium (50ml skimmed milk, 7g glucose and 5ml glycerol) were plated. The biofilms were let to grow for four hours at 37C. In parallel, E.coliand B.subtilis biofilms were grown on microtiter flat and round bottomed plates in different concentrations at 37C for 4h, 24h, 48h and 96h. Additionally, 96- and 24-well plates were used. Untransformed E.coliand B.subtils cells were used as positive control biofilms and Ro water was used as negative control.
Validation of the biofilms by microtiter assay
Advertisement
In order to validate the biofilms, the microtiter assay was performed on each microtiter plate. The medium was discarded by reverse pipetting, using sterile tips and water was added to each individual well. Afterwards, the water was discarded and the plates were incubated in a fume cupboard to dry. When completely dried, the wells were stained with 135L of 0.1% crystal violet solution and incubated at the room temperature for 30 minutes. The crystal violet solution was washed with water by pipetting and the plates were let to dry again. After complete drying, 125L of 30% acetic acid were added to each well and the plates were analysed using a Thermofisher plate reader. The solutions in the 24-well plates were transferred into 96-well flat bottom plates in order to be read. All the plates were read at 595 nm (O’Toole 2011). A microtiter plate containing 30% acetic acid was used to blank the instrument as a negative control. Microtiter plates containing E.coliand B.subtilis biofilms were used as positive controls.
Validation of the biofilms adhesion on titanium
The adhesion of the biofilms on the titanium chips was analysed photometically by comparing the absorbance of LB broth in which titanium chips were submerged before and after four washes. The absorbance was read using a Thermofisher spectrophotometer at 595 nm and water was used to blank the spectrophotometer.
Silk analysis using Attenuated Total Reflectance technique
Threads of synthetic silk provided from Amazon were partly solubilised by submerging in a calcium chloride:ethanol:water solution in 1:2:8 molar ratio at 37C overnight and natural silk from a common house spider was dissolved in 10% sodium hydroxide at 37C overnight. All the silk samples were let to dry overnight in the fume cupboard. After complete drying, all the silk samples were analysed with Thermofisher FT-IR (Fourrier Transform Infrared Reflectance) apparatus using the ATR (Attenuated Total Reflectance) mode by placing a small amount of the sample on the ATR diamond and run the instrument. Samples of undissolved silk threads were analysed in the same fashion (Coventry University 2018). The obtained spectra were compared to the instrument’s IR (Infrared) spectra library. Atmospheric air as a sample free run was used to blank the instrument and also as a negative control.
Results
The experiment was carried out as described in the methods section. Colonies of successfully transformed cells were observed in the blue/white colour selection for recombinants and non-recombinants. White colonies represent the successfully
transformed plasmids and the blue colonies are the negative control. They can be observed in Figure 5 below.
Figure 5. Blue/white colonies selection of E.coli
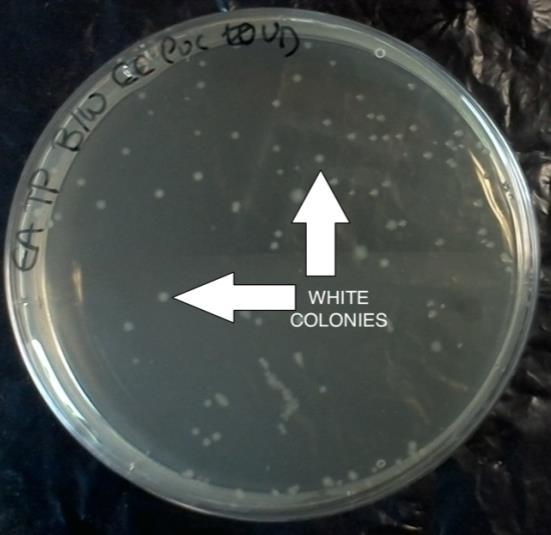
Table 2 presents a summary the number of colonies in the agar plates containing the host bacteria and the plasmid vector insertion, undiluted or diluted 10 -1 , 10 -2 and 10 -3 .
Plate no. Bacteria Vector Dilutions White colonies
1 E.coli pBluescript Undiluted 66 II SK(-) 2 E.coli pBluescript 10 -1 0 II SK(-) 3 E.coli pUC19 Undiluted 118 4 E.coli pUC19 10 -1 6 5 E.coli - (H2O) Undiluted 0 6 B.subtilis pBluescript Undiluted 0 II SK(-) 7 B.subtilis pBluescript 10 -1 0 II SK(-) 8 Control - - 0
Table 2. Number of colonies in the blue/white screening Blue colonies
0
0
0 0 1 0
0
0
A free cell agar plate was used as a negative control, which did not show any colony growth. Two plates containing water and the plasmid vectors were also used as negative controls yet did not produce colonies either. Competent E.coliand B.subtilis without the insert were plated as a positive control. As previously observed, E.coli without any insert produced a blue colony, whilst competent E.coli with insertions of plasmid vectors pBluescript II SK(-) and pUC19 produced white colonies as successful recombinant plasmids. Undiluted E.coli with the vector pBluescript II SK(-) produced 66 white colonies and the viable competent cells can be calculated as follows 66(colonies) x 10ml= 660 = 6.60 x 10 2 cells/ml. On the plate containing






