Using Large Language Models for Assertion Detection in Medical Notes
Cynthia Zhao
Dr. Simona Doboli
Abstract
It is important for healthcare professionals to accurately and efficiently understand clinical notes and determine the presence, absence, or possibility of certain medical problems that may exist in their patients To address this issue, the task of categorizing different assertions, statements expressing facts, can be employed This is also known as clinical assertion detection, and it has been explored using non-generative transformer models to identify assertions classes of medical entities in unstructured clinical notes The main objective of this project is to test the use of generative large language models (LLMs) in the problem of clinical assertion and compare the performance of the generative versus non-generative models While the generative model's outcome may not surpass those of the non-generative model, it nonetheless demonstrates promising potential for application within the healthcare field.
Intro/Background
Clinical assertion detection is known to be a challenging task due to the unstructured nature and free-text format of clinical notes (Chen, 2019) Understanding the valuable information hidden in these clinical notes can be used for various applications such as clinical decision support, disease modeling, medical risk evaluation, medication reconciliation, and quality measurements (Chen, 2019) Previous methods have used non-generative transformer models such as BERT (van Aken et al 2021) to label assertions of medical terms from text
Methods
We used the pretrained model Mistral-7B (Jiang et al 2023) and first tested out different prompts to see which prompt type will elicit the desired response We then finetuned Mistral using LoRA (Hu et al , 2021) and tested out the performance using an unseen i2b2 testing dataset
Dataset Description
The 2010 i2b2 Assertion Task dataset provides a corpus of assertions, concepts, and relations for clinical discharge summaries (Uzuner et al , 2011) We will only be using the present, absent, and possible assertion categories
Format:
c=”concept text” offset||t=”concept type”||a=”assertion value”
Example:
c=”diabetes” 2:14 2:14||t=”problem”||a=”absent”
Figure 1: i2b2 Clinical Assertion Annotated File Format and Examples
Fine-tuning
Methods
Training Data Testing Data
Present Cases 4622 8622
Absent Cases 1596 2594
Possible Cases 309 652
Total Cases 6527 11, 868
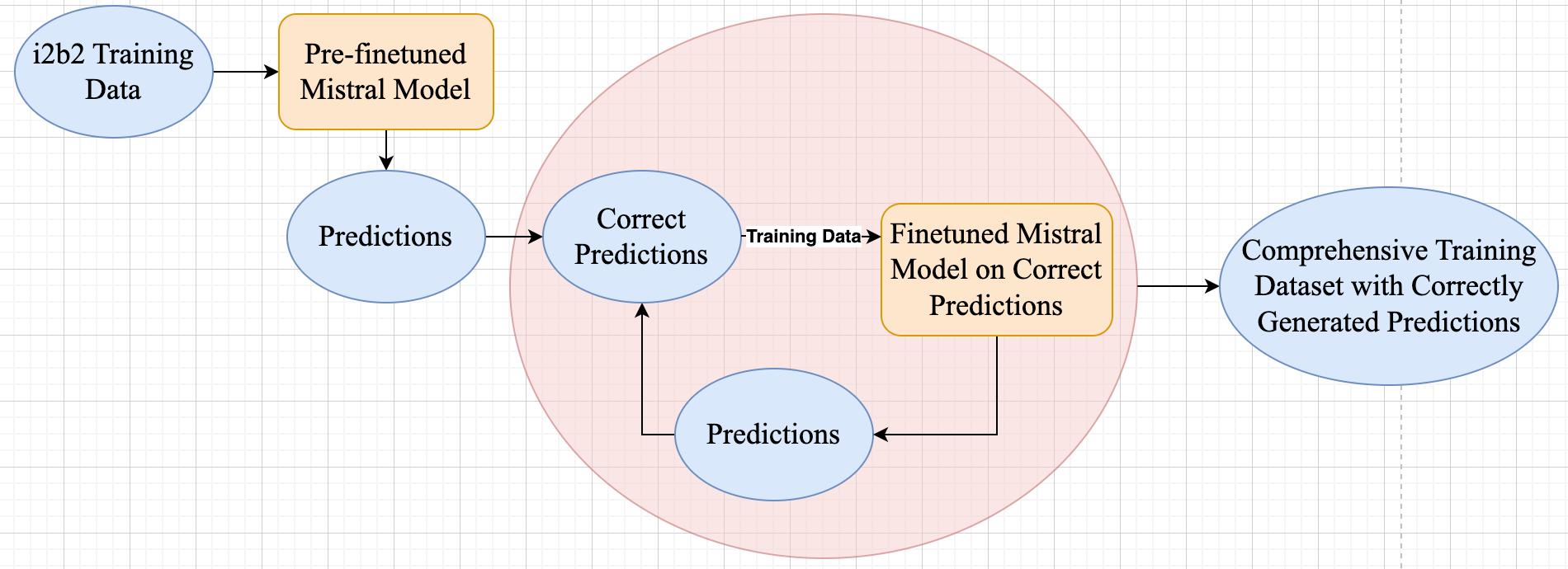
The i2b2 dataset lacks answers containing both assertions and justifications, so we needed to create the desired response format that is necessary for finetuning our model We had to adopt an iterative strategy which involved generating answers with the pre-finetuned model on the i2b2 training dataset and using the correctly generated responses to improve the model's performance on the dataset incrementally This iterative process continued until we compiled a comprehensive training dataset containing accurately formatted responses Once this training dataset was established, we could then apply the model to make predictions on the provided i2b2 testing dataset
Prompt Engineering
We found that the generated answers were greatly dependent on whichever prompt format that was used We used the prompting styles in Table 2 to create our prompt format as shown in Figure 4 We found that the prefinetuned model generated the best responses when using one-shot prompting (providing the prompt with an example of each case: present, absent, and possible) We used zero-shot prompting for our finetuned model.
Table 4: Prompting Styles and Definitions
Prompting Style Definitions
Multiple Choice
Prompting (MCP)
Chain-of-Thought
Prompting (CoT)
Provide multiple choices with each answer associated with “A”, “B”, “C”, etc (Robinson et al., 2023).
Encourage the model to generate solutions with step-bystep reasoning by adding phrases like “Let’s think step by step” before the answer (Kojima et al., 2022).
One-shot Prompting Ask the model a question and provide an example (or shot) of “question-explanation-answers triplets” in the prompt (Liévin et al., 2023).
Zero-shot Prompting Ask the model a question without providing any examples.
<s> [INST]
Patient Note: "lungs : breath sounds clear and equal bilaterally , no retractions "
Considering the information in the patient note, which of the following is the most suitable classification for determining the patient's association with retractions?
A Present B Absent C Possible
Let’s derive the differential diagnosis step by step
Answer: [/INST]
Based on the patient note, the most suitable classification for determining the patient's association with retractions would be "Absent" The patient's breath sounds are clear and equal bilaterally, and there are no retractions present Therefore, among A through C, the answer is B Absent </s>
Results
Training and Validation Loss over Epochs
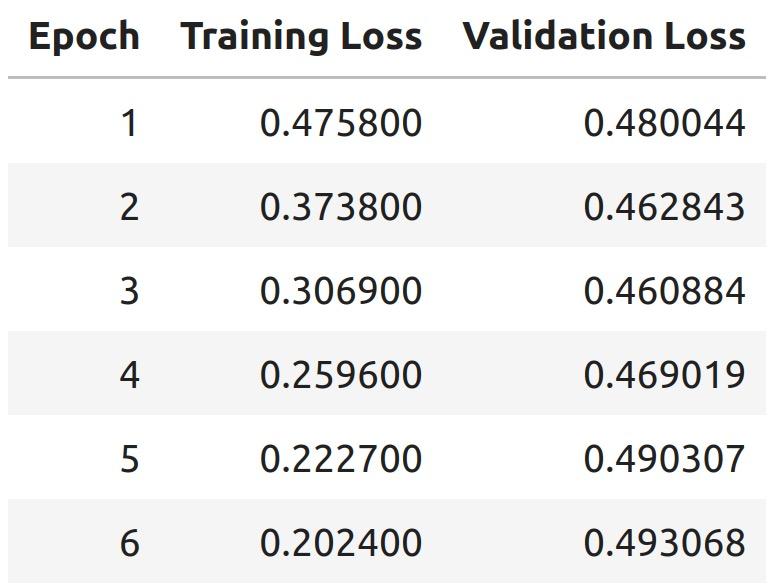

We trained on 6 epochs and used a learning rate of 2e-5 Our comprehensive training dataset was generated using a checkpoint at epoch 3 because it had the lowest validation loss (how well the model is able generalize unseen data)
Precision: the accuracy of positive predictions. Recall (sensitivity): the model's ability to identify all relevant instances.
F1-score (micro): a single value that balances the precision and recall metrics. The micro-average gives equal weight to each prediction

F1-score (micro): 0.89
Present (4622 cases): precision: 0.97, recall: 0.87
Absent (1596 cases): precision: 0.93 recall: 0.94
Possible (309 cases): precision: 0.33, recall: 0.86
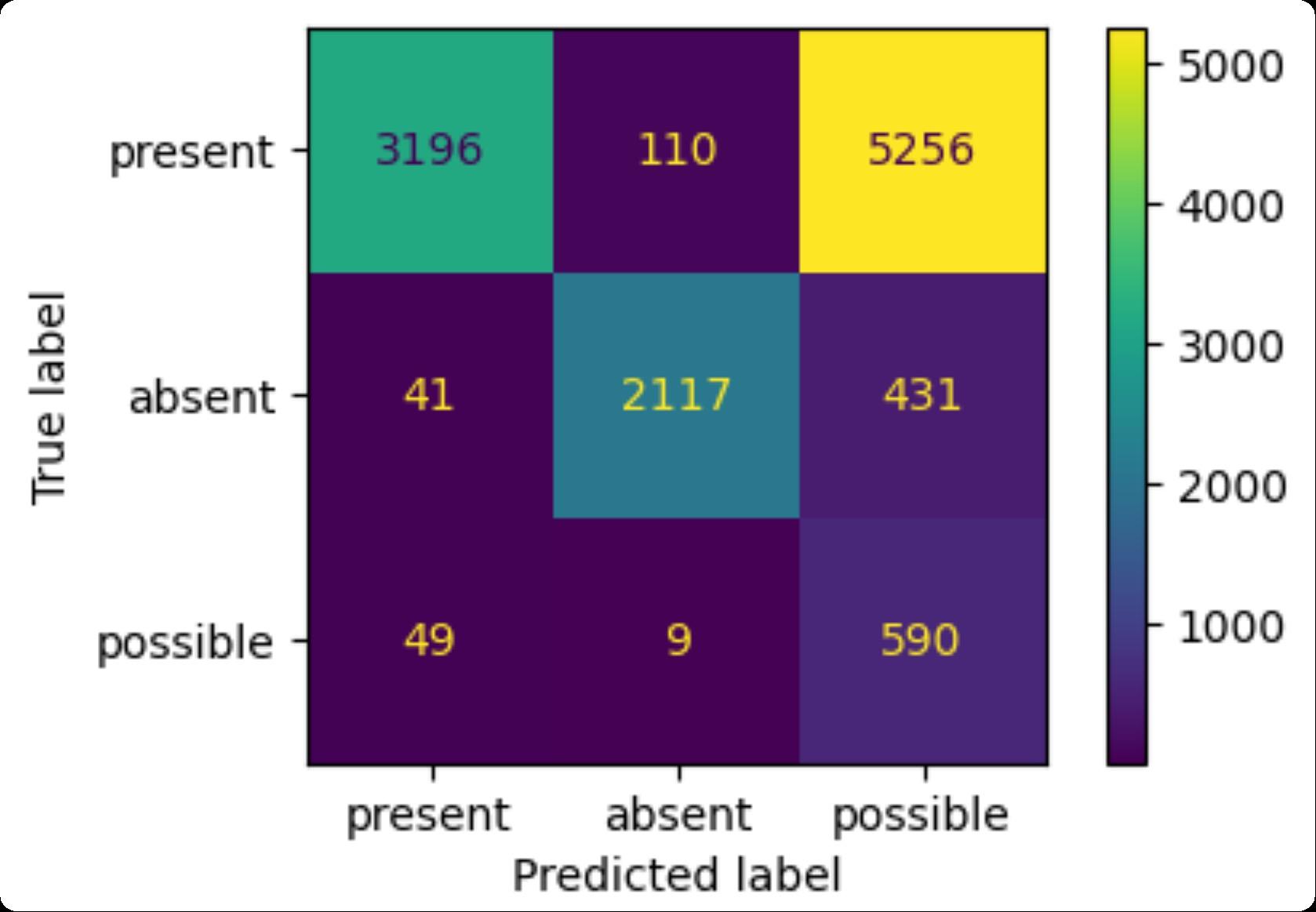
F1-score (micro): 0.50
Present (8622 cases): precision: 0.97, recall: 0.37
Absent (2594 cases): precision: 0.95, recall: 0.82
Possible (652 cases): precision: 0.10, recall: 0.91
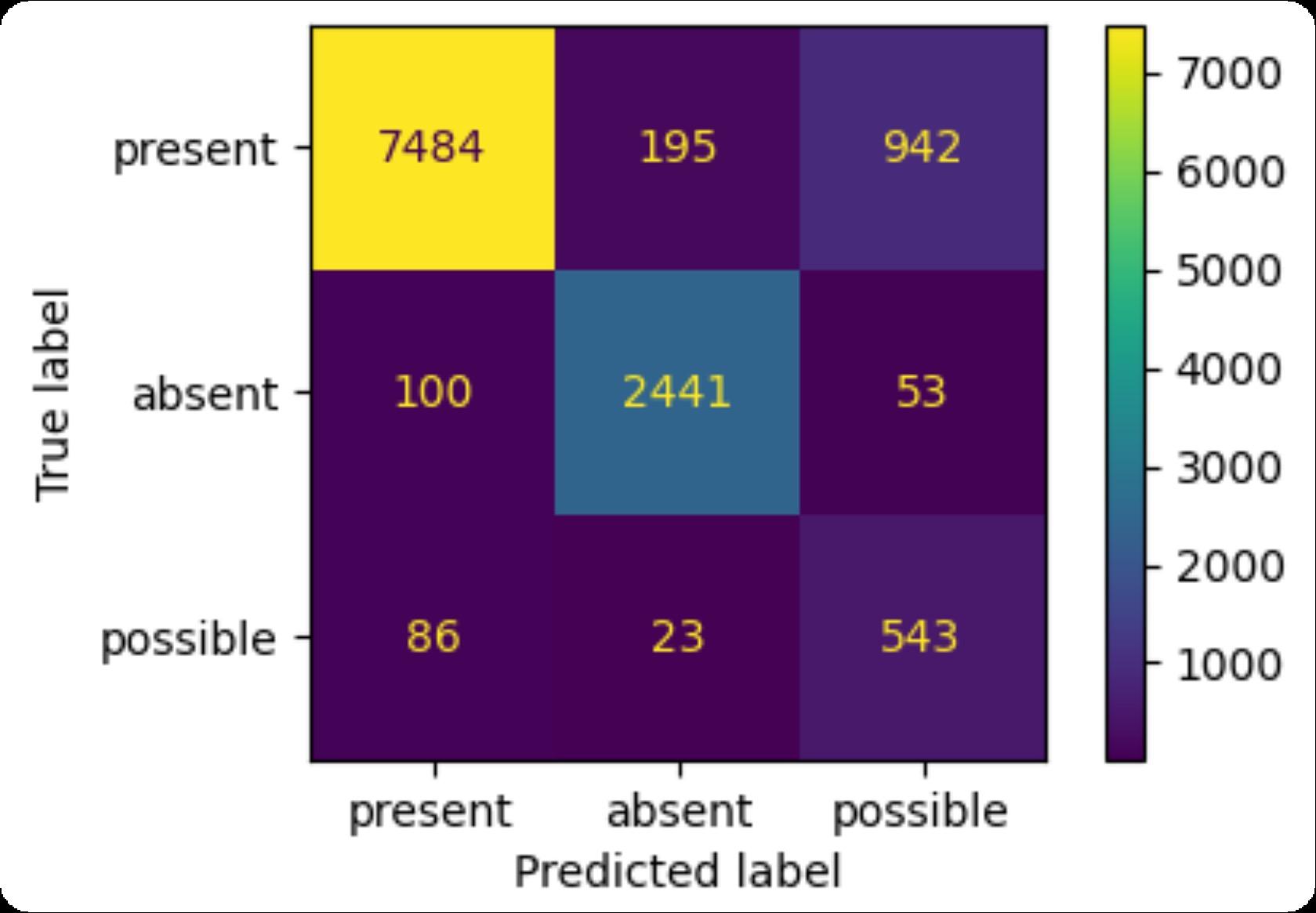
F1-score (micro): 0.88
Present (8622 cases): precision: 0.98, recall: 0.87
Absent (2594 cases): precision: 0.92, recall: 0.94
Possible (652 cases): precision: 0.35, recall: 0.83
BioClinicalBERT Confusion Matrix and Metrics
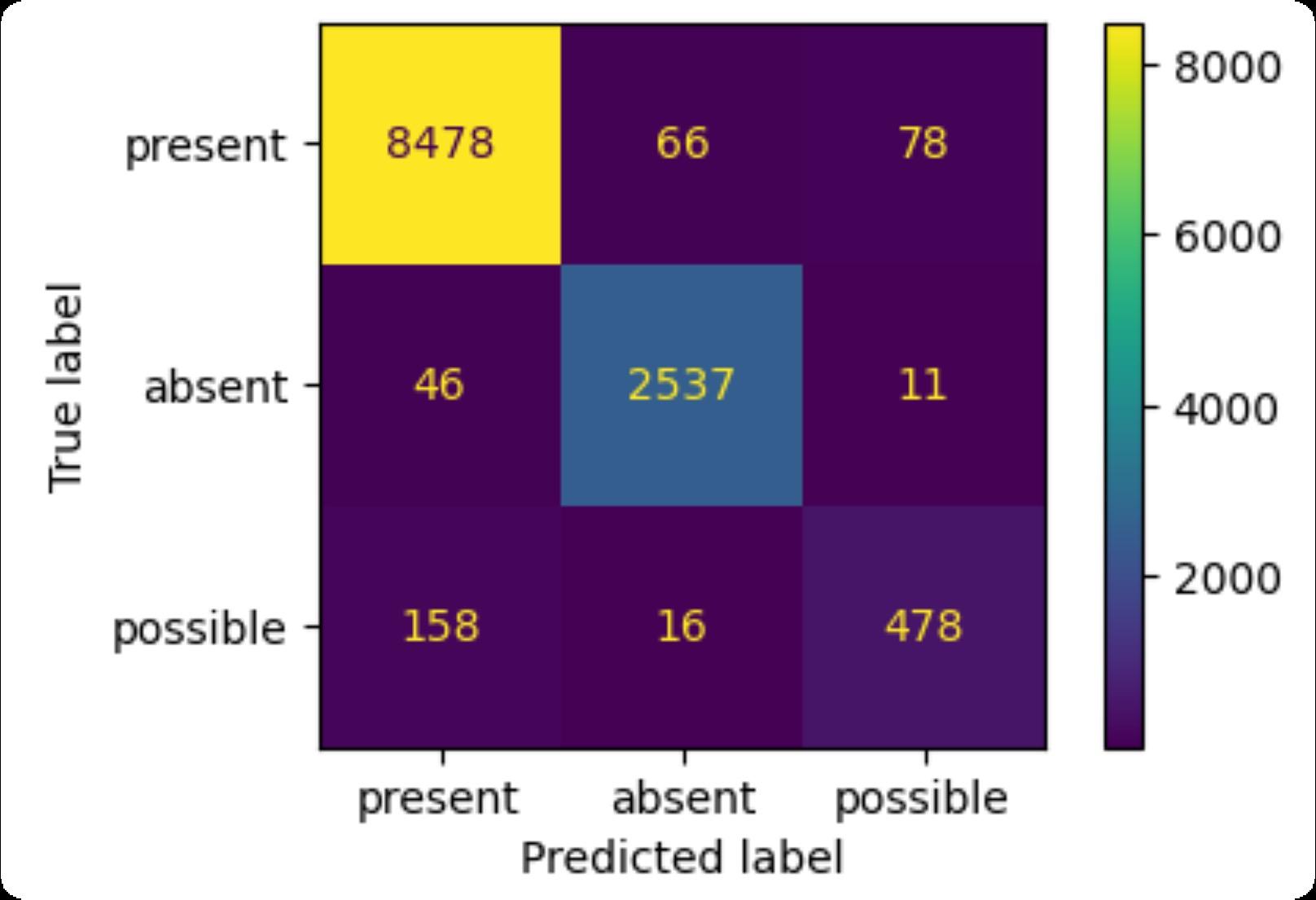
F1-score (micro): 0.97
Present (8622 cases): precision: 0.98, recall: 0.98
Absent (2594 cases): precision: 0.97, recall: 0.98
Possible (652 cases): precision: 0.84, recall: 0.73

Conclusions
Although there were tremendous improvements in the finetune generated predictions, the model continues to struggle with the accuracy in identifying positive possible cases It mistakenly classifies Present and Absent cases as Possible (low precision) However, the model rarely mistakes actual possible cases as Present or Absent (high recall) Our results do not outperform those of the BERT model, primarily because of the increased complexity involved in generating justifications for the predicted assertions Unlike our approach, the BERT model can utilize the entire provided i2b2 training dataset, providing it with a large set of data points for fine tuning This limitation restricted us to using only correct model generated responses for fine-tuning, resulting in fewer data points Despite these challenges, our results are promising and show the potential of generative models in the healthcare field.
Future Work
Further research and exploration into medical question and answer prompt engineering and automatic prompt generation to generate the optimal prompt format could result in better performance Additionally, fine tuning Mistral with other clinical datasets as well as, fine tuning other generative models on medical clinical notes could be areas to be further explored It would be insightful to compare the results of how various datasets can influence the performance of Mistral It would also be interesting to compare the performances of different generative LLMs with the task of clinical assertion

References
www.PosterPresentations.com
Chapman, W W Bridewell, W Hanbury, P Cooper, G F & Buchanan, B G (2001) A simple algorithm for identifying negated findings and diseases in discharge summaries Journal of biomedical informatics, 34(5), 301–310 https //doi org 10 1006/jbin 2001 1029 Chen, L (2019) Attention-based deep learning system for negation and assertion detection in clinical notes International Journal of Artificial Intelligence & Applications, 10(01), 1–9 https://doi org 10 5121/ijaia 2019 10101 Hu, E Shen, Y Wallis, P Allen-Zhu, Z Li, Y Wang, S & Chen, W 2021 LoRA Low-Rank Adaptation of Large Language Models https //doi org 10 48550/arXiv 2106 09685 Jiang, A Q Sablayrolles A Mensch, A Bamford, C , Chaploy D S Casas, D de las, Bressand, F Lengyel, G Lample G , Saulnier, L Lavaud, L R Lachaux, M -A Stock, P , Le Scao T Lavril, T Wang, T Lacroix, T & El Sayed, W (2023) Mistral 7B https://doi org/10 48550/arXiv 2310 06825 Liévin V Hother C E Motzfeldt A G & Winther O 2023) Can large language models reason about medical questions? Patterns, 5 3 https //doi org 10 1016/j patter 2024 100943 Perera S Sheth A Thirunarayan K Nair, S & Shah, N 2013 Challenges in understanding clinical notes Proceedings of the 2013 International Workshop on Data Management & Analytics for Healthcare - DARE 13 https://doi org 10 1145 2512410 2512427 Robinson, Rytting C M & Wingate, D (2023) Leveraging Large Language Models for Multiple Choice Question Answering https //doi org https://doi org/10 48550/arXiv 2210 12353 Sergeeva, E Zhu, H Prinsen, P & Tahmasebi, A (2019) Negation Scope Detection in Clinical Notes and Scientific Abstracts A Feature-enriched LSTM-based Approach AMIA Joint Summits on Translational Science proceedings AMIA Joint Summits on Translational Science, 2019 212–221 Wang, S Tang, L Majety A Rousseau, J F Shih, G Ding, Y & Peng, Y 2022 Trustworthy assertion classification through prompting Journal of biomedical informatics, 132, 104139 https://doi org 10 1016/j jbi 2022 104139 Qian, Z Li, P Zhu, Q Zhou, G Luo, Z & Luo, W 2016 Speculation and negation scope detection via Convolutional Neural Networks Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing https //doi org 10 18653/v1/d16-1078 van Aken B Trajanovska I Siu, A Mayrdorfer M Budde K & Loeser A (2021) Assertion detection in clinical notes: Medical Language Models to the rescue? Proceedings of the Second Workshop on Natural Language Processing for Medical Conversations https //doi org 10 18653/v1/2021 nlpmc-1 5 Uzuner Ö South, B R Shen, S & DuVall, S L 2011) 2010 2b2/VA challenge on concepts, assertions, and relations in clinical text Journal of the American Medical Informatics Association JAMIA, 18 5), 552–556 https://doi org/10 1136/amiajnl-2011-000203 Zhong, R Lee, K Zhang, Z , & Klein, D (2021) Adapting language models for zero-shot learning by meta-tuning on dataset and prompt collections Findings of the Association for Computational Linguistics EMNLP 2021 https://doi org 10 18653/v1 2021 findings-emnlp 244 Acknowledgement This work could not have been possible without the guidance and support from my advisor Dr Simona Doboli
Assertion Category Definition Medical Entity Clinical Note Context Present Problems associated with the patient can be present. This is the default category for medical problems and it contains information that does not fit the definition of any of the other assertion categories increasing weight gain the patient has had increasing weight gain Absent The note asserts that the problem does not exist in the patient. This category also includes mentions where it is stated that the patient HAD a problem, but no longer does. pain patient denies pain Possible The note asserts that the patient may have a problem, but there is uncertainty expressed in the note. Possible takes precedence over absent, so terms like “probably not” or “unlikely” categorize problems as being possible just as “probably” and “likely” do. asthma exacerbation This is very likely to be an asthma exacerbation
Table 3: Number of Each Case for i2b2 Training and Testing Dataset
Table 1: i2b2 Clinical Assertion Definitions and Examples
Mistral Pre-finetuned Results on i2b2 Testing Data (one-shot prompt)
Mistral Post-finetuned Results on i2b2 Testing Data
Mistral Post-finetuned Results on i2b2 Training Data
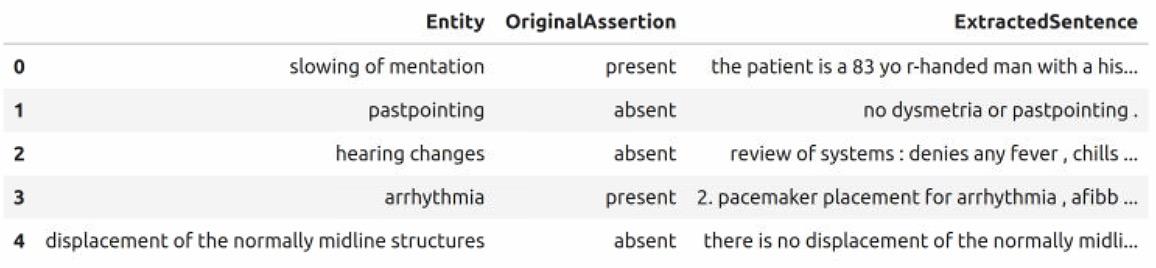
Table 2: Data Frame with Extracted Medical Entity, Assertion, and Context
Figure 3: Our Desired Prompt and Model Generated Response Format
Figure 2: Iterative Strategy to Generate Training Dataset










