Authorbios
Dr.MojtabaAghajaniDelavarisaResearchFellowatAthabascaUniversity.Hereceived hisB.Sc.inMechanicalEngineeringfromAmirkabirUniversityofTechnologyin2001, M.Sc.,andPh.D.inMechanicalEngineeringfromMazandaranUniversityin2003and 2010,respectively.HeworkedattheBabolNoshirvaniUniversityofTechnologyinIranas anassociateprofessorfrom2010until2019.ThenhejoinedprofessorWang’sresearch groupatAthabascaUniversity.Dr.Delavarhasover20yearsofexperienceinmodeling variousindustrialandbiologicalsystemsusingdifferentmodelingschemesincludingmathematicalandnumericalanalysisandsimulation.Hehasauthored/coauthoredover100papersincludingmorethan60peer-reviewedjournalpapers.
Dr.JunyeWangisaProfessorandtheCampusAlbertaInnovationProgram(CAIP) ResearchChairatAthabascaUniversity,Canada.HereceivedhisM.Sc.inThermo-physics fromHarbinShipbuildingEngineeringInstituteandPh.D.inChemicalandMechanicalEngineeringfromEastChinaUniversityofScienceandTechnologyin1989and1996, respectively.ThenhejoinedShanghaiJiaotongUniversityasanassociateprofessorin 1996.From1999till2013,heworkedattheUniversitiesofSheffield,Greenwich,and Loughborough,andScottishCropResearchInstitute,andRothamstedResearch,UK,asa researchassociate,researchscientist,andprincipalresearchscientist,respectively.Dr. Wanghasover30years’experienceinmultiscaleandmultidisciplinarymodelingandisan internationallyrecognizedleaderinmicrobiology,bioenergy,andenvironment.Hehas authored/coauthoredover160papers,includingover120refereedjournalpapers,and servesasassociateeditorandeditorialboardmemberonseveralinternationaljournals.He isalsoareviewerofpapersforover130internationaljournals.
Introduction
1.1Background
Microorganismsareoneofthemostextensivelyspreadandsuccessfulformsoflifeon earth,livinginnaturalhabitats,industrialequipment,andclinicaldevices(Stoodleyetal., 2002).Innaturalhabitats,mostmicroorganismsthriveincommunitiesratherthan planktoniccells.Microorganismstendtogrownotonlyasasinglespeciescommunityof pathogensandnonpathogensbutalsoasmultispeciescommunities(Donlan,2002; Kragh etal.,2016; Melaughetal.,2016).Biofilmscanadheretovarioussolid liquid, air liquid,orliquid liquidinterfaces,suchasimplantedmedicaldevices,livingtissues, industrialorpotablewaterpipes,anddevices,ornaturalaquaticsurfaces.Itiswidely acceptedthatbiofilmsaretheprimarylifestyleofbacteria,withauniquephenotypein termsofgenetranscriptionandgrowthrate.Itwasdiscoveredthat99.9%ofallbacteria adheredtoanaqueoussurfaceandproliferatedtogetherastheircrucialsurvivalstrategy (Geeseyetal.,1977; DonlanandCosterton,2002).Insomesituations,biofilmscanform asswimmingclustersinactivatedslimesinanaquaticenvironmentwheretheyarenot adherenttoasolidsubstratum.Theratioofepilithictoplanktonicmicroorganismsis greaterthan1000 10,000:1(https://www.cs.montana.edu/webworks/projects/stevesbook/ contents/chapters/chapter001/section003/green/page002.html).
Thecharacteristicsofbiofilmsinnaturalhabitatscandiffersubstantially,asevidencedby scanningelectronmicrographsofbiofilmsfromanindustrialwaterdeviceandariver stream(Fig.1.1)(Donlan,2002; Suarezetal.,2019).Asmulticellularcommunitiesof microbialcells,biofilmsareimmersedinaself-producedmatrixofextracellularpolymeric molecules(EPS).Fluorescenceinsituhybridization(FISH)analysesofbiofilm cryosectionsshowedthattheZ400biofilmwaslikelystratified,where Nitrospira was moreabundantinthemiddleofthebiofilmandtheanaerobicanammoxbacteriapresented inthedeeperlayerswhile Nitrosomonas biovolumewasthesamealongthedepthgradient (Fig.1.1AandB)(Suarezetal.,2019).InthethinZ50biofilms,nostratificationwas observedastheammoniaoxidizingbacteria(AOB)andtheoxygenatedwaterandnitrite oxidizingbacteria(NOB)populationswerelocatedsidebyside.Manymicrobial communitieshavecelldensitiesrangingfrom108 to1011 cellspergramofwetmass, comprising1millionto100billioncells(ByrneandDrasdo,2009; Gebreyohannesetal., 2019).Inhoneycombconfigurations,streambiofilmscancohabitwithalgae,EPS,and

Figure1.1
Physicalstructureand“buildingblocks”ofbiofilms:(A)Fluorescenceinsituhybridization(FISH) imagesofaZ400biofilmcryosectionwiththewater biofilminterfaceonthetop(Green:
diatomcells(Battinetal.,2003).Althoughthechemistryandphysiologyofbiofilms mightdifferduetodifferentbacteriaandtheirsurroundingenvironment,thebiomassof biofilmscontainsaround90%EPS,whichaddstotheresemblanceofthemushroom-like structure(Jamaletal.,2018; StewartandFranklin,2008).
Biofilmsinpopulationsaretypicallymadeupofnumerouscoloniesofvariousbacterial species(Vidakovicetal.,2018).Wastewatertreatmentsystemsconsistofthousandsof operatingtaxonomicgroupsatthespecieslevel(Lawetal.,2016; Saundersetal.,2016). Inthenaturalenvironment,biofilmscontainover750distinctspecies(Leyetal.,2006), andoralbiofilmscontainavarietyofthousandsofmicroorganismsasanembraceof bacteriaandeukaryotes.
Microorganismsthrivinginacommunallifestylehavesomebenefitsoverplanktoniccells: (1)increasedrigiditytoerosionbystreamflowstress;(2)enhancedresistanceto antimicrobials;(3)higherbiomassdensityforthetreatmentofvariousinorganicand inorganicsubstratesinbioengineering;(4)improvedcell-to-cellinteraction,genedelivery, andmetabolicworkloadsharing;and(5)providedheterogeneousstructuresfordiffusion andconsumptionofnutrientsbenefits(WangandZhang,2010).
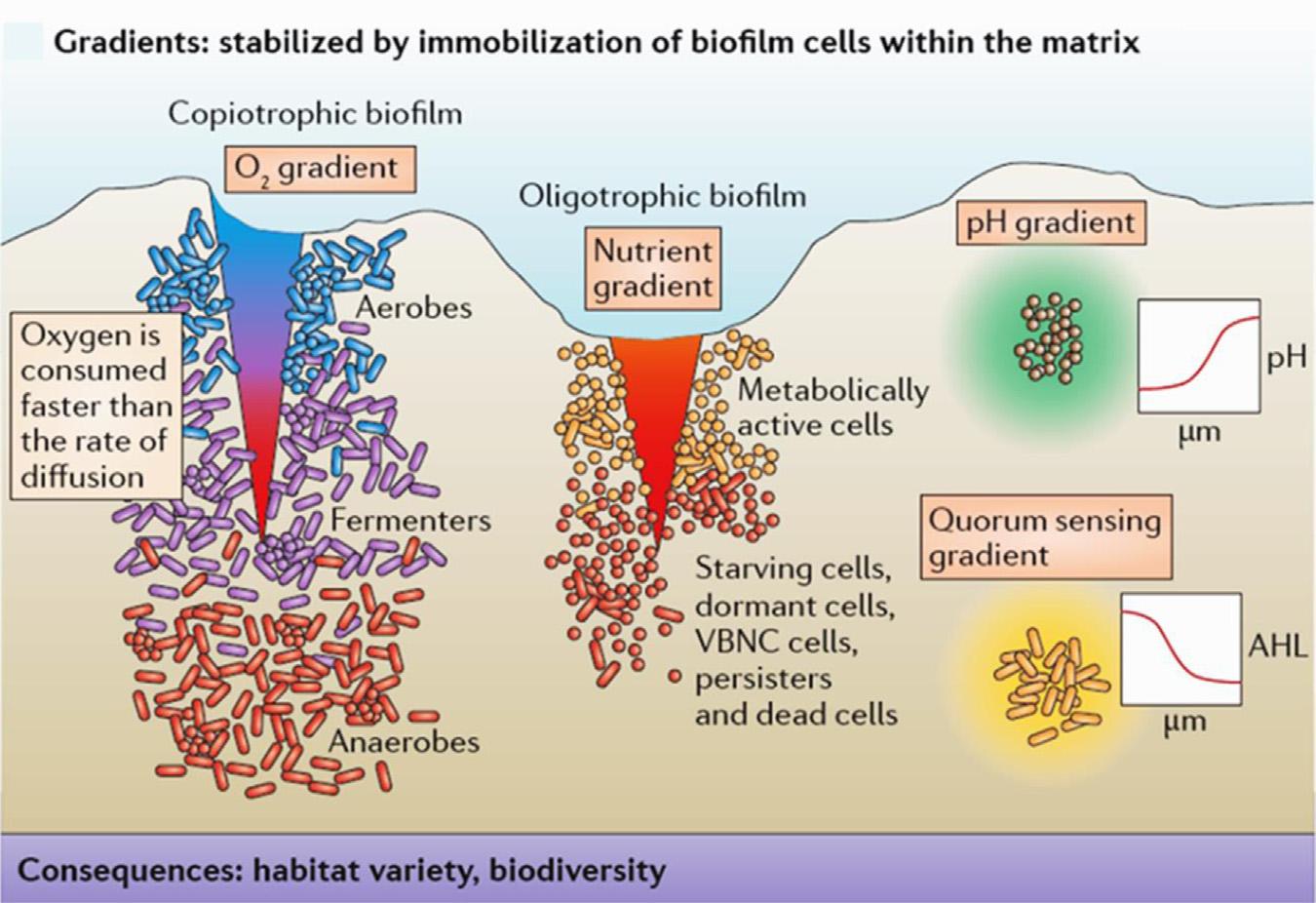
Fig.1.1AandB depictstheheterogeneityoftheEPSmatrixdevelopment(Suarezetal., 2019).Deterministicassemblyinbiofilmsdependsonspecificmechanisms.Forexample, structuredmicroenvironmentsmaylimitthediffusionofelectrondonorsandacceptorsin biofilms,resultinginformsteepgradients.Thethicknessoftheliquidboundarylayerthat limitsdiffusionofsolublesubstrates,includingdissolvedoxygen(DO),fromthebulk liquidtothebiofilmwasdeterminedbycomparingtheammoniumoxidationrates calculatedbythemodeltothosemeasuredduringthenitrogentransformationactivitytests (Suarezetal.,2019).Microorganismsthatliveinsoilaggregatesorhabitatscanadapt theiractivityandcompositioninresponsetothechangesinnutrientandenvironmental circumstances(YoungandCrawford,2004).Thepersistentgradientswerecreatedto enabledistinctlocalhabitatsonafinescale.Microorganismsinanaerobiccopiotrophic biofilmstratifyintermsofoxygenavailability.Becauseoxygenisextractedbyaerobic organismsinthehigherlayersofthebiofilm,alayerofanaerobesdevelopsinthelower layersofthebiofilmduetooxygendepletion.Thiscanbeattributedtothefactthatthe rateofoxygenconsumptionexceedstherateofdiffusion.
Nitrosomonas,Red: Nitrospira.Yellow: Nitrotoga.Blue: Brocadia.Gray:SYTO),(B)FISHimagesofa Z50biofilmcryosectionwiththewater biofilminterfaceonthetop(Green: Nitrosomonas,Red: Nitrospira.Yellow: Nitrotoga.Blue: Brocadia.Gray:SYTO)(Suarezetal.,2019),and(C)SEM (scanningelectronmicrograph)imageofanativebiofilmformedonlowcarbonsteelintersurface inanindustrialwatersystemoveran8-weekperiod,scalebar,20 mm(Donlan,2002).
Flemmingetal.(2016) showedthatbacteriaintheupperlayersofaerobicoligotrophic biofilmsarelikelytoconsumethemajorityofthenutrients,causingstarving microorganismsinthebottomlayers(Fig.1.2).Thesemicrobesadapttosluggishgrowth states,suchasinactivecells,orevencelldeath.Othergradientsinbiofilmsproducedby heterotrophicmetabolismincludepHgradientsandgradientsofsignalingchemicalsthat varywithdistancefromgeneratingcells.
Mostelementsinwater,soil,sediment,andsubsurfaceenvironmentsarecycled biogeochemicallybymicroorganisms(Battinetal.,2008; EhrlichandNewman,2008; Bhanjaetal.,2019; BhanjaandWang,2020, 2021; Meckenstocketal.,2015).The metabolismofterrestrialorganiccarboninfreshwaterenvironmentscontributes significantlytotheemissionofcarbondioxideintotheatmosphere(Battinetal.,2008).
Biofilmsthroughwell-definedinteractionsforspeciallydesignedpurposeshavebeenused inarangeofbioindustries,suchasimprovedsafetyoffoods(Stiles,1996),biosynthesis andbioremediation(Tsoietal.,2019),andinhibitionofpathogenicgrowthin nonfermented,refrigeratedfoods(Gombas,1989).
Biofilmsarefoundnumerouslyinawiderangeofinfectiousdiseasesinbothclinicaland publichealthsettings(Donlan,2002).Theycanpersistentlygrowonbothbioticand abioticsurfaces,suchasahumantoothorlung,acow’sintestine,orrockimmersedina fast-movingstream.Biofilmscancolonizeonvariousmedicaldevices,including
Figure1.2
Biofilmheterogeneitycharacteristics,andsocialcompetitionandcooperation(Flemmingetal., 2016).
intrauterinecontraceptivedevices,prostheticmedicaldevices,catheters,heartvalves, implantdevices,dentalmaterials,andcontactlenses,resultinginavarietyofdeviceassociatedillnesses.Clinicalinvestigationshaveemphasizedthesignificanceofbiofilmsin producinghumanillnesses,accountingforupto60%ofallinfections(ChenandWen, 2011).Biofilmshaveasubstantialinfluenceontheindustrialenvironment,including biofouling,biocorrosion,oilfieldsouring,andeffluent(KlapperandDockery2002).
1.2Historyofbiofilmsstudies
1.2.1Biofilmandbioaggregates
Mostofthehistoryofmicrobiologyhasclassifiedmicrobesasplanktonic,freelyswinging cellsinanutritionallyabundantaquaticenvironmentdependingontheirgrowth environment.However,themajorityofmicrobesinaquaticenvironmentsarenotfreesuspendedmicroorganismsbutinsteaddwellonimmergedsurfaces,wheretheyform organizedcoloniesknownasbiofilms.Untilthe17thcentury,biofilmswerefirst characterizedbyVanLeeuwenhoekfromDelft(1632 1723),whoobservedmicrobial coloniesonshavingsofplaquefromhisteethwithhiscrudemicroscope(Dobell,1932). Thegenestranscribedbybiofilmbacteriadifferfromthosedonebyplanktonic counterparts(Henrici1933).Asearlyas1933,theword“biofilm”wastermedintechnical andenvironmentalmicrobiology(Henrici,1933; ZobellandAllen,1935).
Earlierstudiesofbiofilmsweremainlyinwastewaterfiltering,industrialequipment biofouling,anddentalplaque.LouisPasteur(1822 95)discoveredandsketchedbacterial aggregationastheoriginofaceticwine(Hoiby,2014, 2017).Forbiofilmdescription,much ofthebiofilmstudiesdependonthedevelopmentofinstruments,suchasscanningelectron microscopy(SEM)ortraditionalmicrobialculturetechniques.Twokeyadvancesinbiofilm researchoccurredinthepastfewdecades:thefirstonewastheuseoftheconfocallaser scanningmicroscopetoanalyzebiofilmultrastructure,andthesecondwasthestudyofthe genesrelatedtocelladhesionandbiofilmdevelopment(Battinetal.,2007).
RobertKoch(1843 1910)wasapioneeringmicrobiologistwhoattemptedtoisolateand characterizemicrobesfromtheirnaturalhabitats.Thepureculturetechniqueallowsthe highlevelofreplicationandhandlingtobetterunderstandhowmicroorganismsculture respondtodifferentsituations.Inmicrobiology,thisledtothe“GoldenAge”sinceit providedanopportunitytostudyorganismsinthelabinconsiderabledetailundertightly controlledsettings(suchasgrowthconditions)orwithgeneticmodifications.Because mostbacteriaexistasmultispeciescommunities,pureculturecannotdescribemultispecies communityphenomena.Becauseofthis,biofilmresearchincreasinglyemploysmethods previouslyreservedforstudyingpopulations,suchasmeta-omics-basedmethodologies andhigh-resolutionscanning(Battinetal.,2007).
Thebiofilmresearchwasrevivedinthe1970and1980s.Microorganismsthatgrewona surfacewereconsideredasflatanduniformfilmsofcellsencasedinslime.Thenatureof biofilmsinthehumanhostcanbesignificantlydifferentfromthatatthesurfacesexposedto theenvironment.Inthemid-20thcentury,scientistsdiscoveredthatbiofilmsareprimarily composedofdiversepopulationsofbacteria(Kraghetal.,2016; Melaughetal.,2016).
Environmentalchangesorthemetabolismandmigrationofothercommunitiescause dynamicchangesinnutritionalgradientsforeachpopulation.Thereforeacertainbiofilm community’ssuccessrateishighlydependentontheothermembers’performanceaswell asregulationandcontrolofquorumsensing(QS).Inmedicalmicrobiology, Nickeletal. (1985) introducedtheconceptof“biofilm”growthintheirpioneerresearchintothe physiologicalandbiochemicalfeaturesofbacteria.Theyfoundthatbiofilm-growing bacteriaexhibitedgreaterresistancethanplanktonicallydevelopingbacteria.
Theterm“film”isinsufficienttodescribebacteriallife;hencethename“biofilm”is somehowmisleading.Biofilmcellsdonotjustpileupontopofeachother;instead,they createcomplex,self-organizedstructuresfromthebottomup.Thereareawiderangeof biofilmsthatdifferfromonemicroorganismpopulationtoanother,dependingonthetype ofbacterialcommunityandenvironmentalconditions,suchas“streamers,”“columns,” “mushrooms,”“bioclusters,”“microbialmats,”“microcolonies,”and“bioaggregates” (Atkinsonetal.,1967).Apparently,becausethesetermsarealreadywidelyusedintheir respectivefields,whetherthetermofbiofilmsneedstobereplacedoriscontroversial (Baveye,2020; Flemmingetal.,2021).Asaresult,theword“biofilm”herereferstothe extensivebacterialattachmentonsurfacestodifferentiateaggregatedbacteriafromfree suspended“planktonic”bacteria.Biofilmsrepresentmulticellularmicrobialaggregatesthat arenotlimitedtomicrobialfilmsonsurfaces.
1.2.2Biofilmmodeling
Modelingofbiofilmsdatesbacktothe1970s.Biofilmsweredescribedashomogeneous biomassofasinglemicrobialspecies(AtkinsonandDavies,1974; Williamsonand McCarty1976; Harremoes,1976; RittmannandMcCarty,1980).Thefirst-generation approacheswerequickandeasytoimplement,sometimesusingasimplespreadsheet. However,theydescribeonlyuniformgrowthandcannotcaptureallnonevengrowth. Followingthat,stratifieddynamicmodelsweredevelopedassecond-generationmodels (WannerandGujer,1986).Theselayeredmodelsweredevelopedtorepresentinteractions betweenmultiplesubstratesandspeciesinsidethebiofilm.Theywere,however,unableto accountforthetypicalstructuraldiversity.Therefore,thethird-generationmodelsare developedtorepresentthecharacteristicstructuralheterogeneityinsideabiofilm.
Inthethird-generationmodels,thecoupledmodelofbiomassgrowthsubmodels(e.g., detachment,decay,biomassdivision,andspreading),andtransportsubmodels(e.g.,flow,
substratetransport,andreactions)aresolvedusingthefinitedifferencemethod(FDM)or thefinitevolumemethod(FVM)(Picioreanuetal.,2004).TheNavier Stokesequations, andreactivetransportequationshavebeendevelopedtosimulatebiofilmgrowths (Picioreanuetal.,1998a,b,2001, 2004; Eberletal.,2001; Pizarroetal.,2001; Laspidou andRittmann,2004; Xavieretal.,2005).Bothcontinuummodelsanddiscretemodelsfor biofilmgrowthstartedtobedevelopedtodescribetheformationofmultidimensional biofilmmorphologyinthe1990sandcarrytotodayasthethird-generationmathematical models.Inthecontinuummodels,biofilmgrowthisdescribedusingatransportequation (Eberletal.,2001)andcoupleswithhydrodynamicsandsubstratetransportprocesses.The term“discrete”denotesthatthesystem’sspace,time,andcharacteristicscanonlyhavea finitenumberofstates.Intheearlystage,adomainspaceisdiscretizedintoregulargrid elements,establishingalattice.Thelarge-scaledynamicsofmasstransportand hydrodynamicsaresolvedusingtheFDMortheFVMwhilethebiofilmstructuresaredone usingcellularautomata(CA)orindividual-basedmodel(IbM).
Avarietyofdiscreterulescanresultinfundamentallydiverseandratherrandomstructures forCA.Therulesemployedtosimulateinteractionsatthelocallevelcanbeinspiredonly bybiologicalprinciples,asopposedtoanalysisaccordingtoamathematicalandphysical framework(Alpkvistetal.,2006).IbMwasappliedtodescribebiofilmgrowth(Picioreanu etal.,2004; Yanetal.,2017).Asaresultoftheactionsaswellasinteractionsofthe biomassunitswithoneanotherandtheirsurroundings,thecomplexmorphologyofbiofilms evolves.ThoughCAissimpletobuild,theincreasingbiomasscanonlytravelinarestricted numberoflatticedirections.Additionally,biofilmsaresupposedtobelivingsystemsthat areintrinsicallystochastic.TheCAmodeldescribesstochasticity(Laspidouetal.,2010).
Thethird-generationbiofilmmodelsincludehydrodynamics,masstransfer,and transformations(Eberletal.,2001).Therefore,theyareabletorepresentthe2Dor3D microbialbiofilmmorphologies(Picioreanuetal.,1999; Nogueraetal.,1999a).EPSand QSprocesseshavebeenconsideredinthethird-generationmodels.Biofilmbiomassis madeupofbiologicalgelEPSandwater,accordingtothebiofilm’sconceptualmodelby CoganandKeener(2004).Followingtheconcept, Winstanleyetal.(2011, 2015) developedaconceptualmodeltostudybiofilmblockageinsingleporespace.Later, Kreft andWimpenny(2001) introducedtheIbMtoapproximateEPScreationandspread.The synthesisofEPSwasstoichiometricallyconnectedtothegrowthofbacteria.TheEPS functionedasthefirstlineofprotectionagainsterosionandinvasionofthebacterialcells. Togetherwiththebacterialcells,theexcretedagentEPSwillbeinvolvedintheshoving mechanism(KreftandWimpenny,2001).
QSisacell-to-cellcommunicationfunctionthatbacteriacoordinategeneexpressionand behaviorinthecommunities(Fredericketal.,2011).QSmodelingisrelativelyrecent. Ueckeetal.(2014) developedanIbMforQSsimulationconsideringfluidflow.Ordinary
differentialequationstorepresenttheQSmoleculardiffusionwerecoupledwiththe exteriorflowfieldtosimulatethetimeevolutionoftheinnercells. ZhaoandWang(2017) developedanumerical3DmodelforsimulationsofQSandantibacterialpersistencein heterogeneousmultispeciesbiofilms.
ThelatticeBoltzmannmodel(LBM)forbiofilmmodelinghasbeendevelopedinthepast fewdecades(DelavarandWang,2021a,b).LBMhasitsrootsinstatisticalphysicsand latticegascellularautomata(LGCA)(Frischetal.,1986).LBMusesdistributionfunctions torepresentparticleparceldynamicsandismoresuitableforcouplingwith nonequilibriuminterfaceswithCAorIbM.InLBM-basedframeworks,thebiomassis describedasdiscretequantitiesusingCAorIbM,assignedeithertonumericalgridblocks ortoparticles.Thisallowsthebiofilmmatrixtospreadinmorethanonedirection. Continuummodelsareusedforthetransportofsubstratesusingreactivetransport equationsorLBM. Eberletal.(2001) indicatedinherentdrawbacksinthediscrete approach,suchasthesmallsetofspreadingdirections,biofilmgrowinginauniform environment,adhoc,orsomewhatarbitraryrulesforpriority.However,becauseofthe fundamentalkineticnatureofLBM,itsinterfaceprocessescouldbecoupledeasily throughthedistributionfunctionsinthemesoscaleincomparisonwithtraditional computationalfluiddynamics(CFD)usingFDMorFVM.Therefore,theLBM-based modelshaveseveraladvantages:(1)nonequilibriumdynamics,suchasdiscontinuous interfaceprocesses,multiphaseflow,andflowinporousmediaorcomplexgeometries,(2) easyparallelization,and(3)norequirementofsolvingPoisson’sequationof pressure velocitycouplingthatiscomputationallyexpensive(DelavarandWang,2020).
Becauseofthesecharacteristics,LBMisagoodchoiceformodelingmicroscopic/ mesoscalebiofilmformationusingsimplifiedkineticmodels.Afewhybridtheoretical frameworksthatlinkedparticle-basedandcontinuum-basedmodelsseamlesslyhavebeen developed(DelavarandWang,2021a):(1)integratedLBMandIbM(LBM-IbM)(Graf vonderSchulenburgetal.,2009; TianandWang,2019)and(2)integratedLBMandCA (LBM-CA)(Picioreanuetal.,1999; Tangetal.,2013; Knutsonetal.,2005; Beniougetal., 2017,2019; DelavarandWang,2020, 2021b).Thesefourth-generationmodelsareusually effectiveinstrumentsformodelingbiofilmsindifferenttypesofsituations(Delavarand Wang,2021a).However,iflarge-scaleheterogeneoussystemsaresimulated,thelevelof detailinbiofilmdescriptioncanbealimitation.Mostnotably,LBM-IbMsneed substantiallygreatercomputerresourcesthanthebiomass-basedmodelsunderthesame spatialresolution.
1.3Problemsandobjectivesofbiofilmresearch
Biofilmsareanaturalphenomenonoftheearth’secology,justasbacteriaandfungiare. Manybiofilmsaredetrimentalbecausetheycausemetalcorrosionorwoundinfections.
Everyyear,biofilmscausethelossesoftheUnitedStatesliterallyhundredsofbillionsof dollarsduetoenergylosses,devicecorrosion,foodpoison,biofouling,andclinical infection.Largeseagoingships’fuelconsumptionisexpectedtoriseby30%asaresultof theviscousdragincreasescausedbybiofouling(deCarvalho,2018).Otherbiofilmsmay beusefulforresolvingmajorissues,particularlyinwastewaterpretreatmentsystemsand bioremediationofcontaminatedsoilswhilenaturalbiofilmscanbeharmfultohuman healthandtheenvironment.Forexample,industrialbiofilmsaremoreefficientandstable intermsofbioconversionsandpollutantdegradation.
Microbialsystembehaviorsresultfromintricateinteractionsbetweenalargenumberof cellsandtheirbioticandabioticsurroundings.However,biofilmsdevelopedonsubmerged sedimentareverydifferentfromthatonourteethorfromtheirplanktoniccousins.Itisa keytopichowbiofilmsservemultipleactivitiesofmicroorganismsandbestowthemwith specificfunctionalitiesfordiverseapplicationsthatareusefultocompaniesandthe environment(Vasudevan,2014).Understanding,controlling,andengineeringbiofilmsare allgoalsdrivenbyscientificandpracticalconsiderationssuchasbioremediation,corrosion andbiofoulingprevention,medicalhealthandpharmaceuticals,andecosystemrespiration.
Becausebiofilmsarepresentthroughoutareasofindustrialsystems,theenvironment,and humanhealth,biofilmresearchnowextendsfromthenotionofbiofilmdevelopmentto biofilmbasetechnologiesandtheirengineeringapplications.Biofilmformationiscriticalin boththeearlyphasesofbiofilmdevelopmentandthelatterstagesofthematuredbiofilm lifecycleviacelldispersalinavarietyofindustries.Anenhancedunderstandingofthe emergentpropertiesofbiofilmsandtheirlifestyleallowsanappreciationofbiofilms’ ecologicalandindustrialsuccess,aswellasthepotentialvalueinecologicalandindustrial applicationstodoenormousgoodforourworldasawholeortocontrolandremovebad biofilmsofindustrialandclinicalconcern(Coganetal.,2016).Itwillbefeasibleto manipulatethegrowthandformationofbiofilmsifthebiofilmfunctioncanmechanistically relatetobiotechnologies.Asaresult,scientistsandengineerscanreconsidertheir techniquesinhandlingbiofilmproblemsandsolutionsthatwerepreviouslyneglectedornot handledeffectivelyoncethepathwaystobiofilmgrowthareunderstood.Theycanmodify theirdevicesorbioprocessesinclinical,environmental,orindustrialapplicationstoblock toxicbiofilmsorstimulatefavorableones(Moonsetal.,2009).
Becausebiofilmactivityandbehaviorarecomplexinteractionsbetweenphysical, chemical,andbiologicalprocesses,theinherentlyinterdisciplinarynaturenecessitates researchcontributionsfromdisparatedisciplinessuchasbiochemistry,engineering, mathematics,andmicrobiology,withapplicationsrangingfrombiofoulingtosoil microbialecologytobioremediationtowastewatertreatmenttochronicbacterial infections(Flemmingetal.,2021).Biofilmsarenotviewedaseitherbeneficialorharmful inthiscontext,butratherasacrucialcomponentoftheecosystemsthatsurroundus.Asa
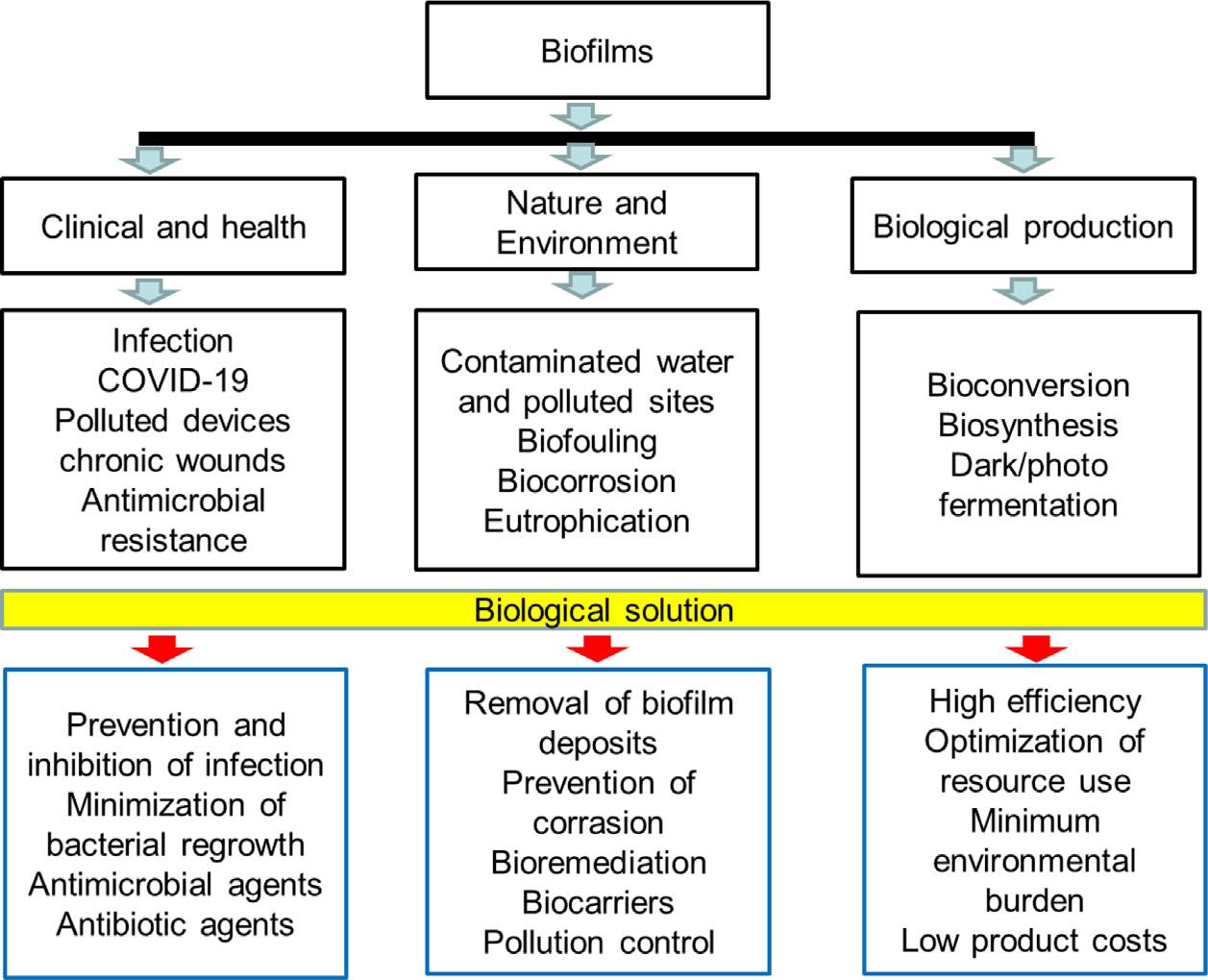
result,understandingbiofilmsrequiresresearchfrommultipleindustries,environments, anddisciplinesonhowbacteriainteractwiththeirsurroundingsandhowtheyimpactthe creationorbreakdownofawiderangeofchemicals. Fig.1.3 depictsthethreemajor domainsofbiofilmsinscienceandengineering.
Clinicalandhealth: Biofilmscanbeharmfultohumanhealthduetomicroorganism infectionsinpatients.Everyyear,millionsofpeoplesufferfromchronicdiseasessuchas cysticfibrosispneumonia,long-standingwounds,chronicinflammation,andimplant-and catheter-associatedinfections,andmanyofthemloseone’slifeasaresult(Bjarnsholt 2013; Sonderholmetal.,2017; TorrettaandPignataro,2018).Indeed,numeroushost environments,suchasblood,tears,saliva,intervascularfluid,andrespiratorysecretions, canbesuitableforbacteriagrowth.Microbialcorrosion,medicaldevice-relatedillnesses, andpersistentwoundsareallcausedbybiofilmsembeddedinhostmaterialastiny aggregates.Foodcontaminatedbybiofilmbacteriaisalsoaglobalpublichealthconcern (Sreyetal.,2013).Asaconsequence,biofilmsareregardedastheprimarycauseof infectioninpublichealth.
Microorganismsmaybenaturallyresistanttoantimicrobialsandantibiotics.Biofilmassociatedorganisms,inparticular,candevelopresistancedeterminantstowithstand antibiotics.Microorganismsinabiofilmare1000 1500timesmoreresistanttodrugsthan
Figure1.3
Problemsandobjectivesofthreemainareasofbiofilms.
theirplanktonicmicroorganisms(SocranskyandHaffajee,2002).Antibioticresistanceisa criticalconcernduetorisingmorbidity,mortality,andhealthcarecosts.Antimicrobial resistancecanalsosignificantlyinfluencehealthcaresettingsandhumanhealthbycauseof contaminationofindwellingmedicaldevicesandthesubsequentspreadofinfections relatedtothesedevices.Themechanismsofthisincreasedresistancedifferfromspeciesto species,antibioticstoantibioticsindifferenthabitats.Thisantibioticresistanceinbiofilms isconsiderablyinfluencedbytheirnutritionalconditions,species,temperature,pH,and exposuretimetoeffectiveconcentrationsofantimicrobialagents.Furthermore,bacterial speciesinabiofilmthatarelesssusceptivetoantibioticsleadtoslowergrowth.Biofilms canbeimpervioustothediffusionofantibioticsandchangethetranslationaland therapeuticpotentialduringinfection.Forexample,thebiofilmcanbeusedasanion exchangerinremovingsuchmoleculesfromthesolutionbecausestronglychargedor chemicallyhighlyreactiveagentsmayberesistedtoreachitsdeeperzones.Asaresult,it isstillunknownhowtoelucidatetheuniquegeneexpressionofbiofilm-associated organisms,evaluatevariousantimicrobialagentsandantimicrobiallocks,andanalyzethe efficiencyofnewhandlinginpreventingorremediatingbiofilmcolonizationofmedical devices.Biofilmsinmedicaldevicesareextremelydifficulttodealwithsincetheirgrowth involvesavarietyoffeatures,phases,andreducedantimicrobialsusceptibility.Hence,new tacticsforcontrollingbiofilmformationinmedicaldevicesarenecessary,suchasthe functionofbiofilmsinantimicrobialresistance,biofilmsasahostforpathogenic organisms,andbiofilmsasacauseofchronicdiseases.
Weseektofindawayhowtoeffectivelyinhibitorregulatebiofilmgrowth,whichissafe andsimpletobeimplementedinclinical,food,water,andenvironmentalmicrobiology. Thekeytosuccessinbiofilmpreventionandcontrolmaybeabetterunderstandingof whatdistinguishesthebiofilmphenotypefromtheplanktonicphenotype(Donlan,2002). Hence,themostpromisingtechniquesaretounderstandhowtopreventbiofilmgrowthas wellashowtodealwiththeseverediseasesrelatedtobiofilminfections(Nogueraetal., 1999b).Forexample,incomplexenvironments,theminimumeffectiveinhibitory concentrationofanymedicineshouldbeidentifiedineliminating,preventing,or destroyingbiofilmswhilebeingresistanttomicrobialresistance(Vasudevan,2014).New treatments,suchastheuseofantiadhesionchemicalsortheutilizationofbacterially createdsignalstopromotebacterialdispersal,shouldbeinvestigatedforexcitingfastacting,effective,andbioavailablemethods(Kostakiotietal.,2013).Bacteriacanalsobe inhibitedbyenvironmentalfactors(OfekandDoyle,1994).Understandingthesignificance ofbiofilmsininfectionshouldimproveclinicaldecision-making(Donlan,2001).Thebest clinicalpracticeswillbeincludedinregulatoryscienceandchoicesbecauseofthe comprehensivescientificexpertconsultprocess,educationaltraining,andstandardization. Thisreducesthelikelihoodofinfectionandencouragestheformationofbiofilm-inhibiting products.
Environmentalprotection: Industrialandagriculturalproductionraisessevereworries aboutcontaminatedsoilandwater.Livestockproductionisthemainsourceof eutrophicationduetophosphorusrunoffanddepletionoftheoxygenintheaquatic systems.Biofilmsareimportantinself-purificationprocessesinsoil,water,andsediments, aswellasinthenaturalattenuationoforganicmatterandpollutants,suchas contaminants,phenols,andspilledoils(LofflerandEdwards,2006).Thetechnologiesof biofilmshaveilluminatedtheirutilityintheremovalofnutrients,pollutants,andheavy metalsfromwastewater,waterdetoxification,andspilledoildegradationbypromoting naturallyoccurringorimportedmicrobiomes.Sincetheendofthe18thcentury,biofilms havebeenusedforbioremediationincontaminatedsitesbybreakingdownundesired materialcreatedbydeadfishandplantresiduals,andbyabsorbingheavyionsfromthe wastewaterwithoutreducingtheoxygenlevel(Nicolellaetal.,2000).Asaresult,biofilms haveenormouspotentialforcleaninguphazardouswastesites,filteringmunicipaland industrialwaterandwastewater,buildingbiobarrierstoavoidpollutionofsoiland groundwater,andmaintainingecologicalbalance.Furthermore,soilmicrobiomesin biofilmsalsoplayacriticalroleinecosystemrespirationandsoilcarbonsequestrationin climatechange(Bhanjaetal.,2019; BhanjaandWang,2020, 2021).Microorganismscan contributefavorablytotheecologicalbalancetoconferthedetrimentalconsequencesof theecosystemandclimatechange.
Microbiologyhastraditionallyconcentratedonsinglespeciescommunities,whichhas tremendouslyaidedourunderstandingofmicrobialgrowth,adaptability,andbiofilm formation.However,inthenaturalenvironment,mostbiofilmsaremultispecies communitiesratherthansinglespeciescommunities.Biofilmsystemsarenotrandomly distributed,butrathertailoredtocertainniches(EliasandBanin,2012).Interspeciescan collaborateandcompetewithoneanother.Interspeciescooperationhasbeendemonstrated toincreasetolerancetohazardouscompoundssuchasantibiotics,antimicrobials,chlorine, anddetergents(Costertonetal.,1994).Multispeciescommunitiesinbioremediationare recognizedtobemoreadaptivetomorecomplexconditionsindecontaminatingpolluted environments.Indeed,bioremediationofcontaminatedwaterandsoilhasbecomean appealingmethod,asorganicpollutants,suchasphenols,areconvertedtoinnocuous compoundsandsecondarymineralwastesbytheprocess.Theeffectivenessof bioremediation,ontheotherhand,isdependentonthesurvivalandpersistenceof microorganismswithinamultispeciesbiofilm.Cooperationandcompetitionamongthese complexcommunitiesarecriticalforincreasingtheresilienceandsurvivalofthevarious species(Moonsetal.,2009).Althoughresearchonmultispeciesbiofilmsisstillinitsearly stages,ithasbeendiscoveredthattheadditionofawell-selectednutritionsupplyor additionalspeciesmicroorganismscansteerthecommunitiestowardmoreefficient bioremediation.Forexample, Juangetal.(2008) showedthatphenolmaybedegraded moreeffectivelyinthepresenceoftetrasodiumpyrophosphatebyutilizingbiofilmsof
Pseudomonasputida BCRC14365.Furthermore,thepresenceofironpromotesthe degradationoftheisothiazolinonebiocide(5-chloro-2-methyl-4-isothiazolin-3-one).Asa result,biofilmtechnologyhasenormouspotentialinremediatinghazardoussites,filtering municipalandindustrialwastewater,buildingbiobarrierstoavoidpollutionofsoiland water,andmaintainingecologicalconservation.
Bioengineeringandtechnology: Theabilitytopromotethegrowthofgoodbiofilmsor inhibitthegrowthofbadbiofilmshasbeenwidelyrecognizedinthefieldsofclinic treatment,biologicalproduction,andenvironmentalprotection.Bioproducts,soluble organicmolecules,andEPSsubstancesconvertedbybacteriaareexamplesofmicrobial productsofinterest(Nogueraetal.,1999b; DelavarandWang,2021a).Many bioconversiontechnologies,includinganoxic/oxygenicphotosynthesisanddark/photo fermentation,havebeendevelopedforbiofuelandbiohydrogenproduction(Delavarand Wang,2021a,b),aswellaswastewatertreatment(DelavarandWang,2020).Theyallare dependentoninteractionsbetweenmicrobialmetabolismandphysical chemical processes.Theprimarygoalsofbioengineeringandbiotechnology,suchasbioconversion, biosynthesis,bioremediation,carbonsequestration,andwastetreatment,aretoincrease efficiency,optimizeresourceutilization,decreaseenvironmentalload,andlowerproduct costs.Variousbioreactors,suchasmovingbedbiofilmreactors(MBBRs),integratedfixedfilmactivatedsludge(IFAS)processes,andmembrane-supportedbiofilmreactors (MBfRs),aretheprimarymeansofconstructingmicrobiomesfortheuseofbiofilmsin recoveringvaluableresourcesfromwastewatertreatmentorsynthesizingbioproducts (Mccartyetal.,2011; Torresietal.,2016).Biofilmformationsinthebioreactorsarethe primarymechanismfororganicpollutantbioconversion(e.g.,organicmatter,nitrogen,and phosphorus).However,usingorganismsinbiofilmreactorsmustbemorecost-effective.To designabioreactororacompleteplant,inwhichbiofilmsconvertsubstrates,exact knowledgeofthebiofilmgrowthprocessesismandatorytoensurethegreatestefficiency andthelowesttotalcosts.Newmetabolicpathwaysorbioreactorstructureshavebeen exploredtoimprovetheefficiencyofbiofilmreactorsandtoincreasetheireconomic potential.Forexample, Roeselersetal.(2008) discoveredtheanaerobicammonium oxidation(anammox)processinapilot-scaledenitrifyingfluidizedbedbiofilmreactor.A significantlyenrichedmicrobialcommunitycollectedfromthisenvironmentwas dominatedbyasingledeep-branchingplanctomycete, CandidatusBrocadia anammoxidans (Roeselersetal.,2008).Weneedtoimprovemicrobialprocessesformore efficientbioconversion,biosynthesis,orbiodegradation,orweneedtofindstrategiesto combatbacteriatoproducecleanenergy.
Microorganismsofmultiplespeciescancontributetohighermetabolismthroughtwoto fourspeciesthansinglespecies,buttheyaresubstantiallymorecomplicatedsystems.A numberofemergentfeatureshavebeenobservedinbothdefinedandundefined populations,includingimprovedstresstolerance,greaterbiomassyield,QSsignaling,and
metaboliccooperation(Tsoietal.,2019).Thehighestprocessingcapacitiesaregenerally consideredasconversionratespertotalreactorvolume,andthespatialstructurehasa significantimpactonsubstratedistribution-relatednutrienttransferandavailabilityfor bacteriauptake(Battinetal.,2016).Naturalhabitatsorindustrialbiofilmshavespatial variabilityrangingfromnanoscalesignalstomicroscalenutritionalgradientstomillimeter structuralsurfacecharacteristicsthatdrivepopulation-levelactivities(HolandDekker, 2014).Thesinglespeciesandhomogenousevaluationsmayexploretimelynoveltraits connectedwithmultispeciescommunities.Furthermore,smartbioengineeringwillbe essentialforenhancingefficiency.Togetherwithspeciesselection,geneticengineeringcan increaseunderstandingofbiofilmformationbetweendiverseorganismsandimprove reactorperformance(Liuetal.,2008).Asaresult,thegenuinepotentialexistsinmixedspeciescommunitiestoachievehighefficiencyonawiderangeofsubstrates.Research datafrompilot-scaleplantsandmodelingcanbehighlybeneficialforassessingthe performanceofprojectedfull-scaleplantsfordesigningandtestingnewtypesofbiofilm processesorreactorstructures.Evenifthereisnofullunderstandingofbiofilmprocesses forsuchacomplexsystemofbioreactors,biofilmresearchmaybecriticalindetermining howabiofilmformsordevelopsinbioreactorsandwhetheranadequatedesignwould encouragethestudyofcertainbiofilmactivities.
1.3.1Objectivesofbiofilmmodeling
Thegoalsofbiofilmmodelingaretogainabetterknowledgeofbiofilmprocesses,reduce experimentaltesting,andscaleup.Amathematicalmodelisasystematictesttotransfera real-worldsystem’sconceptualknowledgeintoamathematicalformulaandlinkvarious processesandweightheirrelativecontributionstostructuralformationandgrowthof biofilms(Wanneretal.,2006).Amathematicalmodelcanbeusedtocombinemany mechanismsthatoperateatdistinctspatialandtemporalscales,suchastransport, metabolic,chemical,mechanical,andgeneticactivitieswithinbiofilms.Biofilmmodeling canhelpresearchersbetterunderstandbiofilmstructure,function,andpopulation dynamics,aswellasthetransition(structureandbehavior)ofbiofilmsgrowingunder variousenvironmentalconditions,potentiallyopeningupnewbiotechnologyapplications (Nogueraetal.,1999b).Innovativesolutionscanbedevelopedbycombiningscientific discoveryinscienceandengineeringforsustainablenaturalresourcemanagementand humanandanimalhealth.
Scale-upisabottleneckinbioprocessdesignanddevelopment.Optimalbiofilmcontrolis achallengeincomplexclinicaltherapy.Thebiomass,structures,andthicknessinbiofilms interactwithhydrodynamics.Ifonepartischanged,itispossiblethattheotherpartswill bechanged.Theinteractionofhydrodynamicsandbiofilmchangecanresultinerratic performance,whenachievingapseudo-steadystate,ataspecifiedtimeandspatialscales.
Theabilitytomonitor,modify,andsimulatebiofilmsfromfirstprinciples,frommolecules toindividualstopopulations,communities,andecosystems,maygomuchbeyondthe explanationoftheecologyandevolutionarytheory.Fromsamplesinshakeflaskstothe finalproductandclinicaltreatment,screeningtrialsinearly-stagedevelopmentof microbialprocessestakealongtime.Nevertheless,itischallengingtoconnectthe naturallyestablishedmultispeciescommunitieswiththefinalproduct.Modelingisto analyzequalitativelyandquantitativelytheeffectsofvariousfactorsonbiofilmformation andgrowth.
Whenexploringavarietyofbiofilm-relatedphenomena,biofilmmodelscanbeappliedfor qualitativeandquantitativeassessmentofexperimentaldataonhowpopulationsfunctionin theirnaturalconstructedconditions(Nogueraetal.,1999b).Athoroughanalysisof experimentalresultsprovidesaninsightintoreal-timecontrolofbiofilmactivitiesandthe potentialtomodifybiofilmformandfunction(Chevalieretal.,2018).Thisusually decreasesthenumberofexperimentaldesignsandteststhatallowtheunderlyinghypothesis tobetested,aswellasextractconceptualandinnovativeknowledgethatpromotes microbiomeresearch,leadingtonovelbiologicalprocessdesigns.Themodelingapproach, inparticular,allowsonetodefinetherelationshipsunderinvestigationandquantifiesthe relativequantitiesofspeciesinthepopulation,individualversusgeneralactivity,andsoon. Themodelingapproachalsoexaminesifbiofilmfunctionsaddresspracticalconcernsthat areintractablethroughfieldobservationsofcommunitiesoflargerspecies.Indeed,studies ofsocialbehaviorssuchascooperationandcompetitionareshiftingfromsingle-species systemstoincreasinglycomplexmultispeciessystems,suchasthoseinvolving host microbeinteractionsandmicrobialconsortia(DelavarandWang,2020).
Finally,despitetheuncertainties,theuseofmechanisticbiofilmmodelsinbiofilmreactor designandmanufacturingisubiquitousinconsultation,anditiscurrentlyhighlybeneficial inbioengineering.Forcomplexsystemswithnumerousinteractingphenomena,modelingis oneofthemostsuccessfulapproachesinacceleratingbothscientificadvancementand translationintonovelsolutions.Modelingcanbeappliedtoexplorenovelprocessdesigns without/atlowexpense,effort,orriskofimplementingallthestrategiestofindoptimal ones,comparedtoexperimentaltesting.Therefore,modelingallowsresearchersand practitionerstoefficientlyscreenalargenumberofdesignoptionstodiscardthosethatfail toachieveperformanceobjectives,suchasdesignedoperatingconditionsandnovel therapeutictechniques.Inbioengineeringandbiotechnology,aniterative design build test learn(DBTL)cyclecanbeusedformicrobiomedevelopment(Lawson etal.,2019).ThisDBTLcoversthelifecycledevelopmentofamicrobiome,fromaninitial microbiomedesigntoengineeringgoals,modelsystems,andexpectedoutcomes.The microbiomeiscreatedsothatitsfunctioncanbetestedagainstasetofmetricstoexamine ifthedesign buildoutcome(s)meetsthedesigngoal(thatis,establishcausation).Thiswill allowexaminingwhatworkedandwhatdidnot(andwhy).Thisisutilizedtoassist decision-makinginsubsequentDBTLdesigncycles.Thismethodcouldacceleratethe
developmentofproductsfromdesignconceptsforharnessingmicrobiomestorealproducts bygivingnovelsolutionsandexpandingscientificunderstandingthroughmodeling.
Biofilmmodelsarealsousedintheeducationandtrainingofprospectivescientistsand engineers(Nogueraetal.,1999b).Modelingapproachesallowstudents,researchers,and scientiststounderstandthemechanismofbiofilmdevelopmentanditsfunctionality,andto examinetheimpactofbiofilmformation,inducedcorrosion,andfoulingonsurfacesof devices.Understandinghowbiofilmsformandfunctionthroughsimulationisoneofthe mostefficientlearningsandtrainingwaysforachievingtheintendedresults.A combinationoffundamental(mechanistic)anddata-oriented(empirical)biofilmreactor designmodelsisrequiredforlearningandtrainingalthoughpredictingtheinteraction amongrelevantscalesisstilladifficulty.Environmentalcharacteristicssuchaslocalized gradientsinpH,dissolvedoxygen,redoxpotential,nutrient,andmetabolicproduct concentrationscanallbeinvestigatedtoexaminetheireffectsonbiofilmformationand development,whetherthebiofilmisalreadybuiltorstillintheconceptualstage(Noguera etal.,1999a; Flemmingetal.,2021).
Thereareseveralprioritiesthatrequiretobeconsideredwhileemployingmodeling approaches:(1)microorganismactivitywithinabiofilm,(2)elucidationofbiofilm deposition,formation,anddetachmentmechanisms,(3)assessmentofthemechanical propertiesofEPSandpreservationofextracellularenzymes,(4)identificationofkeydrivers andparameters,andecologicalinteraction(s)amongdifferentmicroorganisms,(5)increased resistancetoantibioticsandbiocides,(6)geneticexchange,recombination,thereunionof nucleicacidsinintercellularcommunication,andcollectivebehaviorsuchascooperation andcompetitioninresponsetothestressfulandchangingenvironment,(7)waterretention andpreventiontodehydration,and(8)real-timecontrolofbiofilmsystemsforthemost effectivebiologicaltreatmentmethods(Nogueraetal.,1999a; Flemmingetal.,2021).
References
Alpkvist,E.,Picioreanu,C.,vanLoosdrecht,M.C.,Heyden,A.,2006.Three-dimensionalbiofilmmodelwith individualcellsandcontinuumEPSmatrix.Biotechnol.Bioeng.94(5),961 979.
Atkinson,B.,Swilley,E.L.,Busch,A.W.,Williams,D.A.,1967.Kinetics,masstransferandorganismgrowth inabiologicalfilmreactor.Trans.Inst.Chem.Eng.45(6),T257 T264. Atkinson,B.,Davies,I.J.,1974.Theoverallrateofsubstrateuptake(reaction)bymicrobialfilms.PartI-A biologicalrateequation.Trans.Inst.Chem.Eng.52,260 268.
Battin,T.J.,Kaplan,L.A.,Newbold,J.D.,Cheng,X.,Hansen,C.,2003.Effectsofcurrentvelocityonthe nascentarchitectureofstreammicrobialbiofilms.Appl.Environ.Microbiol.69(9),5443 5452.
Battin,T.J.,Sloan,W.T.,Kjelleberg,S.,Daims,H.,Head,I.M.,Curtis,T.P.,Eberl,L.,2007.Microbial landscapes:newpathstobiofilmresearch.Nat.Rev.Microbiol.5(1),76 81.
Battin,T.J.,Kaplan,L.A.,Findlay,S.,Hopkinson,C.S.,Marti,E.,Packman,A.I.,Sabater,F.,2008. Biophysicalcontrolsonorganiccarbonfluxesinfluvialnetworks.Nat.Geosci.1(2),95 100.
Battin,T.J.,Besemer,K.,Bengtsson,M.M.,Romani,A.M.,Packmann,A.I.,2016.Theecologyand biogeochemistryofstreambiofilms.Nat.Rev.Microbiol.14(4),251 263.
Baveye,P.C.,2020.Soilbiofilms:misleadingdescriptionofthespatialdistributionofmicrobialbiomassin soils.SoilEcol.Lett.2,2 5.
Bhanja,S.N.,Wang,J.,Shrestha,N.K.,Zhang,X.,2019.Modellingmicrobialkineticsandthermodynamic processesforquantifyingsoilCO2 emission.Atmos.Environ.209,125 135.
Benioug,M.,Golfier,F.,Fischer,P.,Oltean,C.,Bue ` s,M.A.,Yang,X.,2019.Interactionbetweenbiofilm growthandNAPLremediation:Apore-scalestudy.Adv.WaterResour.125,82 97.
Benioug,M.,Golfier,F.,Olte ´ an,C.,Bue ` s,M.A.,Bahar,T.,Cuny,J.,2017.Animmersedboundary-lattice Boltzmannmodelforbiofilmgrowthinporousmedia.Adv.WaterResour.107,65 82.
Bhanja,S.N.,Wang,J.,2020.Estimatinginfluencesofenvironmentaldriversonsoilheterotrophicrespiration intheAthabascaRiverBasin,Canada.Environ.Pollut.257,113630.
Bhanja,S.N.,Wang,J.,2021.Influenceofenvironmentalfactorsonautotrophic,soilandecosystem respirationsinCanadianborealforest.Ecol.Indicat.125,107517.
Bjarnsholt,T.,2013.Theroleofbacterialbiofilmsinchronicinfections.Apmis121,1 58.
Byrne,H.,Drasdo,D.,2009.Individual-basedandcontinuummodelsofgrowingcellpopulations:a comparison.J.Math.Biol.58(4),657 687.
Chen,L.,Wen,Y.M.,2011.Theroleofbacterialbiofilminpersistentinfectionsandcontrolstrategies.Int.J. OralSci.3(2),66 73.
Chevalier,M.,Ranque,S.,Pre ˆ cheur,I.,2018.Oralfungal-bacterialbiofilmmodelsinvitro:areview.Med. Mycol.56(6),653 667.
Cogan,N.G.,Keener,J.P.,2004.Theroleofthebiofilmmatrixinstructuraldevelopment.Math.Med.Biol.21 (2),147 166.
Cogan,N.G.,Harro,J.M.,Stoodley,P.,Shirtliff,M.E.,2016.Predictivecomputermodelsforbiofilm detachmentpropertiesin Pseudomonasaeruginosa.mBio7(3),e00815 e00816.
Costerton,J.W.,Lewandowski,Z.,DeBeer,D.,Caldwell,D.,Korber,D.,James,G.,1994.Biofilms,the customizedmicroniche.J.Bacteriol.176(8),2137 2142.
deCarvalho,C.C.,2018.Marinebiofilms:asuccessfulmicrobialstrategywitheconomicimplications.Front. Mar.Sci.5,126.
Delavar,M.A.,Wang,J.,2021a.LatticeBoltzmannmethodinmodelingbiofilmformation,growthand detachment.Sustainability13(14),7968.
Delavar,M.A.,Wang,J.,2021b.NumericalinvestigationofpHcontrolondarkfermentationandhydrogen productioninamicrobioreactor.Fuel292,120355.
Delavar,M.A.,Wang,J.,2020.Modelingcombinedeffectsoftemperatureandstructureoncompetitionand growthofmultispeciesbiofilmsinmicrobioreactors.Ind.Eng.Chem.Res.59(37),16122 16135.
Dobell,C.,1932.AntonyvanLeeuwenhoekandHis“LittleAnimals”:BeingSomeAccountoftheFatherof ProtozoologyandBacteriologyandHisMultifariousDiscoveriesinTheseDisciplines.Harcourt,Barace andCompany,NewYork.
Donlan,R.M.,2001.Biofilmformation:aclinicallyrelevantmicrobiologicalprocess.Clin.Infect.Dis.33(8), 1387 1392.
Donlan,R.M.,2002.Biofilms:microbiallifeonsurfaces.Emerg.Infect.Dis.8(9),881.
Donlan,R.M.,Costerton,J.W.,2002.Biofilms:survivalmechanismsofclinicallyrelevantmicroorganisms. Clin.Microbiol.Rev.15(2),167 193.
Eberl,H.J.,Parker,D.F.,Vanloosdrecht,M.C.,2001.Anewdeterministicspatio-temporalcontinuummodelfor biofilmdevelopment.Comp.Math.MethodsMed.3(3),161 175.
Ehrlich,H.L.,Newman,D.K.,2008.Geomicrobiology,fifthed.CRCpress. Elias,S.,Banin,E.,2012.Multi-speciesbiofilms:livingwithfriendlyneighbors.FEMSMicrobiolRev36(5), 990 1004.
Flemming,H.C.,Wingender,J.,Szewzyk,U.,Steinberg,P.,Rice,S.A.,Kjelleberg,S.,2016.Biofilms:an emergentformofbacteriallife.Nat.Rev.Microbiol.14(9),563 575.
Flemming,H.C.,Baveye,P.,Neu,T.R.,Stoodley,P.,Szewzyk,U.,Wingender,J.,Wuertz,S.,2021.Whoput thefilminbiofilm?Themigrationofatermfromwastewaterengineeringtomedicineandbeyond.NPJ BiofilmsMicrobiomes7(1),1 5.
Frederick,M.R.,Kuttler,C.,Hense,B.A.,Eberl,H.J.,2011.Amathematicalmodelofquorumsensing regulatedEPSproductioninbiofilmcommunities.Theor.Biol.Med.Model.8(1),1 29. Frisch,U.,d’Humieres,D.,Hasslacher,B.,Lallemand,P.,Pomeau,Y.,Rivet,J.P.,1986.Latticegas hydrodynamicsintwoandthreedimensions(No.LA-UR-87-2524;CONF-8610281-2).LosAlamos NationalLab.,NM(USA);ObservatoiredeNice,06(France);EcoleNormaleSuperieure,75-Paris(France). Gombas,D.E.,1989.Biologicalcompetitionasapreservingmechanism.J.FoodSaf.10(2),107 117. Gebreyohannes,G.,Nyerere,A.,Bii,C.,Sbhatu,D.B.,2019.Challengesofintervention,treatment,and antibioticresistanceofbiofilm-formingmicroorganisms.Heliyon5(8),e02192.
Geesey,G.G.,Richardson,W.T.,Yeomans,H.G.,Irvin,R.T.,Costerton,J.W.,1977.Microscopicexamination ofnaturalsessilebacterialpopulationsfromanalpinestream.Can.J.Microbiol.23(12),1733 1736. GrafvonDerSchulenburg,D.G.,Pintelon,T.R.R.,Picioreanu,C.,VanLoosdrecht,M.C.,Johns,M.L.,2009. Three-dimensionalsimulationsofbiofilmgrowthinporousmedia.AIChEJ.55(2),494 504. Harremoe ¨ s,P.,1976.Thesignificanceofporediffusiontofilterdenitrification.J.WaterPollut.ControlFed. 377 388.
Henrici,A.T.,1933.Studiesoffreshwaterbacteria:I.Adirectmicroscopictechnique.J.Bacteriol.25(3), 277 287.
Høiby,N.,2014.Apersonalhistoryofresearchonmicrobialbiofilmsandbiofilminfections.PathogensDis. 70(3),205 211.
Høiby,N.,2017.Ashorthistoryofmicrobialbiofilmsandbiofilminfections.Apmis125(4),272 275. Hol,F.J.,Dekker,C.,2014.Zoomingintoseethebiggerpicture:microfluidicandnanofabricationtoolsto studybacteria.Science346(6208),1251821.
Jamal,M.,Ahmad,W.,Andleeb,S.,Jalil,F.,Imran,M.,Nawaz,M.A.,etal.,2018.Bacterialbiofilmand associatedinfections.J.Chin.Med.Assoc.81(1),7 11.
Juang,R.S.,Chung,T.P.,Wang,M.L.,Lee,D.J.,2008.Experimentalobservationsontheeffectofadded dispersingagentonphenolbiodegradationinamicroporousmembranebioreactor.J.HazardMater.151 (2 3),746 752.
Klapper,I.,Dockery,J.,2002.Fingerformationinbiofilmlayers.SIAMJ.Appl.Math.62(3),853 869.
Knutson,C.E.,Werth,C.J.,Valocchi,A.J.,2005.Pore-scalesimulationofbiomassgrowthalongthetransverse mixingzoneofamodeltwo-dimensionalporousmedium.WaterResour.Res.41(7).
Kostakioti,M.,Hadjifrangiskou,M.,Hultgren,S.J.,2013.Bacterialbiofilms:development,dispersal,and therapeuticstrategiesinthedawnofthepostantibioticera.ColdSpringHarb.PerspectMed.3(4),a010306.
Kragh,K.N.,Hutchison,J.B.,Melaugh,G.,Rodesney,C.,Roberts,A.E.,Irie,Y.,Bjarnsholt,T.,2016.Roleof multicellularaggregatesinbiofilmformation.mBio7(2)e00237-16.
Kreft,J.U.,Wimpenny,J.W.,2001.EffectofEPSonbiofilmstructureandfunctionasrevealedbyan individual-basedmodelofbiofilmgrowth.WaterSci.Technol.43(6),135.
Laspidou,C.S.,Kungolos,A.,Samaras,P.,2010.Cellular-automataandindividual-basedapproachesforthe modelingofbiofilmstructures:prosandcons.Desalination250(1),390 394.
Laspidou,C.S.,Rittmann,B.E.,2004.Modelingthedevelopmentofbiofilmdensityincludingactivebacteria, inertbiomass,andextracellularpolymericsubstances.WaterRes.38(14 15),3349 3361.
Law,Y.,Kirkegaard,R.H.,Cokro,A.A.,Liu,X.,Arumugam,K.,Xie,C.,Williams,R.B.,2016.Integrative microbialcommunityanalysisrevealsfull-scaleenhancedbiologicalphosphorusremovalundertropical conditions.Sci.Rep.6(1),1 15.
Lawson,C.E.,Harcombe,W.R.,Hatzenpichler,R.,Lindemann,S.R.,Loffler,F.E.,O’Malley,M.A., McMahon,K.D.,2019.Commonprinciplesandbestpracticesforengineeringmicrobiomes.Nat.Rev. Microbiol.17(12),725 741.
Ley,R.E.,Harris,J.K.,Wilcox,J.,Spear,J.R.,Miller,S.R.,Bebout,B.M.,Pace,N.R.,2006.Unexpected diversityandcomplexityoftheGuerreroNegrohypersalinemicrobialmat.Appl.Environ.Microbiol.72 (5),3685 3695.
Liu,Z.,Li,H.,Liu,J.,Su,Z.,2008.Effectsofinoculationstrategyandcultivationapproachontheperformance ofmicrobialfuelcellusingmarinesedimentasbio-matrix.J.Appl.Microbiol.104(4),1163 1170.
Loffler,F.E.,Edwards,E.A.,2006.Harnessingmicrobialactivitiesforenvironmentalcleanup.Curr.Opinion Biotechnol.17(3),274 284.
McCarty,P.L.,Bae,J.,Kim,J.,2011.Domesticwastewatertreatmentasanetenergyproducer canthisbe achieved?Environ.Sci.Technol.45(17),7100 7106.
Meckenstock,R.U.,Elsner,M.,Griebler,C.,Lueders,T.,Stumpp,C.,Aamand,J.,vanBreukelen,B.M.,2015. Biodegradation:updatingtheconceptsofcontrolformicrobialcleanupincontaminatedaquifers.Environ. Sci.Technol.49(12),7073 7081.
Melaugh,G.,Hutchison,J.,Kragh,K.N.,Irie,Y.,Roberts,A.,Bjarnsholt,T.,Allen,R.J.,2016.Shapingthe growthbehaviourofbiofilmsinitiatedfrombacterialaggregates.PLoSOne11(3),e0149683.
Moons,P.,Michiels,C.W.,Aertsen,A.,2009.Bacterialinteractionsinbiofilms.Crit.Rev.Microbiol.35(3), 157 168.
Nickel,J.C.,Ruseska,I.,Costerton,J.W.,1985.TobramycinresistanceofcellsofPseudomonasaerogenosa growingasabiofilmonurinarycathetermaterial.Antimicrob.AgentsChemother.27,610 624.
Nicolella,C.,VanLoosdrecht,M.C.M.,Heijnen,J.J.,2000.Wastewatertreatmentwithparticulatebiofilm reactors.J.Biotechnol.80(1),1 33.
Noguera,D.R.,Pizarfo,G.,Stahl,D.A.,Rittmann,B.E.,1999a.Simulationofmultispeciesbiofilm developmentinthreedimensions.WaterSci.Technol.39(7),123 130.
Noguera,D.R.,Okabe,S.,Picioreanu,C.,1999b.Biofilmmodeling:presentstatusandfuturedirections.Water Sci.Technol.39(7),273 278.
Ofek,I.,Doyle,R.J.,1994.Principlesofbacterialadhesion.BacterialAdhesiontoCellsandTissues.Springer, Boston,MA,pp.1 15.
Picioreanu,C.,VanLoosdrecht,M.C.,Heijnen,J.J.,1998a.Mathematicalmodelingofbiofilmstructurewitha hybriddifferential-discretecellularautomatonapproach.Biotechnologyandbioengineering58(1), 101 116.
Picioreanu,C.,vanLoosdrecht,M.C.,Heijnen,J.J.,1998b.Anewcombineddifferential-discretecellular automatonapproachforbiofilmmodeling:Applicationforgrowthingelbeads.Biotechnol.Bioeng.57 (6),718 731.
Picioreanu,C.,VanLoosdrecht,M.C.M.,Heijnen,J.J.,1999.Discrete-differentialmodellingofbiofilm structure.WaterSci.Technol.39(7),115 122.
Picioreanu,C.,Kreft,J.U.,VanLoosdrecht,M.C.,2004.Particle-basedmultidimensionalmultispeciesbiofilm model.Appl.Environ.Microbiol.70(5),3024 3040.
Picioreanu,C.,VanLoosdrecht,M.C.,Heijnen,J.J.,2001.Two-dimensionalmodelofbiofilmdetachment causedbyinternalstressfromliquidflow.Biotechnol.Bioeng.72(2),205 218.
Pizarro,G.,Griffeath,D.,Noguera,D.R.,2001.Quantitativecellularautomatonmodelforbiofilms.J.Environ. Eng.127(9),782 789.
Rittmann,B.E.,McCarty,P.L.,1980.Modelofsteady-state-biofilmkinetics.Biotechnol.Bioeng.22(11), 2343 2357.
Roeselers,G.,VanLoosdrecht,M.C.,Muyzer,G.,2008.Phototrophicbiofilmsandtheirpotentialapplications. J.Appl.Phycol.20(3),227 235.
Saunders,A.M.,Albertsen,M.,Vollertsen,J.,Nielsen,P.H.,2016.Theactivatedsludgeecosystemcontainsa corecommunityofabundantorganisms.ISMEJ.10(1),11 20.
Socransky,S.S.,Haffajee,A.D.,2002.Dentalbiofilms:difficulttherapeutictargets.Periodontology28(1), 12 55.
Sønderholm,M.,Kragh,K.N.,Koren,K.,Jakobsen,T.H.,Darch,S.E.,Alhede,M.,Bjarnsholt,T.,2017. Pseudomonasaeruginosa aggregateformationinanalginatebeadmodelsystemexhibitsinvivo-like characteristics.Appl.Environ.Microbiol.83(9),e00113 e00117.
Srey,S.,Jahid,I.K.,Ha,S.D.,2013.Biofilmformationinfoodindustries:afoodsafetyconcern.FoodControl 31(2),572 585.
Stewart,P.S.,Franklin,M.J.,2008.Physiologicalheterogeneityinbiofilms.Nat.Rev.Microbiol.6(3),199 210. Stiles,M.E.,1996.Biopreservationbylacticacidbacteria.AntonieVanLeeuwenhoek70(2),331 345.
Stoodley,P.,Sauer,K.,Davies,D.G.,Costerton,J.W.,2002.Biofilmsascomplexdifferentiatedcommunities. Annu.Rev.Microbiol.56(1),187 209.
Suarez,C.,Piculell,M.,Modin,O.,etal.,2019.Thicknessdeterminesmicrobialcommunitystructureand functioninnitrifyingbiofilmsviadeterministicassembly.Sci.Rep.9,5110.
Torresi,E.,Fowler,S.J.,Polesel,F.,Bester,K.,Andersen,H.R.,Smets,B.F.,Christensson,M.,2016.Biofilm thicknessinfluencesbiodiversityinnitrifyingMBBRsimplicationsonmicropollutantremoval.Environ. Sci.Technol.50(17),9279 9288.
Tang,Y.,Valocchi,A.J.,Werth,C.J.,Liu,H.,2013.Animprovedpore-scalebiofilmmodelandcomparison withamicrofluidicflowcellexperiment.WaterResour.Res.49(12),8370 8382.
Tian,Z.,Wang,J.,2019.LatticeBoltzmannsimulationofbiofilmcloggingandchemicaloxygendemand removalinporousmedia.AIChEJ.65(9),e16661.
Torretta,S.,Pignataro,L.,2018.Theroleofbiofilmsinupperrespiratorytractinfections.In:Infectionsofthe Ears,Nose,Throat,andSinuses.Springer,Cham,pp.31 43.
Tsoi,R.,Dai,Z.,You,L.,2019.Emergingstrategiesforengineeringmicrobialcommunities.Biotechnol.Adv. 37(6),107372.
Uecke,H.,Muller,J.,Hense,B.A.,2014.Individual-basedmodelforquorumsensingwithbackgroundflow. Bull.Math.Biol.76(7),1727 1746.
Vasudevan,R.,2014.Biofilms:microbialcitiesofscientificsignificance.J.Microbiol.Exp.1(3),00014.
Vidakovic,L.,Singh,P.K.,Hartmann,R.,Nadell,C.D.,Drescher,K.,2018.Dynamicbiofilmarchitecture confersindividualandcollectivemechanismsofviralprotection.Nat.Microbiol.3(1),26 31.
Wang,Q.,Zhang,T.,2010.Reviewofmathematicalmodelsforbiofilms.SolidStateCommun.150(21 22), 1009 1022.
Wanner,O.,Eberl,H.J.,Morgenroth,E.,Noguera,D.R.,Picioreanu,C.,Rittmann,B.E.,Van Loosdrecht,M.C.M.,2006.MathematicalModelingofBiofilms.IWAScientificandTechnicalReportNo. 18IWATaskGrouponBiofilmModeling.
Wanner,O.,Gujer,W.,1986.Amultispeciesbiofilmmodel.Biotechnol.Bioeng.28(3),314 328. Williamson,K.,McCarty,P.L.,1976.Amodelofsubstrateutilizationbybacterialfilms.J.WaterPollut. ControlFed.9 24.
Winstanley,H.F.,Chapwanya,M.,McGuinness,M.J.,Fowler,A.C.,2011.Apolymer solventmodelof biofilmgrowth.Proc.R.Soc.Math.Phys.Eng.Sci.467(2129),1449 1467.
Winstanley,H.F.,Chapwanya,M.,Fowler,A.C.,O’Brien,S.B.G.,2015.A2Dchannel-cloggingbiofilmmodel. J.Math.Biol.71(3),647 668.
Xavier,J.B.,Picioreanu,C.,VanLoosdrecht,M.C.,2005.Aframeworkformultidimensionalmodellingof activityandstructureofmultispeciesbiofilms.Environ.Microbiol.7(8),1085 1103.
Yan,Z.,Liu,C.,Liu,Y.,Bailey,V.L.,2017.Multiscaleinvestigationonbiofilmdistributionanditsimpacton macroscopicbiogeochemicalreactionrates.WaterResour.Res.53(11),6898 8714.
Young,I.M.,Crawford,J.W.,2004.Interactionsandself-organizationinthesoil-microbecomplex.Science304 (5677),1634 1637.
Zhao,J.,Wang,Q.,2017.Three-dimensionalnumericalsimulationsofbiofilmdynamicswithquorumsensing inaflowcell.Bull.Math.Biol.79(4),884 919.
Zobell,C.E.,Allen,E.C.,1935.Thesignificanceofmarinebacteriainthefoulingofsubmergedsurfaces. J.Bacteriol.29(3),239 251.
Furtherreading
Gerlach,R.,Cunningham,A.B.,2010.InfluenceofBiofilmsonPorousMediaHydrodynamics.PorousMedia: ApplicationsinBiologicalSystemsandBiotechnology,pp.173 230.
Hajishengallis,G.,2015.Periodontitis:frommicrobialimmunesubversiontosystemicinflammation.Nat.Rev. Immunol.15(1),30 44.
Hibbing,M.E.,Fuqua,C.,Parsek,M.R.,Peterson,S.B.,2010.Bacterialcompetition:survivingandthrivingin themicrobialjungle.Nat.Rev.Microbiol.8(1),15 25.
Horn,H.,Lackner,S.,2014.Modelingofbiofilmsystems:areview.Prod.Biofilms53 76.