scıentific Carolina
SPRING 2023 | Volume 17 | Issue 2
—THE FIGHT TO CURE CANCER— full story on page 58


SPRING 2023 | Volume 17 | Issue 2
—THE FIGHT TO CURE CANCER— full story on page 58



Check out all of our previous issues at issuu.com/uncsci.





As the organization continues to grow, we would like to thank our Faculty Advisor, Dr. Lillian Zwemer, for her continued support and mentorship.









Mission Statement:
Founded in Spring 2008, Carolina Scientific serves to educate undergraduates by focusing on the exciting innovations in science and current research that are taking place at UNC-Chapel Hill. Carolina Scientific strives to provide a way for students to discover and express their knowledge of new scientific advances, to encourage students to explore and report on the latest scientific research at UNC-Chapel Hill, and to educate and inform readers while promoting interest in science and research.
Letter from the Editors:
Another beautiful spring semester at Carolina is coming to an end, but research here on campus goes on. At the core of any research is curiosity and you, as the reader, may also be curious as to what exciting research many scientists, faculty, and students at UNC-Chapel Hill engage in. Carolina Scientific works to make such projects and scientific information more accessible to the campus-wide community. In the process, we also aim to foster scientific writing, creative designing, and critical thinking skills among students. Here, we hope to quench your curious minds through this Spring 2023 issue, and we greatly hope you enjoy learning about the fascinating research happening right here at Carolina.
- Megan Bishop and Sarah (Yeajin) Kim
Editors-in-Chiefs
Megan Bishop
Sarah (Yeajin) Kim
Design Editor Cassie Wan
Copy Editor Gargi Dixit
Managing Editor Isaac Hwang
Treasurer
Publicity Chair
Fundraising Chair
Associate Editors
Ambika Bhatt
Sarah Giang
Heidi Cao
Meitra Kazemi
Neil Sud
Jasmeet Singh
Maddy Stratton
Online Content Manager Sreya Upputuri
Faculty Advisor Lillian Zwemer, Ph.D.
Staff Writers Copy Staff
Whitney Abed
Esha Agarwal
Klodia Badal
Kruti Bhargav
Julia Boltz
Taylor Born
Samantha Breen
Estella Calcaterra
Kaitlyn Campbell
Shu Chen
JR Cobb
Grayson Coleman
Ciara Daly
Charisma Daniel
Morgan Davis
Anooshka Deshpande
Reagan Gulledge
Emily Harper
Jessica Hoyt
An introduction to the Lineberger Comprehensive Cancer Center and the research going on.
Full story on page 58. Image courtesy of the National Institute of Cancer via Unsplash
carolina_scientific@unc.edu

carolinascientific.org
instagram: @carolinascientific facebook.com/CarolinaScientific
Torrance Jenkins
Ashwath Kapilavai
Lintao Li
Simran Malik
Rahul Mehta
Lyssa Menendez
Isha Mistry
Nat Moody
Mahika Nagaradona
Hari Patel
Ria Patel
Yifei Pei
Andrew Phan
Matthew Rodzen
Julia Sallean
Vina Senthil
Skye Scoggins
Natalie Travis
Ashley Villanueva
Layla Williams
Rujula Yete
Karen Zhu
Sneha Adayapalam
Quinten Curtis
Daniela Danilova
Corinne Drabenstott
Natalie Druffner
Nastia Hnatov
Lily Hohn
Alexander Kinrade
Sprihaa Kolanukuduru
August Little
Alacia McClary
Claire Nolan
Rashmi Ramanujam
Nihith Ravikanti
Tsehai Ricketts
Arora Rohrbach
Avery Sallean
Arushi Vaish
Rachitha Vijayakumar
Sophia Vona
Anna Vu
Kelly Yun
Heidi Cao
Estella Calcaterra
Tanisha Chourdhury
Srinithi Gali
Clara Lord
Katrina Murch
Caroline Norland
Emily Ormond
Skyler Peterson
Heidi Segars
Amelia Spell
Sharon Wang
Kelly Yun
Bhavika Chirumamilla
Tanisha Choudhury
Heidi Cao
Jessica Hoyt
Tia Robinson
Emily Ormond
The
Counting Catepillars!
Jessica Hoyt
The “Dark Matter” of Biology
Ashwath Kapilavai
Science Behind the Magic
Kruti
Negative
Julia Boltz
Out With the Old, In With the New
Taylor Born
The Untapped Potential of the Endocannabinoid System
Kaitlyn Campbell
Sugar-Sweetened Beverage Tax: No “Sweet Deal”
Anymore
Shu Chen
The Fight to Cure Cancer
Grayson Coleman
My Body My Test: Cervical Cancer Self-Screening
Emily Harper
Nitric Oxide: The Future of Diabetes Managment
Torrance Jenkins
The Pulse of Inequality
Simran Malik
Research of Great Valve-ue
Lyssa Menendez
Developing Flies, Scientists, or Both?
Nat Moody
The
Julia
“Staying
Vina Senthil
Pruning the Human Blueprint
Karen Zhu
Physical Sciences
Molecular Builders
Ciara Daly
Gravitational Waves and Gateways to Science
Rujula Yete
Social Sciences
Abuzz About the HHIVE Lab
Esha Agarwal
Self-Regulated and Active Learning
JR Cobb
Advances in Health Communication
Charisma Daniel
Thorugh the W(ring)er
Mahika Nagaradona
Fueling the Fight Against Type 1 Diabetes
Hari Patel
Optimal Pain Relief-Without the Side Effects
Ria Patel
Brain Circuitry Underlying Substance Abuse
Anooshka Deshpande
Antecedent Predictors of Suicidality and Depression
Yifei Pei
Breaking the Chains of Substance Use Disorder
Matthew Rodzen
Are You Feeling Hangry?
Skye Scoggins
Practicing Parenting with Virtual Reality
Natalie Travis
Of Mice
Ashley Villanueva
The Pink Elephant
When thinking about greenhouse gasses and climate change, people often picture exhaust from cars waiting in traffic or formidable factories releasing dark smog into the atmosphere. Nevertheless, rivers and streams also produce these same greenhouse gasses due to a variety of unknown factors. Dr. Amanda G. DelVecchia, biogeochemist in the Department of Geography at UNC Chapel Hill, aims to understand the processes and interactions that may influence this production across freshwater systems in the U.S. using recent data collected by the National Ecological Observatory Network (NEON). More specifically, she hopes to uncover the correlation between factors such as dissolved oxygen concentrations and stream slope with carbon dioxide, methane, and nitrous oxide levels. With this information, she can help others create more accurate
models of greenhouse gas emissions, mark their change over time, and potentially attribute gas production to specific interactions and physical factors.1
Despite only starting as a UNC professor in the fall, DelVecchia first became interested in conducting this environmental research during her undergraduate career in Chapel Hill. As an Environmental Science major, she researched carbon sequestration, the storage of carbon dioxide from the atmosphere, in South American mangroves. Inspired by this work, she studied biogeochemical interactions within the Nyack floodplain aquifer in Montana to complete her

PhD. DelVecchia became specifically interested in ecology when she noticed that “many ecologists care deeply about what they’re doing, and they make this effort to not only collect results that are novel and important for science, but things that are relevant to the local community.”2 DelVecchia eventually came back to Chapel Hill to teach students about freshwater systems and conduct research pertaining to greenhouse gasses because of their current global importance.
DelVecchia’s study provides extensive information about greenhouse gasses in freshwater streams across the U.S. The research can cover many locations, variables, and types of greenhouse gasses

due to the data collected by NEON, an organization whose goal is to provide scientists with ecological data from around the country.1 She notes that, “Typically, datasets are limited to maybe a particular greenhouse gas, a particular set of environmental variables, or oftentimes the summer because it is far more fun to sample in the summer. NEON is unique because it not only expands this range of sites to twentyeight sites across the U.S., but all three greenhouse gasses are collected, along with aquatic data, terrestrial data, and imagery.”2
Using this data, DelVecchia looked at the correlation between mean air temperature, stream slope, dissolved oxygen, total nitrogen concentrations, etc. and greenhouse gas production across the U.S. to determine which variables might be drivers and how the values varied across regions. Understanding the correlation between these variables and greenhouse gas production is the first step in potentially explaining the processes that create variation among these gasses.
DelVecchia discovered that working with NEON data, however, is not always easy. Since the data is collected by many different people, the way that it is formatted varies frequently and can be challenging to work with. The information given by NEON is in its raw form, requiring many stages of filtering to end up with a result that can be used by scientists. DelVecchia and her team spent a lot of time figuring out which data they wanted to incorporate into the study and which they felt should be left out due to potential error. Once they developed this criterion, the process became much easier.2
After going through the tedious steps of cleaning data, DelVecchia completed her analysis. She found that physical aspects such as temperature, watershed land cover, dissolved oxygen, channel slope, and precipitation all correlate with carbon dioxide and methane production. Warmer, lowgradient rivers that only flow part of the year also released the highest gas concentrations and contained the most variation.1 One surprising piece of information found from this analysis was the amount of methane that was released by these rivers and streams. Rivers are often considered well-oxygenated systems because the water is always moving and can exchange gasses with the atmosphere. Slow, stagnant systems such as ponds that are not as well-oxygenated often produce methane because of the anaerobic processes that occur in those conditions. DelVecchia’s study found that the lowgradient, warmer rivers and streams
released more methane than expected, showing that anaerobic processes might have a greater impact on these systems than previously thought.2 Nitrous oxide, on the other hand, did not reveal similar trends and instead only varied with the total nitrogen concentration of the system.1 Of course, correlation does not equal causation, but the study gives scientists a reason to believe that specific interactions might be driving greenhouse gas variation and that this topic should be further investigated.

Many aspects of this research excite DelVecchia, as she believes the information can change the way scientists manage rivers and think about environmental change. For example, knowing what drives greenhouse gas production can help scientists predict how carbon and nitrogen cycles will be altered by climate change and what systems will be most affected. This information will also be useful when determining the effects of land use change or modifying a river network.2 In the future, she hopes to collaborate with other researchers to expand on the paper and create a better picture of the processes that make up freshwater systems and their ecosystems. DelVecchia imagines her research continuing in a variety of different ways to better understand the relationship between greenhouse gasses and freshwater systems.2 Greenhouse gases are a predominant topic in many minds today as their influence on the world becomes clearer with climate change research. This study shows that there is still a long way to go in understanding how these gasses work and their potential consequences.
1. DelVecchia, A.G.; Rhea, S.; Carter, A.M.; Aho, K.S.; Hotchkiss, E.; Stanley, E.H.; Bernhardt, E.S. L&O. 2022.
2. Interview with Amanda G. DelVecchia, Ph.D. 1/27/23.
“Greenhouse gases are a predominant topic in many minds today as their influence on the world becomes clearer with climate change research.”
Leopard sharks and remora fish have the most known ocean-based symbiotic relationship, but the focus may not be small enough. Symbiosis is the interaction between two different organisms within the same area, where both organisms benefit from the actions of the other. On one hand, these interactions can be mutualistic, when both organisms benefit from each other. However, on a micro (to avoid repeating
small) scale, cyanobacteria, algae, and plants also use photosynthesis, converting sunlight, carbon dioxide, and water into oxygen that ends up in the atmosphere. Diatoms, a type of phytoplankton, are single-celled photosynthetic eukaryotes that are the primary producers in polar oceans and the base of the polar food chains. Even though they are small in size, they supply a great amount of the Earth’s oxygen. Marine bacteria also have many roles, including assimilating and decomposing the majority of the organic carbon fixed by the diatoms (Figure 2).1

According to Alecia Septer, Ph.D, diatoms require bacteria to allow them to grow. The diatoms release carbon molecules that the bacteria make to create vitamins, such as vitamin B12. This is a mutualistic symbiotic relationships as the compound exchange helps each organism grow.1


Dr. Alecia Septer is an associate professor at the Earth, Marine, and Environmental Sciences department at the University of North Carolina at Chapel Hill. Dr. Septer’s Lab investigated the relationship between diatoms and bacteria alongside Dr. Marchetti’s Lab, which studies how phytoplankton are affected by and adapt to their environment. Her research primarily focuses on microbial interactions and how microbes cooperate and compete with each other for various resources in the environment (Figure 3).2
The main experiment Dr. Septer conducted for her
research investigated the relationship between the diatom P. subcuryata and various associated bacterial strains in the Southern Ocean such as Sulfitobacter sp. SA1 and Olleya sp. A30. Dr. Septer cultivated the bacteria attached to P. subcurvata, and Dr. Marchetti isolated and studied the P. subcurvata diatom.2
While the initial experiment was ongoing, the researchers faced the hurdle of COVID-19 affecting the project’s production. The diatoms they study are extremely sensitive and need to be maintained in culture. If the diatoms had died, it would have been nearly impossible to obtain more during the lockdown, especially from the Southern Ocean. The team had to get permission from the university to do experiments under heavy restrictions, but they managed to get them completed.2

Data from these experiments indicated that the naturally associating bacterial community occurring in P. subcurvata cultures is required for their survival in the stationary phase. Thorough investigation of the results indicated that the relationship between bacteria and diatoms is complex, and that the roles of each differ based on environmental conditions and physiological capabilities.1
A new issue to tackle is how symbiotic association may be impacted by future climate conditions. Dr. Septer’s team used a model to predict how global warming would impact photosynthetic organisms like bacteria. Results showed that these groups would decrease, lowering oxygen production levels and thus hurting other forms of life like plants and animals.1 When asked about the issue of climate change and how it will affect these relationships in the Southern Ocean, Dr. Septer responded by stating that the relationship between climate change and symbiosis is an understudied area that needs more consideration in future research.2 In polar systems, specifically, water temperature is extremely low and microorganisms are sensitive to change. She suggests that future testing of diatombacteria interactions should be done under possible future climate change scenarios to see how they are affected.
One of the most important outcomes from the collaboration was the discovery of a new approach to cultivate and culture the bacteria stuck to the phytoplankton cells. Dr. Septer’s team attempted two methods that resulted in four of the five bacterial isolates that, when added back to the phytoplankton cells, stuck to those cells. As Dr. Septer explains how this research will be used in fields beyond her own, she comments, “We think that our co-cultivation approach will be broadly applicable to other researchers that work in other habitat systems. If they want to try to cultivate bacteria that are physically associated with phytoplankton cells, they can use our method because it is very general” (Figure 1).2
When questioned about how important collaboration was in the making of this paper, Dr. Septer replied, “Collaboration was huge and there was no way I would have done this with just my lab, and I think there was no way the other lab would do this on their own.”2 Cooperation not only happens in a lab between the microorganisms being studied but is also important between the researchers studying them.
References
1. Andrew, S., Wilson, T., Smith, S., Marchetti, A., & Septer, A. N. ISME Communications 2022 2, 97. https://doi. org/10.1038/s43705-022-00181-w
2. Interview with Alecia N. Septer, Ph.D. 1/27/23
“We think that our co-cultivation approach will be broadly applicable to other researchers that work in other habitat systems.”

 by Samantha Breen
by Samantha Breen
Prepare to wrap your brain around this. Within a single mouse brain exists nearly 100 million cells. It would take at least 10 years to count these cells manually– but there is hope. Dr. Jason Stein and his team of researchers have developed a tool called NuMorph, which can automatically recognize a nucleus within an entire mouse brain. This same tool may hold the key to understanding exactly how our genetics influence brain structure.
Dr. Jason Stein is the principal investigator of the Stein Lab, a neurogenetic research lab in both the Department of Genetics and the UNC Neuroscience Center at the UNC School
of Medicine. Beginning in January 2016, the Stein Lab has been investigating how the human genome influences brain structure and development, and how this may lead to the risk of psychiatric disorders.
In his most recent study, Dr. Stein and his team, along with Dr. Lei Xing, used a mouse model of a disease called neurofibromatosis type one (NF1) -- a condition characterized by the formation of tumors within the nervous system. This mouse model has the same genetic mutation found in people with NF1, allowing the researchers to directly see the changes in the brain. Within these models, Dr. Stein and his team performed a technique called tissue clearing, which removes lipids and sends light through the brain, enabling them to take a full 3D picture. With this technique, they were able to see every cell – 100 million of them – in the brain with the help of NuMorph. Short for nuclear morphometry, NuMorph helps the lab understand these differences in cell counts across wild-type mice with no genetic mutation, as well as knockout mice with modulated genetics similar to a patient with the disorder. Wild-type mice have genes that are unchanged and in their natural form, while the knockout mice have both copies of their NF1 gene removed within neural progenitor cell nuclei. These progenitor cells give rise to cells of the Central Nervous System.2 Numorph can essentially “train” a computer to count the differences in cell types between the wild-type and knockout mice. (Figure 1). In order to achieve this, it took nearly a month for graduate student Oleh Krupa and four graduate research assistants to manually count and annotate cells to configure the computer to automatically recognize a nucleus. This tedious process proved worthwhile as it led the researchers to great success.
The team’s goals, however, were not accomplished alone. The Stein Lab works closely with other researchers in the UNC Neuroscience Center. Dr. Mark Zylka, the principal investigator of the Zylka Lab, generated a mouse model with a particular mutation in the mouse genome using CRISPR, a genome-editing technique. When the mouse genome was modified, breeding was performed to get heterozygous mutants and wild types. Wild types have the phenotype generally
found in the population, whereas heterozygous mutants have the defective NF1 gene and the NF1 phenotype. By finding genetic mutations that exist in humans with particular diseases, researchers can generate a similar mutation in the mouse and see how it changes the brain.
Several new neurotechnologies are also designed to count cells within the brain. So, what makes NuMorph different? For one, NuMorph has a higher accuracy for the specific task of counting nuclei. Another algorithm called ClearMap, which is similarly used in whole mouse brains, was also tested for this task. ClearMap is designed to count very sparse objects or objects that are very far apart, which makes it easier to count them. However, when objects are closer together like the nuclei in a mouse brain, a challenge arises.
“NuMorph is trained for the specific purpose of counting nuclei, whereas ClearMap was designed to count sparse labels. So, it’s not really designed for the same task,”1 Dr. Stein notes. He also emphasized the value of whole-brain imaging techniques rather than other forms of methodology to study the brain. Usually, to find structural differences, researchers take an individual “slice” of the brain. With this method, however, researchers are unsure if their particular “slice” has the answers they are looking for. In a sense, they have to be “lucky” that the slice chosen has the structural differences that exist, according to Dr. Stein. On the other hand, with whole-brain imaging, researchers can find any structural differences that exist, avoiding the issues posed by taking an individual slice of the brain (Figure 2). However, a downside of this thorough technique is the sheer size of the data. Dr. Stein mentions that just one image of a mouse brain has nearly a terabyte of data which, to put into perspective, is
equivalent to nearly 75 million pages of text.3 Human brains are each worth about a petabyte in size, 1000 times more than a mouse brain, which poses a challenge since the Stein Lab hopes to study human brains in the future.

“You have to create really efficient computational code to handle extremely large data sets,” says Stein. He also hopes to analyze even larger data sets as he studies human brains. “The future is to move towards human brains by using inherent genetic variation -- the natural genetic differences that exist among the population -- rather than inducing a mutation.”1 Knowing the region or cell type can introduce bias by leading researchers to only look for specific characteristics within a particular region. Dr. Stein hopes to utilize computational approaches like these because they allow for more unbiased approaches. Rather than targeting a specific region or cell type, this method measures every single region. He notes that this lack of bias leads the researcher to continue proceeding throughout the study without a specific hypothesis, which has led to a higher level of reproducibility in other fields like genetics. Dr. Stein’s future goals lie in exploring other mouse models for psychiatric disorders. The lab’s next project involves another mutant mouse model to investigate the CHD8 gene associated with autism risk, generated by the Zylka Lab. Additionally, the Stein Lab would like to work with human brains using postmortem human tissue. With this method, the researchers can use inherent genetic variation to study how genetics influence the human brain.
Dr. Stein and his team have made significant strides with NuMorph, clarifying the relationship between the human genome and the development of psychiatric disorders. The Stein Lab is paving the future of neurogenetic research.
1. Interview with Jason Stein, Ph.D. 02/06/23.

2. Cerdeno, V. M. & Noctor, S.C. Frontiers in Neuroanatomy. 2018, 12, 1662-5129. Neural Progenitor Cell Terminology.
3. Krupa, O.; Fragola, G.; Hadden-Ford, E.; Mory, J. T.; Liu, T.; Humphrey, Z.; Rees B. W.; Krishnamurthy, A.; Snider, W. D.; Zylka, M. J.; Wu, G.; et al. Cell Reports 2021, 37(2): 109802. NuMorph: tools for cortical cellular phenotyping in tissue cleared whole brain images. (accessed March 23rd, 2023)
4. Paceadmin. Putting Data into Perspective - Pace Technical. Pace Technical - Managed IT Services Toronto, 7 Mar. 2012, https://pacetechnical.com/112-putting-data-into-perspective/#:~:text=A%20single%20terabyte%20can%20 store,the%20form%20of%20ard%20drives. (accessed March 23rd, 2023)

 By Estella Calcaterra
By Estella Calcaterra
Acritical missing puzzle piece in understanding cell regulation and communication is hidden in plain sight: the lipid droplet (LD) organelle. Most biology students learn that organelles are specialized parts of a cell, each with a unique function. Most familiar is the mitochondria, which produces the cell’s energy, and the endoplasmic reticulum, which produces molecules. However, a deeper look into cell physiology reveals that organelle communication is equally as important, enabling the cell to adapt and maintain a consistent internal environment.1,2,3,4
Initially, LDs were assumed to simply be fat storage within the cell because lipids (fats) are incompatible with water. LDs also have a unique singlephospholipid layer (Figure 1), unlike most bi-layered organelles. Phospholipids are barrier lipids enclosing the cell or organelle from the environment.4 Due to their irregular structure and contents, LDs physically travel to other organelles to complete their functions.
LD functions are divided into two main categories: lipid regulation and lipid signaling.2,4 Lipid regulation includes controlled storage and release of lipids for energy. The second category of LD function, lipid signaling, refers to lipid mediators which regulate processes like lipid transportation, immune
system response, and both cell and inter-organelle communication.2,3 Both functions are critical to every aspect of cell function, as researchers like Dr. Sarah Cohen at the University of North Carolina at Chapel Hill are now discovering.
Dr. Cohen and her lab utilize advanced imaging methods to observe LD organelle movements.4 One of her lab’s specialties is multispectral imaging. Each organelle is marked with a different wavelength-correspondent fluorescent marker of the light spectrum. The
fluorescence is recorded over time and clarified by sorting each fluorescent point into a different color depending on its wavelength with prisms, which, Dr. Cohen explains, “…split the light onto [an] array of detectors to collect spectra in each pixel of the image, and then we use math to computationally separate the different colors.”4 This light-microscopy method observes patterns of LD movement near organelles (Figure 2).4
Light microscopy methods like multispectral imaging are powerful, but
the specific moment of contact between LDs and other organelles. Dr. Cohen states that the dimerization-dependent fluorescence uses, “… a [weakly] fluorescent protein, … unless it touches another protein, it’s dimer,”4 When the protein on an LD finds an identical protein on an organelle, they bind to form a dimer and release a bright fluorescence. This indicates contact between the organelles at the membrane contact site. Dr. Cohen reveals that the dimer bond is reversible, so when the dimer bond breaks as the organelles move away, the fluorescence darkens.4 Using these imaging methods, Dr. Cohen outlined 3 main projects in her lab’s current research: identifying proteins at LD-organelle membrane contact sites, LD communication in cell differentiation or development, and LD presence in neurodegenerative disease(s).
Dr. Cohen describes protein identification as similar to dimerizationdependent fluorescence. Instead of dimers, her lab can select certain proteins at membrane contact sites with enzymes, which are proteins that regulate chemical reactions. She explains the process of selecting proteins with enzymes as “targeting”, where half an enzyme is placed onto the lipid droplet and half on the other organelle.4 When the LD and organelle meet at the membrane contact site, the enzyme tags the selected proteins.4 The tag on a protein acts as a “handle” to pull the protein out of the cell, which can later be observed in a mass spectrometer, a tool used to identify substances via molecular weight.4 By recording the proteins at the membrane contact site, her lab can better understand the mechanisms of how communication occurs or what is being communicated between the two organelles.
Cell differentiation is the process of cell development from a
new to a specialized cell. Dr. Cohen points out that LDs may be the key to understanding stem cell specialization during development due to the LD’s role in signaling pathways. She emphasizes that, “…we find that the pattern of contact between different organelles to be a really key feature of a given cell type… the shape of the organelles and the communication between the organelles is really different.”4 Currently, cell differentiation is a complex process not yet fully understood. Further study into patterns of contact between LDs and organelles, as well as which pathways are activated will unveil unknowns with cell differentiation.
Her lab’s third area of study focuses on the connection between LD presence and disease. Presence of LDs has been implicated in lipid metabolism disorders such as diabetes or atherosclerosis, as well as neurodegenerative disorders like Alzheimer’s or dementia. Dr. Cohen explains, “…there is this emerging theme that contact sites are implicated in multiple neurodegenerative diseases: Alzheimer’s, Parkinson’s, [and] amyotrophic lateral sclerosis.”4 She notes that Alzheimer’s was of particular interest. When the disorder was classified, one characteristic was “adipose saccules” or lipid bubbles, which Dr. Cohen theorizes may be the accumulation of LDs. She
states, “This is something that was observed early on, but … has gotten a lot less attention.”4 She adds that, “…there are defects in lipid metabolism in other neurodegenerative diseases too.”4 She notes the increase of LDs in certain brain cells, and whether LDs are beneficial or a symptom of disease is not yet known.4
Dr. Cohen mentioned many other LD processes, such as lipid protein localization, innate immunity, immune response, LD antibacterial properties, and misfolded protein removal.4 She emphasizes that further LD research is necessary, and that scientists need to fundamentally understand the complicated LD before any use in medicine. However, Dr. Cohen believes there are many possible therapeutic applications in the future and is excited to continue uncovering the mysteries of our cells via further exploration and reexamination of the previously overlooked LD.4
References
1 Walther, T.C.; Chung, J.; Farese, R.V. Jr.; Annu Rev Cell Dev Biol. 2017, 33, 491-510.
2 Cohen, S.; Int Rev Cell Mol Biol. 2018; 337, 83-110.

3 Jarc, E.; Petan, T.; Biochimie 2020, 169, 69-87.
4 Interview with Sarah Cohen, Ph.D. 2/8/2023.

 By Morgan Davis
By Morgan Davis

North Carolina is known for its exceptional oysters. NC oysters have a distinct, complex flavor that reflects the diversity of NC water environments. Although delicious, the consumption of raw oysters can be risky. Doing so can lead to the development of foodborne illnesses, which can cause gastrointestinal pain, nausea, and even death. Unfortunately, there is no way of knowing if raw seafood is safe to eat by sight alone. However, Dr. Rachel Noble and her team at the Noble Lab
are dedicated to finding ways to safely consume raw seafood through the study of shellfish, vibrio pathogens, and water quality.
Between 2001 and 2002, Dr. Rachel Noble and Denene Blackwood, the current lab manager, noticed a blind spot in water quality research. While the coasts of North Carolina and Dr. Noble’s previous residence of California both bring millions of tourists a year, Dr. Noble saw that the quality of North Carolina’s rural areas and waterways were only being given a fraction of the attention. Hence, with perseverance and a few passionate research technicians, the Noble Lab was born. After its establishment, the lab quickly began working with North Carolina natives in rural towns on water quality topics. Twenty years later, this lab is at the forefront of their field in making things that appear complicated— such as water quality—more streamlined using cuttingedge biological technology.

According to Dr. Noble, “[the
Noble Lab] has always pushed the envelope on using the newest tools to quantify the things that cause illness.”1 Today, the Noble Lab’s research encompasses everything from applied environmental microbiology to studying marine microbial food webs. Holding true to their roots, they use their expertise in water quality research to investigate the clammy issue affecting NC’s trademark oysters: vibrio pathogens and their impact on public health and shellfish harvesting. Vibrio is a genus of bacteria that can cause foodborne illnesses associated with eating undercooked seafood. According
to the CDC, vibrio bacteria are naturally present in warmer coastal waters.2 Since shellfish are filter-feeders, they consume a lot of water, and can act as indicators of how much of a pathogen is present in a waterway and where the contaminated water is going. Thus, examining shellfish allows for a more complete understanding of not only how pathogens get into shellfish, but the risks associated with the pathogens to human health (Figure 2).3
Dr. Noble’s interest in shellfish and vibrio pathogens has led to the creation of several fascinating studies, including Dr. Noble’s 2016 Oyster probiotic study. For humans, taking probiotics helps the body maintain a healthy community of microorganisms and eliminate pathogens. This concept, theoretically, could work for shellfish as well. Similar to how humans eat bacteria-rich foods like kimchi and yogurt, the Noble Lab could add specific types of good bacteria to reduce or replace the bad bacteria in shellfish. Dr. Noble and her lab were able to prove their hypothesis right by giving oysters probiotics that would kill pathogens inside them.
After concluding her research on the oyster probiotic study, Dr. Noble still wanted to find ways to reduce the risks associated with raw shellfish consumption. Therefore, she created the Underdock Oyster Cultivation (UDOC) and Revolutionizing Rapid Molecular Diagnostics (MDx) Studies. The purpose of the UDOC study was to compare the riskiness of recreational oyster cultivation to commercial. Funded by the NC Sea Grant, the Underdock Oyster Project gave citizens the opportunity to grow oysters off their docks. However, docks
can be filled with toxins, like boat waste and run-off from nearby residences. Additionally, the water around docks is stagnant, sediment-rich, and muddy; an environment in which vibrio thrives. Through determining the concentration of vibrio pathogens in UDOC oyster farms, Dr. Noble’s study found UDOC oyster farms to be significantly riskier than commercial oyster farms.
While the NC Underdock Program ended in 2019, there is a possibility that citizens are still growing UDOC oyster farms for personal consumption. As stated earlier, there is no way to know if a raw shellfish is safe to eat through sight alone. To fix this issue, Dr. Noble created a new study focused on revolutionizing Rapid Molecular Diagnostics. It aimed to create tools that are rapid, cost-effective, user-friendly, and could be used to test if raw seafood is safe for human consumption. Before this study, testing seafood involved an incubation process that would take 18-24 hours. Now, the new rapid MDx has reduced this window to 2-5 hours!
Additionally, the rapid MDx created from this study can test for six different pathogens at once. “It’s kind of like making cookies,” described Dr. Noble. “Once you make the cookie batter [rapid testing tool], what ingredient [pathogen] you add to it to make it that specific type of cookie is irrelevant.”1 Developing the rapid MDx process was complex, but the Noble Lab’s efforts have made pathogen testing as straightforward as baking cookies.
Currently, Dr. Noble is collaborating with a company on the rapid MDx study to develop a gastrointestinal panel that could test for bacteria, like Salmonella, Norovirus, and Campylobacter. Collaboration is a concept the Noble Lab is well acquainted with. Dr Noble enjoys building industry partnerships, as it allows her and her partners to develop new, effective tools to mitigate community issues and teach the local community about water quality through hands-on experience. As an ongoing project, Dr. Noble has partnered with Dr. Liz DeMattia and the Duke Community Science Initiative to develop a water quality app for high school students with the aim of inspiring them to connect more with their outdoor environment.
The work of the Noble Lab shows how water quality research is much larger

than only looking at oil spills. Water quality impacts our everyday lives, from the water we brush our teeth with to the raw oysters we eat on a beach vacation. The next time you eat sushi, mussels, or clams at a nice dinner, perhaps Dr. Noble’s research helped with making sure your food was safe to eat!
1. Interview with Dr. Rachel Noble, Ph.D. 02/01/2023
2. Center for Disease Control & Prevention (2019). Vibrio Species Causing Vibriosis. Retrieved from https://www.cdc.gov/vibrio/faq.html
3. Froelich, B.A., B. Phippen, P. Fowler, R.T. Noble, and J.D. Oliver. 2016. Differences in total Vibrio spp. V. vulnificus, and V. parahaemolyticus abundance between clams and oysters in North Carolina. Applied and Environmental Microbiology. doi: 10.1128/AEM.02265-16



It’s the little things that matter most in life. Although invisible to the naked eye, water sources all over the globe like oceans and lakes are full of tiny microbes. Also known as microorganisms, microbes including bacteria are essential to numerous environmental processes. One example is the carbon cycle. This cycle is a series of biogeochemical processes where carbon is transferred from the atmosphere into the Earth, and back again. One of the ways that carbon can reenter the atmosphere is through the degradation of organic matter, where these dutiful microbes come into play.
At UNC-Chapel Hill, Dr. Carol Arnosti is currently working in her lab to further understand how these environmental processes work and their causes. Dr. Arnosti is a professor in the department of Earth, Marine, and Environmental Sciences at UNC, and a principal investigator of the Arnosti Lab. She has a bachelor’s degree in Chemistry from Lawrence University and a Ph.D. in Chemical Oceanography, jointly from MIT and the Woods Hole Oceanographic Institution. Dr. Arnosti’s lab focuses on microbial biogeochemistry and oceanography, with many ongoing projects geared towards one main focal point: microbial-driven carbon cycling in the ocean.1
The Arnosti Lab conducts research both in a laboratory setting and out in the
By Reagan Gulledgefield to gain a more extensive knowledge base of unknowns in oceanic microbial ecosystems. “I basically study how things rot; processes that happen in compost heaps where organic matter is broken down also occur in the ocean,” Dr. Arnosti explained.1 The organic carbon that is naturally produced in the ocean is also, for the most part, naturally recycled by
bacteria. Heterotrophic microbes use extracellular enzymes to break down the organic matter, a process which eventually transfers matter back into the atmosphere. Much of marine primary productivity (the production of organic material from aquatic or atmospheric CO2) is processed by microbes, and the rate of their degradations is vital to the carbon cycling in marine systems.1 In the ocean, photosynthesis only happens in surface waters, which is where the sunlight penetrates, while dissolved organic carbon resides everywhere.
Even though microbes are incredibly small, they have a large impact on a global scale. The carbon cycle in the ocean has an enormous influence on the global carbon cycle. The location
of bacteria in the ocean and how they decompose organic matter affects the primary productivity of the ocean. Even though nitrogen and phosphorus are returned to the water when organic matter is degraded, it is important to know when and where this degradation occurs to determine phytoplankton growth. Phytoplankton are organisms that form the base of the marine food web and are an essential component to the other half of the carbon cycle: photosynthesis. Both the carbon cycle and photosynthesis influence each other based on their separate productivity. The depth at which organic matter is eaten helps control how much carbon dioxide (CO2) the ocean can take up from the atmosphere.2
“Even though microbes are incredibly small, they have a large impact on a global scale.”Dr. Carol Arnosti
Dr. Arnosti has embarked on 30 separate research voyages around the globe and has collaborated with scientists internationally through her years of research.3 One of her current projects even has a field location on Svalbard, a Norwegian archipelago in the Arctic Sea. This research is focused on Svalbard because most carbon cycling investigations are carried out in temperate environments, not polar climates. However, the Arctic is also particularly vulnerable to rapid rates of climate change, so it is important to record microbial productivity rates at this type of location. The experiments that the Arnosti Lab carries out, examines carbon cycling in water columns and sediments.3 These two types of data are essential for understanding how carbon is consumed.


During her time as a lead researcher, Dr. Arnosti and her team have faced many hurdles, the biggest one being funding. Being in a lead position in her lab means that she has to write numerous grant proposals in order to keep her projects funded and team members paid. This task is exceedingly time consuming and stressful because people do not get paid without it. “Bad weather at sea and unexpected stuff, those are normal and don’t bother me. I never worry about the weather because it’s going to happen, and you should have plans A-G.” expressed Dr. Arnosti.1
Another growing aspect of the
Arnosti lab is the time that she and her student-assistants are spending at the campus’ Be-A-Maker Space to build new prototypes.3 The prototypes are being used to try and find ways to better visualize happenings at the microscopical level in the sediments. “Sediments are complicated, and there are many steps to organic matter degradation. By building these cells we are trying to spatially separate some of the steps out that normally happen together so that we can isolate different things and determine how they’re structurally being changed”.1
As a result of the extensive research, there have been many exciting and unexpected findings in Dr. Arnosti’s lab. “Mother nature sometimes comes knocking on your door and tells you, ‘You’ve been all wrong in your assumptions about how some aspects of carbon cycling works, and you just have to rethink what you thought.’”1 She and her team have discovered new ways
in which the microbial communities function in marine environments. Additionally, some of the findings regarding the bacterial carbon uptake processes, as well as some of the tools they stumbled into serendipitously, are being used in medical and biomedical research. Specifically, the research has applications in the identification of which organisms can absorb certain types of organic carbon in the gut. Although the current knowledge of the carbon cycle in marine environments is limited, Dr. Arnosti is one of the leading researchers in the field, adding valuable knowledge to the subject as the world becomes more cognizant of the true extent of climate change.
References:
1. Interview with Carol Arnosti, Ph.D. 02/01/2023
2. Arnosti, C. Ann. Review of Marine Science 2011, 3, 401-425.
3. Arnosti Lab. https://arnostilab.web. unc.edu/
“However, the Arctic is also particularly vulnerable to rapid rates of climate change, so it is important to record microbial productivity rates at this type of location.”
 By Jessica Hoyt
By Jessica Hoyt
Have you ever turned over a leaf and found a bug crawling around underneath? Some may be frightened by this critter, while others may wonder about it’s travels. Maybe they would consider how many leaves this little guy has consumed in his curt lifespan — or even what adaptations it has developed to survive predatory attacks or harsh climates. Or, perhaps, you’d find the closest shoe to smash him to pieces. Most people avoid bugs at all costs, but not Dr. Allen Hurlbert. The phenomenon of studying organisms and their interactions within the environment is known as ecology.1
As the late winter season ushers in warm days, animals and plants alike are left befuddled by early spring temperatures and react accordingly: confused. If the temperature reaches a particular threshold, plants begin to bloom, and insects emerge. Why would this pose an issue for anyone? Wearing shorts in February may seem fabulous to us, but our environment becomes stressed if thrown off balance.1
Birds may migrate back to their southern homes during a pseudo-spring but become bombarded by possible late weather storms. When insects arrive too early, there is not enough food to survive.2 On the same token, if birds arrive too late, they miss out on the bug buffet and their offspring suffer as a result.2 Every living thing has a domino effect on another.1
When these animals arrive at different times, the imbalance creates what is known as a phenological mismatch.2 Phenology is a study within ecology that observes timed events, such as bird migration, flowering patterns, and hibernation. Typically, all these natural cycles work in tandem with one another: the sun warms the earth, plants bloom, bugs emerge and eat plants, and birds migrate back and eat the bugs. Furthermore, bug populations are controlled and plant population is relegated,
birds pollinate the plants, and their offspring are able to eat.
Dr. Allen Hurlbert, a biology professor at UNC, studies the significance of these climate-driven mismatches, and has led a citizen science project known as ‘Caterpillars Count!’2 Hurlbert’s research depends on many people to collect data, as insect and arthropod populations far outnumber humans. By thumbing through tree branches and bushes, anyone can help supply useful data to monitor patterns in ecosystem health. Survey participants can follow instructions found on the Caterpillars Count! webpage and enter data via mobile app or website form.2 The data is later subsampled after compilation by Dr. Hurlbert’s team for research purposes.2

The project uses several methods to observe these insects: beat sheets, visual surveys, and frass collection.3 Beat sheets are blocks of stretched white fabric that allow observers to easily find bugs that may be camouflaged.2 Participants beat branches and observe the organisms that fall off on the beat sheets.3 Regular visual surveys of lower tree branches are also collected.3 Two groups collect this data: citizen scientists and trained scientists.3 Any possible discrepancies between the quality of the data compiled by either group were compared and found to be reliable.3
“We are essentially measuring bird food,” Dr. Hurlbert said, “because if we can monitor the phenology of arthropods, we are able to utilize existing datasets to compare overlapping
results and determine mismatches.”2
The research participants are looking for far more organisms than caterpillars. The possibilities range anywhere from spiders and ants to beetles and aphids. What is the purpose of counting all these crawling and flittering insects? When analyzing the data, different population numbers helps Dr. Hurlbert and his team understand more about ecosystem health. Populations too large in regards to bird populations communicate a mismatch has occured.
A mismatch occurs when there are imbalances in the climate — particularly in the spring months — signaling hibernating insects to emerge from overwintering. Insects dine on mostly vegetation to survive, making them herbivores. Birds eat these herbivores and then later feed their young the catch of the day.3
In his research, Hurlbert outlines the differences between the two collecting groups. Apart from the regular surveys, trained scientists use a method of data collection known as ‘frass monitoring.’3
“Frass is just caterpillar poo and is used to understand bug abundance,” Dr. Hurlbert explained. “Surveying on the ground can only tell you a small fraction of what lives in the trees, so we use beat sheets to collect frass. Since all caterpillars make frass, it is a helpful method in determining their seasonal population.”2

Imagine a caterpillar in the winter months. As the skies darken and the air becomes frigid, it digs a safe burrow in the soil to hibernate. After only a few months, there is a week of abnormally warm weather. It’s February, but they didn’t bring their caterpillar calendar with them. They can’t check a weather app or look at the forecast either. The coast seems clear and they writhe their way to the surface, ready to feast on a flush buffet of leaves and greenery.
Animals are much different than humans. They rely on environmental cues to perform important life processes. Examples of environmental factors include temperatures, light cycles, diet, humidity and mineral deficiencies — all can affect the behavior of any animal. Disparities in any can be cause for disaster in a weak biome. The consequences spillover into daily human life as well — in agricultural settings, insect overpopulation, and cause a continually crumbling trophic
hierarchy.2 Luckily, the first steps are found in citizen science projects like Hurlbert’s.
Hurlbert was able to create a curious project to tackle a greater issue than an individual can comprehend. As more and more data is collected, one will be able to illustrate this problem more clearly. Follow the progress or become a part of the solution at https://caterpillarscount.unc.edu/ and download the app.
Thanks to the consistent work of Dr. Hurlbert and his army of research participants, phenological issues have been identified in local environments. Without identifying the cause, the problems may become more widespread and harm other populations within nature. The growing issue of climate change sparks little fires throughout the natural world, but Dr. Hurlbert is working to combat each flame, one caterpillar at a time.

https://caterpillarscount.unc.edu/
1. Prather, R M; Dalton, R M; Barr, B; Blumstein, D T; Boggs, C L; Brody, A K; Inouye, D W; Irwin, R E; Martin, J G A; Smith, R J et al. Proc. Royal Soc. B 2023, 290: pp 2-22
2. Zoom Interview with Allen Hurlbert, Dr. 02/02/23
3. Hurlbert, A; Hayes. T; Mckinnon, T; Goforth, C. CSTP 2019, 2: pp 2-22
Mysterious, yet known – check. Partially understood, yet complex – check. Always on the move – check. Found in every living organism – also check. Ribonucleic acid, or RNA, is a key molecule for protein synthesis, and thus, the creation and maintenance of life – yet it is involved in many other bodily systems that scientists are still working to find out. One of those leading the charge is Dr. Qi Zhang, an associate professor here at the UNC School of Medicine.
Dr. Zhang received his PhD in chemistry from the University of Michigan in 2007, where he developed novel experimental techniques for visualizing RNA motions. However, he was only an undergraduate sophomore when his interest in molecular biology was sparked.1 At the time, Dr. Zhang found the molecular motion of proteins fascinating and began creating his own computer programs to simulate protein folding – over 20 years ago.1 Since then, he completed his postdoctoral training in the structural biology of RNA at UCLA and started the Zhang Research Group at Chapel Hill in 2011-2012. His lab is researching the unknown properties of RNA – what do its “noncoding” parts do?1
RNA is primarily known as the molecule that is responsible for protein synthesis. It works with its more famous counterpart DNA by “transcribing” DNA’s code and relaying that information to the protein-building parts of a cell.2 These proteins become the building blocks for all of life’s processes. The three types of RNA involved are extremely well-known and documented: messenger (mRNA), transfer (tRNA), and ribosomal (rRNA).3 More specifically, mRNA is referred to as “coding” RNA. Noncoding RNA, which does not code for proteins, has a diverse array of functionality, including gene suppression, regulation, and even slowing down tumors.3 In fact, Dr. Zhang cites the relative explosion of RNA research over the last 30 years as the reason behind some major advancements in the field of biology and medicine. A shining example is the successful creation of the mRNA COVID-19 vaccines.4
“Scientists have considered RNA as a ‘dark matter’ in biology for a very long time,” says Dr. Zhang. “We know what
DNA does, and what proteins do, but with RNA we did not have much of a clue.” Only around 2% of a human genome transcribed by RNA actually codes for proteins, with the rest of our genome’s use left unknown.5 Dr. Zhang notes the inherent difficulty in studying the many structures and complex folding patterns of RNA – its mobility. RNA is a constantly moving and shapeshifting molecule, which makes it extremely difficult to obtain high-resolution pictures of its structures for understanding the mechanism of their functions.1 One of the goals of the lab is to take a ‘nano-video’ of RNA. This essentially means having a dynamic representation of real RNA molecules interconverting among many varying states, so that their mobile functions can be better understood.1
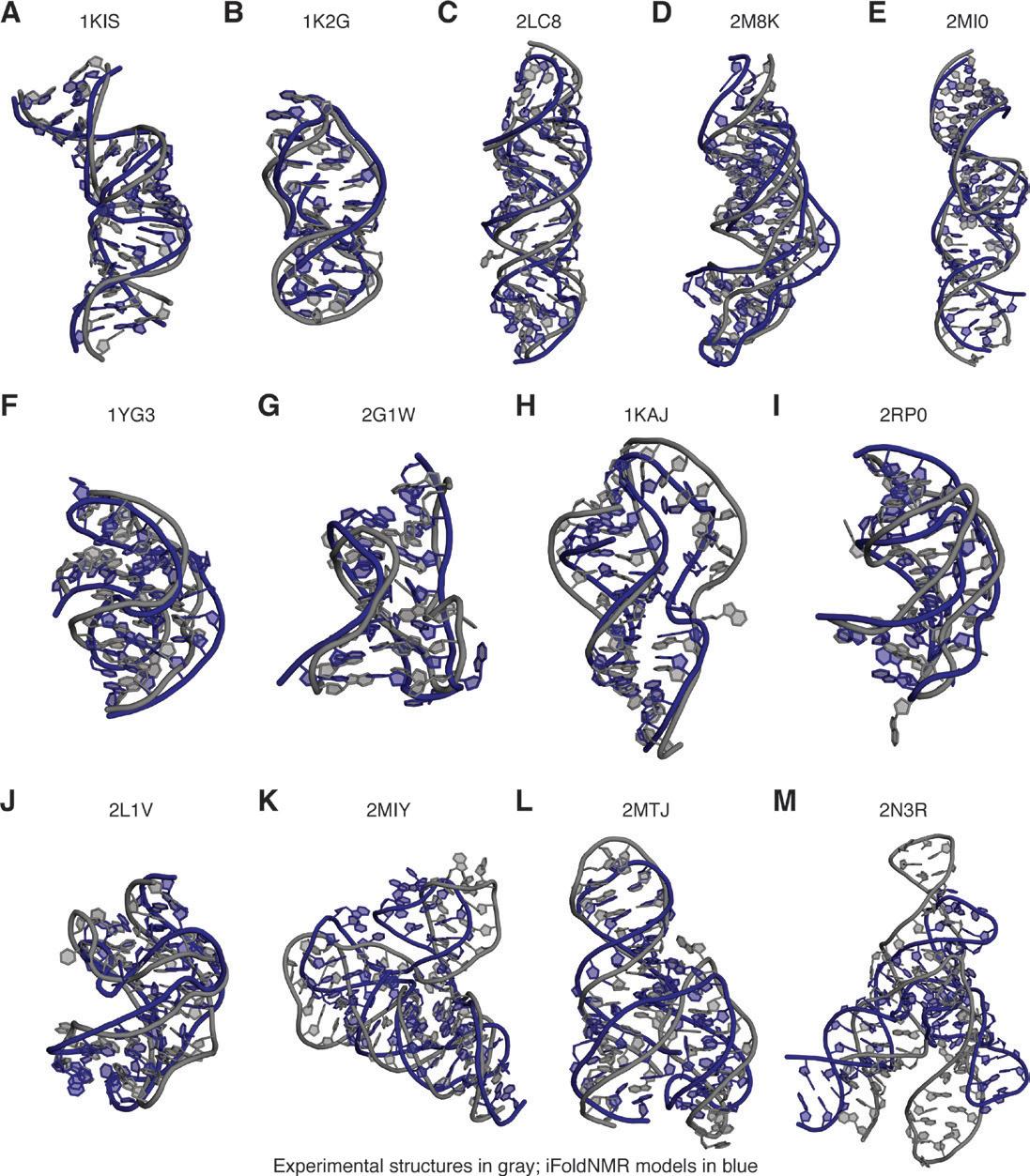
In 2017, Dr. Zhang assisted in developing a software known as iFoldNMR to model complex RNA structures accurately and quickly. The software is novel in its much simpler requirements of experimental data for creating an accurate model compared to others of its kind. It uses constraint data taken from nuclear magnetic resonance (NMR) experiments used to determine certain structures of RNA. From the data, iFoldNMR performs simulations that consider many of the different base-pair interaction biases to create a model for the structure in question.6 Figure 1 shows the visual differences in the structures generated by iFoldNMR and the experimental structures. They noted that specific structures were reproduced far better than other computational methods, including pseudoknots with base triples, kissing dimers, and RNA–ligand complexes. Comparisons were evaluated using a root mean square deviation metric. Overall, the software provides an Dr. Qi Zhang


innovative way to better observe RNA structures that researchers may uncover during NMR experimentation.6
The Zhang Lab has several ongoing projects dedicated to uncovering functionalities of nucleic acids and nucleic-acid sensing proteins. One such project concerns the role of a newly discovered DNA sensor, cGAS, in the innate immune response. The activation of this protein by viral and bacterial DNA can give rise to the synthesis of a small RNA molecule that triggers a chain reaction of pathways to alert the innate immune system. Understanding the ways that this protein affects biological pathways can add another piece to the puzzle of RNA’s capabilities.7 Additionally, another project is rewinding the clock to discover how protein synthesis came about. Dr. Zhang describes the experiment as “researching the origin of life.” The project is sponsored by a grant from the W.M Keck Foundation8 and is partly undertaken by sophomore Ethan Meyerhoffer. “We are trying to figure out a potential mechanism for protein production using just RNA molecules, something that would make monumental advancements for the explanation of prebiotic life,” he explained. Essentially, the project is looking for evidence to support the idea that RNA could have been the precursor to all life on Earth.9
When asked about getting into research as an undergraduate, Dr. Zhang offered strong words in favor of starting early. He cited the work of his mentors at both Michigan and UCLA as the inspiration for his work, and his undergraduate research as the spark. The Zhang Lab and many others alike in the School of Medicine are always looking for highly motivated undergraduates to participate in research roles. These include mentored work as well as the potential to
start their own projects that align with the lab’s goals. Together, they work towards shedding more “light” on the “dark matter” that is ribonucleic acid. We have already seen RNA research being used to create the COVID-19 vaccine in record times. It is no secret that uncovering the untapped potentials of RNA and its interactions with proteins can bring major advancements to fields like immunology, virology, genetics, biophysics, and drug discovery. You may wonder what propels RNA researchers into exploring these uncharted waters of molecular biology. Dr. Zhang referenced the two words that he felt best described his motivations thus far: “staying curious.”
1. Interview with Qi Zhang, Ph.D. 02/01/2023.
2. Wang, D., Farhana, A., (2022, May 8). Biochemistry, RNA structure. In StatPearls. StatPearls Publishing. Retrieved February 9, 2023 from https://www.ncbi.nlm.nih.gov/books/NBK558999/
3. Encyclopædia Britannica. RNA. https://www.britannica.com/science/RNA (accessed February 9, 2023).
4. Centers for Disease Control and Prevention. Understanding how covid-19 vaccines work. https://www.cdc.gov/coronavirus/2019-ncov/vaccines/different-vaccines/how-they-work.html#:~:text=First%2C%20mRNA%20COVID%2D19%20vaccines,is%20 called%20the%20spike%20protein (accessed February 9, 2023).
5. Lee, H., Zhang, Z., & Krause, H. M. Trends in Genetics 2019, 35(12), 892–902.
6. Williams, B., Zhao, B., Tandon, A., Ding, F., Weeks, K. M., Zhang, Q., & Dokholyan, N. V. Nucleic Acids Research 2017, 45(22), 12638–12647.
7. Interview with Sanath Yeduri, Undergraduate. 02/07/2023.
8. University of North Carolina at Chapel Hill. Carolina scientists earn prestigious Keck Award - University of North Carolina at Chapel Hill: The campaign for Carolina.. https://campaign.unc.edu/ story/carolina-scientists-earn-prestigious-keck-award/ (accessed February 9, 2023).
9. Interview with Ethan Meyerhoffer, Undergraduate. 02/09/2023.

 By Lintao Li
By Lintao Li
When telling people about his work on proteins, Dr. Kuhlman usually gets the slightly humorous response of, “what do you make, better meat?”1 However, proteins are more important than just food prioritized by gymgoers. From transmitting signals to serving as building blocks for the body, proteins are large molecules with a vast array of cellular functions in our bodies.2 To perform these activities, proteins must fold into a specific 3-D shape, making the process of protein folding a crucial area of study.2 Dr. Brian Kuhlman, a researcher at the University of North Carolina at Chapel Hill, is at the forefront of this field, using a combination of experimental and computational methods to uncover the mysteries of protein folding and advance our understanding of these macromolecules.1

Proteins are complex molecules that play essential roles in nearly all cellular processes. Made up of long chains of small molecules called amino acids (molecules with a nitrogen and an acid group), each protein sequence is synthesized through a process known as translation, where small cellular machines called ribosomes read the genetic code contained in genetic material called messenger RNA. The ribosomes assemble the appropriate sequence of amino acids based off the messenger RNA to form the final protein. The many functions of proteins cannot be understated.2 To give just a few examples, structural proteins such
as collagen provide the underlying framework for cells and tissues, while enzyme proteins act as catalysts that speed up chemical reactions. Transport proteins like hemoglobin carry oxygen and other essential molecules throughout the body, while signaling proteins such as hormones and neurotransmitters regulate cell behavior and communication.3 Due to these many functions, proteins are the targets of many drugs and treatment plans. The malfunction or misregulation of proteins can result in diseases such as cancer, while pathogens act based on protein-protein interactions. Therefore, understanding the structure, function, and regulation of proteins is a critical aspect of modern biology and medicine.4
One of the main tools Dr. Kuhlman’s work uses is Rosetta, a computational platform that performs a variety of proteinrelated functions. This tool has become an indispensable resource for Dr. Kuhlman’s lab. The power of Rosetta lies in its ability to consider all possible shapes of a protein and identify the most favorable one based on complex interactions between the different amino acids.5 After a model of the protein is created, its associated amino acid sequence is created and its structure and function are analyzed. If something doesn’t conform to how the protein was expected to function, Rosetta is used again to recalculate a more favorable sequence. This back-and-forth computational biology allows Dr. Kuhlman to improve existing proteins and even make new functional proteins.1
One recent task the Kuhlman lab is trying to tackle with this technique is to create a vaccine for de ngue, a mosquitoborne disease caused by the dengue virus (Figure 1). Almost 4 billion people are at risk of infection, with hundreds of millions of infections and around 20,000 deaths each year.1 The underlying cause for the disease’s widespread prevalence is the lack of a universally safe vaccine; Dr. Kuhlman even states that “it’s been harder to create a good vaccine for dengue than for Covid.”1 One way he has tackled this is through Rosetta by modeling a Dr.
stabilizing protein dimer (two proteins bound together) similar in structure to the proteins found on the dengue virus, paving the path towards a more robust vaccine.1

Another project Dr. Kuhlman is involved in is the development of bispecific antibodies. Antibodies are Y-shaped proteins produced by the immune system in response to the presence of foreign substances, such as viruses, bacteria, and toxins (Figure 2). They protect the body against infection and disease by recognizing and binding to antigens: specific parts on the surfaces of harmful invaders. Every antibody is highly specific, with 2 binding sites in a unique shape that allow it to recognize and bind to a particular antigen. The binding can have several effects, such as neutralizing pathogens, marking antigens for destruction by other components of the immunesystem, or triggering an immune response.6 The two binding sites on antibodies produced by our bodies bind to only one specific antigen. However, a bispecific antibody, as the name suggests, binds to two different antigens, allowing for more complex interactions.1 Such interactions are the target for Dr. Kuhlman’s collaboration with the pharmaceutical company Eli Lilly to create bispecific antibodies that can bind both cancer cells and the body’s
immune cells. This takes advantage of the body’s own immune system to fight off cancer in a process known as immunotherapy. Eli Lilly is currently producing these antibodies with promising results, and clinical trials are actively being conducted.1

Computational protein design has vast applications, and the future of creating proteins is bright. With newer and more powerful methods in the future such as DeepMind’s AI-driven protein structure predictor AlphaFold (Figure 3), Dr. Kuhlman is hoping to develop novel enzymes and better protein switches.1 Developing enzymes will increase the efficiency of all fields of work, from agriculture and food processing to pharmaceutical production, while protein switches can help us develop better drugs, such as diabetic drugs that only activate when insulin levels are high enough.1 The application of Rosetta and protein folding understanding cannot be overstated. With each new application comes a host of treatments and techniques to improve healthcare and other cellular interactions.
References:
1. Interview with Brian Kuhlman, Ph.D. 01/30/23
2. What are proteins and what do they do?: MedlinePlus Genetics. (2021, March 26). MedlinePlus. https:// medlineplus.gov/genetics/understanding/howgeneswork/ protein/
3. Protein function. (2014). Learn Science at Scitable. https://www.nature.com/scitable/topicpage/proteinfunction-14123348/
4. Protein misfolding and degenerative diseases. (2014). Learn Science at Scitable. https://www.nature.com/ scitable/topicpage/protein-misfolding-and-degenerativediseases-14434929/

5. Kaufmann KW, Lemmon GH, Deluca SL, Sheehan JH, Meiler J. Practically useful: what the Rosetta protein modeling suite can do for you. Biochemistry. 2010;49(14):2987-98. doi: 10.1021/bi902153g. PMID: 20235548; PMCID: PMC2850155.
6. NHGRI. (2023, February 9). Antibody. Genome.Gov. https://www.genome.gov/genetics-glossary/Antibody
 By Rahul Mehta
By Rahul Mehta
In the next twenty-four hours, about seventeen individuals will die due to the lack of a satisfactory organ to replace a dysfunctional one. Behind these nearly two dozen people is a list of over 100,000 U.S. citizens whose lives hinge upon the chance that an essential organ replacement becomes available.¹ In a new era of medicine where vaccines for novel diseases can be crafted within a year and numerous options exist for cancer treatment, one may attempt to surmise what options exist for those awaiting transplants. In recent media, UMD cardiac surgeons attempted the modification of a pig’s heart using CRISPR gene editing techniques. By making edits to relevant DNA sequences, researchers made the heart more suitable to function within the human body. Although the result filled a hole in the patient’s chest for more than six months, this branch of CRISPR implementation is still in its rudimentary stages. The recent passing of this individual was met with deliberation about the future of such technology and its ethical implications; however, numerous relevant professionals have also acknowledged the immense potential of this multidisciplinary scientific feat.

Since the early 1990s, scientists have been attempting to exploit stem cells, a special set of cells with a unique ability to divide consistently due to their expression of the enzyme telomerase. The significance of telomerase lies in its ability to block natural signals for cellular self-destruction, known as apoptosis. During the simultaneous replication of two DNA strands, one
strand is not completely copied. DNA can only be duplicated in one direction and the ends of each parent strand are opposing. Consequently, one replicate is copied in short segments, called Okazaki fragments. For DNA replication machinery to fill in the gaps, RNA needs to attach to these Okazaki fragments, but at the end of one strand, there is nothing to attach to. The result is DNA shrinking in size following each replication. The premise of apoptosis lies in eliminating nonfunctional cells from the body after excessive DNA shortening. However, when telomerase is expressed, it extends the protective telomere caps of essential DNA, protecting cells from being eliminated due to functional DNA damage. Controlled telomerase expression allows for stem cells to divide continuously without infliction of damage upon DNA. Though present in small numbers within adults, stem cells’ most configurable forms are most prevalent within embryos (embryonic stem cells). In other words, these embryonic stem cells have more flexibility in which
cell type they become. The issue with studying embryonic stem cells lies in ethical considerations of their use given that their experimentation means that scientists would be experimenting with human cells that could have eventually differentiated into functional humans. Today, scientists are attempting to work around this ethical impediment, but the end goal remains clear—creating organs in vitro, or organogenesis.
The formal definition of organogenesis is the “formation of organs from stem cells involving multiple stages including proliferation, differentiation into specialized cell types, and the formation of organs.”² Stem cells need to multiply to express proteins specific to their organ, and ultimately come together to form a completed organ. Once the cells have differentiated, they begin morphogenesis, the process of organizing themselves into the proper structures that make up the organs. This process is essential for developing many organs, including the heart, lungs, and kidneys during embryonic development. Despite the
contemporary nature of creating organs in dishes separate from the human body, recent advancements have conveyed prospects of making significant contributions to medical science in the near future. By understanding organogenesis, scientists and doctors can open doors for patients in an industry saturated by demand. At the University of North Carolina at Chapel Hill, Dr. Ronit Freeman is attempting to merge the forefronts of engineering, computer science, and biology to craft a novel approach to this intricate process.
Dr. Freeman’s approach consists of combining proteins and nucleic acids at the molecular level; in this case, peptides and nucleotides represent the most fundamental units, or monomers, of the two aforementioned macromolecules, respectively. She attempts to determine whether the structural formation of “cellular skeleton” (spindle fiber) is
affected by complementary peptide DNA monomers, corresponding DNA sequences with different peptides attached with the ability to form complex junctions with various geometries and stabilities. The creation of “cellular skeleton” components in the form of peptide-DNA monomers could help provide a tool for creating complex and functional tissues that closely mimic natural biological structures. Tissues are complex structures that require specific environments at the microscopic level, such as the presence of specific cell types, proteins outside of the cell, and signaling molecules, such as ligands, to maintain their function. Crafting spindles using peptide-DNA nanotechnology offers a way to create these microscopic level environments with precision as they can be designed to mimic the structure and function of natural protein fibers.
One of the key advantages of the peptide-DNA spindles is that they can be functionalized with other molecules to control their assembly and properties. Dr. Freeman mentioned that she could “attach different types of ligands (small signaling molecules) to the peptides or DNA strands” to control the assembly of the spindles; these molecules can also be used to target the assembly specific cellular structures or molecules.³ This opens up a range of possibilities for the development of new technologies for

controlling cell division and other cellular processes.

In order to use peptideDNA spindles for studying the function of the mitotic spindle, Dr. Freeman also emphasized the need to be able to mimic the complex dynamics of the spindle. This is challenging because the mitotic spindle is a highly dynamic structure that undergoes constant reorganization during cell division. However, recent advances in live-cell imaging and highresolution microscopy have enabled researchers to track the movements of individual molecules and visualize the structure of the spindle at high resolution. Using these tools, Dr. Freeman has been able to show that the peptide-DNA spindles can form highly ordered arrays that resemble the microtubules in the mitotic spindle. The peptide-DNA spindles can also capture and align chromosomes in dividing cells, and undergo dynamic changes in response to different stimuli, such as changes in pH or temperature.
In the future, Dr. Freeman hopes to implement her advancements in hopes of revolutionizing the field of in vitro organogenesis. By using synthetic spindle to the degree of control achieved by Dr. Freeman, one may be able to guide the growth and development of cells; if done with precision, this process may pave the path to creating functional organs that can be used for organ transplantation as well as other life-saving medical purposes.
1. Organ donation statistics. https:// www.organdonor.gov/learn/ organ-donation-statistics (accessed February 27th, 2023).
2. Encyclopædia Britannica, inc. Organogenesis. https:// www.britannica.com/science/ organogenesis (accessed February 27th, 2023).
3. Interview with Dr. Ronit Freeman, Ph.D. 03/17/2023.


 By Isha Mistry
By Isha Mistry
Blood vessels are widely known for transporting blood throughout our body, so that the necessary nutrients get sent to our tissues and organs. But blood vessels also have another role that is not as obvious. Dr. Victoria Bautch1 studies the development of blood vessels to assess their role in certain cancerous diseases. Dr. Bautch and her team’s main goal is to understand the process of blood vessels developing normally to be able to grasp what occurs abnormally in the developmental process that leads to certain diseases.
Blood vessels can be involved in diseases in two ways: directly and indirectly. In the case of cancer, blood vessels are indirectly involved, and there are no specific mutations in the blood vessels themselves. Instead, the different environment associated with cancerous tumors changes the way blood vessels function. Research shows that “tumors induce vasculature through the unbalanced, local expression of a small number of growth factors [stimulate growth of blood vessels].”2 In other cases, blood vessels are affected directly via a specific mutation in the blood vessels’ cells themselves. Out of the many different types of blood vessel cells, Dr. Bautch and her lab studies the building block of blood vessels: the endothelial cell (Figure 1). Dr. Bautch’s research allows for the better understanding of how abnormalities in the development and functions of endothelial cells are
associated with certain diseases, which is a vital first step to understand cures and prevention.
One way to understand the role of blood vessels is to study the pathways associated with them. Specifically, studying the negative regulators involved in the pathways can lead to a better explanation of how disease-causing abnormalities may occur. Negative regulators suppress cellular responses to certain signals in a variety of ways.
The research done by Dr. Bautch along with her colleagues, in the paper published in 2021, “SMAD6 Transduces Endothelial Cell Flow Responses Required for Blood Vessel Homeostasis,”3 explores angiogenesis (Figure 2), which is the formation of new blood vessels dependent upon chemical signals.
The specific pathway Dr. Bautch and her team investigated was the Bone Morphogenetic Proteins, BMP, pathway. They wanted to comprehend how the endothelial cells themselves can regulate the BMP signals received by assessing the effect of these negative regulators in the BMP signaling pathway. She suspected that abnormalities occur and normal functioning ceases when the cells either get overwhelmed by a spike in signals, or the negative regulators become overactive. Dr. Bautch and her team were able to determine the important role of negative regulators by manipulating the expression of negative regulators and altering the signals received by the endothelial cells.
To investigate the BMP signaling pathway, Dr. Bautch and her team used

an in vitro system, which involves directly experimenting on cells. Dr. Bautch and her lab looked at endothelial cells lying flat on a dish and examined how the negative regulators affected the cellular responses.
The signaling in the BMP pathway occurs by the BMP protein, but since this protein is unavailable during in vitro experiments, Dr. Bautch and her team used mechanical signals on the cells. The signal was generated by the flow of fluid over the cells that made up the blood vessel in the petri dish, and the force of the fluid that mimicked blood led to a signal in those blood vessels that led to a cellular response. The machine they used pumped fluid through the cells for about three days, and they varied the level of force going through the cells. The variation of force allowed them to connect blood perfusion to the development of certain cancerous diseases. For example, “when that force isn’t uniform, but when it’s very oscillatory, that often happens in areas that are prone to developing atherosclerosis [the buildup of substances on the walls of blood vessels].”1 Dr. Bautch and her team also explored what happens when cells lack the presence of a negative regulator. They found that the normal response of the cells to fluid flow across them is altered. Normally, the endothelial cells align with the flow of blood, but “when we took away this negative regulator, the cells didn’t look like they were doing that at all.”1 She found that the cells were simply sitting flat, as if there was no flow going across them. Her publication outlines that “reduced Notch [a negative regulator] signaling…
prevented endothelial cell alignment under homeostatic laminar flow.”3 This helped to highlight the importance of negative regulators and their role in the response from cells. The unaligned cells affect the endothelial cells’ response to the signals around them, potentially leading to cancerous diseases.

set up a quantitative PCR amplification, in which the amount that was amplified reflected the amount of RNA for the negative regulator that was present.
Dr. Bautch and her team have gained insight about the BMP pathway which can help us better recognize why some diseases may occur when there is mutation in this pathway. From this research, they were able to grasp the true significance and importance of negative regulators on this pathway and their effect on cellular response in blood vessels. For example, patients with mutations often develop abnormalities in their blood vessels called arteriovenous malformations (AVMs) (Figure 3), or tangles in their blood vessels that disrupt normal blood flow. The better understanding of this BMP pathway will allow us to recognize possible ways to treat these AVMs.
Dr. Bautch and her team encountered a few hurdles while developing this research, the largest of which was how to monitor the expression of the negative regulator because it was hard to track since it was a protein. To ensure the presence of the negative regulator in some cells was lowered, the researchers turned to measure RNA through quantitative reverse transcriptase PCR (qRT-PCR). They isolated the RNA from endothelial cells to
“What I think characterizes our research the most is trying to use different ways to understand how blood vessels form and function,”1 said Dr. Bautch. She has many research projects in which she studies the development of blood vessels, both in vivo (in life) and in vitro, and all these projects combine to advance our understanding of ways in which abnormalities contribute to disease. She has collaborations within UNC with a lab in the Biomedical Engineering Department to extend her research done in this paper to cells that are in a tube, rather than flat in a dish as they are in this research project, to mimic the blood vessel of an organism. She hopes to continue learning about how this pathway works.
1. Interview with Victoria L. Bautch, Ph.D. 1/30/23.
2. Nagy J A; Chang S-H; Dvorak A M; Dvorak H F. Why are Tumor Blood Vessels Abnormal and Why is it Important To Know 2009, 1
3. Dana L. Rutler; Ziqing Liu; Kimlynn M. Ngo; Shaka X; Allison Marvin; Danielle B. Buglak; Elise J. Kidder; Victoria L. Bautch. SMAD6 transduces endothelial cell flow responses required for blood vessel homeostasis 2021, 1, 24:387-398
“From this research, they were able to grasp the true significance and importance of negative regulators on this pathway and their effect on cellular response in blood vessels.”
Remember the good ol’ days back when we were just one cell? Neither do I. We all were, at once, just single cells with a long road ahead of us. Thanks to the wonderful processes of the cell cycle and DNA replication (Figure 1), animals and organisms, such as ourselves, develop into fully-grown adults with cells that perform various functions. The cell cycle, the processes by which cells divide and reproduce, is critical in human development and general health. In fact, according to Dr. Bob Duronio, “regulating the progress of the cell cycle is one of the most fundamentally important things that cells do.”1 Despite its importance, research regarding the cell cycle among multicellular organisms is less expansive than its unicellular counterpart.
After completing his Ph.D., Dr. Duronio, now Chair of the Department of Biology, was struck with curiosity and interest in research regarding the cell cycle of Drosophila (fruit flies). Having always been interested in the fundamental mechanisms of biology, Dr. Duronio found and still does find the topic worth researching. “What if a deeper
understanding of the cell cycle and the mechanisms that regulate it is the key to our scientific understanding of cellular development and cancer?” Questions like these are the focus of interest and curiosity of the Duronio Lab.

Dr. Duronio has dedicated his career to researching the cell cycle and the potential applications of such research. When considering Duronio’s research, a reasonable question to ask is “why fruit flies”? Not only is the use of

fruit flies in lab settings well-established with over 100 years of use in labs, but the reproduction of these flies is also fast and cheap in comparison to other common organisms used in labs. Additionally, fruit flies offer specific advantages to Dr. Duronio’s research. The life span of a fruit fly is a developmental system, meaning it goes through stages including larvae and embryonic phases (Figure 2). This allows the lab team to research the cell cycle at different stages, finding possible differences between the stages. Lastly, and possibly most importantly, the machinery that controls the cell cycle is evolutionarily conserved in fruit flies.1 This means that many of the genes and DNA involved in the cell cycle in fruit flies are similar or maintained in humans. This connection makes fruit flies a popular target for studies regarding human development and disease like cancer.
The process the Duronio Lab uses to explore the cell cycle is largely on the basis of trial and error. Given the microscopically small nature of the subject of the research, you are safe to say that Dr. Duronio and his lab spend lots of time under the microscope. Using a technique known as CRISPR, the lab is able to pinpoint individual genes and make specific mutations to them. By doing so, Dr. Duronio is able to determine the function of the gene. It is through these sorts of experiments that he explores the impact that certain genes and pathways can have on cell development, a prominent topic in cancer research. In addition to aiding the understanding of cancer, the research done in the Duronio Lab also deals with topics of cell regeneration. Duronio’s research has aided in understanding the “varying degrees of regenerative potential after tissue damage”.2 This
research helps the scientific community understand the cell cycle mechanisms that allow some organisms to regenerate large amounts of tissue while others cannot. Salamanders, for example, are able to regrow whole limbs while other organisms lack the capacity for regrowth. Using CRISPR and other analysis tools to observe the impact that specific genes have on cell development and replication, the Duronio Lab is contributing to the scientific pool of knowledge (Figure 3).

Like all scientific labs, data must be collected. In this case, it can be both numerical and solely observational. The lab collects data by “making a mutation”1 with CRISPR and observing how the change in the gene affects the development of the cells and flies. Quantitative data can then be derived from the observations, including what percent of the flies or cells survived or to what point in development the organism reached. This data serves as scientific evidence that can be used to significantly report the findings of the lab.
While Dr. Duronio is dedicated to his research and unwavering in his academic contributions to the scientific community, his work extends beyond the number of publications he has achieved. Rather, being both a mentor and a mentee is very important to Dr. Duronio. Seeing that his interest in science largely stems from a high school biology teacher that sparked his curiosity, mentoring the students in his lab is of foremost importance to Duronio. After all, he claims that curiosity is “the most important feature of being a scientist.”1 Not only does Duronio mentor his students, but he also recognizes that being a scientist means being someone who is continuously growing. Through not only his research into the impacts of gene editing in Drosophilia, but also his dedication to science mentorship, the Chair of the Biology Department sets an example for the future of what it means to be a scientist.

References
1. Interview with Robert Duronio, Ph.D. 01/27/2023.
2. Meserve, Joy; Duronio, R.J; Developmental Biology 2018, 444, 43-49
“[Dr. Duronio] claims that curiosity is ‘the most important feature of being a scientist.’”
Scars tell a story of the past, but to someone with a scarred eye, it becomes a story shrouded by blindness. The anatomy of the eye is fascinating. From the colorful iris to the dark, mysterious pupil, it is no wonder why the human eye is so alluring. However, overlooked is the transparent part of the eye that acts as a line of defense and helps focus on what is needed – the cornea.
Throughout life, the cornea is under constant renewal supported by stem cells, which are sometimes called the “master cells of the body” because of their ability to develop into a variety of cells. These corneal-renewing stem cells—appropriately named limbal stem cells (LSCs)—are located in an area called the corneal limbus, between the cornea and the sclera, the white part of the eye.1 Deficiency of these essential stem cells is one of the leading causes of blindness, specifically blindness in the cornea, where the only current treatments are costly and often inaccessible transplantations.1,2
At the UNC-Chapel Hill School of Medicine, Dr. Hua Mei works in the Ophthalmology Department tackling corneal blindness. Working towards her Ph.D. from the University of Cambridge, Dr. Mei developed an interest in LSCs and grew a passion for translational eye research. She later joined Dr. Sophie Deng’s lab as a post-doctorate continuing her studies with LSCs at the University of California, Los Angeles (UCLA), where she also assisted in establishing a national limbal stem cell bank with the UCLA Broad Stem Cell Research Center. Now researching as a UNC faculty member, Dr. Mei has extended her interests into investigating corneal wounding and healing to understand the properties underlying corneal blindness.
Selecting the mice model, Dr. Mei investigated the trajectory of LSCs in a recent paper on distinguishing active and inactive LSC populations.2 However, an intriguing question arose. If the cornea was wounded, which highly expressed genes would be present as healing occurred? To answer this, her research team wounded the cornea, and as the eye healed and

was analyzed, a single gene stood out.
The results showed that the neuroblastoma suppressor of tumorigenicity I (NBL1) gene, a founding member of the neuroblastoma family, was most expressed as the body’s natural response to a nerve injury. Neuroblastoma is the uncontrollable growth of young nerve cells that ultimately become cancerous.3 Although NBL1 typically suppresses nerve cell growth, it was most expressed when cell growth was expected to replace the wounded nerve cells. To understand this phenomenon, Dr. Mei studied another leading cause of corneal blindness: excessive corneal scarring.1 Dr. Mei hypothesized that NBL1 was responsible for acting as the stop signal toward excessive scarring, or fibrosis, and that adding this gene to wounded corneas would prevent fibrosis development. This hypothesis presented an exciting and promising therapeutic to corneal blindness.
To determine the specific role NBL1 played, her research team set up experimental and control groups with the mice model. In the experimental group, her team created corneal

wounds in mice cornea and injected the gene in an anesthetized eye. When the lab received the gene, it was bonded with a biological tag called the Fc protein. Therefore, to reaffirm that the Fc protein played no function in corneal wound healing, a control group was created where the protein was solely injected. Another control group was then treated with chemically inactive substances. The control groups were to ensure that the treatment effects were from the NBL1 gene instead of the Fc protein or experimental procedures.
Fourteen days after treatment, her team analyzed the experiment with different methods. First, using optical coherence tomography (OCT), which is a popular non-invasive imaging test that creates pictures of the back of the eye, the lab found that the NBL1-treated group indicated smaller scar areas than the control groups (Figure 1). Second, her research team evaluated the treatment of NBL1 by protein expression. When the body heals wounds, cells like myofibroblasts promote wound closure. After wound closure completes, myofibroblasts will typically degrade. However, recent studies have found that many scarring conditions result from excessive myofibroblasts failing to undergo their programmed cell death.6 Results of the NBL1-treated group indicated reduced myofibroblast expression, showing that NBL1 gene treatment led to an antiscarring effect (Figure 1).
Dr. Mei’s team then applied their findings from the mice model to the human model to determine if the gene showed similar anti-scarring effects.5 Through a collaboration with the non-profit eye bank “Miracles in Sight,” her lab obtained donated human corneas, allowing research on eye tissue with more responsive cells compared to the animal model.5 Left and right human corneas from the same donor were treated with NBL1 and the Fc protein, respectively. The experiment determined that the control group with Fc protein showed normal myofibroblast formation, while the experimental NBL1treated cornea indicated reduced myofibroblast formation similar to the mice model data. This project confirmed that the NBL1 application presented sufficient anti-scarring effects in mouse and human models.
Discovering the presence and effects of the NBL1 gene is promising given its potential as an anti-scarring therapeutic. Although the body already produces this gene naturally, NBL1 is only produced in the later stages of the healing process, creating a time lag that leads to more scarring.5 With earlier application of the anti-scarring effects or with a wound-healing-inducing drug, NBL1 could effectively bypass this time difference and prevent unnecessary scarring.
Interestingly, the cornea’s anatomy and development are very similar to the skin, where many cuts and scars form. Anti-scarring drugs exist, but they pose long-term risk factors. Dr. Mei emphasizes that there are “no real [FDA approved] drugs to prevent scar formation on the cornea or skin.” With the anatomical similarity between the eye and skin, NBL1 could be applied to the
skin, increasing the possibility of an FDA-approved NBL1 drug. Currently, NBL1 is patented, and Dr. Mei’s group is collecting the necessary pre-clinical data required for clinical trials. Due to the easy enrollment of skin-scar patients, the team may focus on skin-scar trials first, allowing later corneal scar trials.
In summary, Dr. Hua Mei’s research has contributed significantly to the understanding of corneal wound healing and scarring. Her discovery of the NBL1 gene’s anti-scarring effects is a promising step towards developing more accessible and affordable treatments for corneal blindness. In the future, NBL1 may be seen not in the lab but as an ingredient in topical ointments or in an eyedrop after a minor scratch, preventing the lingering signs of past stories called scars.
1. Oliva, M. S.; Schottman, T.; Gulati, M. Turning the tide of corneal blindness. Indian J Ophthalmology, 2012, 60, https://doi. org/10.4103/0301-4738.100540
2. Song, Z.; Chen, B.; Tsai, C.-H.; Wu, D.; Liu, E.; Hawkins, I.S.; Phan, A.; Auman, J.T.; Tao, Y.; Mei, H. Differentiation Trajectory of Limbal Stem and Progenitor Cells under Normal Homeostasis and upon Corneal Wounding. Cells 2022, 11, https://doi.org/10.3390/ cells11131983
3. Nolan, K.; Kattamuri, C.; Luedeke, D. M.; Angerman, E. B.; Rankin, S. A.; Stevens, M. L.; Zorn, A. M.; Thompson, T. B. Structure of neuroblastoma suppressor of tumorigenicity 1 (NBL1): insights for the functional variability across bone morphogenetic protein (BMP) antagonists. The Journal of biological chemistry, 2015, 290, https:// doi.org/10.1074/jbc.M114.62841
4. Gupta, N.; Tandon, R.; Gupta, S. K.; Sreenivas, V.; Vashist, P; Burden of corneal blindness in India. Indian journal of community medicine : Official Publication of Indian Association of Preventive & Social Medicine, 2013, 38, https://doi.org/10.4103/0970-0218.120153
5. Interview with Hua Mei, Ph.D. 02/02/23.

6. Darby, I. A.; Laverdet, B.; Bonté, F.; Desmoulière, A. Fibroblasts and myofibroblasts in wound healing. Clinical, Cosmetic and Investigational Dermatology, 2014, 7, 301–311. https://doi. org/10.2147/CCID.S50046 Dr. Hua Mei


We’re taught from an early age that DNA is a static force in each of our cells. It controls everything about us, but it doesn’t change with time—and it certainly doesn’t have memory. Until recent history, this theory appeared to be right. But there’s an emerging field called epigenetics that has overturned everything we thought we knew about DNA, unlocking an entire new world of genetics for us to explore.
Relatively new to the world of science, epigenetics is focused on the components of genetics located just above or attached to the DNA. A genetic record of our environment and our choices, epigenetics explores not only the sequence of DNA, but how it’s expressed. The entirety of our DNA exists in each one of our cells, but the activation of different genes results in different expression, individualizing a kidney cell from a liver cell.1 Expression is the driving force behind cell differentiation—and it’s what makes each cell unique. But how does this expression work?
Each of us have enough DNA inside our body to reach from the sun and back three hundred times once stretched out.1
Not all of that can fit normally in each cell, so instead, the DNA is wrapped tightly around hockey-puck shaped histone complexes called nucleosomes, like beads
on a string.2 Once coiled, the DNA forms chromatin, which will later condense into chromosomes. Gene expression, then, is determined by how available the DNA

is for transcription, as seen in Figure 1.1 This accessibility depends largely on the unwinding and winding of DNA around the histones.3
Dr. Greg Matera is delving into the specific forces behind what causes changes in DNA coiling in the genomes of the Drosophila fruit fly, which was chosen as the lab’s model organism due to its unique genome.4 According to Dr. Matera, the fruit fly keeps its histone genes in the same proximity, making it possible to study the whole gene family simultaneously—a feat that is impossible in more scattered mouse or human genomes.3 The goal behind Dr. Matera’s research is to study these histone genes and the effects of mutations on them. His research aims to discover how epigenetics is regulated by the factors that influence histones.
These factors are less well known than histones, though they play just as important a role. They are called posttranslational modifications, or PTMs. These PTM’s come in many distinct types, such as methylation, acetylation, and phosphorylation. While the mechanics of each differ slightly, they all attach to tails on the histone proteins and add chemical groups. Consequently, the DNA reacts to the change, restricting or uncoiling enough to allow gene expression.3
In Drosophila, these PTMs are the focus when studying histone proteins. Dr. Matera’s research

aims to discover their effects on histone genes by creating mutations that alter the effect of PTMs. To accomplish this, in collaboration with the Duronio, McKay, Strahl, and Marzluff labs, the “UNC Histone Replacement Group” has developed a genetic platform that allows manipulation of the histone gene family, as seen in Figure 2.4 Genetically engineered gene clusters are isolated, manipulated, and then put back into the embryos of fruit flies. The physical expression of these genes is then studied to distinguish the effects of mutant genes from the wild type.3

The data gained from this research will expand upon current knowledge of epigenetics and the post-translational modifications that affect them. The results could show the extent that these PTMs affect gene expression and provide insight into the evolving world of epigenetics. Progress in this research promises many exciting opportunities, though it takes time and effort for each step of the process. Oftentimes, comparison of gene mutations to wild type genes yields only correlations and not causations, which require further examination. The microcosm of epigenetics only seems to grow larger with each discovery, and often, observed data opens more doors than it closes as we discover more information about genetics that are is yet understood. But as Dr. Matera states, this is not cause for discouragement. In his words, “Something was an unknown unknown and now we’ve turned it into a known unknown—and that’s how science gets done”. Through more research, other possibilities can steadily be chipped away until causation remains, expanding current knowledge of epigenetics and how it affects all of us.
Current research asserts that epigenetics is largely influenced by individual actions and environmental factors, like diet and sleep. Personal decisions hold more power over the DNA than previously thought, and through research like Dr. Matera’s, science will be one step closer to cracking the epigenetic code. In the
meantime, reconsider how much of your DNA you really believe is set in stone and remember that in the end, the expression of your DNA is changing as you change, too.
1. Anthony T. Annunziato, Ph.D. Nature.https://www.nature.com/ scitable/topicpage/dna-packagingnucleosomes-and-chromatin-310/ (Accessed 02/13/11)
2. BioInteractive.https://www. biointeractive.org/classroom-resources/ how-dna-packaged.(Accessed 03/13/23)
3. Interview with A. Gregory Matera, Ph.D. 2/02/23
4. Meers MP, Leatham-Jensen M, Penke TJR, McKay DJ, Duronio RJ, Matera AG. Methods Mol Biol. 2018;1832:309-325.
“Something was an unknown unknown and now we’ve turned it into a known unknown. And that’s how science gets done.”
- Dr. Matera
The human genome is composed of approximately 3 billion nucleotides. However, humans only have about 20,000 protein-coding genes–a mere 2% of their entire genome. Scientists refer to the remaining 98% of the human genome as “non-coding DNA.” Though non-coding DNA comprises most of the genome, scientists have yet to determine the various functions this genetic material may serve in the context of human development and disease. Dr. Douglas Phanstiel, an assistant professor of cell biology and physiology at UNC-Chapel Hill, and the researchers in his lab are among the scientists and researchers investigating the elusive non-coding DNA–specifically, how its role in the regulation of gene expression affects human development and disease.

Enhancers are regulatory elements of DNA that activate higher levels of DNA transcription. They can regulate the expression of genes, even those that are hundreds of thousands or even millions of base pairs away. This regulation can occur through DNA looping: when proteins and protein complexes simultaneously bind the DNA and cause the intervening DNA to form a loop structure. DNA loops bring enhancers in closer proximity with genes, therefore allowing enhancer-mediated gene regulation to occur (Figure 1). According to Dr. Phanstiel, alterations in the regions connected by DNA looping or in the loops themselves may have serious effects on human development and disease.1 The Phanstiel lab maps DNA loops and other genomic events across time courses of human macrophage and microglia activation, human megakaryocyte

development, and other processes. A macrophage is a white blood cell that kills microorganisms in the body. Megakaryocytes are a type of hematopoietic stem cell--a cell that gives rise to different types of blood cells--that convert to platelets. Through the analysis of DNA loops, the researchers can understand how the loops form, the impact they have on gene expression, cellular phenotype, and human disease.1
The Phanstiel lab is currently undertaking four different projects to understand chromatin looping and assess its role in human disease.1 The first project involves studying the mechanisms of chromatin looping from a basic biology standpoint, while the other three projects directly examine the relationship between chromatin looping and disease. One project investigates how a new type of chromatin loop can cause acute myeloid leukemia (AML), while another is evaluating chromatin loops and other epigenetic features as a

genetic basis of Alzheimer’s disease. Epigenetics is the study of how an individual’s behaviors and environment can alter their genes. In the fourth project, the lab maps chromatin loops in primary human chondrocytes to understand the genetic underpinnings of osteoarthritis since chondrocytes are the primary cells found in cartilage.
Figure 3. IDRs/chromatin-associating fusion proteins, such as NUP98, bind chromatin at enhancer sequences and gene promoters and bring loci together to form condensates dependent on phase separation. Such condensates activate oncogenic transcription. Figure courtesy of Quiroga et al.

Dr. Phanstiel developed an interest in 3D chromatin structure and loops while completing his postdoctoral research at Stanford University a decade prior.1 “I was excited by it, because so little was known about 3D chromatin structure at the time,” he said.1 Through organic interactions with the P.I.’s of other labs at UNC, Dr. Phanstiel realized different labs could collectively apply their respective areas of expertise to learn more about debilitating human diseases such as AML and Alzheimer’s disease.1 From there, his lab delved into studies of 3D chromatin structure through the lens of human disease.1
The Phanstiel lab has also taken an interdisciplinary approach to their research into 3D chromatin structure and disease, through the implementation of software. Most of the lab’s work relies on DNA sequencing technology, which has the capacity to produce billions of data points. The sheer amount of data necessitates the use of software to analyze, interpret, and communicate the data to others.1 The lab is working on software that can reduce the complexity of the data by converting the billions of data points into smaller, digestible quantities of information. To visualize and communicate the data to people with efficiency, the lab has developed softwares, including Plotgardener, a genomic data visualization package that can create precise figures in R, BedtoolsR, an R package that allows for genomic data analysis and manipulation, and CoralP, which can visualize human phosphatome data (Figure 2).3 “Science is only really valuable if you can communicate it to other scientists and people,” Dr. Phanstiel said.1
In investigating chromatin looping and the onset of AML, the Phanstiel lab, in collaboration with Dr. Gang (Greg) Wang, a professor in UNC’s Department of Biochemistry and Biophysics, has discovered a new type of chromatin looping that is driven by phase separation.1 Phase separation is the separation of a homogenous mixture into two distinct phases. In this context, phase separation refers to liquid-liquid phase separation (LPPS) occurring in a chimeric fusion protein called NUP98-HOXA9.5
Fusion proteins are encoded by genes that result from chromosomal translocations, rearrangements of chromosomes that result in chromosomes breaking and a fragment of the chromosomes affixing to different chromosomes.6 NUP98 is a
Dr. Douglas Phanstiel
protein that is constitutively expressed by cells, or is constantly being produced.6 HOXA9 is a transcription factor that enables cell proliferation and differentiation during embryogenesis, the process of embryo development from a zygote.6 Transcription factors are proteins that bind to genes, controlling the rate of transcription.
LPPS of NUP98-HOXA9 increases the transcription of leukemogenic (relating to leukemia) genes.5 NUP98-HOXA9 binds to various regions of DNA and pulls them into close physical proximity via LPPS, activating transcription of these genes (Figure 3).5 This process involves the formation of abnormal 3D chromatin structures—loops that “rewire connections between enhancers and target genes.”5 Further analysis revealed that these loops contained the proto-oncogenes, genes involved in normal cell growth, PBX3, and HOX that were being upregulated or expressed at increasing levels.6 When proto-oncogenes are upregulated, normal cell growth can become dangerously proliferative.
The Phanstiel lab is exploring strategies to disrupt these loops in order to slow or even reverse oncogenesis.1 To do this, they are taking an even deeper look into the exact mechanisms through which the loops form. However, Dr. Phanstiel says it’s difficult to know where to start to prevent the formation of the newly discovered loops.1 To tackle this hurdle, the lab is screening numerous small molecules that could potentially inhibit the loops.1 Through this translational research, Dr. Phanstiel and his team hope to identify small molecule drugs that could inhibit the loops, curbing the development of cancer.1
1. Interview with Douglas Phanstiel, Ph.D. 02/07/23

2. Gene Expression. https://www.nature.com/scitable/ topicpage/gene-expression-14121669/ (accessed March 14th, 2023)
3. Phanstiel Lab. http://phanstiel-lab.med.unc.edu/index. html (accessed March 4th, 2023)
4. Kramer, N.E.; Davis, E.S.; Wenger, C.D.; Deoudes, E.M.; Parker, S.M.; Love, M.I.; Phanstiel, D.H. Bioinformatics, Volume 38. 2022, 2042-2045. https://doi.org/10.1093/ bioinformatics/btac057
5. Ahn, J.H.; Davis, E.S.; Daugird, T.A.; Zhao, S.; Quiroga, I.Y.; Uryu, H., Li, J.; Storey, A.J.; Tsai, Y.; Keeley, D.P. et al. Nature 2021, 591-595. https://doi.org/10.1038/s41586-021-03662-5
6. Quiroga, I.Y.; Ahn, J.H.; Wang, G.; Phanstiel, D. Current Opinion in Genetics and Development. 2022, Volume 74. https://doi.org/10.1016/j.gde.2022.101901

The way each person’s life unfolds is unique. From a biological perspective, one way to think about everyone’s different life stories is to fit them within the context of genetic diversity. Genes are the raw material basis for how an individual becomes who they are in conjunction with social factors of upbringing, education, culture, and life experiences. Individuals are unique because a nucleotide (the building block of DNA) at a particular locus (a location along a sequence of DNA) of a gene varies from person to person. While genetic diversity gives rise to the multiplicity of life, it does the same to human diseases and disorders. For a given disorder, two people may exhibit different symptoms with varying degrees of severity due to genetic variance. Neuro-disorders are an example of such, often characterized as a spectrum because symptoms are vastly distinct and specific to each individual.
Dr. Hyejung Won is a professor in the Department of Genetics and Neuroscience Center at the UNC-Chapel Hill School of Medicine. She studies nucleotide variants from Genome-Wide Association Studies (GWAS) of neurological disorders. GWAS is a study approach that scans the DNA of individuals affected with a disorder and the DNA of unaffected individuals and then compares differences in nucleotide sequences between the two groups. GWAS

has uncovered thousands of possible disorder-associated loci that often contains dozens of genetic variants.1 However, not all the uncovered variants are causal for a disorder.2 The current challenge for scientists is to determine from that immense pool of possible variants which ones are causal. Confirming the causal variant is difficult because of linkage disequilibrium (LD), which is the inheritance of two or more nucleotide variants that are located near each other and therefore, inherited in sets.2 Thus, a causal variant might be in proximity to unrelated variants of the disorder. The difficulty is to separate the sets of variants identified by GWAS and to see which ones are truly causal. GWAS only tells which set of those alleles are commonly found in disorderaffected individuals, and thereby tentatively marks disorderassociated, but not which specific variant causes the disorder.⁵ As a Ph.D. student, Dr. Won characterized mouse models of autism spectrum disorder (ASD). In each mouse model, she mimicked a genetic mutation found in individuals with ASD.⁵ However, the limitation in doing so is that researchers were testing one variant at a time to see if the chosen variant caused ASD-like symptoms in mice.⁵ Mouse models are not high-throughput; they are costly, slow, and inefficient in simultaneously testing hundreds to thousands of possible variants. “So that made me think that

I have to do something, to find out what those thousands of variants are doing in a more systematic fashion, rather than just do it one by one, and wish for the best,” said Dr. Won.⁵ Hence, she adopted Massively Parallel Reporter Assays (MPRA) for simultaneously characterizing thousands of genetic variants associated with neurodisorders. By introducing genetic variants in neural cell types, MPRA can validate the gene regulatory impacts of thousands of variants in the neuronal context in a single experiment.1
Dr. Won first studied schizophrenia risk variants using MPRA. Using the variants identified by GWAS as a blueprint, Dr. Won’s lab inserted each of those variants separately into small circular rings of DNA derived from bacteria called plasmids that can be utilized as a vessel to transfer genes from one organism to another. The plasmid contains a promoter, which is a sequence of DNA serving as a marker telling the relevant molecules in a cell to come and read the nucleotide variants to transcribe RNA for making the proteins that initiate the processes for exhibiting a trait (in this case, the symptoms of schizophrenia). Another piece of DNA inserted is a Green Fluorescent Protein gene (GFP), used as a visual indicator for tracking where the RNA strands the entire plasmid construct codes for end up in a cell. Lastly, the lab inserted a unique barcode in the plasmid. This barcode is an artificially constructed DNA sequence like a tag assigned to each specific variant. The final completed plasmid is placed inside a benign virus used as a transporter for injecting the plasmid into human neural progenitors (HNPs) extracted from developing brains, which are neuron stem cells that can be specialized into a variety of working cells in the brain. The HNPs were cultured, allowing them to transcribe the plasmid into RNA. Since HNPs are not fully specialized, culturing them mimics neural development in the brain given a known injected nucleotide variant of interest. Two weeks later, the lab team extracted RNA from the HNPs and then sequenced them to backtrack to the DNA barcode sequence that coded for it. The number of barcode sequences backtracked from RNA is quantified using polymerase chain reaction (PCR), a method that makes copies of the DNA of interest from the original sample size.1
variants GWAS identified.1

However, there is still more pruning left to do. Further experimental testing is required to verify the significance of those 439 variants contributing to schizophrenia symptoms. What follows is the challenge of figuring out the respective functions and impacts each confirmed variant wields on a disorder’s molecular pathway. The molecular pathway is the process by which a trait develops, beginning from the genes to the networks of proteins that manifests a trait. “I think the ultimate goal is can we identify the molecular pathway underlying a disorder and can we reverse that,” says Dr. Won.⁶ The reversal of the molecular pathway is a possible research direction in the future toward possible treatments addressing the genetic basis of disease. There is still a long way to go before the day treatments come about since a marathon of pruning remains. Dr. Won thinks that her lab “is the first 1% of that journey,”.⁶ Genetics is very complex and there is so much yet unexplored. Nevertheless, 1% is a start. The eagerness to discover and undaunted spirit of scientists like Dr. Won, is a perpetual, dynamic force expanding the knowledge of the human blueprint.
A typical variant contains two groups of alleles that are tested using MPRA in the experiment: risk alleles that are more frequently observed in schizophrenic individuals and protective alleles that are less frequently observed in schizophrenic individuals. If the quantity of barcode expression from HNPs injected with a risk allele differs from the quantity of barcode expression from HNPs injected with the corresponding protective allele, this indicates that a particular variant may contribute to schizophrenia risk by altering gene expression. In using MPRA, Dr. Won was able to narrow down to 439 schizophrenia risk variants from the original 5,173 schizophrenia-associated
1. McAfee JC, Lee S, Lee J, Bell JL, Krupa O, Davis J, Insigne K, Bond ML, Phanstiel DH, Love MI, et al. 2022 Sep 17. Systematic investigation of allelic regulatory activity of schizophrenia-associated common variants. doi:https:// doi.org/10.1101/2022.09.15.22279954.
2. McAfee, J. C.; Bell, J. L.; Krupa, O.; Matoba, N.; Stein, J. L.; Won, H. Focus on Your Locus with a Massively Parallel Reporter Assay. Journal of Neurodevelopmental Disorders 2022, 14 (1). https://doi.org/10.1186/s11689-022-09461-x.
3. Won, H. MPRA and GWAS Used for SNPs Identification; 2022.
4. Won H. 2022. MPRA Construct.
5. Won H. 2023a. MPRA 1st Interview.
6. Won H. 2023b. MPRA 2nd Interview.
“Genetics is very complex and there is so much yet unexplored. Nevertheless, 1% is a start.”
Have a headache? Take some Advil or Tylenol. Allergies? Benadryl or Zyrtec. Bacterial infection? Antibiotics. To the typical patient, pharmaceuticals appear a deceptively simple solution: swallow the pill and feel better. For Professor Jeff Aubé at the Eshelman School of Pharmacy at UNC Chapel Hill, one of the scientists designing the drugs we take for granted, pharmaceuticals are a much more complex issue. Dr. Aubé has dedicated his career to developing small molecule drugs tackling a vast portfolio of diseases including prostate cancer, tuberculosis, and opioid addiction.
Developing a small molecule drug, a compound with a small molecular size that is designed to inhibit a certain protein or disease-causing agent, is an overwhelming task (National Cancer Institute).1 From the moment a drug is ingested, it has countless biochemical hurdles to jump over before taking effect, with the human circulatory, digestive, and immune systems actively working against it.
When a consumer ingests a pill and swallows it, it first reaches the stomach, and then the liver after being absorbed into the gut; the first hurdle is posed - the compound must survive this initial pass of metabolization without decomposing but remain soluble enough to be absorbed into the bloodstream. When the drug finally reaches each cell in the body, which occurs a mere two minutes after it hits the bloodstream, it needs to be able to enter through the cell membrane and navigate the cytoplasm to find the steroid receptors in cells and bind to them. The
drug needs to be selective - if it also binds to other receptors that are not being targeted, it will have unintended effects that could range from neutral to harmful. Finally, after it has achieved its job and mitigated the symptoms of disease, the body must be able to excrete the drug without any side effects of toxicity.

For a professional moleculebuilder like Professor Aubé, the difference between success and failure can be a careful balancing act between how hydrophilic (attracted to water) and how hydrophobic (repelled by water), the drug is. If a drug is too hydrophilic, it will dissolve completely and then dissociate in the many solutions it encounters in the human body, rendering it useless. If it is too hydrophobic, it will instead be attracted to nonpolar fats and sit immobile in the body’s fat cells, never reaching its intended targets. However, the molecule’s composition is not the only necessary consideration; before the drug ever reaches the patient, it must be capable of being packaged into pill form and sitting stably on the shelf without

decomposing from UV radiation, heat, or other factors that may compromise the efficacy of a drug. Due to the complexity of these criteria, very few successful drugs are void of side effects; it is an inherent tradeoff between the severity of disease and magnitude of the side effects of the treatment, where chemists aim to maximize efficacy and minimize complications.

To determine which compounds may be up to the task, Aubé and his lab members begin by running an assay, a term for sets of chemical tests to determine molecular properties, with a wide variety of chemicals to determine which options may be effective. Hydroponic screening is a process during which a chemist runs a bioassay on a target and a collection of molecules with diverse chemical structures to find a starting point for medicinal chemistry. The target of these assays can range from enzymes, pathogens, mutated gene products, or receptors. Targets are typically identified by biological scientists, experts on the respective disease for which treatment is being explored, collaborating with the medicinal chemistry team. Another method to begin the process is a phenotypic screen, which screens not against a target, but instead for a compound that can perform a particular molecular task which is predicted to inhibit the pathogen or disease.
The collaborative relationships and drug design journeys that follow these initial investigations vary. Professor Aubé became involved in researching pharmaceuticals to treat prostate cancer when a colleague, structural biologist Dr. Emily Scott, began using X-ray crystallography to determine how a
metabolizing enzyme in prostate cancer cells was inhibited by a commonly used small molecule cancer drug. Dr. Scott sought Aubé’s chemical support to help assemble the enzyme binding site. Their work uncovered that the binding model previously assumed between the enzyme, CYP-17-CapA1, and the drug, Abiraterone, was incorrect.2 Their discovery sparked an effort between Aubé and Scott that has led to the improvement of Abiraterone as an inhibitor of CYP-17-CapA1; an updated and more precise version of the drug is currently in a clinical trial.
Another exciting project began when a coworker connected with a Tuberculosis researcher on a tour of
research forward. Aubé credits these successful relationships and projects to scientists who are willing to “check their ego at the door” and show genuine interest in the work of their collaborators. To build these crucial relationships, Aubé advises, “even as a young scientist early in your education, appreciate the importance of collaborating and team science and developing the curiosity of what people do to make it happen.”4
the Great Wall of China at a research conference. The two began discussing the possibility of Aubé’s lab developing a library of potential compounds to inhibit the beta lactams pathway, which is toxic to tuberculosis bacteria.
The chemical support Aubé ’s lab provides does not stop once a potential small molecule drug is identified. In the Aubé lab, “hand-off” is a forbidden term when discussing projects; medicinal chemistry can be thought of as a constant feedback loop between the chemistry and biology team leading to additional testing, improvement, and collaboration. 3
Dealing with such a wide range of diseases and chemicals can seem overwhelming, but for Professor Aubé and his team, the relationships they have built with collaborators tie everything together and propel the
1. National Cancer Institute, https:// www.cancer.gov/publications/ dictionaries/cancer-terms/def/smallmolecule-drug
2. Fehl, Charlie, et al. https://doi. org/10.1021/acs.jmedchem.8b00419
3. Interview with Jeffery Aube, Ph.D. 2/7/2023
4. Interview with Jeffery Aube, Ph.D. 2/7/2023
“All these required properties for a successful small molecule drug ultimately depend on chemical structure”
What do Cerro Tololo (Chile), Perth (Australia), and Chapel Hill (USA) have in common? They are all home to at least one special telescope less than 40 inches in diameter. The Skynet Robotic Telescope Network is a widespread group of small-scale, ground-based telescopes used primarily for visual astronomy. Directed by Dr. Dan Reichart at the University of North Carolina at Chapel Hill, Skynet observations have contributed to countless discoveries, with a paper using Skynet data being published about every 20 days. Initially, the Skynet telescope network was meant to observe gamma ray bursts. These are bright, energetic explosions occurring in distant galaxies that release gamma rays, the highest energy light in the electromagnetic spectrum. In 2005, Dr. Reichart and his team set a world record by observing a gamma ray burst from 12.9 billion years ago, which was the oldest gamma ray burst observed at that time. This was incredible since the universe itself is only 13.7 billion years old. As astronomers around the world saw Skynet’s potential, more telescopes were integrated into the system. Now, Dr. Reichart is using this network to search for visual counterparts to gravitational waves.

When massive explosions occur in the universe, such as the collision of black holes or the death of a giant star, the fabric of spacetime itself is affected. In 1916, as a part of his theory of relativity, Albert Einstein theorized that a ripple in spacetime created by such events would travel as a gravitational wave. Gravitational waves are invisible to us - they travel at the speed of light by stretching and compressing space. The first
 By Rujula Yete
By Rujula Yete
time a gravitational wave was detected was in 2015, when the Laser Interferometer Gravitational-Wave Observatory (LIGO) in California detected a ripple in spacetime 1000 times smaller than the nucleus of an atom.1 As LIGO continues to detect gravitational waves from cosmic collisions, Dr. Reichart and his team search for the visual counterparts to such events. Specifically, they are analyzing the emissions in the electromagnetic spectrum that come from events that produce gravitational waves. Once the location of the source of a gravitational wave is found, Skynet telescopes can be pointed towards that area, allowing them to capture any optical changes that occur. In 2017, Skynet telescopes contributed to the detection of the first visual counterpart to gravitational waves. A large international team of scientists collaborated to analyze the electromagnetic data that resulted from the collision of two binary neutron stars. Neutron stars are formerly massive stars that run out of fuel and collapse, leaving behind a very dense core of neutrons, a particle found in the nucleus of an atom. The data from the study showed
various changes in visible, ultraviolet, infrared, x-ray, and radio emissions throughout the collision process.2 Using this electromagnetic data, the scientists were able to calculate the Hubble Constant, which indicates how fast the universe is expanding.3 This value provides insight into the age of the universe and how things might change in the future. This research will not only help us better understand how gravitational waves work, but also how cosmic collisions impact our universe.
As Skynet continues to pioneer discoveries in small-telescope astronomy, Dr. Reichart has another key goal with the system: make astronomy accessible to students. Much of his time is dedicated to developing the Astrophotography (ASTR 110) course at UNC, where students use Skynet telescopes to capture and process their own images of space. Dr. Reichart and his team of astrophysicists, programmers, and educators received a $3 million grant from the US Department of Defense to create this curriculum and give students direct experience with the same astronomy tools used by researchers to make discoveries about our universe.4 Dr. Reichart emphasizes that teaching astronomy has far greater implications than just increasing the number of astrophysicists in the world. “Astronomy can be a gateway drug to get people hooked on science,” he says. By exposing students to real-world tools that help them explore the universe, he not only hopes to make students excited about science, but also to help them believe they all have a place in the world of scientific discovery.
Science helps us navigate our world and continues to drive forward our future. By investing in science education
research, we can ensure that science is accessible, interesting, and fun for future generations. “Of all the sciences, [astronomy] is the one that’s easiest to get excited about,” Dr. Reichart says. Astronomy helps spread appreciation and understanding for science, something Dr. Reichart believes is much needed in society. “People might not go into science, but you want them to be pro-science,” he says. He and his team continue to collaborate across the world to observe the deepest ends of the universe, all while leaving a positive impact for students here at home.
1. Interview with Daniel E. Reichart, Ph.D. 1/31/23.
2. Reichart, D. Gravitational Wave Counterparts https:// www.danreichart.com/gwcounterparts (accessed February 24th, 2023).
3. Reichart, D. Skynet and PROMPT https://www. danreichart.com/skynet (accessed February 24th, 2023).
4. NASA. “Neutron Stars, Pulsars, and MagnetarsIntroduction.” Nasa.gov, Mar. 2017, imagine.gsfc.nasa.gov/ science/objects/neutron_stars1.html.
5. LIGO Caltech. “What Are Gravitational Waves?” LIGO Lab | Caltech, 2019, www.ligo.caltech.edu/page/what-aregw.


6. Abbott, B. P., et al. “Multi-Messenger Observations of a Binary Neutron Star Merger.” The Astrophysical Journal, vol. 848, no. 2, 16 Oct. 2017, p. L12, https://doi. org/10.3847/2041-8213/aa91c9.
7. “A Gravitational-Wave Standard Siren Measurement of the Hubble Constant.” Nature, vol. 551, no. 7678, 16 Oct. 2017, pp. 85–88, https://doi.org/10.1038/nature24471.

 By Esha Agarwal
By Esha Agarwal
Despite what the name may suggest, the HHIVE lab is not a place invested specifically in plant science and biology. Rather, this UNC-Chapel Hill research facility, led by Drs. Jordynn Jack, Jane Thrailkill, and Kym Weed, is a powerhouse dedicated to research in the health humanities, a field that is surprisingly understudied among aspiring healthcare providers and others along STEM career paths. Let’s delve deeper to understand what the health humanities truly is. Health humanities is a discipline that uses research methods, tools, and insights from the humanities in order to study and improve human health and wellbeing practices. Health humanities focuses on creating new language and perspectives in STEM fields to limit miscommunication between patients and healthcare providers.1,2 The health humanities utilizes both epistemology—theories of knowledge and ways of knowing— and empirics—verifiable and data-driven observations—and therefore uses research methods that meld together bioethics, anthropology, rhetoric, and wellness.1,2 In a changing society pushing through pandemics, political and racial reckonings, climate change, and more, health humanities has the capability to provide vital new perspectives to these multifaceted issues. Yet, this discipline is relatively unheard of, and all three professors followed unconventional paths on their way to this field.
Interested in the rhetoric of science, Dr. Jack had long focused on the language of science before shifting to health and medicine, investigating how not only science communication but scientific enterprise is rooted in persuasion. She is leading a collaboration between HHIVE and the Digital Literacy and Communications (DLC) department on a pilot study on perand polyfluoroalkyl substances or PFAS chemicals in NC and their dumping in the Cape Fear River in NC. PFAS chemicals, are those that are resistant to heat, oil, grease, and water and are used to coat a variety of commodities.3 PFAS chemicals are

concerning because they do not break down naturally over time, can contaminate drinking water sources, and bioaccumulate in wildlife.3 While many have investigated the greater impact and spread of PFAS, less research has been done on how the public understands the problem, how it is communicated, and the public’s experiences with it.2 The pilot study aims to conduct oral history interviews to provide new perspectives on public opinion on PFAS and, using the tools of the DLC, will create a public-facing archive to allow general access to gathered stories.
Like so many UNC-Chapel Hill students, Dr. Thrailkill started out as a pre-med major, but had always known and acknowledged her interest in English and Philosophy as well. She eventually switched her major to English but focused on medical issues and medical journalism.1 After earning her PhD in English, she continued building on the works of late UNC-Chapel Hill professor Lilian Furst, in medicine, literature, and their intersections. Dr. Thrailkill is now working on two separate projects: first, writing an article on William James, one of the fathers of American psychology, and how reading a psychology textbook for its literary and persuasive style can provide a miniature experiential psychology immersion.1 Her second project is an essay about the agony of empathy, in which she expands on both the importance of and difficulties with teaching empathy in medical schools.
Similarly, Dr. Weed began her undergraduate career on the pre-med track; her family had long been in the medical field, so it “was the obvious choice” and her “legacy.”1 However, she soon realized that it was in her literary classes that she was able to ask the most meaningful questions. Still, she pursued a biochemistry and molecular biology degree and worked in pharmacology before earning a master’s degree and, later, a PhD in English. Dr. Weed is now continuing to utilize all facets of her skill set as part of the Global Convening Program. Through this program, she collaborates with interdisciplinary teams around the world using stories, language patterns, and narratives to increase conversations and awareness about antimicrobial resistance in order to find solutions to this issue.1 Another project Dr. Weed is contributing to is focused on breast cancer research. In a recent study, Drs. Ebonee Butler and Melissa Troester, principal investigators in the Carolina Breast Cancer Study, determined they had the correct instruments and tools to measure barriers and racial bias in healthcare but found that participants were not able to provide the most accurate and precise answers with these scientific scales.1 They approached the HHIVE Lab to help them in developing and utilizing health humanities and
oral history research methods to aid in asking better questions to create new vocabulary and new instruments in the area of breast cancer research.
“Be open to the meandering path.”1 – Dr. Weed Throughout the conversation, each of the professors emphasized the importance of open-minded thinking, listening, and curiosity. While many students enter college with the fixed idea of pursuing medicine, Drs. Thrailkill and Weed highlight the significance of taking one’s time, enjoying the circuitous route when possible, and learning in-depth about the career path one intends to pursue to truly understand both its advantages and disadvantages. Dr. Jack notes that having the ability to listen and genuinely take in what one is saying, often gained from humanities courses, is a necessary life skill.2 Focused on rhetoric, narrative, and disciplines interwoven in science, the Health Humanities finds that “if you can change the narrative, you can change the world.”1
The HHIVE Lab and its endeavors embody the interdisciplinarity promoted by UNC,-Chapel Hill, taking daring next steps and delving into areas of science and humanities that no other institution on campus has been able to tackle. They found that many researchers and students in their lab did not know of the lab previously and had joined through cold emails – they had been working on research projects and discovered obstacles that could not be resolved when confining themselves to a singular field.1
“I almost want fewer people to know about this field, because once people know about it, they want it!,” Dr. Thrailkill joked.1
“[The lab] is an incredible powerhouse because it runs on no resources!” she explained.1 She and Dr. Weed continued, stating that while their department has supported them “beautifully,” the health humanities is a rapidly-growing field and a lack of resources has prevented them from reaching their full potential of research projects, and that it is the enthusiasm of students and faculty in the lab that really fuels their work as of now.1 They also mention that the addition of 2-3 more faculty and a greater show of university funding could help them take their research and findings to the next level, as well as help expand their lab and influences to students across campus— many of whom are unaware of the lab’s existence—so more can enjoy the “magic of the HHIVE.”1,2
1. Interview with Jane Thrailkill, PhD. and Kym Weed, PhD. 02/03/23


2. Interview with Jordynn Jack, PhD. 02/08/23
3. US Department of Health and Human Services. Per- and Polyfluorinated Substances (PFAS) Factsheet. https://www. cdc.gov/biomonitoring/PFAS_FactSheet.html (accessed February 2023).
Many undergraduate students walk into an introductory STEM class, like Biology 101 or Calculus 1, and feel overwhelmed. For them, these classes will be the first time they experience large lecture settings and such fast-paced content. The courses are difficult, especially when students have not yet had the chance to form review groups or develop their own study methods. This can lead many students to ask themselves how they could succeed in such a difficult environment. In the School of Education at the University of North Carolina at Chapel Hill, Dr. Jeffrey Greene searches for the answer.
From a young age, Dr. Greene has been interested in how people think. As an undergraduate at Carleton College in Minnesota, he studied psychology. He took an interest in the field of educational psychology, as he found it rewarding due to the field’s optimism. As he put it, the field tries “to find ways for everyone to learn more effectively and achieve what they want to achieve.”2 Now, at UNC, he continues to research critical
thinking, self-regulated learning, and digital literacy.

One of the psychological phenomena Dr. Greene studies is self-regulation. This term describes how one controls shortterm behaviors to stay on track for long-term superordinate goals.2 Dr. Greene mentions that it has been shown that a student’s application of self-regulation to their learning can help them succeed in daunting introductory STEM courses.2 As he puts it, studying self-regulated learning “involves investigating how people pursue goals, often in the face of adversity.”2 While clear-cut definitions of self-regulated learning are often clunky, Dr. Greene further explains that people who exhibit self-regulation often have several strategies for motivation and information retention, and they actively employ these strategies when challenged.2 Tying back to why he was first interested in educational psychology, Dr. Greene remains optimistic about self-regulated learning since “the capacity to self-regulate is something that basically everyone can acquire,” which allows more students to become effective learners.2

A study published last January by Dr. Mladen Raković, one of Dr. Greene’s former post-doctoral fellows, sought to investigate the acquisition of self-regulated learning skills.3 The research focused on how employing self-regulated learning strategies early on could boost performance and make large STEM classes more effective for learning (Figure 1). Dr. Raković selected a class of this type at a large southeastern university. After the first exam, two key events took place. First, all students were asked to reflect on their preparation for and execution of the test. Second, students who scored below the C+ range were provided with resources to implement self-regulated learning skills to improve their trajectory in the class.3 The results showed two notable trends. The students who mentioned adapting their behavior in their reflection were more likely to improve, without explicitly committing to change.3 Additionally, those who actually did change their behavior almost always improved.3
The students who experienced these academic improvements are textbook examples of self-regulated learning. They reflected on an outcome, asked questions about why the outcome happened, and planned for the future accordingly. This scenario embodies students owning their learning: a key facet of self-regulated learning. However, the other side of this coin is that instructors ought to own their teaching, too. Dr. Greene mentions the “unspoken agreement” between students and professors, wherein professors lecture and students absorb information, most of the time passively.2 Researchers at Carnegie Mellon University’s Human-Computer Interaction Institute, among others, have shown that this setup may not be optimal.4 Instead, they find that active learning often engages students more.4
Active learning is a set of principles “used by instructors as pedagogies,” as Dr. Greene puts it.2 Though formally defining it is elusive, it often entails pushing students to analyze, interpret, and apply their learning, essentially moving to higher levels of understanding than just absorption of information (Figure 2).1 Bloom’s Taxonomy, named for its
creator, late psychologist Benjamin Bloom, organizes these levels of understanding into a hierarchy.1 Employing active learning techniques can push students higher up the taxonomy, implying higher levels of learning.1
A combination of self-regulated learning and active learning pedagogies could redefine how students learn.3,4 However, to be effective, it will require effort on both sides of the lectern. Dr. Greene notes that students are often reluctant to engage in active learning since they wonder why changes to the aforementioned “unspoken agreement” are necessary.2 The of effort will be made by professors proactively informing their students about the structure and benefits of active learning. Then, students would have to accept such information and put effort into that new active learning setup. Dr. Greene emphasizes that self-regulated learning is a choice: students must decide to engage meaningfully with their classes in order for them to reap the benefits.2 If both sides of a class can uphold their commitments to these new learning principles, then students may gain the deeper understanding seen in the higher levels of Bloom’s Taxonomy.1,4
It is worth noting, though, that researchers are being “appropriately humble” about their findings.2 There is a growing body of evidence that highly structured STEM courses with active learning styles help students succeed, but Dr. Greene adds that more research is still needed on how effective this type of class is in the humanities.2 Not everyone must employ these techniques, and it’s important not to generalize. In fact, Dr. Greene notes that traditional lecture-style classes, where students must simply “absorb” information, are still important.1,2 Students must know what they’re doing before they can travel up Bloom’s Taxonomy, meaning that memorization and understanding will remain crucial.2 Providing structures to best to support students in early STEM classes may prove just as complex as the courses themselves, but Dr. Greene’s research into self-regulated learning could pave an important step along the way.
1. Armstrong, P. (2010). Bloom’s Taxonomy. Vanderbilt University Center for Teaching. Retrieved 13 February 2023 from https://cft.vanderbilt.edu/guides-sub-pages/bloomstaxonomy/.

2. Interview with Jeffrey Greene, Ph.D. 01 February 2023.

3. Raković, M; Bernacki, M.L.; Greene, J.A.; Plumley, R.D.; Hogan, K.A.; Gates, K.M.; Panter, A.T. Contemporary Educational Psychology. 2022 68.
4. Yannier, N.; Hudson, S.E.; Koedinger, K.R.; Hirsh-Pasek, K.; Golinkoff, R.M.; Munakata, Y.; Doebel, S.; Schwartz, D.L.; Deslauriers, L.; McCarty, L.; Callaghan, K.; Theobald, E.J.; Freeman, S.; Cooper, K.M.; Brownell, S.E. Science 2021 374 (6563), 26-30.
 By Charisma Daniel
By Charisma Daniel
While every generation has its public health challenges, the world’s newest generation faces another rendition of a much older threat: cigarettes.
Vape products and e-cigarettes have become popular amongst teens and adolescents. While many members of the public perceive vaping as a safer alternative to smoking traditional cigarettes, recent research shows that the latent threats of e-cigarettes and vaping can be just as dangerous. There has been a rise in cases of EVALI (E-cigarette Vaping Associated Lung Injury) amongst adolescents.2 Correcting misperceptions and increasing messaging on the dangers of vaping is imperative to help youth make healthy decisions.
Dr. Seth Noar has recently launched the Communicating for Health Impact Lab (CHI) at the University of North Carolina at Chapel Hill. Dr. Noar’s driving question was, “How can we reach individuals and populations with messages to encourage people to adopt healthy behaviors?” While his newly launched research lab is growing, he has a long road to reach this point.
After receiving his Ph.D. from the University of Rhode Island, Dr. Noar began a postdoctoral position at the University of Kentucky. Dr. Noar liked the applications of his education at the University of Kentucky. He stated, “It wasn’t just theoretical. It wasn’t just articles that I would write or something I was interested in. [Health communications] had a practical application to have some positive impact out in the world, and I got really interested.”
In 2011 Dr. Seth Noar joined the UNC Hussman School of Journalism and Media as a professor. During his time at UNC, Dr. Noar worked with student groups and

conducted research. In 2020, the CHI Lab received funding from the National Institute of Health (NIH) in partnership with the Food and Drug Administration. He stated that the grant helped him hire a project manager and give structure to his research projects.
The FDA uses a Perceived Messages Effectiveness scale (PME) to help gauge how well audiences are receiving their Real Cost Campaigns. However, the PME scale was created to measure effectiveness on adults rather than youth ages 13 -17. It was also developed before the advent of vaping and e-cigarette usage, which has become prevalent in today’s generation. Due to these limitations, the current PME scale is not sufficient today to measure youth and vaping. The CHI Lab endeavors to fill these knowledge gaps with their research specifically involving youth, vaping, and e-cigarette products.
Dr. Noar’s research campaign Advancing Perceived Message Effectiveness: A New Measure for Youth Prevention Media Campaigns, aimed to create a new PME for youth vaping. Dr. Noar’s team created an item pool using 48 youth subjects. An

item pool is a collection of test items used to make individual tests to distribute to a larger population. His team conducted a scale development study on 800 youths across the nation. The scale development study will be used to further refine the questions used for the PME scale.
In the next stage of research, Dr. Noar’s team will determine whether their designed PME scale actively predicts the impacts of the selected advertisements on youths’ intentions to smoke e-cigarettes. The CHI Lab will sample 1,028 adolescents who will be exposed to either The Real Cost e-cigarette ad conditions or control ad conditions. Participants will view 1 to 3 ads weekly and will then be evaluated on the third week for two factors. The first factor is their intention to smoke, and the second factor is whether they believe in the dangers of smoking.
In addition to their endeavors to refine the PME scale for youth and vaping, Dr. Noar’s team is investigating new ways to deliver their messages to youth. Dr. Noar stated, “One tool, that, to my knowledge, the FDA is not using is text messaging.”
receive daily text messages for 20 days, and three months after the study, there will be a follow-up evaluation. A study will be conducted prior to the text messaging campaign to identify the themes for the text messages and to trial them with adolescent focus groups.
The insights gained from the CHI Lab will help adjust the knowledge gap needed to communicate with youths and regulate e-cigarette and vape products effectively.
Lastly, Dr. Noar states, “... increasingly, the diseases we see in our country, and even the world, are caused by modifiable, changeable lifestyle factors.” Strategically crafting messages is a lot more elaborate than people would initially believe. This is why health communication is developing into such a large scientific field with an increased need for research.
Dr. Noar recognizes that while communication techniques are not the only way to influence public health, it is an essential tool to modify behaviors at the population level and to prevent diseases, helping people lead happier and healthier lives.
During the early stages of the CHI Labs research, they hypothesized that youth would favor flashy modern forms of communication such as emojis or gifs. Surprisingly, Dr. Noar’s team discovered that the youth preferred a much more down-to-earth approach with simple text statements. This ironic discovery further highlights the importance of health communications research.
This fall, the CHI Lab will implement its research campaign, Impact of e-cigarette prevention messages on adolescents, which investigates how text messaging can affect vaping dissuasion among youth. In this study, 506 youths will

1. Interview with Seth Noar, Ph.D. 02/07/23
2. UNC Health and UNC School of Medicine Newsroom. “UNC Researchers Tackle the E-Cigarette or Vaping Product Use–Associated Lung Injury (EVALI) Epidemic.” 3 Jan. 2023
3. Advancing Perceived Message Effectiveness: A New Measure for Youth Prevention Media Campaigns (2019-2023)
4. Impact of E-Cigarette Prevention Messages on Adolescents (2020-2025)

“...while communication techniques are not the only way to influence public health, it is an essential tool to modify behaviors at the population level and prevent diseases, helping people lead happier and healthier lives”Figure 2. Impact of vaping prevention advertisements on US adolescents. Image courtesy of Dr. Noar.
Pulmonary fibrosis, or the accumulation of scar tissue in the lungs, is one of the most malignant and challenging diseases to treat in the medical world (Figure 1). It can arise due to several factors ranging from genetics to environmental exposures such as radiation or infection. Scarring can also occur in patients who have rheumatology conditions such as scleroderma or lupus, rheumatoid arthritis, those who receive cancer treatments, or even severe COVID-19. However, no matter the cause, fibrosis is onset by activating the fibroblasts—cells that make scar tissue. While in healthy people these fibroblasts can successfully heal scarring and remain silent, their activation into myofibroblasts is what leads to fibrosis, which has proven to be fatal. For instance, idiopathic pulmonary fibrosis, which is potentially one of the most lethal and difficult-to-treat forms of lung fibrosis, affects about 40,00060,000 people in the US; many die within a few years of diagnosis. Existing treatments can slow this process down, but it doesn’t reverse or treat the fibrosis. However, exciting work is currently underway in the lab of Dr. Jim Hagood at the UNC School of Medicine in an effort to reverse lung fibrosis and

 By Kruti Bhargav
By Kruti Bhargav
reinstate normal lung function.
According to Dr. Hagood, he has been interested in the formation of scar tissue in the lungs for a long time due to the lack of understanding and therapies available. Attempts to reverse fibrosis greatly depend on understanding its course of action; as a result, the lab largely focuses on trying to understand what factors specifically regulate fibroblast activation and how they produce high amounts of collagen and scar tissue. Over the past few years, the lab has focused on exploring Thy-
1—a molecule that is typically present in normal fibroblasts but is decreased or silenced in activated fibroblasts that form scar tissue. In order to better understand the production, function, and therapeutic abilities of this molecule, Dr. Hagood’s lab is currently studying tissue culture fibroblasts within mouse models of lung fibrosis and tissues from human patient samples. At present, Dr. Hagood and the team feel as though they have a strong understanding of the different mechanisms of Thy-1 fibrosis suppression and therefore are working
towards utilizing this data to develop peptides that may bring about new treatments for pulmonary fibrosis.

While this research mainly relies on a biological and medical framework, the lab collaborates broadly with bioengineers, chemists, and various people with medical backgrounds over the course of this research. Moreover, Dr. Hagood and his team have built many relationships with researchers not only in the US but throughout the world as well. According to Dr. Hagood, “These collaborations have been a very rewarding process of the job.”1 Working with investigators who have different skills and expertise compared to those in the lab is essential to achieving scientific breakthroughs. Currently, the lab is collaborating with an applied physical scientist from UNC as well as computer and data scientists for single-cell work.

This exciting research may have far-reaching applications in the future within the medical field. Due to the severity and prevalence of idiopathic pulmonary fibrosis, Dr. Hagood believes novel treatments are crucial to battling this aggressive disease. According to him, the most notable use of this research would be to utilize the data for the development of new therapies or drugs to treat or reverse lung fibrosis rather than simply slow it down. However, drug development is a very challenging area of research. For instance, there are thousands of potential new drugs that are developed through basic science work yet very few make it through to the market as viable patient drugs. Nevertheless, the team has received a lot of guidance from experts who have worked on drug development in the past and are enthusiastic to undertake the task. Additionally, this research can allow for a greater understanding of how cell identity is regulated, what markers cause cells to differentiate along a specific pathway, and whether the differentiation is reversible.
However, no major research is exempt from obstacles. At times, Dr. Hagood states that it is essential to revisit one’s hypothesis and adjust methods in accordance with the data. Additionally, other challenges may include managing time to pursue all the different ideas discussed, ensuring there is sufficient funding to support the work being done, and getting published in scientific journals. However, as difficult as these challenges may be, Dr. Hagood suggests they are just as exciting and rewarding to address.
Moving forward, the lab is hoping to extend this research into investigating other forms of lung fibrosis as well as fibrosis in other organs. So far, they have collaborated with experts who research liver and kidney fibrosis and believe what they’re working on can be applied to a wide range of different chronic diseases in the future. In the end, Dr. Hagood encourages people with a scientific interest to keep asking questions, as we need more people with bright ideas and enthusiasm to enter the research field. With greater research into the development of novel treatments, we may progress towards a world where the threat of fibrosis may not seem as alarming as it does today.
1. Interview with Dr. Jim Hagood, M.D. 02/13/2023
2. Mayo Clinic. “Pulmonary Fibrosis.”
https://www.mayoclinic.org/diseasesconditions/pulmonary-fibrosis/ symptoms-causes/syc-20353690. (Accessed: 02/13/2023)
“...the most notable use of this research would be to utilize the data for the development of new therapies or drugs to treat or reverse lung fibrosis rather than simply slow it down.”Dr. Jim Hagood
 By Julia Boltz
By Julia Boltz
In the early years of the early human race, people spent their days on their feet, hunting, farming, and gathering food in order to survive.1 However with the rise of modern technology and globalization, the need for a highly active lifestyle is no longer common, potentially contributing to the rise in cardiovascular disease, or illness related to the heart and blood vessels, in many nations.1 In the United States, many Americans spend a large part of their day in what is known as a sedentary behavior, defined as very low-intensity behaviors in a seated, reclined, or supine posture.1 While it is likely that our human ancestors spent time engaged in sedentary behavior, it is believed that most of this behavior involved squatting, kneeling, or sitting with good posture.1
Modern aspects of contemporary life, including transportation, entertainment, and many professions, actively encourage individuals to be sedentary. The global COVID-19 pandemic only increased the prevalence of sedentary behavior (Figure 1).1 The decrease in physical activity and increase in sedentary behavior in America, and globally, has prompted Dr. Lee Stoner and colleagues to investigate what effects this kind of behavior has on human cardiometabolic health, and what policies can be implemented to mitigate these effects. Cardiovascular disease is the leading cause of death worldwide, which makes understanding risk factors and contributors to this deadly disease critical for the betterment of society.
A primary aspect of Dr. Stoner’s research involves determining how certain cardiometabolic indicators are impacted by a sedentary lifestyle. One indicator of health that is critical to examine is blood pressure.2 In a meta-analysis done in 2022, it was found that sitting a single bout of prolonged, uninterrupted sitting increases blood pressure.2 Over time, the repeated increase in blood pressure detrimental impacts the cardiovascular system, including stiffening of blood vessels and

and his associates found that sitting for 3 hours led to increased arterial stiffness.3 Stoner has hypothesized that this increased arterial stiffness is caused by hemodynamic, autonomic, hormonal, and metabolic changes (Figure 2).3 Hemodynamic changes, or changes in the movement of blood through the body, are likely caused by limited muscle activity when sitting,3 which leads to blood pooling in the veins in the lower body and a subsequent decrease in blood returned to the heart.3 A decrease in blood return to the heart leads to decreased stroke volume, which is the blood pumped out of the heart with each heartbeat. The decrease in stroke volume leads to decreased blood flow through the aorta – the big artery emanating from the heart. Blood flowing through the aorta provides a stimulus that is important for maintaining the health of the aorta and other arteries (much like dumbbell curls supplies a stimulus to the biceps). When the important blood flow-induced stimulus is lessened, the arteries lose their protective mechanisms and they become stiffer.3 Overall, it is evident that sitting for long periods of time can prevent our cardiovascular system from working optimally.

Through his research, Dr. Stoner has been able to establish that a sedentary lifestyle is harmful but translating this knowledge into policy is equally as necessary as the research itself. Current U.S. policy advocates for getting 150 minutes of moderate exercise per week, yet there is limited policy to guide sedentary behavior reduction (Figure 1).1 Dr. Stoner deprescribes sedentary behaviors are “multidimensional and very complex”, which makes designing policy around cardiometabolic health difficult.5
It is crucial that the American public be not only aware of how sedentary behaviors can cause negative health outcomes, but understand the best practices for reducing this behavior.5 To better understand sedentary “behavior”, has begun to
incorporate both quantitative and qualitative research to better understand the intersecting aspects of modern lifestyles and how to design policy around it.5 One of the ways Dr. Stoner goes about collecting this data is through a method called ecological momentary analysis, which studies the movement and location of people at various, random moments. To do this, Stoner uses a phone app, paired with an accelerometer, or a device that tracks movement, that prompts people at random times of the day to better understand the 24-hour activity cycle, which incorporates physical activity, sleep, and sedentary behavior across the day.5 Through this methodology, Stoner hopes to understand behavior at a more granular level.5
Dr. Stoner feels that policy should consider the interactions with the 24-hour activity cycle. He is quoted saying, “In 20 years, I want to be helping to lead the policy about how we should be engaging in activity behaviors throughout the day, including sleep, sedentary behavior, and physical activity”.5 In order to do this, Stoner hopes to help design something resembling 24-hour clock for the U.S. public to use that advises individuals on what behaviors they should be engaging with at different points in the day.5 Developing this clock proves difficult, as there is much about sedentary behavior or sleep, and even less about hour 24-hour activity behaviors interact.5 It is essential that public policy advises the public not only about how long to sleep or how long to be active, but also how to ensure that the behaviors are high quality. For example, it is important to consider not only how long someone sleeps, but also the quality of the sleep (Figure 1).5 Dr. Stoner views it as the job of scientists to understand the complexities of 24-hour activity behaviors in relation to cardiometabolic health, and translate this research into simple messages that can be understood and implemented by the public.5 The health of the American public relies greatly on public policy and research, which is why the work done by Dr. Stoner and others on how everyday behavior may be causing cardiovascular harm is crucial in saving lives.
References
1. Higgins S; Pomeroy A; Bates LC; Paterson C; Barone Gibbs B; Pontzer H; Stoner L. Front Physiol 2022, 13
2. Adams, NT; Poles, J; Paterson, C; Stoner, L. Medicine & Science in Sports & Exercise 2022, 54(9S)
3. Kelsch E; Diana JC, Burnet K; Hanson ED; Fryer SF; Credeur DP; Stone KJ; Stoner L. J Appl Physiol 2021, 131(1)
4. Shirwany NA; Zou MH. Acta Pharmacol Sin 2010, 31(10)
5. Interview with Lee Stoner, Ph.D. 01/30/2023 Dr. Lee Stoner

Every ten minutes, the number of people on the national transplant waiting list grows by one person; however, only a fraction of a percent of people on this waiting list accounts for the number of Americans who undergo an organ transplant (Figure 1). An organ transplant is an invasive procedure that had its first successes within the last seventy years. There are many essential steps to ensure that the new organ does not get rejected by the patient and that it finds stability within its new home. A drug called Mycophenolate mofetil (MMF), is a commonly prescribed immunosuppressant in transplant recipients. This drug reduces the effectiveness of the patient’s immune system, which prevents their immune system from attacking its own tissue and rejecting transplanted organs. However, severe gastrointestinal toxicity limits the effectiveness of MMF, and research on how to reduce this toxicity is gaining traction. Dr. Matthew Redinbo, a Kenan Distinguished Professor of chemistry, biochemistry, microbiology, and genomics at the University of North Carolina at Chapel Hill, has been recognized with awards for his research, teaching, and mentoring. Working alongside his team at Redinbo lab, Dr. Redinbo uses genetic material and proteins to identify gut bacterial enzymes that can reactivate MMF to ensure that transplanted organs are not

rejected. Understanding the function of gut bacterial enzymes can be lifesaving for not only organ transplant recipients, but also cancer patients because GI toxicity reduces the efficacy of critical drugs.

With poop, Dr. Redinbo and his team began this potentially lifechanging research. Collecting fecal samples from five renal transplant recipients taking MMF, as well as four healthy individuals, marked the beginning of their research. The Redinbo Lab did fourteen experiments using these samples. The first experiment consisted of reviewing the genetic material that was recovered from the fecal samples. To do this, the team used shotgun metagenomic sequencing, a technique that examines thousands of microorganisms at once. This technique determined the exact

sequences for all the genes present in the fecal samples, most of which were from bacteria (Figure 3). They found that transplant recipients had bacteria Bacilli, Gammaproteobacteria, and Erysiplotrichia in their flora, whereas healthy individuals had only Actinobacteria and Verrucomicrobiae (Figure 2). Having discovered these distinctions, in addition to knowing the total translated protein sequences, the team then performed an experiment to detect the genes that produce Betaglucuronidase (GUS) enzymes. These enzymes are responsible for generating the GI toxicity of Mycophenolate because they convert an inactive drug substance that is needed for metabolism into an active drug, which hurts the GI tract. Once they identified GUS genes that were either unique or entirely absent in transplant recipients, they then attempted to explain the range in mycophenolic acid (MPA) reactivation using their knowledge about which bacteria and GUS genes were present or absent. This next step seemed logical because these methods have long been used to connect candidate genes to functional results. However, in this case the team was unable to find any correlations between the MPA reactivation rates and specific GUS metagenomic features which could have pinpointed the cause of GI toxicity. Nonetheless, the Redinbo Lab pivoted their approach to find a better solution.
This time, the team used proteomics which is a method to examine exactly which proteins, rather than genes, are present within our gut. Since very few genes are expressed into proteins, genes reflect genetic potential, as opposed to genetic reality. Using proteomics, the team was able to prove that MMF-treated transplant recipients have a greater abundance of GUS enzymes. Higher levels of GUS enzymes reflected faster rates of reactivation of MPA when compared to healthy individuals. This implied that GI toxicity would be due to the fast reactivation of MPA caused by an increase in GUS enzymes following an organ transplant. Overall, the data and findings reinforced the importance of our microbiome in MPA-induced toxicity. Due to Dr. Redinbo and his team, blueprints now exist for researchers to identify transplant recipients at risk for MPA-induced gut damage, which could help alleviate GI toxicity that limits essential drug therapies.

The University of North Carolina at Chapel Hill is home to one of the nation’s leading medical research centers, making it the perfect place for Dr. Redinbo. His research on the microbiome is proving to be impactful in not only biology, but
microbiology, toxicology, and immunology. Understanding, targeting, and being able to block GUS enzymes would change drug therapies worldwide. This would help improve medications for cancer, pain, and now also improve immunosuppressant medications. The data Dr. Redinbo and his team has collected indicates that microbial GUS enzymes

are a primary factor influencing the toxic side effects of drugs. There may not be a cure yet, but there is a dedicated team of researchers working tirelessly to find the answer. No ten minutes go by without this team getting one step closer.
1. Interview with Matthew R. Redinbo, Ph.D. 01/30/2023
2. Learn How Organ Allocation Works - OPTN. Organ Procurement and Transplantation Network. (n.d.). Retrieved February 13, 2023, from https://optn.transplant. hrsa.gov/patients/about-transplantation/how-organallocationworks/#:~:text=Every%2010%20minutes%20 another%20person,donor%20can%20save %20eight%20 lives.
“No ten minutes go by without this team getting one step closer.”
 By Kaitlyn Campbell
By Kaitlyn Campbell
Cannabis could be the key to preventing neurodegenerative diseases such as HIV, Alzheimer’s, or Parkinson’s. Well, not exactly cannabis, but rather endocannabinoids. Cannabis attaches to a specific receptor in the body, giving the stereotypical feelings associated with getting high. But getting someone high isn’t cannabis’s only skill: the form of cannabinoids found naturally in mammalian bodies, endocannabinoids, may be the key to preventing neurodegenerative diseases such as HIV, Alzhiemer’s, or Parkinsons. However, there are enzymes already in the body that also bind to these receptors known as endocannabinoids. The cannabis enzymes in plants, phytocannabinoids, differ from the endocannabinoids found in the mammalian endocannabinoid system. Endocannabinoids are the neurotransmitter receptors found in the brain cells as part of the central nervous system, and they play a crucial role in the functioning of our brain, endocrine, and immune tissues. Dr. Sylvia Fitting, a researcher in the Department
of Psychology and Neuroscience, is working to determine how the endocannabinoid system can prevent neurological damage caused by the HIV1 protein. She is hopeful she can extend this research to other diseases.2
Before coming to UNC, Dr. Fitting worked to get her Bachelor’s and Master’s degrees in Germany and completed her graduate work at the University of South Carolina.1 During her work at South Carolina, Dr. Fitting initially worked in the field of experimental psychology but then transferred to behavioral neuroscience with a focus
on individuals exposed to HIV and as a response to the infection their body generates certain proteins that can have a detrimental effect on neurons.1 During her postdoctoral position at Virginia Commonwealth University, she focused on cell firing concerning HIV exposure.1 When she arrived at UNC in 2015, she directed her focus to the cannabinoid system and its involvement with HIV neuron degeneration.1
Currently, Dr. Fitting is attempting to determine the effect of enzyme inhibitors on the endocannabinoid system in relation to HIV proteins, using cell cultures from the prefrontal cortex of a mouse.2 Generally, an enzyme inhibitor works by binding to a specific receptor, which can either block or slow down the effect of the target enzyme. In these cell cultures, Dr. Fitting was able to see how the HIV protein affects neuronal responses by hindering a specific type of cell called glial cells.1 The main player most people know about in the brain is neurons, which are responsible for conveying information through chemical signals in the brain, but a less

known, but just as vital cell type are glial cells. Glial cells in the endocannabinoid system protect the neurons and increase the intensity of chemical signals sent between cells, increasing overall communication.
When discussing the protective effects of glial cells in the brain, Dr. Fitting said, “These enzyme inhibitors can be protective” in inhibiting the neuronal degradation commonly associated with HIV exposure.1 This discovery has allowed Dr. Fitting to establish a more direct study of the endocannabinoid system.
Furthermore, Dr. Fitting has been able to show that the cannabinoid system can be protective against the damage caused by the HIV-1 protein through behavioral studies involving the prefrontal cortex of the brain.2 The animal subjects, specifically mice, were tested through what is known as a choice task: one of five holes in a contraption is selected. If the animal remembers which hole is selected and itself “selects” the same hole by poking it, the mouse will receive a reward.1
To determine if exposure to the HIV1 protein alters attention span, mice exposed to the HIV1 protein then were put into a chamber where they had to perform the choice task. They were controlled to a set of control mice which had no exposure.1 Through these tests, Dr. Fitting was able to observe that there was an attention deficit in the mice who received exposure to HIV1 proteins.1 To expand on these findings, drugs known as corrected. Her results showed evidence that endocannabinoid enzyme inhibitors could decrease the attention deficits caused by HIV-1 exposure.1 This signifies that the enzyme inhibitor was likely binding to the endocannabinoid receptor, which could potentially reverse the damage done by the HIV-1 protein.
The discovery that enzyme inhibitors prevent damage to glial cells in mice had a substantial impact in the field of neuroscience. By acknowledging that there is a direct relationship between endocannabinoid enzyme inhibitors and neuronal damage caused by HIV-1, researchers could take steps to establish treatments for HIV neuronal degeneration that had the potential to influence large groups of people. Dr. Fitting is currently extending her research through what is called a working memory task.1 In this method, scientists monitor the mice while they are making decisions, yet their reward is delayed. This delay can trigger neuronal firing in the brain, which scientists are able to observe through neuronal imaging. This new research method could allow for the discovery of more details concerning the duration of the attention span, not just the existence of an attention span as previously.

This monumental work will not only help others expand upon the relationship between endocannabinoid enzyme inhibitors and brain function in infected mice, but will impact fields beyond its own.1 Developments in this area can lead to more cannabinoidbased medication and eventually changes in policy and community outlook on the use of cannabinoidbased drugs. Dr. Fitting is hopeful that “more work can be done with genetics facilities” to focus more on the live virus and its effect on the central nervous system, which varies significantly from current research on the periphery and microbiome.1
It would be a bold assumption to say that this research will find a cure for Alzheimer’s or Parkinson’s, yet such developments are not entirely out of the question. Dr. Fitting’s endocannabinoid research will open many doors for
new experiments involving complex neuronal imaging and lead to discoveries surrounding neurological degeneration.
1. Interview with Sylvia Fitting, PhD. 1/24/23
2.Yadav-Samudrala BJ, Fitting S. Mini-review: The therapeutic role of cannabinoids in neuroHIV. Neurosci Lett. 2021 Apr 17;750:135717. doi: 10.1016/j.neulet.2021.135717. Epub 2021 Feb 12. PMID: 33587986; PMCID: PMC7994193.

“Dr. Fitting has been able to show that the cannabinoid system can be protective against the damage caused by the HIV-1 protein through behavioral studies involving the prefrontal cortex of the brain.”
Each year, there are about 422 million people diagnosed with diabetes, the majority from low- and middleincome regions. One of the major culprits of this common illness is sugar intake, which is potentially caused by sugarsweetened beverage (SSB) consumption. In April 2018, South Africa became one of the first countries to implement taxes on sweetened beverages, which successfully reduced SSB consumption. Aiming to investigate factors related to the reduction of purchasing the taxed drinks—such as people’s intentions towards reducing sweetened beverage intake—Dr. Michael Essman led a team to answer the question of how exactly the tax led consumers to alter their purchasing and consumption behaviors.1 Dr. Essman and his team are leading one of the first studies examining what factors may change the influence of the health policy on the public. These findings may serve as an example for other countries to resolve concerns about diabetes and other health consequences of a high-sugar diet.

Dr. Essman finished his Ph.D. studies in the Global Food Research Program at University of North Carolina at Chapel Hill in 2020. He had a mission of evaluating food and nutrition-related policies around the world in hopes of improving people’s daily diets and preventing diet-related diseases. Previously, he focused mainly on the effect of the SSB tax in South Africa by examining people’s dietary changes
before and after the tax and the function of “news media” during this tax period.2,3 His work helped build an evidence base for the effectiveness of South Africa’s health program. His most recent study is based off previous results that the tax did reduce people’s purchase of taxed sweetened beverages. The study aimed to address three main research questions: how certain psychological constructs were associated with how much consumers drink the taxed beverage, whether such constructs themselves changed from pre- to post-tax, and how these constructs changed the influence of the tax on sweetened beverage intake. Specifically, there are four main psychological constructs in Dr. Essman’s study: awareness of the tax, intentions to reduce sugar-sweetened beverage (SSB) intake, perceived health risks of SSB, and SSB-related knowledge. All these constructs could potentially result in the decline of people’s purchase of the taxed sweetened beverage.
Dr. Essman and other researchers conducted interviews focused on twenty-four-hour diet recall with about 2500 participants between the ages of 18-39 in the lower income Langa township of South Africa. They prompted interviewees to

recall the food and beverage consumed as precisely as possible. Researchers then categorized this recalled data as either taxed or untaxed. After that, Dr. Essman and his team gave participants a survey on the four aforementioned psychological constructs and scored each participant’s response for further analysis. Dr. Essman underscored two things making this study unique. The first is the study’s demographic characteristics: lower income groups are always understudied but suffer from a great rate of diabetes. The second distinctive factor is interviewing and surveying such a large number of participants, which significantly reduced interviewer and questionnaire bias.4
Next, Dr. Essman and his team employed different analytical approaches to quantify important variables in the study. First, in order to estimate participants’ beverage consumption, they used a two-part model that links the probability of a consumer purchasing the sweetened beverage to the amount consumed. The two-part model incorporates two questions: “Did you buy the sweetened beverage?” and, “If so, how much did you purchase?”. Second, the team utilized both logistic and linear regression models to predict the relationship between
two variables to examine whether and how the four psychological constructs changed in pre-and-post-tax periods.
Eventually, Dr. Essman’s team found that after the tax implementation, only the intention to reduce the SSB intake was positively associated with taxed beverage consumption. In other words, people who expressed the intention to decrease their sweetened beverage consumption were more likely to consume less beverage. As for the second research question about changes in the constructs over time, three out of four constructs (tax awareness, SSB-related knowledge, and risk of SSB perception) slightly increased in the post-tax period compared to the pre-tax, while the intention to reduce SSB intake decreased significantly after the tax. Lastly, SSB-related knowledge and intention to reduce sweetened beverage intake did change the effect of the tax on the taxed beverage. That is, changes in both the knowledge of and the intention towards purchasing sweetened beverages may reduce the impact of such tax.
In light of the results, Dr. Essman points out that the behavioral change is most likely due to the price sensitivity of lower-income individuals. That is, people’s reduced consumption of the taxed SSB are unlikely to be explained by tax awareness, SSB-related knowledge, or risk of SSB perception. He also suggests that because of the small increase of the three constructs in the post-tax period, more widespread media and communications may seek to further improve people’s awareness and understanding of this tax policy. Dr. Essman infers that the sweetened beverage tax not only increased price, but also led people to have a deeper understanding of the health implications of the taxed beverages.
Dr. Essman spoke about a hurdle encountered during the study: “the biggest thing that stands out to me is that this type of research requires a really large team effort.”4 Indeed, collaboration is the key to making a study of this magnitude successful. Fortunately, in his current study, Dr. Essman largely worked with
other interdisciplinary scientists, such as Dr. Francesca Dillman Carpentier from the UNC Hussman School of Media and Journalism as well as other economists and psychologists. “Understanding the complicated ways [in which] populationlevel measures, like sugary drink levies, affect the entire food system, does really require a multidisciplinary viewpoint,” Dr. Essman emphasized.4
Dr. Essman and his team’s results not only provide proof of the influence brought by the current tax on sweetened beverages in South Africa but give suggestions about how to increase the tax’s effectiveness in reducing sweetened beverage purchases and lowering the rate of related diseases. More broadly, Dr. Essman’s mission in doing this type of research is to contribute to building a scientific evidence base for healthrelated policies. “As more evidence accumulates, the stronger strength a certain policy will have, and other governments [will be] more encouraged to do the same effective thing.”4 In other words, Dr. Essman and his team are using their research to serve both a political and health goal: to maximize the function of any health policy. He hopes future studies can build on the current approach, but collect more data beyond the four
psychological constructs measured currently in order to better understand people’s behavioral change under altered policy.
1. Essman, M.; Zimmer, C.; Carpentier, F.D. et al. Int J Behav Nutr Phys Act. 2022, 19, 136.

2. Essman, M.; Taillie, L.S; Frank, T.; Ng S.W; Popkin B.M; Swart E.C. PLOS Med. 2021, 18,5, e1003574.
3. Essman, M.; Stoltze, F.M; Carpentier, F.D. et al. BMC Public Health. 2021, 21, 454
4. Interview with Dr. Essman, Ph.D. 02/06/23.
“Understanding the complicated ways [in which] populationlevel measures, like sugary drink levies, affect the entire food system, does really require a multidisciplinary viewpoint”
 By Grayson Coleman
By Grayson Coleman
UNC is a nexus of groundbreaking research and innovative therapeutics in the world of oncology. UNC Lineberger Comprehensive Cancer Center in conjunction with UNC Hospitals have repeatedly made headlines for their advancements in cancer treatment and management. Being nationally ranked as a premier research institution at the forefront of the fight against cancer, dozens of labs concentrated in niche disciplines uniquely suited to tackle particular queries related to cancer are included under the center’s expansive title. Virology, Epidemiology, Genetics, Immunology, Cell Biology, and more are all “Lineberger” departments. In February 2023, Chief Science and Technology Advisor and White House Office of Science and Technology Policy Director Arati Prabhakar came to campus to observe the work being done at UNC’s Lineberger Comprehensive Cancer Center. From Dr. Pengda Liu’s cGAS-STING pathway mapping to Dr. Jean Cook’s cell cycle investigations to Dr. Gianpietro Dotti’s CAR T cell therapies, PhDs and MDs alike are making incredible strides against the heterogenous disease that claims the lives of over half a million each year in the United States alone. As the second leading cause of death in the United States, cancer has brought brilliant minds from across the globe together in their efforts to find a cure to such a multifaceted disease. Different researchers have approached this question in various ways using their respective expertise to foster a collaborative atmosphere.
What makes cancer such a challenging disease to manage is both its heterogeneity and its adaptivity. Cancer is heterogenous in the sense that each cancer varies from patient to patient and can even behave differently or inconsistently within a single patient. Tumors are constantly evolving and mutating, allowing them to metastasize, while advancing and evading the immune system. The latter is what makes tumors adaptive or “plastic.” The plasticity of cancer refers to its ability to adjust its strategies and defenses in response to the body’s natural opposition. These characteristics make cancer a formidable foe and demonstrate why curing it is such a complex task. However, these challenges have only strengthened Lineberger’s resolve.
Dr. Pengda Liu, one of Lineberger’s lead principal investigators, has found his home within the Department of Biochemistry and Biophysics. He has honed his research in cancer biology to focus primarily on the malformation of certain proteins and the signaling cascades that precede them. These molecular errors contribute incredibly to the tumorigenesis – the creation of cancerous tumors – in several forms of cancer. His research tracking signaling cascades has taken him on a long and winding road through the human genome. The mTOR signaling pathway particularly piqued Dr. Liu’s curiosity, as it has for many other biochemists within his field. The mTOR or “mechanistic target of rapamycin” signaling pathway regulates protein translation, cell proliferation, and immune cell maturation. Its activation has been implicated in the metabolism of several chronic conditions including cancer. Dr. Liu’s research on the mTOR pathway has been characterized by his desire to develop better inhibitors and therapeutics to impact the lives of affected patients.
Presently, Dr. Liu and his team are investigating the cGASSTING pathway – a valuable asset to the body’s cells that is a part of the innate immune system. The innate immune system


is the body’s first line of defense and is defined by its immediate and nonspecific responses. It uses the cGAS-STING pathway to detect and alert the immune system to foreign DNA found within a cell’s cytosol. This works incredibly well in identifying viral DNA that has been inserted into idle human cells and in detecting mutations such as those found in tumors. This pathway, however, seems to be dysregulated in many tumors, thus minimizing its efficacy in cancerous cells. By conducting loss of function experiments on cultured human cells, Dr. Liu and his collaborators turn off expression of certain genes in a systematic fashion to map a pathway. Using these genetic and biochemical approaches, the lab can track changes in the cells due to their manipulations and determine a rough cascade. By learning how this pathway should work, they can better understand why it is failing in their patients.
The third and most personal prong of Dr. Liu’s career interests is his investigation into Ewing sarcoma – an extremely rare pediatric bone cancer which currently has no targeted therapies and receives minimal research interest from the greater cancer community. Afflicting fewer than 200 Americans each year, Ewing sarcoma is caused by a fusion oncogene, where two unrelated genes are conjoined by an odd mutation. Deubiquitinases such as OTUD7A, which encourage cell proliferation and have been found to be highly expressed in Ewing sarcoma tumors, stand out as possible progressors of this illness. Dr. Liu and other scientists hypothesize that inhibitors to OTUD7A might help shrink these tumors in patients. He and his team have utilized the system AtomNet, a virtual drug screening platform established by Atomwise, Inc, to find potential new therapies which might aid patients identified as having OTUD7A overproduction.
Ultimately, Dr. Liu aims to direct his research towards developing new therapeutics. Cancer treatment has historically been one-dimensional, limited to monotherapies such as chemotherapy, radiation, and surgery. Monotherapies, though, are ineffective, as cancers always seem to develop a resistance or engineer a new workaround. Instead, clinicians and scientists alike have begun to focus on suppressing oncogene expression to stop tumor growth, later shifting to activate natural tumor suppressive functions to eliminate the cancer if possible. This is what Dr. Pengda Liu calls the “gas and brake” system. First, the physician must release the “gas” of cancer development. By destabilizing molecules that are aiding in cancer growth, such as OTU7A and other deubiquitinases, the physician can cut off the resources to the tumor. Then, the physician must step on the “brake” and halt further cancer growth. This can be accomplished by restoring dysregulated immune functions such as cGASSTING to attack the tumor and initiate normal endogenous defenses against the cancer.
Dr. Jean Cook and her research team are located at the Genetic Medicine Building on the UNC-Chapel Hill School of Medicine campus. Her research has been dedicated to understanding the molecular mechanisms which govern cell proliferation and DNA replication. Genetics is the foundation of

tumorigenesis and the discoveries made by Dr. Cook and her team of undergraduates, graduate students, and postdoctoral scholars directly integrates with the translational work done in the Lineberger Center to bring the lab to the bedside of patients. Dr. Cook is fascinated with how one cell becomes two, and it is the magic of cell proliferation that drives her lab’s analyses. A study she is particularly fond of was a “technical Tour de Force” by a postdoctoral fellow, Dr. Liu Mei. Dr. Mei was investigating cells via live cell imaging—more on that in a second—and wished to understand more about the chromosomes she was viewing. By treating the cells with certain antibodies, Dr. Mei was able to analyze the geography of these cells’ chromosomes. The importance of this research stems from differences between chromosome segments. Euchromatin refers to segments of a chromosome where the DNA is very loosely packed and easily accessible for proteins to read and replicate. Heterochromatin segments, however, are the incredibly dense and tightly wound strands of DNA which are much harder to replicate. Dr. Mei’s guiding question centered on how cells accessed heterochromatin during the preparation steps of DNA replication to allow cells to proliferate. What she observed following her antibody treatments was that the euchromatin got a head start in replication since it was more accessible to replication proteins, while heterochromatin replication was delayed. This was expected. What was unexpected was that the rate of heterochromatin replication increased such that both populations of DNA finished replicating simultaneously. This discovery helped to answer the question of how such a heterogenous mix of DNA types were able to be replicated. Perhaps even more interesting, however, is what happens when this process is dysregulated. When cells replicate their DNA later than they should, the segments that are skipped are primarily heterochromatin due to them being last in the queue. When segments of DNA are missing in chromosomes, they can become fragile and even break, causing catastrophic cascades that, in many instances, can lead to tumorigenesis. It is crucial that research continues in this area not only to determine why this is occurring and why these failures are not being recognized by the body’s defenses, but also to develop clinically practicable methods of identification and alleviation of these malignant replications.
Another study that has been in the works for over six years now is the enterprise of graduate students Jacob Matson and Dalia Fleifel. Matson, a former graduate student of the Cook Lab, discovered that pluripotent stem cells, an incredibly early developmental cell with the ability to mature into any tissue type in the body, could prepare their chromosomes at a much faster rate than mature, differentiated cells. This tracked logically, as development occurs at an accelerated rate early on and plateaus as one grows up. But lab members became even more intrigued when another lab abroad discovered that scientists could induce a differentiated cell to become a pluripotent cell. This kind of technology could, put simply, revert any adult cell back to its undifferentiated state. This was a remarkable breakthrough for the field of regenerative medicine. The work of Jacob Matson along with this discovery has been passed on to Fleifel, who is taking these two studies a step further by identifying the gene or combination of genes that leads to this acceleration of chromosome preparation. It was found in the pluripotent stem cell paper that only four genes were necessary to revert a mature,
differentiated cell and those same four genes are being experimented with by Fleifel. In a “plug and chug” style assay with these genes, she is attempting to discover why these cells are so much more efficient than their adult counterparts in hopes that these findings can initiate a new avenue of cancer treatments.

Unlike other researchers in the cancer field, Dr. Cook is not solely interested in examining how disorders in the cell life cycle progress into cancer; she is also interested in how these disorders progress towards aging. As she says, aging and cancer are the “flipside of the same coin.” In cancer pathology, cells divide when they should not, while in aging, cells fail to divide when they should. This dual focus in her lab means that every discovery made towards aging can simultaneously impact the field of cancer biology and vice versa.
Behind the headway Dr. Cook is making in understanding the underlying scaffolding of cancer and the numerous publications produced by her lab, there is a zoo of incredible analytical tools. Most fascinating, perhaps, is her live cell imaging approach, which allows her to examine cell activity in real time. This is the same technology previously mentioned in Dr. Liu Mei’s investigations of euchromatin and heterochromatin. Via molecular genetics technology, Dr. Cook can attach GFP (green fluorescent protein) to a protein of interest which will fluoresce upon the fusion protein’s expression. What is able to be seen under the microscope is nothing short of remarkable.

Dr. Gianpietro Dotti is on the front lines of cancer treatment both as an MD treating active cancer patients and as an immunological researcher ushering in a new era of cancer therapeutics. Dr. Dotti and his team in Marsico Hall at UNCChapel Hill are a part of a growing movement in medicine to reengineer patient immune cells in the lab to target tumors. Following a synthetic manipulation in lab, these cells are then infused back into patients, reprogramming the immune system to better combat cancer. The name of this new technology is CAR T cell therapy (standing for chimeric antigen receptor), and it has the potential to turn the world of cancer pharmaceuticals on its head. T cells are a prominent immune cell type which can identify and target foreign antigens via their TCR complex. A TCR complex or T Cell Receptor complex is a protein structure on the surface of T cells which Dr. Dotti and his team have manipulated to target the surface of cancer tumors. By introducing a synthesized TCR complex to a patient’s T cells, Dr. Dotti can program these cells to target certain forms of cancer exclusively. How are these CAR T cells produced, though? What makes a target promising? How is this different than all previous cancer procedures and why is this not the new standard of care? These are all great questions with even more fascinating answers.
The production of a CAR T cell is a complex and advanced process. Dr. Dotti’s CAR T cells are a highly modified version of
patient immune cells with three key adjustments: genes for prolonged IL 15 release, a “suicide” gene, and a CAR TCR complex. IL 15 is a signaling molecule that encourages the proliferation and survival of T cells. Without this genetic addition to constitutively release this cytokine, Dr. Dotti’s CAR T cells would expire before any significant impact could be made within the patient. Dr. Dotti has termed his second genetic addition “the suicide gene.” This remarkable feat of biochemistry is arguably the most ingenious aspect of the CAR T cell manufacturing process. This gene is a failsafe against a negative reaction induced by these cells. Should a patient experience harmful symptoms as a result of this treatment, a specialized drug can be administered which activates the CAR T cells’ suicide gene. Once this gene is activated, the cells commit apoptosis, which is also known as programmed cell death. Within 24 hours, the patient will no longer have these cells in their system nor experience any more symptoms.
Dr. Dotti then determines the appropriate proteins for his CAR T cells to target through several rounds of trial and error. Research is often a grueling process of testing hypotheses until a usable result presents itself. Long hours were put into investigating the literature and analyzing RNA sequencing data on cell surface markers present on their intended cancer types. Even longer hours were then invested into testing these cell surface markers on cancer cell lines. The biggest challenge in this testing phase was finding a cell surface marker which was both highly expressed on tumor cells but also lowly expressed on normal cells. This was necessary to ensure the specificity of these T cells’ target and reduce any autoimmune attacks. To accomplish this result, Dr. Dotti would express the target antigen at varying levels on the cancer cell lines to observe the tolerance of his CAR T cells. His aim was to have CAR T cells that had low affinity for an antigen so it would not attack antigens on normal cells.
One of the greatest drawbacks to traditional cancer treatments is their lack of exclusivity. Treatments such as radiation and chemo are incredibly toxic to patients because they target healthy cells too. CAR T cell therapy could seemingly replace these monotherapies, making cancer treatment more bearable for the patients. Tragically, however, there are three major barriers currently preventing the standardization of CAR T cell therapy: the manufacturing cost, the overhead of cultivating proper facilities, and its limited application. As of 2023, to produce a single round of CAR T cell treatment would cost the patient roughly $400,000. Comparatively, therapies such as radiation and chemo are much cheaper, with annual costs between $5,000
and $10,000 depending on the cancer type. And even these treatments remain unaffordable for many patients. Aside from the costs of manufacturing is the overhead involved in producing these facilities. Expanding these specialized cellular facilities will prove challenging if CAR T cell therapy is to reach clinics internationally. CAR T Cell therapy is also currently limited in the cancer types it can treat. Currently, the CAR T cell therapies that have been developed are only able to attack liquid tumors such as leukemias and lymphomas. This leaves patients with solid tumors such as colon, lung, and brain cancer in an unfortunate gap which researchers are attempting to fill. However, solid tumors have proven to be tricky and pose more challenges than one may anticipate. Solid tumors can adapt at a much faster rate than liquid tumors because they are uniquely able to generate a lymphotoxic microenvironment around them. Like a forcefield, solid tumors can create barriers that make it impossible for immune cells to approach without dying. Not only can they develop these microenvironments, but they can also augment themselves to present different strengths and weaknesses when they metastasize to a new region of the body. This heterogeneity, as has been previously discussed, is an extremely frustrating aspect of cancer pathogenesis. Due to its limited application, the future of this technology is incredibly dependent not just on price and widescale feasibility but also on the ability to use it on all cancer types.
CAR T cell therapy variants are not just new pharmaceuticals being rolled out annually; these “living medicines” are reinventing the industry and redefining personalized medicine. As of 2023, the FDA has approved 5 CAR T cell treatments with many more in active clinical trials. With any luck, CAR T cell treatment will become the standard of care in the near future.
work of Dr. Liu, Dr. Cook, and Dr. Dotti is ongoing, and you have the potential to continue their research as you graduate and move forward in your own career. While no one individual can solve such a complex problem as a cure for cancer on their own, progress is possible when the entire community comes together. By joining the effort to combat cancer, you and others who share this goal can create a collective impact that has the potential to be immeasurable. Certain cancer types still lack adequate funding and research expertise. The cellular and genetic failures that lead to cancer expansion are not yet fully understood. The new field of immunological reengineering treatments is fresh and requires continued attention and refinement. As UNC-Chapel Hill and the extensive international community work together to overcome these challenges, I hope to see you join in this important effort. It will truly be a Good Day To Be A Tar Heel when the cancer diagnosis of tomorrow does not have to carry the same weight as today.
1. “Car T Cells: Engineering Immune Cells to Treat Cancer.” National Cancer Institute, https://www.cancer.gov/about-cancer/treatment/ research/car-t-cells.

2. Choi, Gyeyoung, et al. “Price and Prejudice? the Value of Chimeric Antigen Receptor (CAR) T-Cell Therapy.” International Journal of Environmental Research and Public Health, U.S. National Library of Medicine, 28 Sept. 2022, https://www.ncbi.nlm.nih.gov/pmc/ articles/PMC9566791/.
3. Derewicz, Mark. “UNC Lineberger Hosts White House Chief Science and Technology Advisor: UNC-Chapel Hill.” The University of North Carolina at Chapel Hill, 14 Feb. 2023, https://www.unc.edu/ posts/2023/02/14/unc-lineberger-hosts-white-house-chief-sciencetechnology-advisor/.
4. “Ewing Sarcoma.” Mayo Clinic, Mayo Foundation for Medical Education and Research, 7 Jan. 2022, https://www.mayoclinic. org/diseases-conditions/ewing-sarcoma/symptoms-causes/ syc-20351071?utm_source=Google&utm_medium=abstract&utm_ content=Ewings-sarcoma&utm_campaign=Knowledge-panel.
5. “Financial Burden of Cancer Care.” Financial Burden of Cancer Care, Apr. 2022, https://www.progressreport.cancer.gov/after/ economic_burden.
6. “Gianpietro Dotti.” UNC Lineberger, 28 Apr. 2022, https:// unclineberger.org/directory/gianpietro-dotti/.
7. “Jean Cook Lab.” Jean Cook Lab, https://sites.google.com/site/ cooklabgroup/home.
The purpose of this segment is not only to highlight the groundbreaking cancer research being conducted on UNC’s campus but to also raise awareness for the innovations that still need to be made. The fight against cancer requires the collective efforts of individuals, like you, who are reading this article. The

8. “Ohsu Knight Cancer Institute.” OHSU, https://www.ohsu.edu/ knight-cancer-institute/car-t-cell-therapy-cancer.
9. Pengda Liu Lab, https://unclineberger.org/pengdaliulab/.
10. “An Update on Cancer Deaths in the United States.” Centers for Disease Control and Prevention, Centers for Disease Control and Prevention, 28 Feb. 2022, https://www.cdc.gov/cancer/dcpc/ research/update-on-cancer-deaths/index.htm.
For many, growing up means new freedoms, especially on a college campus. But as the spring semester rolls around, it is becoming more understood that with adulthood comes new responsibilities. Particularly for college women, a new concern regarding reproductive health and the looming idea of a first pap smear appears. Luckily, research shows that for college educated women in North Carolina, 90.9% percent have received a pap smear within the last three years.¹ Receiving a routine checkup with a pap smear every three years is essential for women to detect changes and possible growths that could lead to cervical cancer.
A preventative regimen for cervical cancer is established for many American women from a young age. Beginning around the age of 11 or 12, American girls typically receive the HPV vaccine to prepare for cancer prevention from possible transmission of HPV. If the immune system is unable to completely extinguish the virus in an infected individual, HPV will proliferate inside the body and transform normal cells into cancerous ones, which often grow into cancerous lesions.


Lesions on the cervix are usually detected during a cancer screening procedure that is performed in tandem with a routine pap smear. In America, women are encouraged to begin receiving regular pap smears beginning at age 21. That being said, cervical cancer remains a prominent cause of death for women globally. It may be surprising considering the current preventative procedures, but cervical cancer is a true disease of disparity. In recent years, around 90% of new cases and deaths from the cancer were reported from countries considered to be low and middle income.² Dr. Jennifer Smith, an award-winning researcher working at the UNC- Dr.

Chapel Hill Center for Women’s Health Research, is one of many scientists particularly outraged by this statistic. As she describes it, the greatest obstacle to improving death and disease rates for cervical cancer is to bridge the gap that can be seen in access to preventative services. For women in the United States, higher cervical cancer rates are seen in Black and Hispanic populations, women who are underinsured or uninsured, and especially in women who are living below the poverty line. Typically, higher rates can be linked to rural areas where average household incomes are significantly lower. The trends are seen both domestically and globally. Throughout her time studying HPV spread and cervical cancer, Dr. Smith has fought as part of her mission to spread the message that “local is global, and global is local.” She has led studies of cervical cancer globally, from areas in rural North Carolina to countries such as China, Kenya, and South Africa. While performing research in rural or lowincome areas, Dr. Smith has worked outside of the box to spread messages about the importance of cervical cancer screening. With her team, she has conducted canvassing of the areas using methods like bus advertisements, radio segments, and even approaches as unique as scouring Craigslist. As Dr. Smith explained, much of the heavy lifting is done while attempting to identify individuals with little access to care who are eager to receive aid in both screening and continuity of care.
through the process of home use, where it can then be mailed to labs where the samples are analyzed. Through periods of testing, Dr. Smith has found the use of the MBMT kits to be highly successful at detecting any present cancerous lesions, even compared to the detection rate of samples collected by healthcare workers in a clinic. There has been a recent rise in the use of self-testing kits for a myriad of diseases in national programs across the globe, particularly in Denmark and the Netherlands. Many people have likely even used a similar kit in their own homes, seeing as at-home tests for COVID-19 have become popular in the United States.
As for the future of her work, Dr. Smith explains that she not only aims to focus resources on continuing to promote regular cancer screening, but she also emphasizes the importance of continuity of care. Resources in healthcare in rural and lower income areas pose a threat to many individuals who receive positive test results for diseases such as cervical cancer. Properly equipped medical centers and trained healthcare providers are often few and far between in such areas. At the end of 2022, Dr. Smith was one out of a team of three researchers from the UNC Lineberger Comprehensive Cancer Center to be awarded a $3 million grant to continue essential efforts in fighting for cervical cancer screening and prevention.³ The grant will be used to fund clinical trials and research towards determining further methods and technologies most effective in both detection and treatment of cervical cancer. Dr. Smith explained that, going forward, she is interested in exploring techniques such as utilizing integrated healthcare systems to identify women who are under screened for cervical cancer or at high risk for HPV infection.
Dr. Smith’s groundbreaking work in the field of cervical cancer has emerged from her success in utilizing self-testing kits to perform widespread cancer screening in underserved areas. Aptly named “My Body My Test” (MBMT), the innovative self-testing apparatus comes with instructions to walk women

1. Explore Cervical Cancer Screening in North Carolina | 2022 HWC. (2020). America’s Health Rankings. Retrieved February, 2023, from https://www.americashealthrankings. org/explore/health-of-women-and-children/measure/ cervical_cancer_screen_women/population/cervical_cancer_ screen_women_College/state/NC
2. Cervical cancer. (2022, February 22). World Health Organization (WHO). Retrieved February, 2023, from https://www.who.int/news-room/fact-sheets/detail/cervicalcancer
3. UNC researchers to use $3 million grant to improve cervical cancer screening and treatment - UNC Gillings School of Global Public Health. (2022, December 16). UNC Gillings School of Public Health. Retrieved February, 2023, from https://sph.unc.edu/sph-news/unc-researchers-to-use3-million-grant-to-improve-cervical-cancer-screening-andtreatment/
“It may be surprising considering the current preventative procedures, but cervical cancer is a true disease of disparity.”

Diabetes is one of the most prevalent diseases both in the United States and worldwide, affecting one in ten Americans, and its rates are on the rise. With diabetes, a person’s body either cannot make enough insulin or cannot process it properly, leading to an excess of glucose in the bloodstream. Untreated diabetes can cause serious health conditions,- including vision loss, heart conditions, kidney disease, and nerve damage. While there is no cure for diabetes, the condition can be managed with medication and self monitoring.1 Continuous glucose monitors (CGMs) are implantable sensors that continuously measure the concentration of glucose in the blood (Figure 1), unlike handheld glucometers that require diabetic people to prick their finger multiple times throughout the day. However, the use of CGMs is time-limited as the implantation process elicits a foreign body response (FBR) from the immune system.2
Dr. Mark Schoenfisch, the Peter A. Ornstein Distinguished Professor at UNC-Chapel Hill, and his lab have been working on reducing the FBR caused by sensor implantation using nitric oxide-releasing coatings. The Schoenfisch lab aims to create a “next-generation sensor membrane that stores and releases nitric oxide” to extend the use duration of CGMs.3
The process of implanting the CGM injures the tissue surrounding it, prompting the FBR. Over time,
the body develops a fibrotic response to the site of insertion. During the foreign body response, immune cells accumulate, consume glucose, and ultimately form a thick layer of collagen around the glucose sensor (Figure 2). This process and resulting fibrotic layer greatly decrease the sensor’s performance (i.e., ability to measure glucose) and useable lifetime.
Nitric oxide (NO) is a diatomic free radical (meaning it has an unpaired electron), involved in several vital physiological processes such as wound healing, anti-inflammatory responses, angiogenesis (the formation of new blood vessels), and blood pressure regulation. These roles of NO throughout the body motivated Dr. Schoenfisch and his group to delve deeper into the possibilities of implementing NO donors to reduce the FBR caused by CGMs, thus “harnessing nitric oxide’s therapeutic potential.” 3
In its natural gaseous state, nitric oxide is difficult to apply directly to a biomedical device. Instead, compounds that store and release NO under normal physiological conditions are used to achieve the desired effects of NO. For the implantable glucose sensors, the Schoenfisch lab synthesizes NO-releasing silica nanoparticles (Figure 3). Polyurethane films containing either

“Continuous glucose monitors (CGMs) are implantable sensors that continuously measure the concentration of glucose in the blood”
these NO-releasing nanoparticles or nonNO-releasing nanoparticles (as controls) are coated onto existing CGM sensors. The sensors are then tested in vivo to assess the ability of NO to reduce the FBR. The tissue microenvironment surrounding each sensor is characterized based on the presence of immune cells associated with the foreign body response. The sensors capable of NO release have shown mitigation of both the acute and chronic stages of the FBR.2

One of the greatest challenges of working with NO is the clock. Many NO donors release the molecule for less than 24 hours. However, Dr. Schoenfisch and his lab have designed porous silica nanoparticles that have been shown to release NO for up to 28 days.4 For the duration of the NO release, the sensors fabricated with these nanoparticles showed a decrease in the FBR and in the collagen density surrounding the sensor. Additionally, the analytical performance of these sensors was more accurate than control sensors over this period.

Despite the progress Dr. Schoenfisch and his lab have made thus far, he continues to strive for
improvement. When speaking of the successes of the project, Dr. Schoenfisch acknowledged that “we’ve shown that we can measure glucose more reliably and for longer periods.”3 However, he strives to further prolong sensor performance. In addition to increasing the duration of NO release, combining active mitigation of the FBR through NO with other promising passive strategies, is a path forward. While NO release is limited and will eventually expire, passive FBR mitigation through electrospun fibers, for example, should prove beneficial beyond the release of NO because it relies on the chemical and physical characteristics of the implant surface itself.⁵
Even though continuous glucose monitoring systems have been around for decades, there is much room for improvement. Those with diabetes rely on glucose monitors, and the utility of these monitors relies on measurement speed and accuracy. CGM performance begins to deteriorate immediately after implantation, which can have serious effects for the user. Researchers will continue striving to improve this technology to ultimately develop a long
performing closed-loop system whereby glucose levels are maintained “behind the scenes” with little required input by the patient.
1. What is Diabetes? https://www.cdc. gov/diabetes/basics/diabetes.html (accessed January 11th, 2023).
2. Taylor, J.B; Malone-Povolny, M.J; Merricks, E.P; Wimsey, L.E; Soliman, D; Nichols, T.C; Wallet, S.M; Maile, R; Schoenfisch, M.H. Int. J. Mol. Sci 2022, 23, 11635.
3. Interview with Mark H. Schoenfisch, Ph.D. 1/27/23
4. Malone-Povolny, M.J; Merricks, E.P; Wimsey, L.E; Nichols, T.C; Schoenfisch, M.H. ACS Sens. 2019, 4, 3257-3264.
5. Malone-Povolny, M.J; Bradshaw, T.M; Merricks, E.P; Long, C.T; Nichols, T.C; Schoenfisch, M.H. ACS Biomater. Sci. Eng. 2021, 7, 2444-2452.
Oxygen is the most abundant element in the human body, making up 65% of a person’s body mass.1 It is utilized by the cell for various purposes, but a notable function is oxygen’s role in cellular respiration. Without oxygen, cellular respiration cannot be completed; consequently, human cells would not have an adequate amount of ATP to carry out their important functions. Some organs have a greater demand for oxygen, like the brain, heart, and kidneys. In fact, if the brain’s oxygen supply is interrupted for as little as four minutes, permanent brain damage would occur.2 This is because the brain requires oxygen for the metabolism of glucose3 that influences critical brain functions such as thinking, learning, and memory.4 Due to the importance of high oxygen levels, a measurement device called a pulse oximeter provides insight into the amount of oxygen circulating a person’s blood.
Unfortunately, pulse oximetry suffers from a fatal flaw: melanin, a
substance in cells that is responsible for skin pigmentation, interferes with the accuracy of oxygen level measurements. The presence of melanin can lead to an overestimation of oxygen levels when the actual values are much lower. Many may find it surprising that this inaccuracy exists considering the importance of pulse oximetry in all aspects of healthcare. Many published articles discuss the problems with pulse oximetry, yet much of the public continues to trust the traditional pulse oximeter to deliver accurate measurements. The problem with this inaccuracy is that it only affects certain populations, and it creates a gap in the precision of healthcare across different racial and ethnic groups. These misleading results may cause patients to not receive adequate treatment for their low oxygen levels. Despite this, there seems to be a surprisingly low urgency in the scientific community towards correcting this problem.

Luckily, here at the University of North Carolina at Chapel Hill, Wubin Bai,
Ph.D., and his talented research team are working towards solving this ignored problem and eliminating this medical disparity in pulse oximetry. The Bai Lab aims to use fundamental research and translate it to healthcare or environmental impacts. Dr. Bai enjoys his work as he feels that he is essentially a middle ground

between fundamental researchers and frontline healthcare workers. He says that “such a style of connecting them and starting a conversation [with them]—an easy and smooth conversation—drives me to think that I’m doing something quite meaningful”.6
As an applied sciences lab, Dr. Bai works to understand the principles —exactly what he is doing with his work in pulse oximetry. Current pulse oximetry technology doesn’t work by measuring the amount of oxygen in the body. Instead, it measures the amount of oxygenated versus deoxygenated hemoglobin, the protein in our red blood cells that picks up oxygen and delivers it to the rest of the body’s cells. Pulse oximetry measures the absorbance and transmittance of certain wavelengths by hemoglobin, specifically red and infrared light wavelengths. Oxygenated hemoglobin is better at absorbing infrared light, and it transmits more red light; the opposite is observed for deoxygenated hemoglobin.5 By determining the ratio of infrared light and red light transmitted, pulse oximeters can determine how much oxygenated hemoglobin there is, but as Dr. Bai explains, melanocytes in the skin interfere with the amount of light absorbed and transmitted.6 People with darker skin
pigmentations have a greater number of melanocytes, which leads to greater inaccuracies as the skin pigmentation darkens. Dr. Bai’s lab has found a solution to this problem where they don’t modify the technology altogether, but instead improve upon the previous technology by adjusting the wavelength calibration to account for melanin presence.
Dr. Bai and his research team’s agenda for the pulse oximeter involves more than just correcting the melanindetecting problem itself—they want to create it in a low cost, mass manufacturable way. Currently, traditional pulse oximeters are readily accessible to the public. This is because some people must consistently check their oxygen levels due to certain health conditions, as was especially apparent during the COVID-19 pandemic. Thus, pulse oximeters can be bought at CVS, Amazon, or other outlets for use outside of a medical setting. Since the traditional oximeters are so widely available in the market, it will take a low cost and mass manufacturing approach to replace them with the melanin-detecting pulse oximeters. As for other goals, Dr. Bai’s research lab is hoping to create a comfortable device that’s just as soft, or even softer, than a band-aid. Now, the Bai lab is in the process of approval to enact
clinical trials to test their device.
This melanin-detecting pulse oximeter has the potential to have significant public health impacts, and it can lead to a better delivery of care to different racial groups. “I always thought those issues required policy to solve, but technology also plays a very essential role,” says Dr. Bai, as he reflects on the impact of his work6.of the power of engineering to improve public health.

Dr. Bai largely accredits the ideas on how to correct the pulse oximetry to detect melanin to his students. He says that UNC has a talented student body, and because of this, he hopes to create a collaborative research environment for students in his lab where all ideas are valued.6
1. Shah, R. Elements That Keep Us Alive Also Give Color to Fireworks. https:// biobeat.nigms.nih.gov/2015/07/elementsthat-keep-us-alive-also-give-color-tofireworks/#:~:text=By%20mass%2C%20 about%2096%20percent,including%20 water%2C%20proteins%20and%20DNA (accessed February 12th, 2023).
2. What You Need to Know About Brain Oxygen Deprivation. https:// www.spinalcord.com/blog/whathappens-after-a-lack-of-oxygen-to-thebrain#:~:text=Without%20oxygen%2C%20 the%20brain’s%20cells,to%20power%20 the%20brain’s%20cells. (accessed February 24th, 2023).
3. Sugar and the Brain. https://hms.harvard. edu/news-events/publications-archive/ brain/sugarbrain#:~:text=Brain%20 functions%20such%20as%20 thinking,communication%20between%20 neurons%20breaks%20down (accessed February 24th, 2023).
4. CPR – adult and child after onset of puberty. https://medlineplus.gov/ency/ article/000013.htm#:~:text=Time%20is%20 very%20important%20when,4%20to%20 6%20minutes%20later (accessed February 12th, 2023).
5. Pulse Oximetry Basic Principles and Interpretation. https://medicine. uiowa.edu/iowaprotocols/pulseoximetry-basic-principles-andinterpretation#:~:text=Oxygenated%20 hemoglobin%20absorbs%20more%20 infrared,and%20absorbs%20more%20 red%20light (accessed February 12th, 2023).
“Dr. Bai [...] is essentially a middle ground between fundamental researchers and frontline healthcare workers.”


 By Lyssa Menendez
By Lyssa Menendez
The heart affects every part of our body, yet most people go about their day without its importance crossing their mind! It’s hard to imagine a lifestyle where one is cognizant of their heart throughout everyday life. However, for those with cardiovascular troubles, their heart’s capabilities are as common a thought as what they are going to eat for lunch that day. To help this disadvantaged group, Dr. Boyce Griffith, a professor of Mathematics and Biomedical Engineering (BME) at UNC, conducts research in nontraditional ways to improve current treatments for heart valve issues.
Dr. Griffith and his research team use biomedically engineered computational models to discover ways to improve existing medical devices (Figure 1). To understand why computational models are useful for this research, it’s important to understand the focus of Griffith’s study: valve disease. The heart operates in a specific pattern, with blood
flowing into the right atrium then into the right ventricle through the tricuspid valve. The heart then pumps the blood through the pulmonary valve and into the lungs, oxygenating the blood which will be used by the entire body! From there, it flows through the pulmonary veins into the left atrium, where the blood will be pumped through the mitral valve into the left ventricle. Finally, blood travels through the aortic valve to the rest of the body before returning to the right atrium.1 The four valves enable this system to thrive by preventing backflow and thus allowing a consistent flow of blood from right to left (Figure 2). Therefore, defective valves decrease the effectiveness of the entire system. Since the left side of the heart pumps blood to the entire body instead of just the lungs (which the right side does), it must withstand higher pressures, causing the mitral and aortic valves to have higher likelihood of developing problems.
There are many other reasons why these valves can begin to fail. For instance, contraction of rheumatic fever can cause valve issues in younger patients, and some individuals are even born with disorders like bicuspid aortic valve disease, which results in the aortic valve only having two leaflets, causing significant malfunctions in proper closing and blood flow regulation. Dr. Griffith specifically focuses on the valve issues in the elderly caused by typical aging. While defective valves may seem like a rare issue, about ten percent of people seventy years or older have some form of valve disease.²
A healthy valve can open well when necessary and stay tightly closed at all other times. The mitral and tricuspid valves even have cords that help them close tightly! When these cords become less effective, the leaflets become weaker and allow more irregularity with blood flow. This happens more commonly in elderly patients, putting them at a greater risk of not having blood constantly pumped to all parts of their body. To improve valve functionality in these patients, valves can be surgically repaired, but are often replaced by either a chemically-treated pig heart valve or a mechanical valve. Mechanical valves are mostly reliable but sometimes produce blood clots, forcing patients with artificial valves to take blood thinners called anticoagulants. Another option, pig valves or similar prosthetics, are better at preventing clots and do not require to take anticoagulants. However, these devices only last about ten to fifteen years. Given this trade-off, Dr. Griffith and his research team hope to use biomedically-engineered computational heart models to derive solutions for these design issues in valve replacements. Using several mathematical systems, Griffith’s team has been able to make digital models of the heart, which include valves and simulated blood flow. However, this system is very hard to replicate through medical imaging because closely approximated models provide a much more detailed view. Using

mathematics and imaging of anatomical features specific to each patient, useful models of the heart can be personalized. Dr. Griffith and his colleagues hope to make these models accurate enough in terms of blood flow and clotting to predict the effectiveness of varying medical devices for different patients.3 They also aim to determine characteristics of replacements that will make replacement valves more effective overall. Dr. Griffith recognizes that different placements of the valve replacements or new surgical methods may prove to be more reliable than others. Specifically, he focuses on how metal valve replacements are usually compressed extremely tightly in order to fit through a patient’s actual valve (Figure 3). There is some speculation that this squeezing may damage the artificial leaflets. While he says it is too early to tell, Dr. Griffith hopes to use his computational models to figure out how to replace valves without damaging parts of the heart or of the inserted device.
In addition to the quest to improve the design and practice of valve repair and replacement surgery, Dr. Griffith also seeks to increase the credibility of computational models. His team is working with researchers from the Food and Drug Administration (FDA) to improve their process of making computational models. Although the FDA has been reluctant to see these models as sufficient evidence for a variety of findings (ranging from evidence in support of a new drug to research on blood flow through faulty valves), Griffith believes that working together could help the FDA understand the validity and value of digital designs in medical fields. Therefore, Dr. Griffith hopes that his work will not only increase knowledge of heart repairment practices but also further validate the use of computational models in medicine, creating new possibilities for several other disciplines.
1. Cleveland Clinic. 4 Heart Valves: What They Are and How They Work. https://my.clevelandclinic.org/health/articles/17067-heart-valves (accessed Month Dateth, 2023).

2. Emery, Erin. After Age 65, 3 Things to Ask Your Doctor about Heart Valve Disease.https://www.uchealth.org/today/ heart-valve-disease-and-older-adults/ (accessed Month Dateth, 2023).
3. Interview with Boyce E. Griffith, Ph.D. 02/06/23.
“Dr. Griffith hopes that his work will not only increase knowledge of heart repairment practices but also further validate the use of computational models in medicine, creating new possibilities for several other disciplines.”
 By Mahika Nagaradona
By Mahika Nagaradona
When one thinks of drug delivery, the methods that come to mind are through swallowing, inhalation, or simply through injection. Now through new technology, a new method has surfaced in the form of intravaginal rings (IVRS). Though it may seem odd to imagine that medicine is able to enter your body through rings that are inserted in your vagina, this method is one that has already been used successfully for many years. It continues to slowly improve in the medical market through research and innovation. Traditional IVRs have many product limitations as a platform technology for women’s health because they are limited in terms of size, feel, and look; which causes drug delivery innovation to be limited as well. Here at The University of North Carolina at Chapel Hill, S. Rahima Benhabbour, MS. Ph.D., is an Associate Professor at the University of North Carolina at Chapel Hill-North Carolina State University Joint Department of Biomedical Engineering and the Founder and Director of AnelleO, Inc. She holds a different idea on how to make IVRs more efficient for drug delivery and personalized for all women. Could 3D printing technology be used to re-engineer rings in a way that just wasn’t possible with traditional injection modeling or extrusion methods? Inspired by her former mentor, Dr. Benhabbour has started to explore this idea through research and innovation in her lab.
Currently, the idea of using 3D printing to increase drug delivery efficiency through intravaginal rings has had major success. 3D printing allows the surface area of

the ring to be enhanced, which gives Dr. Benhabbour and her team the opportunity to experiment with how the drug will be released, how long the release will last, and the quality of the drugs in the rings.2 Traditional IVRs cannot give the flexibility in manufacturing needed to allow for adjustments. Another aspect of the new manufacturing methods comes from the way the drug is held inside the rings. Dr. Benhabbour and her team incorporate the drug post-fabrication, as it allows for a specific control on how they release the drugs, the dosage control, and how many drugs can be released at once. Not only has 3D printing created flexibility in manufacturing, but it has also increased efficiency. This allows for faster prototyping, which authorizes focus group studies and talking to the end users about their opinion on the final product. Talking to the prototype users can determine how they feel about the color, size, shape, or even the feel of the ring.1

The new manufacturing methods permit the changes of the prototypes in real-time without compromising the efficiency of the drug release. The most important part of this research to Dr. Benhabbour is to create a technology for women that supports what a woman needs. “As a woman engineer, I look at it as an opportunity to make an impact in a field that is not quite served by women who can actually engineer devices for unmet medical needs for women’s health,” said Dr. Benhabbour.1

The way IVRs works is that the medicine is embedded within the entire matrix of the ring, while the design inside of the rings are there to enhance the surface area and to streamline the diffusion of the drug throughout the ring. The designs that were put within the ring are in the form of incisions of different shapes and sizes on the ring itself.2 The designs would allow for easier diffusion of the drug to the vagina and control how much of the drug can be incorporated in the ring and how it diffuses out of the ring, through the enhancement of the surface area. The latest ring that Dr. Benhabbour is working on is called ‘Generation Four’ which has been re-engineered to hide the medicinal design, so it looks like a smooth ring. The drug is put within the ring and is not visible to the naked eye. The small incisions and lines within the ring are there to aid with the drug delivery and release process. Diffusion is the main mechanism that is used to release the drug from the matrix. The diffusion is controlled by the distance that the drug has to move out of the matrix. The interaction between the environment and the matrix is what allows for tweaking by changing the design within the ring. The injection molded rings are not able to do this process because the cross-section is solid and the drug will just diffuse until it can no longer diffuse into the environment. This is the reason the traditional IVRs are not able to release the entire drug load and can only release up to 45%, however, the

new technology can achieve up to a 100% drug release.1
Intravaginal rings are used for various different drug delivery methods ranging from HPV prevention to hormone injection. Dr. Benhabbour lab’s main focus is prevention of HIV, other STIs, and unplanned pregnancies. The lab is mainly developing the rings as a multipurpose prevention technology with two different multipurpose prevention technology (MPTs) that are ongoing at the moment which includes an anti-HIV and a contraceptive and the other is an anti-HIV, anti-herpes, and a contraceptive. Dr. Benhabbour (Founder & Director) and her coFounder, Rima Janusziewicz, of the start-up company, AnelleO (https://anelleo.com/) are using the 3D printing technology to develop IRVs that will be used for infertility treatment. In both her lab and at her start-up company, Dr. Benhabbour works with unmet needs in women’s health. “This is a platform technology that can be developed to address unmet medical needs where there is a sizable market by addressing gaps and/or limitations in the current standard of care, or to introduce a new treatment option for an unmet need,” said Dr. Benhhabour.1
The future of this research and IVRs is a very promising one. The next step for the IVR technology is clinical trials since the product is still undergoing testing. This is important because this is a platform technology and the drug incorporation is disassociated from the manufacturing process. Once this technology is approved, the sky is the limit and it sets the stage for the next innovations to happen quicker. Dr. Benhabbour emphasizes the importance of making an impact with the work she does. “The next step would be to partner up with a large pharmaceutical company with interest in women’s health, so we can advance this technology quicker.”1 The experiences and exposure that have been given to Dr. Benhabbour and her team have allowed her to continue innovating and researching different unmet needs that she believes would significantly improve the quality of health for women all
world.
References
 By Hari Patel
By Hari Patel
Diabetes is an autoimmune illness with life-altering conditions and far-reaching impacts on every aspect of a person’s life. As the 7th leading cause of death in the United States, this illness affects 34 million people in the country and can lead to a host of serious complications.1 Yet, despite its prevalence and effects, type 1 diabetes is often overlooked by the medical research community. Furthermore, patients with this disease are victims of health inequity, as there are disproportional rates of treatment and survival worldwide among various races, ethnicities, and ages.2 It is critical that researchers continue to explore how type 1 diabetes affects the body, as well as its underlying causes.
This disease primarily affects the pancreas but then spirals into a plethora of issues throughout the body. The pancreas is a gland located behind the stomach that plays an important role in regulating blood sugar levels. Normally, it releases insulin into the bloodstream in response to rising blood sugar levels, which helps transport glucose into the cells where it can be used for energy. With type 1 diabetes, the body’s immune system mistakenly attacks and destroys the insulin-producing beta cells in the pancreas, leading to hyperglycemia, or a spike in blood sugar levels, as there is no insulin to transport the glucose. These elevated levels of sugar can damage blood vessels, disrupting the oxygen supply to organs and leading to several illnesses such as heart attacks, strokes, and blindness. Though the exact cause of type 1 diabetes is unknown, the disease is strongly linked to factors such as genetics and viruses in the environment.3
While there are many important elements that come
into play when regulating this disease, including physical activity and hormonal balance, arguably the most important one is nutrition. As Dr. Elizabeth Mayer-Davis, Distinguished Professor of Nutrition and Medicine and Director of UNC’s Nutrition Obesity Research Center, puts it, “type 1 diabetes mainly has to do with disruptive energy metabolism,” or an inefficient use of energy in the body, strongly influenced by what food is digested.4 Because this disease is not prevalent in the literature and research, Dr. Davis, along with her colleagues from Stanford University and the Translational Research Institute for Metabolism and Diabetes, decided to form a team with the goal of discovering effective ways of managing the weight and blood glucose levels of patients with type 1 diabetes. Dr. Davis started her work as a registered dietitian and took an interest in patients with diabetes and the health equities associated with the disease. As a result, she went on to obtain a Master’s in Public Health and PhD in Epidemiology.
Complementing
Dr. Davis’s research on the prevalence of diabetes among young adults, the consortium was awarded 1.9 million dollars to continue research on diabetes and more effective ways of managing it. Their latest

research analyzes the most optimal diets for various people with type 1 diabetes regarding weight loss and decreased blood sugar levels. The three studied diets consisted of two hypocaloric plans and a Mediterranean based provision. The first was a low carbohydrate, hypocaloric diet with only 15% - 20% of calories coming from carbohydrates. In addition to this, less than 10% of the total calories could come from saturated fat, and at least 37% of the total calories had to come from monosaturated fat. The second diet was similar to the first, except that fat intake was limited to 30% of total calories consumed. The third diet differed from the rest, as it had alterations to the foods consumed but no caloric restriction: only healthy Mediterranean items were allowed for consumption, including olive oil, fruits, vegetables, fish, and legumes, and other foods such as butter, alcohol, and pastries were restricted from consumption. These three diets were able to be prescribed, as they are generally recommended by the American Diabetes Association and have worked in the past with patients facing type 2 diabetes.
In the study, each participant was analyzed to obtain information including race, weight, age, BMI, blood analysis, and health history. These measurements were made in an attempt find any correlations between these factors and a specific diet. After this, the participants were split into three groups, one for each nutrition plan. Every three months, the participants consulted with registered dietitians for a checkup. If a participant’s current plan was not working well in terms
of reducing weight and blood sugar levels, a swap to another diet was made. The study was planned to include nine months of data collection, but was cut short to three due to COVID-19 restrictions. Despite this interruption, the results were intriguing: it was found that whether the diet reduced calories or altered food consumption, participants found decreases in weight while maintaining or lowering blood sugar levels.5,6 These outcomes exemplified that making nutritional changes can be beneficial to patients with type 1 diabetes.
This study also served as a cornerstone in type 1 diabetes nutrition plans, as more research can be done to further examine and optimize diets for patients with this condition. Through further experimentation, researchers could analyze individuals responding to low-carbohydrate or low-fat diets in addition to exploring potential correlations between responses.
In conclusion, type 1 diabetes is overlooked in research, and Dr. Davis’s team has made tremendous contributions towards discovering optimal solutions for weight and blood sugar management. Diabetes rates are slowly increasing each year in the United States, and research like this is pivotal for millions of individuals. New research continues to show that it is important for people with diabetes to be physically active and to follow a healthy nutritional plan to handle the disease effectively.
1. CDC. “Diabetes Fast Facts”. https://www.cdc.gov/diabetes/ basics/quick-facts.html (accessed February 13th, 2023).
2. CDC. “Diabetes: Advancing Health Equity”. https:// www.cdc.gov/diabetes/health-equity/index.html (accessed February 13th, 2023).
3. CDC. “What is Type 1 Diabetes?”. https://www.cdc.gov/ diabetes/basics/what-is-type-1-diabetes.html#:~:text=If%20 you%20have%20type%201,builds%20up%20in%20the%20 bloodstream (accessed February 13th, 2023).
4. Interview with Dr. Elizabeth Mayer-Davis, MSPH, Ph.D, 02/09/23
5. Corbin, Karen, et al. “Design of the Advancing Care for Type 1 Diabetes and Obesity Network energy metabolism and Sequential Multiple Assignment Randomized Trial Nutrition Pilot Studies: An Integrated Approach to Develop Weight Management Solutions for Individuals with Type 1 Diabetes” 2022, 177, 2. https://doi.org/10.1016/j.cct.2022.106765
6. Igudesman, Daria, et al. “Weight Management in Young Adults with Type 1 Diabetes: The Advancing Care for Type 1 Diabetes and Obesity Network Sequential Multiple Assignment Randomized Trial Pilot Results” 2022, 35, 688699. https://doi-org.libproxy.lib.unc.edu/10.1111/dom.14911

"it was found that whether the diet reduced calories or altered food consumption, participants found decreases in weight while maintaining or lowering blood sugar levels."
 By Ria Patel
By Ria Patel
Addiction, numbness, overdose, respiratory depression, drowsiness, and nausea: the various side effects of opioids. Opioids are a class of drugs that produce morphine-like effects in humans, including the infamous drugs like heroin, fentanyl and oxymorphone. Ironically, as some of the best synthetic and semi-synthetic pain relievers, opioids are known to come with some of the worst side effects. Why is this? Scientists attribute this phenomenon to the lack of a strong, molecular understanding of the interplay between opioid small molecules and their receptors in the field of pharmacology. Opioid receptors have a high, distinctive selectivity for our body’s naturally occurring pain-relieving peptides (which are very short chains of amino acids). To mimic that exact function and structure to achieve optimal pain relief without any side effects seems to be quite impossible. Or at least, until now!
For an ambitious graduate student at UNC-Chapel Hill’s Department of Pharmacology in the School of Medicine, Dr. Jeff DiBerto, this so-called impossible problem was merely the beginning of his journey as a researcher. As an undergraduate student who attended UNC, Dr. DiBerto knew he had had a strong interest in drug actions but was unsure about his plans beyond university.1 With this interest, Dr. DiBerto approached Dr. CJ Malanga during his time in undergrad, who introduced him to the world of research and structural biology. More specifically, he focused on behavioral pharmacology, and including the muopioid receptor.1 After his successful experience, Dr. DiBerto went on to pursue a PhD in Pharmacology at UNC, where he was able to expand on his research interests in molecular pharmacology. He eventually helped publish detailed structures of the entire human opioid receptor family—a publication that could be used to help guide the creation of better pain medications.2
Dr. Diberto led this project under the UNC School of
Medicine lab of Bryan Roth and in collaboration with Dr. Eric Xu’s CAS Key Lab of Receptor Research in China. By using cryogenic electron microscopy (cryoEM) technology, the Xu lab provided Dr. DiBerto with a detailed view of the interactions between the delta-, kappa-, mu- and nociception opioid receptors and their respective peptides.3 The cryoEM technology revealed peptide-
Figure 1. Results of the biochemical assays for delta-, kappa-, and mu-opioid receptors. The top half shows how incompatibilities between peptides and receptors results in their subtype-selectivity. Immediately below that are binding curves, and finally below that are functional curves. Figure courtesy of Dr. DiBerto.

Figure 2. A graphical abstract of the results of this research. It shows the conserved message motif between all four opioid receptors and their unique extracellular recognition sequences. Additionally, it shows where the receptors can be found in the brain and their respective peptides. Figure courtesy of Wang et al.

bound opioid receptor structures in complex with their G protein effectors, or simply put, opioid receptors in their active states.2
Structures of the activated G protein-coupled opioid receptors, which were novel to the field, provided a tremendous wealth of knowledge to Dr. DiBerto as previous research on opioid receptors was only able to show receptors in either inactive or active-like states, with one exception being the mu-opioid receptor that had already been solved in complex with G protein bound to the semi-synthetic peptide, DAMGO.1
With the aforementioned structures, Dr. DiBerto was then able to perform successive structure-guided biochemical studies to understand the mechanisms that guide peptidereceptor selectivity and drug signaling.2 Dr. DiBerto and others in the Roth lab used the structures to design mutant, or artificially made, opioid receptors. These were then tested in cellular biochemical assays in cells to assess how they could alter receptor signaling. With the mutant receptors, Dr. DiBerto and others measured incompatibilities between peptides and receptors, binding curves that depict receptor and peptide interactions, and functional curves that measure the ability of the receptors to activate G proteins in response to peptide binding. Results of the experiments for the delta-, kappa-, and mu-opioid receptors can be seen in Figure 1.1
Collaboration between Dr. DiBerto, the Xu Lab and the Roth Lab was able to reveal various conserved mechanisms of activation and recognition for all four opioid receptors as well as key differences in peptide recognition that could be used to create receptor-specific drugs. For starters, the conserved message motif “YGGF” on the N-terminal (beginning) of peptides was found to drive receptor activation.3 Furthermore, sequential variations at the C-terminal (end) of opioid peptides were found
to aid in receptor selectivity. In addition, extracellular loops on the receptors themselves were found to be critical for the selective recognition and filtering of specific opioid peptides (Figure 2).
By providing such results, Dr. DiBerto and his colleagues hope to inspire the creation of more specifically structured opioids that can bind to receptors in the same way our naturally made peptides do. The current goal is to effectively stabilize and synthesize opioids which have the ability to bind perfectly to receptors to elicit pain relief via specific transduction pathways, all while avoiding triggering mechanisms that lead to severe side effects and overdosing. Thanks to Dr. DiBerto, we are now one step closer to developing safer opioids!
1. Interview with Jeff DiBerto, Ph.D. 01/30/2023

2. Derewicz, M. (2023, January 12). Scientists take another step toward creating better pain medications. Newsroom. Retrieved February 13, 2023, from https://news. unchealthcare.org/2023/01/scientists-take-another-steptoward-creating-better-pain-medications
3. Wang, Y., Zhuang, Y., DiBerto, J. F., Zhou, X. E., Schmitz, G. P., Yuan, Q., Jain, M. K., Liu, W., Melcher, K., Jiang, Y., Roth, B. L., & Xu, H. E. (2023). Structures of the entire human opioid receptor family. Cell, 186(2). https://doi.org/10.1016/j. cell.2022.12.026
“The current goal is to effectively stabilize and synthesize opioids [...] while avoiding triggering mechanisms that lead to severe side effects and overdosing.”

 By Anooshka Deshpande
By Anooshka Deshpande
Substance use disorders claim about 11.8 million lives worldwide every year.1 They are characterized by persistent drug use despite social, health, and behavioral consequences.
Teenagers and young adults in their twenties are most vulnerable to developing them. Additionally, certain individuals can be genetically predisposed to substance use disorders if their parents were diagnosed with them. Dr. McElligott, an assistant professor in psychiatry at UNC-Chapel Hill, investigates the development and manifestation of psychiatric illnesses. Her overarching goal is to improve human health and develop treatments for these illnesses by studying how environmental factors, such as chronic stress and drug exposure, can modify neurobiological pathways in the brain.
The brain contains billions of nerve cells called neurons. Neurons that perform a certain function are grouped together in clusters called nuclei. Although nuclei are localized to specific regions of the brain, their neurons can extend up to several feet in length and contact other neurons that perform a different function. The path that a neuron takes to a different region of the brain is called its projection. Thus, when neurons with different functions project onto each other and release various messengers, they can yield a variety of behavioral effects.
Neurons communicate with each other by releasing two types of chemical messengers - neurotransmitters and neuropeptides - that govern our thoughts, feelings, and behavior (Figure 1). They can be excitatory, inhibitory, or both. As their name implies, excitatory messengers tend to excite a nucleus, causing the neuron to fire and spread its message to adjacent neurons. On the other hand, inhibitory messengers tend to
prevent the neuron from firing and spreading its message. Dr. McElligott’s lab studies the expression of a neuropeptide called neurotensin (Figure 3). Current research has established that the central amygdala - a brain structure that is activated when we learn to fear an object or situation - is involved in developing addictions and substance abuse. One of her lab’s objectives is to identify the specific types of neurons in the central amygdala and their projections that contribute to substance abuse. In order to study this, the lab used a technique called optogenetics. In this technique, a light-sensitive protein called opsin is inserted into a neuron. Then, blue light is shone onto the neuron, causing it to fire (Figure 2). This enables scientists to observe the neuron’s projection path and its effect on behavior.
Figure 1. Neurons communicate with each other by releasing chemical messengers. Top neuron communicates with bottom neuron by releasing neurotransmitters and neuropeptides that bind to receptors on bottom neuron’s membrane. Figure from Wikimedia Commons.

The lab inserted a gene that codes for a protein called channelrhodopsin, a type of opsin, into neurons residing in the central amygdala. Channelrhodopsin causes these neurons to fire in the presence of blue light. The lab discovered that a group of neurons in the central amygdala express a neuropeptide called neurotensin. These neurotensin-expressing neurons then send inhibitory signals to the parabrachial nucleus, an evolutionarily old part of the brain that is stimulated in life-threatening situations. In order to study this newly discovered pathway projecting from the central amygdala to the parabrachial nucleus in mice, the lab created a situation in which mice could self-stimulate their central amygdala. Mice were placed in a box that contained two compartments. Whenever they would enter the second compartment, a computer would turn on a laser that would shine light onto neurotensin-expressing neurons, leading to inhibition of the parabrachial nucleus. The lab observed that mice spent more time in the second compartment. They found that optogenetic stimulation of this pathway conferred positive valence, or reward-like behavior.2 This implies that the mice wanted there to be minimal activity in their parabrachial nucleus.


Next, the lab observed whether the mice were willing to perform an operation in which they could completely inhibit their parabrachial nucleus. Mice were placed in another box that contained two holes. Whenever they poked their nose through one hole, an infra-red beam would optogenetically stimulate the pathway of neurotensin-expressing neurons projecting from the central amygdala to the parabrachial nucleus. The other hole served as a control where no stimulation occurred. The lab encouraged the mice to spend time in the control by putting a fruit loop around the hole. The lab found that mice still learned and spent more time poking their nose in the hole that optogenetically stimulated the pathway of
neurotensin-expressing neurons from the central amygdala to the parabrachial nucleus. This was an important discovery that could elucidate the neural circuits responsible for substance abuse in humans. A question that Dr. McElligott hopes to follow up on in this study is why some mice learned to poke their nose faster than others in the optogenetically stimulating hole. She is also curious whether mice would work extra hard to stimulate this pathway while experiencing drug withdrawal.3
Initially, the opioid epidemic sparked Dr. McElligott’s curiosity about substance abuse. Decades ago, street drugs did not contain the lethal opioid fentanyl. These days, however, most street drugs contain a fentanyl analog, which consumers unknowingly ingest. She states that “the changing landscape has made us think of how we need to change our approaches in the lab to address the crisis in various ways.”3 Dr. McElligott has faced numerous successes and obstacles in her research career. Nearly all of the research grants she applied for at the start of her career were rejected. Her perseverance carried her through this obstacle. She received an important research grant in the summer of 2020 which she states was “a cornerstone to her success.”3
Deaths from substance abuse are surging in regions where drugs are easily accessible. However, we can curb deaths by developing drugs targeting neural circuits that drive substance abuse, which Dr. McElligott hopes to accomplish. Wouldn’t it be incredible if future generations experience lower mortality rates from drug addictions? Dr. McElligott strives to make this a reality.
1. Drug Use. Our World In Data, https://ourworldindata.org/ drug-use.
Figure 2. Optogenetics - A technique that scientists use to examine a neuron’s function. A gene that codes for a lightsensitive protein called channelrhodopsin is added to a neuron’s DNA sequence. When light is shone onto the neuron, the neuron is activated and performs its function. Figure from Wikimedia Commons
2. Manipulation of central amygdala neurotensin neurons alters alcohol consumption. Maria Luisa Torruella-Suarez, Jessica R Vandenberg, Gregory J Tipton, Brennon R Luster, Kedar Dange, Gunjan K Patel, Jenna A McHenry, J. Andrew Hardaway, Pranish A Kantak, Nicole A Crowley, Jeffrey F DiBerto, Sara P Faccidomo, Clyde W Hodge, Garret D Stuber, Zoe A McElligott.
3. Interview with Zoe McElligott, Ph.D., 01/16/2023.
“This was an important discovery that could elucidate the neural circuits responsible for substance abuse in humans.”
 By Yifei Pei
By Yifei Pei
Suicidal thoughts and behaviors are more common amongst adolescents than most may assume. Suicide was the second-leading cause of death among those aged 10-24 in 2020, and many more adolescents have suicidal thoughts or attempt suicide.1 If researchers can predict causes of depression and suicidal thoughts/behaviors, they can make a difference by reducing the risks.

Funded by the National Institute of Mental Health, Dr. Girdler is working on a longitudinal, observational study designed to look at the predictors of suicidal thinking and behaviors, as well as other mood symptoms among female adolescents. The objective of this project, according to Dr. Girdler, is to address an urgent public health problem that has increased dramatically over the past year—teenage suicide attempts.2 Through a recent meta-analysis, Dr. Girdler and her team found that suicidal thoughts and behaviors (STBs) exponentially rise during the adolescent transition (Figure 1).1

In this study, Dr. Girdler and her team investigate how exposure to stress and fluctuations in estrogen levels over a period of two months may predict mood disorders and SBTs in pre-pubescent (or preadolescent) girls over a 4-month followup period. The study only includes female adolescents as it aims
to investigate the relationship between hormonal fluctuation brought by periods and changes in mood, which has been confirmed in older women.
“We have already known for decades,” says Dr. Girdler, “that stress is an independent predictor of depression and suicidality.”2 The uniqueness of the study is that it accounts for Dr.
hormonal sensitivity when investigating the effect of stress exposure (Figure 2). “The majority of us do not show depression or suicidal thoughts within a menstrual cycle, as triggered by hormonal change,” Dr. Girdler pointed out. “But for the percentages of females who do have a hormone sensitivity profile and have been exposed to stress, interventions may also include hormonal aspects that could be implemented.”2 Research on older women found that perimenopausal women have similar hormonal fluctuation as peripubertal girls. During transitional phases such as puberty or menopause, women go through hormonal changes that are not observed during other phases of the reproductive life cycle. Physiological methods such as progesterone to stabilize the estrogen level among older women are taken in some extreme cases. These methods have been shown to be highly effective at preventing depressant activities in a number of randomized control trials conducted using animals.3 Therefore, hormone stabilizers may be effective in certain high-risk and vulnerable adolescent females.
Every day for 70 days, adolescent female participants complete surveys on their smartphones. “Have you thought about killing yourself today?”, “What has been your urge to do so?”, and “What is the level that you feel you can resist this urge?” are among key questions asked in surveys. A urine sample is required on an everyday basis, from which estrogen and progesterone levels are measured to quantify the association between hormonal fluctuation and mood. “This study is very statistically powerful in measuring hormone sensitivity—how sensitive are girls in terms of mood changes and suicidality changes with that hormone changes,” said Dr. Girdler.2
The research process is difficult and sometimes heartbreaking for researchers and study clinicians. In the early stages of the project, the clinician was frequently contacting the kids after detecting mood changes. Questions like “It looks like your mood has worsened today compared to yesterday, could you tell me what is going on?” were asked, and this intervention prevented accurate reporting of suicidality by the participant. “We are changing their behaviors by reaching out to them,” according to Dr. Girdler, “but this is supposed to be an observational study.” In light of this, they have set very strict rules regarding contact, making it explicit to participants that immediate intervention will occur only if a direct threat to their life is evident. “We just have to accept the fact that we cannot and should not help,” said Dr. Girdler, as “the goal of the study, ultimately, is to develop interventions for future research.”2 By not intervening, the study will help more kids in the future.
If the results of this study are significant, changes in estrogen levels can predict worsening mood before the onset of clinical depression and self-harm. “If we know what the antecedent predictors are,” Dr. Girdler clarifies, “we have targets for intervention.” There can be behavioral interventions that help kids cope with stressors like school rejection. For girls who show high sensitivity to hormones, hormone stabilizers can be used to reduce associated mental health risks.
The study uses ecological momentary assessment (EMAs), which is a survey of 25 questions detailing feelings toward suicidal ideations in an individual’s normal environment.
1. America’s Health Rankings. Public Health Impact: Teen Suicide. https://www.americashealthrankings.org/explore/ health-of-women-and-children/measure/teen_suicide (accessed February 13, 2023).
2. Interview with Susan Girdler, Ph.D. 02/02/23
3. Frye C.A. J Psychopharmacol 2010, 25(3), 421–428.
“If the results of this study are significant, changes in estrogen levels can predict worsening mood before the onset of clinical depression and self-harm.”
 By Matthew Rodzen
By Matthew Rodzen
Substance use disorder (SUD) is a complex public health crisis that affects millions of individuals and communities world-wide. The consequences of SUD can be devastating, hindering personal growth and wellbeing, making it significant to explore and develop innovative solutions for those struggling with SUD. Dr. Daughters, a substance use disorder researcher, is dedicated to studying the nature of SUD and developing effective treatments.
Driven by a passion for psychology research, Dr. Daughters has committed to finding new ways to tackle the complex issue of SUD. One of her most recent projects is the development of the Life Enhancement Treatment for Substance Use (LETS ACT) treatment, which is a smartphone app designed to help individuals with SUD develop new, substance-free rewards in their lives (Figure 1).1 In the words of Dr. Daughters: “LETS ACT aims to help the individual rekindle or develop new substance free rewards in their life and then not only figure out what they are, but engage in them each day. So, there’s positive reinforcement each day for something other than the substance use. The idea being to tip that balance of reward for the substance down and the non-substances up, so it starts to even.”1

The app is an effective tool for individuals to use outside of traditional therapy sessions, making treatment more accessible and affordable. Dr. Daughters aims to provide an effective treatment option for individuals struggling with SUD who may not have access to traditional therapy sessions.1
Dr. Daughters’ research also includes studying the effects of brain stimulation on SUD, which aims to improve oscillation frequencies in the brain.1 Brain stimulation can be used to enhance the effects of therapy sessions, making it a useful addition to traditional treatment options (Figure 2).1 Dr. Daughters envisions a future where her treatment options, including brain stimulation and the LETS ACT app, can be used in combination with therapy sessions to address the multifaceted nature of SUD.1
However, SUD research faces significant challenges, including the need for precision medicine. Personalized treatment, based on an individual’s response to triggers and

cues in the environment, is essential to tailor treatment to the individual. Dr. Daughters sheds light on the current situation: “What happens now is that people with a substance use disorder are treated with one treatment. It’s a one size fits all approach and what we’re trying to do is improve precision medicine where someone comes in and gets an assessment so that we know that maybe this person is having such a hard time recovering because of their response to triggers and cues in the environment that remind them of substance use. And so if that’s the case, then we can provide X treatment for that.”1 Despite the challenges, Dr. Daughters’ work with patients has been rewarding and she emphasizes the importance of continued research and innovation in the field.1
and affordable treatment options for individuals struggling with SUD. Precision medicine and neuroimaging studies are also crucial in developing effective treatment options for SUD patients. Continued research and innovation in the field of SUD are imperative in providing hope for millions of people worldwide struggling with the disorder.


With continued innovation and new discoveries, SUD research has the potential to make a significant impact on public health. Dr. Daughters is determined to find innovative solutions to help individuals break the chains of SUD. The work being done by Dr. Daughters and her peers is invaluable, providing hope for millions of people worldwide who struggle with SUD. Neuroimaging studies also play a critical role in understanding how treatments affect physical structures and pathways in the brain. By conducting these studies, Dr. Daughters and other researchers can develop more effective treatment options for SUD patients (Figure 3).
It is crucial to recognize that SUD is not only a medical issue but also a social issue that affects individuals and communities.2 The consequences of SUD are far-reaching, affecting not only the individual but also their family, friends, and society as a whole. Finding effective treatments for SUD is vital in mitigating these consequences and improving public health outcomes.2 Furthermore, SUD research requires a multifaceted approach, as it is a complex disorder that requires treatment from various angles.
Dr. Daughters’ innovative research into SUD provides hope in the ongoing fight against this debilitating disorder. Her work on the LETS ACT app and brain stimulation highlights the potential for technology to provide accessible
References
1. Interview with Stacey B. Daughters, Ph.D. 2/14/23
2. Daley DC. Family and social aspects of substance use disorders and treatment. J Food Drug Anal. 2013, 21(4):S73-S76.
“It’s a one size fits all approach”
If you have ever felt a little moodier when you are hungry, you are not alone! The Carolina Affective Science Lab, directed by Dr. Kristin Lindquist, researches how hunger is associated with the emotion of anger, causing the infamous phenomenon of “hanger.”

Dr. Kristen Lindquist is a professor and researcher in the department of Psychology and Neuroscience at UNC-Chapel Hill. Dr. Lindquist received her bachelor’s degree and Ph.D. in Psychology from Boston College. She continued her postdoctoral fellowship at Harvard University as a part of their Mind/Brain/ Behavior Initiative.2 Her current research focuses primarily on how bodily sensations, brain functions, and surrounding experiences affect emotion.
As an undergraduate, Dr. Lindquist fell in love with the study of human emotion because she was fascinated by how the brain and body’s connection forms unique emotional experiences. One of her more recent studies, covered in her published paper, “Feeling hangry? When hunger is conceptualized as emotion,” explains why people emotionally react in certain ways based on their bodily sensations.3

To begin this investigation, Dr. Lindquist and her research team conducted a series of studies examining the effect of hunger. . The first was a correlation study, meaning that she observed both hunger and emotion without experimentally controlling these variables. First, participants filled out an online
survey rating their levels of hunger. Next, they were shown various images yet were told to ignore them. Finally, participants were presented with ambiguous stimuli; for example, if someone didn’t speak Mandarin, they were shown a Mandarin symbol and asked to indicate whether the symbol was positive or negative. Interestingly, they found a correlation between negativity and hunger. Those who reported they were hungrier were more likely to assume the ambiguous stimuli were negative if primed with a negative image. Notably, there was no effect on positive stimuli.1
The correlation
motivated Dr. Lindquist to design a causation study by placing undergraduate participants in a controlled social setting. There were two conditions with two different groups. Dr. Lindquist asked one group to fast for five hours preceding the experiment and instructed another group to eat within an hour of the study as a control. Dr. Kristin Lindquist
One condition was presented with the task of telling a story given a face that was either angry, sad, or neutral, which acted as a way for participants to be aware of their emotions. Next, all participants were asked to do a frustrating mental task of counting concentric circles for an extended period. Next, the computer fake crashed, and the research assistant would imply it was the participant’s fault. The assistant told the participant they may have to redo the circle task but, in the meantime, to fill out a quality survey while they talk to their supervisor.
Dr. Lindquist predicted that people who were hungry and unaware of emotional feelings because they did not complete the face-story task, would misattribute their hunger to the world around them. From studying 236 individuals, the lab found that interestingly, people who were feeling hungry and not paying attention to their emotions were more likely to rate their experience as unpleasant and even punish the assistant by rating them negatively.1 For example, participants were more likely to say that the researchers were judgmental and not empathetic. In addition, participants were able to better regulate their emotions when they paid better attention to their feelings.1

The studies by Dr. Lindquist were the first form of evidence to suggest that purely metabolic sensations and processes contribute to how people experience the social
world. Dr. Lindquist’s findings have significant implications for emotional regulation in conditions of famine and food scarcity. Additionally, results suggest that when human metabolic needs are not met, it negatively impacts mental wellness as well as physical health.1
Dr. Lindquist reminds her students: “Part of mental wellbeing and being cognitively on top of things is actually taking care of the metabolic states of your body, which means eating well, getting enough rest, and doing the things that are essential to having a physically healthy life.”1 Commonly, college students forget to eat healthy food and get enough rest, especially during exam seasons, which can impact how people experience their day-to-day lives.
Currently, Dr. Lindquist is following up on how individual sensitivities may interact with emotions. Her current study is expanding on more physiological measures such as heart rate and gastric sensations.1 While these studies will potentially be more difficult to quantify, Dr. Lindquist suggests they will be able to contribute to the findings of how body sensations affect emotions and experiences.
1. Interview with Kristen Lindquist, PhD. 01/26/2023
2. About. Kristen A. Lindquist. (n.d.). Retrieved February 13, 2023, from http://www.kristenalindquist.com/about
3. MacCormack, J. K., & Lindquist, K. A. (2019). Feeling hangry? When hunger is conceptualized as emotion. Emotion (Washington, D.C.), 19(2), 301–319. https://doi.org/10.1037/ emo0000422

 By Natalie Travis
By Natalie Travis
Anew way of learning how to parent is arising in the world of psychology—practicing with Virtual Reality. This is the focus of Dr. Andrea Hussong’s research to optimize VR technology as a tool for improving emotional regulation in children. Dr. Hussong first discovered the pairing of psychology and VR when she came upon a museum exhibit that was discussing how the technology could be used to evoke emotional responses. She began to consider how VR can be used to develop a parenting intervention program. This can be especially useful for parents struggling with substance use disorders to help them improve not only their own emotional regulation but also their children’s. Emotional regulation is often called coping skills and describes the way people deal with difficult feelings.2 There is a pattern in which some children of parents struggling with substance abuse are more likely to experience neglect, house displacements, psychological disorders, and poor social skills— all of which can negatively affect the development of healthy emotional regulation. Dr. Hussong recognized that traditional treatment faculties could do more to prevent relapse for the parents and often fail to reduce child mistreatment. Children can continue to experience neglect after a parent’s treatment, further contributing to problems regarding emotional regulation. This cycle can even increase the children’s risk of developing their own substance problems and addictions later in life. The goal of Dr. Hussong’s research is to help parents struggling with substance use disorders to improve emotional regulation in their children by practicing coping with difficult situations
through VR simulations of a child having a tantrum.
The research behind Dr. Hussong’s advances in VR technology suggests that parents must be able to regulate their own emotions in order to improve their children’s emotional regulation. Like many who are victims of substance abuse, these recovering parents struggle with their own emotional regulation and have high emotions that are “slow to recover.”1 This makes it difficult for these parents to control their emotions and meet their goals. This lack of emotional control becomes an issue when parents need to calm their child down in a moment of misbehavior or tantrum. Tantrums, especially in public, are highly stressful for both the child and the parent. In those stressful times, the parent needs to be able to regulate


their own emotions so they can help the child calm down. Dr. Hussong suggests that practicing with VR could help them be prepared to deal with these situations because children learn how to regulate their emotions by following models set by their parents. The child is then more likely to carry these struggles into adulthood and follow their parent’s path of addiction. This new technology has the potential to help parents regain control over their emotions during stressful times so they can in turn act as a positive model for their children. Dr. Hussong aims to stop substance use disorders proactively by teaching parents to aid in their children’s emotional regulation early on.


Dr. Hussong’s research requires input from other fields, such as social work and computer science, and she often collaborates with community treatment centers for women struggling with substance use disorders. All the women included in the study have children aged 2 to 6, some with behavioral issues. Many of the women are attempting to regain custody of their children and this research may improve their parenting abilities, specifically in regulating both their emotions and their children’s, in order to increase their potential to be seen as fit parents. Dr. Hussong also works with a private tech company called Virtually Better that combines VR technology and psychological interventions, allowing her to translate her ideas into tangible tools for helping those in need.
Many challenges face Dr. Hussong and her team as they explore how to use VR to evoke emotional responses. Her first exploration in combining technology with psychology included large wristbands with digital screens that would ask participants survey questions. Developing this technology was difficult and more complications have come along with the use of sophisticated VR. While the objective of this intervention is to produce a simulation of a child having a tantrum that will evoke a stress response out of the parent, physiological differences between the simulated child and their own, have made it difficult to generate a relatable simulation for every parent. Dr. Hussong is also evaluating evidence that will indicate how to program the VR for each specific parent. If results show that a stronger stress response is evoked from a child that looks like their own, her team would need to hire many child actors of different races, ethnicities, and genders. The easiest option to solve this problem would seem to be using animated children so that parents can make a relatable avatar of their child. However, animated children have been ineffective thus far in evoking the proper emotional responses. Some of the options to portray a child having a tantrum that were tested include a 2D video, 3D animation, and a 3D video with a child actor. Based on Dr. Hussong’s research, the 3D video in the headset evokes the strongest response but obtaining child actors is difficult and
expensive. Adapting to these challenges is Dr. Hussong’s focus of inquiry before further applying VR technology to improve emotional regulation in children. There is much work to be done and Dr. Hussong states that “We’re not there yet, we’re still in the middle of it all.”1 She views the most exciting part of her research as being able to look at people’s entire lifespan and determine the best time to intervene. This is clearly seen in her work, with VR technology as an early intervention being the most recent addition. Dr. Hussong aims to stop substance use disorders at what her research shows to be the source: emotional regulation problems in childhood. Her research in using VR technology to allow parents to practice regulating their own emotions during stressful times of child misbehavior is promising and has the potential to reduce children’s risk of developing substance use disorders.
1. Interview with Andrea Hussong, Ph.D. 2/13/2023
2. Pennequin, V., Questel, F., Delaville, E., Delugre, M., & Maintenant, C. (2020). Metacognition and emotional regulation in children from 8 to 12 years old. British Journal of Educational Psychology, 90(Suppl 1), 1–16. https://doi. org/10.1111/bjep.12305
“[Dr. Hussong] views the most exciting part of her research as being able to look at people’s entire lifespan and determine the best time to intervene.”

 By Ashley Villanueva
By Ashley Villanueva
Social barriers, impulsive behaviors, learning disabilities, and outbursts are common characteristics of autism spectrum disorder (ASD). The traits can prove to be challenging for people diagnosed with ASD and their loved ones who want to make sure that they are treated properly. Caring for those with disabilities often takes guidance, a willingness to adapt, and genuine care from a nurturing community. Many say, “it takes a village”, and this includes a community of neuroscientists, who are currently using their intellectual resources to support these individuals through bridging scientific gaps.
During her postdoctoral fellowship at Stanford University, Dr.
Jessica Walsh, Assistant Professor in Pharmacology and Principal Investigator of the Walsh Lab at UNCChapel Hill made crucial observations that have fueled her current research directions. In addition to her general science research, Dr. Walsh worked with a clinical psychologist as an executive function coach to help adolescents with behavioral deficits learn strategies for achieving optimal success in an academic and social environment. “I am very committed to outreach and personally feel that having exposure to the clinical population can really inform a basic scientist who studies neurodevelopmental and psychiatric disorders,” Dr. Walsh shares, “I have found it invaluable for both directing my
scientific questions and reminding me of why I have chosen this career path. I hope that one day our research can help those in need.”1 Some of this research includes investigating the neural basis
of motivated behavior focusing on neurodevelopmental disorders.1
The identification of neural circuits pertaining to motivated behavior is a daunting task in any brain. Researchers use mouse models as they have similar structural and genetic characteristics to human brains, which allows scientists to probe circuits and create models for human disorders. In one of Dr. Walsh’s experiments assessing social behavior, a test mouse, which harbors a genetic deletion implicated in ASD, is placed in a 3-chamber apparatus and allowed to roam freely for 20 minutes. In one chamber, there is a container where a novel juvenile mouse is placed (social side). In the second chamber, the same container is present, but empty, serving as a novel object (empty side). To assess sociability in mice, Dr. Walsh’s team measures how much time the mouse spent in the chamber with the social target versus time spent in the empty chamber with the novel object. Mice are social creatures by nature and typically will spend more time in the social chamber. Interestingly, Dr. Walsh’s team has demonstrated that mice with distinct genetic mutations spend significantly less time in the social chamber, therefore, exhibiting social deficits similar to those seen in humans diagnosed with ASD.2 But what brain systems are associated with these findings?
The serotonin system plays a critical role in social behavior. Interestingly, dysfunction of this system has been implicated in a host of neurodevelopmental disorders, including ASD. It is not surprising that selective serotonin reuptake inhibitors (SSRIs), which block the reabsorption of serotonin, are commonly prescribed to individuals diagnosed with ASD. However, they do not aid in the relief of the core symptoms, such as social deficits and stereotyped behavior, and have variable efficacy as well as negative side effects.1 The overall goal of the Walsh Lab is to use their science
findings to aid in the development of more targeted therapies tailored to discrete behavioral impairments.
To begin to isolate which brain regions and cell types underlie the behavioral deficits present in mouse models for ASD, the Walsh Lab utilizes whole brain clearing technology and light-sheet microscopy. Their methods provide whole brain images at cellular resolution to measure c-Fos expression, a proxy marker for neuronal activity. ClearMap software then generates maps of altered brain activity for recognition of high-probability target regions. Once researchers identify specific brain regions, they selectively modulate circuits using techniques such as optogenetics and chemogenetics, aka DREADDS. Additionally, using in vivo calcium imaging with fiber photometry, they can measure changes in the population activity of specific cell types. It allows them to observe behaviorally relevant brain activity during discrete bouts of social interaction in freely moving mice.1 The Walsh Lab also utilizes slice electrophysiology to determine the physiological and receptor mechanisms responsible for the social deficits present in distinct mouse models for ASD. Their findings can be later utilized by the pharmaceutical industry for the creation of drugs that target the appropriate ion channels. Although Dr. Walsh and her team have made promising strides in understanding the brain mechanisms of ASD, they face many challenges. While utilizing mice as a model system has many advantages, it can be challenging to properly design behavioral experiments that truly model a human condition. Working with mice themselves proves to be a challenge, as they need to be controlled in the most rigorous ways to ensure reliable experimentation. Additionally, preparation of the mice for optogenetic or fiber photometry experiments can require up to 6 weeks of post-surgery to begin experiments.
Dr. Jessica WalshHowever, the scientific community does not work alone. The brain itself is the most dynamic organ, with branches of science in all directions collaborating to unpack the mechanisms behind human cognition, behavior, and actions. In many ways, this reflects the movements we must embody in learning how to best love and care for others around us who may not share the same neural patterns or responses. As Dr. Jessica Walsh shares, “The greatest discoveries are made in a team”,1 or as many say, “it takes a village.”
1. Interview with Jessica Walsh, Ph.D. 1/23/23


2. Systemic enhancement of serotonin signaling reverses social deficits in multiple mouse models for ASD
https://www-nature-com.libproxy.lib. unc.edu/articles/s41386-021-01091-6 (accessed February 14, 2023)
3.5-HT release in nucleus accumbens rescues social deficits in mouse autism model https://www.nature.com/ articles/s41586-018-0416-4 (accessed March 17, 2023)
“Dysfunction of [the seretonin system] has been implicated in a host of neurodevelopmental disorders, including ASD.”

 By Layla Williams
By Layla Williams
For the next ten seconds, whatever you do, do not think about a pink elephant. Now, I am willing to bet that the first image that crossed your mind was none other than a pink elephant. A thought becomes more intrusive the harder you try to suppress it. As paradoxical as it may seem, research shows that the best way to overcome unwanted thoughts and fears is to face them headon. Obsessive-compulsive disorder (OCD) affects up to 3% of the population and involves unwanted thoughts and fears that lead to compulsive behaviors.1 Exposure and response prevention, also
known as exposure therapy, is a form of treatment for OCD that exposes people to circumstances that provoke their obsessions and the associated distress.2
This has been proven to be an effective method of preventing compulsive responses and improving symptoms of OCD, although improvement rates vary. Therefore, it is important to determine potential predictors of positive treatment outcomes. Dr. Jonathan S. Abramowitz, a professor in the UNC-Chapel Hill Department of Psychology and Neuroscience and an OCD expert, made it his mission to do just that.
Most people probably know someone with OCD, as it is common. Consequently, the general population should be familiar with obsessivecompulsive disorder and treatment methods. “People [with OCD] have fears of everyday things and situations, and then they engage in excessive compulsive rituals that seem like they’d work to alleviate the fears, but they end up making the fears even more intense.”3 There are various types of mental health treatments out there, but as Dr. Abramowitz put it, “most of them are snake oil and placebos.”3 Exposure therapy, on the other hand, is fairly straightforward and has valid,
compelling scientific data that has proven this treatment is extremely effective. Although individual results may differ, people who undergo exposure therapy and commit to the treatment process tend to experience improvements in symptoms. The latest research suggests that around 60% of people who complete exposure therapy see a significant reduction in their symptoms. Reports show that people may stop avoidance behaviors and reduce ritualistic behaviors and may report that they are less afraid of the situations and thoughts that once triggered their compulsive actions.3
Witnessing these successes firsthand is immensely rewarding. Some of Dr. Abramowitz’s earliest experiences in graduate school involved working with people with OCD, using exposure therapy to help them move on with their lives. When he saw how well exposure therapy worked, he knew that he wanted OCD treatment to be the focus of his research and professional career. In Dr. Abramowitz’s lab, the research team studies the predictors of improvement with exposure, as well as ways to optimize the learning that takes place when people utilize the therapy. The researchers work to determine different
ways to vary the delivery of exposure and response prevention to optimize fear extinction.3 A recent study conducted by the team investigated how effectively participants completed their exposure therapy assignments as a predictor of treatment response for all OCD symptom dimensions.
The participants involved in this study were fifty adults between the ages of eighteen and fifty-six, all of whom received a diagnosis of OCD. The participants completed sixteen two-hour sessions of individualized treatment that occurred twice a week at either the University of North Carolina at Chapel Hill or Utah State University. The participants were randomly assigned to receive either traditional exposure and response prevention therapy or exposure, response, prevention, acceptance, and commitment therapy. The latter is an integration treatment in which clients learn to stop avoiding their deeper emotions and recognize that their responses are normal reactions to certain situations that should not stand in the way of them moving on in life.4 Researchers advised participants to avoid engaging in compulsive rituals in between sessions, and prescribed them daily exposure
homework tasks for practice. Typically, the homework involved independently repeating the exposure from that day in various settings. Participants utilized the Yale-Brown Obsessive Compulsive Scale, a standardized rating scale that provides a specific measure of the severity of symptoms of OCD.Self-report measures occurred during a pretest, post-treatment, and a 6-month followup. OCD and depressive symptoms were significantly reduced immediately following treatment and remained at this level when retested at follow-up.5 These findings will be fundamental for future research to better understand the extent to which adherence to exposure therapy results in OCD symptom reduction.
Dr. Abramowitz continues to further the research surrounding OCD and exposure therapy while also preparing a new generation of future experts in the field to carry on this important work. “Given the emerging body of consistent evidence that adherence with [exposure and response prevention therapy] is predictive of changes in OCD symptoms,”5 there is optimism and anticipation for fruitful advancements in the near future. In combination with these factors, the availability of new technology is sure to
lead to “more substantial and durable improvements for individuals with OCD.”5 The groundbreaking work achieved by scholars like Dr. Abramowitz is leading us closer to the discoveries necessary to improve mental health in our society.
1. Mayo Clinic Staff. Obsessivecompulsive disorder (OCD). https:// www.mayoclinic.org/diseasesconditions/obsessive-compulsivedisorder/symptom s-causes/syc20354432 (accessed October 8th, 2022).
2. Carey, P. What Exactly Is Exposure and Response Prevention (ERP) Therapy? https://www.treatmyocd. com/blog/what-is-exposure-andresponse-prevention-therapy (accessed October 8th, 2022).
3. Interview with Jonathan S. Abramowitz, Ph.D. 10/13/2022.

4. Psychology Today Staff. Acceptance and Commitment Therapy. https:// www.psychologytoday.com/us/ therapy-types/acceptance-andcommitmenttherapy (accessed October 17th, 2022).
5. Ojalehto, H. J.; Abramowitz, J. S.; Hellberg, S. N.; Buchholz, J. L.; Twohig, M. P. J. Anxiety Disord. 2020, 72, 102210.
“Research shows that the best way to overcome unwanted thoughts and fears is to face them head-on.”













Are you interested in communicating science to a broad audience?
Do you want to engage in thought-provoking invesitgations?
Does your passion for the sciences extend into the world of research?
Do you want to combine your creative talents with your fascination with science?
Carolina Scientific is always looking for staff writers, designers, and illustrators!
If you are interested, please contact carolina.scientific@gmail.com Follow us on Instagram @carolinascientific Find us on Facebook facebook.com/CarolinaScientific

Follow us on Twitter @UNCSci


Check out our website carolinascientific.org

Spring 2023 | Volume 17 | Issue 2
This publication was funded at least in part by Student Fees which were appropriated and dispersed by the Student Government at UNC-Chapel Hill as well as the Carolina Parents Council.

 Image by Ildar Sagdejev, [CC-BY-SA-3.0].
Image by Ildar Sagdejev, [CC-BY-SA-3.0].
“Everything is theoretically impossible, until it is done.”
- Robert A. Heinlein