https://ebookmass.com/product/sers-for-point-of-care-and-
Instant digital products (PDF, ePub, MOBI) ready for you
Download now and discover formats that fit your needs...
Advanced Nanomaterials for Point of Care Diagnosis and Therapy Sushma Dave
https://ebookmass.com/product/advanced-nanomaterials-for-point-ofcare-diagnosis-and-therapy-sushma-dave/
ebookmass.com
Point of Care Ultrasound 2nd Edition Nilam Soni
https://ebookmass.com/product/point-of-care-ultrasound-2nd-editionnilam-soni/
ebookmass.com
Clinical Psychomotor Skills (5-Point) 7th Edition Joanne Tollefson
https://ebookmass.com/product/clinical-psychomotor-skills-5-point-7thedition-joanne-tollefson/
ebookmass.com
Hematopoietic Cell Transplantation for Malignant Conditions, 1e 1st Edition Qaiser Bashir Md (Editor)
https://ebookmass.com/product/hematopoietic-cell-transplantation-formalignant-conditions-1e-1st-edition-qaiser-bashir-md-editor/
ebookmass.com
Functional Assessment and Program Development 3rd Edition, (Ebook PDF)
https://ebookmass.com/product/functional-assessment-and-programdevelopment-3rd-edition-ebook-pdf/
ebookmass.com
Breakthrough Supply Chains: How Companies and Nations Can Thrive and Prosper in an Uncertain World Christopher Gopal
https://ebookmass.com/product/breakthrough-supply-chains-howcompanies-and-nations-can-thrive-and-prosper-in-an-uncertain-worldchristopher-gopal/ ebookmass.com
How I wrote over 40 books and over 350 articles within 3 years Duodu Henry Appiah-Korang
https://ebookmass.com/product/how-i-wrote-over-40-books-andover-350-articles-within-3-years-duodu-henry-appiah-korang/
ebookmass.com
A novel graph-based framework for state of health prediction of lithium-ion battery Xing-Yan Yao
https://ebookmass.com/product/a-novel-graph-based-framework-for-stateof-health-prediction-of-lithium-ion-battery-xing-yan-yao/
ebookmass.com
Nominalization: 50 years on from Chomsky's remarks Artemis Alexiadou And Hagit Borer
https://ebookmass.com/product/nominalization-50-years-on-fromchomskys-remarks-artemis-alexiadou-and-hagit-borer/
ebookmass.com
Practice Makes Perfect Complete German https://ebookmass.com/product/practice-makes-perfect-complete-germanall-in-one-ed-swick/
ebookmass.com
SERS forPoint-of-careand ClinicalApplications Thispageintentionallyleftblank
Editedby AndrewFales
CenterforDevicesandRadiologicalHealth,U.S.FoodandDrug Administration,SilverSpring,MD,UnitedStates
Elsevier Radarweg29,POBox211,1000AEAmsterdam,Netherlands TheBoulevard,LangfordLane,Kidlington,OxfordOX51GB,UnitedKingdom 50HampshireStreet,5thFloor,Cambridge,MA02139,UnitedStates
Copyright © 2022ElsevierInc.Allrightsreserved.
Nopartofthispublicationmaybereproducedortransmittedinanyformorbyany means,electronicormechanical,includingphotocopying,recording,oranyinformation storageandretrievalsystem,withoutpermissioninwritingfromthepublisher.Detailson howtoseekpermission,furtherinformationaboutthePublisher’spermissionspolicies andourarrangementswithorganizationssuchastheCopyrightClearanceCenterandthe CopyrightLicensingAgency,canbefoundatourwebsite: www.elsevier.com/permissions .
Thisbookandtheindividualcontributionscontainedinitareprotectedundercopyright bythePublisher(otherthanasmaybenotedherein).
Notices Knowledgeandbestpracticeinthis fieldareconstantlychanging.Asnewresearchand experiencebroadenourunderstanding,changesinresearchmethods,professional practices,ormedicaltreatmentmaybecomenecessary.
Practitionersandresearchersmustalwaysrelyontheirownexperienceandknowledgein evaluatingandusinganyinformation,methods,compounds,orexperimentsdescribed herein.Inusingsuchinformationormethodstheyshouldbemindfuloftheirownsafety andthesafetyofothers,includingpartiesforwhomtheyhaveaprofessionalresponsibility.
Tothefullestextentofthelaw,neitherthePublishernortheauthors,contributors,or editors,assumeanyliabilityforanyinjuryand/ordamagetopersonsorpropertyasa matterofproductsliability,negligenceorotherwise,orfromanyuseoroperationofany methods,products,instructions,orideascontainedinthematerialherein.
ISBN:978-0-12-820548-8
ForinformationonallElsevierpublicationsvisitourwebsiteat https://www.elsevier.com/books-and-journals
Publisher: SusanDennis
AcquisitionsEditor: KathrynEryilmaz
EditorialProjectManager: MariaElaineD.Desamero
ProductionProjectManager: KiruthikaGovindaraju
CoverDesigner: GregHarris
TypesetbyTNQTechnologies
StefanoFornasaro,ClaudiaBeleites,ValterSergo andAloisBonifacio
CHAPTER3SERSprobesandtagsforbiomedical
PietroStrobbiaandAndrewFales
CHAPTER4SERSbiosensorsforpoint-of-careinfectious diseasediagnostics. ....................................115
HoanThanhNgoandTuanVo-Dinh Introduction ...................................................................115
CHAPTER5SERS-basedmolecularsentinelnanoprobes fornucleicacidbiomarkerdetection ................135
BridgetCrawford,Hsin-NengWangandTuanVo-Dinh Introduction ...................................................................135
DevelopmentoftheiMSnanoprobeforlabel-freehomogenous biosensing. ....................................................................137
Silver-coatedgoldnanostarsforSERSdetection ............137 DetectionschemeoftheSERSiMSnanoprobe..
DevelopmentofiMSfordetectionofmicroRNA biomarkers ..............................................................141
DetectionofmiRNAbiomarkerswithinbiologicalsamples...144 RNAextractedfromcancercelllines ...........................144
ClinicalevaluationofmiRNAcancerbiomarkerdetection usingiMSnanoprobes. ..............................................146
MultiplexeddetectionofmiRNAbiomarkers... ....................153 Developmentofmultiplexingtechnique........................153
Multiplexdetectionofendogenoustargetsextractedfrom breastcancercelllines. ..............................................157 iMSbioassay-on-chip.. ..............................................158
Conclusion.. ..................................................................162
Acknowledgments... ........................................................162
References... ..................................................................162
CHAPTER6SERSdetectionoforalandgastrointestinal cancers .............................. .......................169
AlexanderCzaja,M.S.andCristinaZavaleta,PhD Introduction. ..................................................................169
Oralcancer.. ..................................................................172 Introduction .............................................................172
Optimizationanddesignconsiderations ........................172 Discussionandfuturedirections... ...............................175
Esophagealcancer.. ........................................................177
Optimizationanddesignconsiderations ........................178 Discussionandfuturedirections... ...............................181
Stomachcancer. .............................................................182
Intestinalcancer .............................................................186 Introduction .............................................................186 Optimizationanddesignconsiderations ........................186 Discussionandfuturedirections... ...............................192
Concludingremarks ........................................................193 References... ..................................................................193
CHAPTER7InvivoimagingwithSERSnanoprobes ..............199
ChrysafisAndreou,PhD,YiotaGregoriou,PhD, AkbarAliandSuchetanPal,PhD Introduction. ..................................................................199
RamanimagingwithSERSnanoprobes ..............................200
InvivoimagingwithSERSnanoparticles multiplexing potential. .......................................................................201
Biologicalbarriersandopportunities.. ................................204
Passivetumortargeting ....................................................204
Opsonization/sequestrationbythemononuclearphagocyte system. .........................................................................207
Sequestrationbasedonphysicochemicalproperties.. ............207
Blood brainbarrier... .....................................................209
Moleculartargeting .........................................................212
ExvivoSERS-basedmolecularimaging.. ...........................212
InvivomolecularimagingwithSERSnanoprobes... ............213
MultimodalimagingusingSERSnanoprobes ......................215
ThefutureofinvivoRamanimaging. ................................218
Imagingdeeper ..............................................................218
SERSandendoscopy. .....................................................219
Spatiallyoffsetoptics. .....................................................220
Imagingfaster. ...............................................................222 Nanoprobeadministration ................................................223
Contributors SaraAbalde-Cela,PhD
InternationalIberianNanotechnologyLaboratory,AvenidaMestreJose ´ Veigas/n, Braga,Portugal
AkbarAli
DepartmentofChemistry,IndianInstituteofTechnology,Bhilai,Chhattisgarh, India
ChrysafisAndreou,PhD
DepartmentofElectricalandComputerEngineering,UniversityofCyprus, Nicosia,Cyprus
ClaudiaBeleites
ChemometrixGmbH,Wolfersheim,Germany
AloisBonifacio
RamanSpectroscopyLab,DepartmentofEngineeringandArchitecture, UniversityofTrieste,Trieste,Italy
BridgetCrawford
DepartmentofBiomedicalEngineering,DukeUniversity,Durham,NC,United States;FitzpatrickInstituteforPhotonics,DukeUniversity,Durham,NC,United States
AlexanderCzaja,M.S.
DepartmentofBiomedicalEngineering,UniversityofSouthernCalifornia,Los Angeles,CA,UnitedStates;USCMichelsonCenterforConvergentBioscience, LosAngeles,CA,UnitedStates
AndrewFales
CenterforDevicesandRadiologicalHealth,U.S.FoodandDrugAdministration, SilverSpring,MD,UnitedStates
StefanoFornasaro
RamanSpectroscopyLab,DepartmentofEngineeringandArchitecture, UniversityofTrieste,Trieste,Italy
YiotaGregoriou,PhD
DepartmentofBiologicalSciences,UniversityofCyprus,Nicosia,Cyprus
HoanThanhNgo
SchoolofBiomedicalEngineering,InternationalUniversity,VietnamNational University,HoChiMinhCity,Vietnam
SuchetanPal,PhD
DepartmentofChemistry,IndianInstituteofTechnology,Bhilai,Chhattisgarh, India
LauraRodrı´guez-Lorenzo,PhD
InternationalIberianNanotechnologyLaboratory,AvenidaMestreJose ´ Veigas/n, Braga,Portugal
ValterSergo
RamanSpectroscopyLab,DepartmentofEngineeringandArchitecture, UniversityofTrieste,Trieste,Italy
MiguelSpuch-Calvar,PhD
CINBIO,UniversidadedeVigo,CampusUniversitarioLagoas,Vigo,Pontevedra, Spain
PietroStrobbia
DepartmentofChemistry,UniversityofCincinnati,Cincinnati,OH,UnitedStates
TuanVo-Dinh
FitzpatrickInstituteforPhotonics,DepartmentofBiomedicalEngineering, DepartmentofChemistry,DukeUniversity,Durham,NC,UnitedStates; DepartmentofBiomedicalEngineering,DukeUniversity,Durham,NC, UnitedStates;FitzpatrickInstituteforPhotonics,DukeUniversity,Durham,NC, UnitedStates;DepartmentofChemistry,DukeUniversity,Durham,NC,United States
Hsin-NengWang
DepartmentofBiomedicalEngineering,DukeUniversity,Durham,NC,United States;FitzpatrickInstituteforPhotonics,DukeUniversity,Durham,NC, UnitedStates
CristinaZavaleta,PhD
DepartmentofBiomedicalEngineering,UniversityofSouthernCalifornia,Los Angeles,CA,UnitedStates;USCMichelsonCenterforConvergentBioscience, LosAngeles,CA,UnitedStates
Editorbiography AndrewM.FalesreceivedhisBSinbiochemistryandmolecularbiologyfromthe UniversityofMaryland,BaltimoreCountyin2010,andhisPhDinbiomedicalengineeringfromDukeUniversityin2016.HisdissertationfocusedonthedevelopmentoftheranosticnanoplatformsusingSERS-baseddetectionstrategies.From 2016to2020,heconductedresearchatFDA’sOfficeofScienceandEngineering Laboratories,studyingemergingbiomedicalopticaltechnologies.In2020,hejoined theFDA’sOfficeofProductEvaluationandQualityasascientificleadreviewer.His researchinterestsincludeplasmonics,pulsedlaser nanoparticleinteractions,and Ramanspectroscopy.
Disclaimer
Theopinionspresentedbytheeditorandauthorsaretheirownandshouldnotbe construedtorepresentthoseoftheFoodandDrugAdministrationortheUSDepartmentofHealthandHumanServices.Thementionofcommercialproducts,their sources,ortheiruseinconnectionwithmaterialreportedhereinisnottobe construedaseitheranactualorimpliedendorsementofsuchproductsbytheUS DepartmentofHealthandHumanServices.
Thispageintentionallyleftblank
DataanalysisinSERS diagnostics 1 StefanoFornasaro1,ClaudiaBeleites2,ValterSergo1,AloisBonifacio1
1RamanSpectroscopyLab,DepartmentofEngineeringandArchitecture,UniversityofTrieste, Trieste,Italy; 2ChemometrixGmbH,Wolfersheim,Germany
Introduction Thischapterfocusesonthesurface-enhancedRamanspectroscopy(SERS)data analysisworkflowinbiomedical/clinicalstudies.Themostcommoncharacteristic forthesestudiesisthattheydealwiththeanalysisofcomplexandinformationrichdatasets,composedoftenstohundredsofobservations(i.e.,spectra),each composedofhundredsorthousandsofvariables(i.e.,wavenumbers).Biofluids (e.g.,blood,plasma,saliva,tears,andurine)aretypicalsamplesusedinbiomedical SERSstudies.Allbiofluidscontainawealthofbiochemicalinformation,whichcan beexploitedbyphysicianstomonitormanyclinicallyrelevantaspects,suchasapatient’sgeneralhealthstatus,theimmunesystem,theclinicalresponsetoacertain drug,orthedeliveryofnutrients(tonamefewexamples).However,extracting theimportantinformationfromthesedatasetsisapuzzlingprocess.Extractingasinglenumberfromeachspectrum(e.g.,theintensityofoneband)sotohaveonlyone variableassociatedwitheachsampleis,frommanypointsofview,veryconvenient. Univariatedataanalysismethodssuchasdifference(e.g.,diseasedminuscontrol) spectralanalysis,orchangesinbandsintensity(orarea)valuescanbeusedtoextract theusefulinformationfromthedataset.Thisapproach,however,isofteninsufficient inSERSspectroscopy,andinparticularin“label-free”SERS,sincecontributions frommultipleanalytesarepresentineachspectrum.
Thus,amultivariateanalysisapproachisoftennecessary.Forthatreason,itisof greatimportancetohavearobustmultistepdataanalysisworkflowavailable,allowingforthecorrectuseandinterpretationofthespectraldata.
OtherspectroscopictechniquesasRamanorIRpresentsimilarchallenges,and canbenefittothesameextentfromtheuseofamultivariatedataanalysis.SERS, however,hassomeuniquecharacteristicswhichmakeitmorechallenging,froma dataanalysispointofview.Differentlyfromothertechniques,aSERSspectrumalwaysoriginatesfromarelativelysmallfractionofthemoleculespresentinasample, whichhaveveryspecificinteractionswithasurface.Infact,itisnowwidely acceptedthatfewmolecules,i.e.,thoselocatedatsurfacepositionswheretheplasmonicamplificationoftheelectromagneticfieldishighest(oftenreferredtoas“hotspots”),areaccountableformostofthesignalconstitutingaSERSspectrum,
SERSforPoint-of-careandClinicalApplications. https://doi.org/10.1016/B978-0-12-820548-8.00002-3 Copyright © 2022ElsevierInc.Allrightsreserved.
whereasthemajorityoftheothermoleculeshaveanegligiblecontribution.Inother words,inSERSthecontributiontothesignalisnotequallydistributedatall.Afirst consequenceofthisfactisthattheintensityofthesignalobservedishighlydependentonthespatialdistributionofthese“hot-spots”ontheSERSsubstratewith respecttothesizeofthelaserspotusedforexcitation.Thisdependencecanlead toremarkableintensityfluctuationsuponprobingdifferentareasofthesamesubstrate,oruponinvestigatingthesamesampleusingtwodifferentsubstrates.Inother words,thisleadstoanimportantsourceofvariabilitywhichneedstobedealtwith. Moreover,thefactthatthesignalalsodependsonthespecificinteractionbetween theanalytesandthesubstratecanleadtoothercomplications:theoverallspectral shapecanbeconcentration-dependent(i.e.,differentbandsareobservedforthe sameanalyte,dependingontheconcentration),andslightdifferencesinthephysicochemicalcharacteristicsofthesurfaceoritsenvironmentcanbringdrasticspectralchanges(e.g.,minordifferencesinpHorionicstrengthcanleadtoquite differentSERSspectra).Thisalsomeansthatsmalldifferencesamongdifferent batchesoftheSERSsubstrateswillfurthercontributetospectralvariability.This analyte metalinteractionisalsoreflectedintheoccurrenceofacharacteristic intenseandbroadbandinallSERSspectrabetween100and300cm 1,which mustbeconsideredwhenconsideringthesubtractionofabaseline.Allthese SERS-specificaspectsfurthercomplicatetheuseofthistechniqueforbiomedical applications,andexalttheimportanceofdataanalysis.
Thegoalofthischapteristointroduceageneralaccountofeachstepofthisprocess,highlightingcentralideasandspecificchallengesrelatedtotheanalysisand interpretationofSERSspectraldata.Webelievethatageneralunderstandingof thekeyaspectsoftheproblemcanbeofgreatervaluethanprovidingsolutions forspecificapplications,sincethesuccessofaspecificanalysisisoftenlimitedto theapplicationforwhichitwasoptimized.Thus,wetrytogivepracticaladvice tothereaderthroughaquickoverviewofthemostrelevanttechniquesfordatavisualization,modeling,classification,andcalibrationinSERSstudies,withan emphasisonboththeircapabilitiesandweaknesses.Theapproachadoptedispurposelyavoidingin-depthmathematicaltreatmentstomakethischaptermorereadablebyabroaderaudience.Equationsarethusavoidedinmostinstances,although theinterestedreadercanfinddetailedmathematicalexplanationsinthesuggested literature.
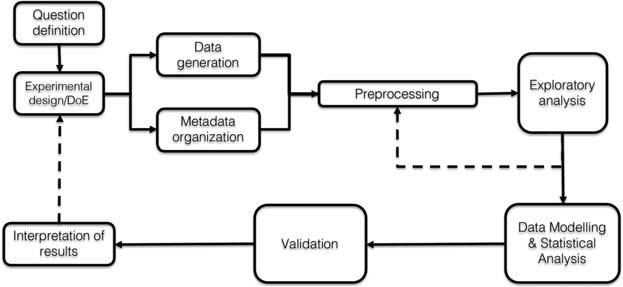
Generaldataprocessingworkflow Theblockdiagram(Fig.1.1)graphicallyrepresentsthegeneraldataprocessing workflowforatypicalSERSexperiment,startingwiththebiologicalquestion, includingdesignofexperiments(DoEs),(meta)datacollection,theselectionand employmentofappropriatetechniquesthatwillextractusefulinformationfrom thesamples,theverification/validationoftheresults,andendingwiththebiochemical,biological,orclinicalinterpretation.Thisinterconnectedpathinvolvesa
GeneraldataprocessingworkflowforSERSstudies.
Credit:StefanoFornasaro.
constantfeedbackbetweenexperimentaldesign,dataprocessing,andstatistical analysis:it’snotunusualtoreturntodatafromanearlierstage(sometimeseven totherawdata)todouble-checkkeyelementsandresolvedataambiguities.Inother words,itisextremelyimportanttochecktheoutcomesofeachdataanalysisoperationstepbystep,andifnecessarilygobacktoapreviousstep,inasortofselfconsistentprocess.
Studydefinitionanddatacollection AtypicalSERSstudybeginswithabiological/clinicalquestion,which,forexample, mightinvolvetwogroupsofindividuals,oftencalledcaseandcontrol(e.g.,patients vs.healthy).Eveninthisverysimplescenario, nesteddatastructures areoftenpresent(e.g.,multiplesamplesperpatient,manyspectrapereachSERSsubstrate).A characteristicofnesteddatastructuresisthatthemeasurementswithineachnesting aremorecloselyrelatedtoeachotherthanthemeasurementsbetweennesting.Here, theSERSintensitiesforeachwavenumberwithinasubstratearemorerelatedor correlatedtoeachotherthantheintensitiesbetweensamples.Andtheintensitiesbetweensamplesaremorecorrelatedtoeachotherthantheintensitiesbetweenpatients.Accordingto TheoryofSampling allthesepropertiesshouldbecarefully consideredbeforedataarecollectedtoensurethatthesamplestobemeasuredreally representtheconditionunderinvestigation[1].Thisisbecausea(biomedical)SERS experimentcannottotallybeundercontrol,sincetherearealwaysunknownvariationsinreplicatesorindividuals.Evenso,thisaspectisoftenoverlookeduntilafter thedataarecollected!Onthecontrary,awell-planned experimentaldesign isthe firstelementofasuccessfuldataanalysis:anadequatenumberofreplicatesatthe relevantinfluencingfactorsfacilitatedataanalysisand,eventually,datainterpretation.Therefore,thesamplesizeshouldbealwayscountedasthenumberofreplicates/individualsratherthanthenumberofspectra.Ifthedatasetiscomposedof
FIGURE1.1
alargenumberofspectrainanesteddatastructure,thespectrabelongingtothetopmosthierarchicalstructure(thepatientinthepreviousexample)shouldbeprocessed together.Theimportanceofthispointcanneverbeoverstatedwhenitcomestoget anhonestassessmentofamodel’spredictiveabilityonnewcases(see“Verification ofresults”section).
DoEisaframeworkforsystematicevaluationofcomplicatedsystemsthatare influencedbymultiplefactors[2].TheobjectiveofDoEistoeffectivelyplanand conductexperimentssothattheexperimentaldomainissystematicallyinvestigated, oftenwithasfewexperimentsaspossible[3].Thekeypointisthatsomevariation cannotbeavoided,butexperimentscanbesetupinsuchawaythatitdoesnotinterferewiththeresults.
Forexample,manySERSstudiesmakeuseofablockingdesigntoaccountfor theeffectoftheuseofdifferentbatchesofAgorAucolloidsassubstrates.In comparingtheSERSresponseofasampletoseveralexperimentalvariablesorinfluencing factors (e.g.,nanoparticlesconcentration,pH,incubationtime),acommon strategyistoperformallmeasurementsusingthesamebatchofSERSsubstrates (e.g.,thesamebatchofmetalcolloid),studyingtheinfluenceofonevariablewhile keepingtheothersatafixedvalue.Then,anothervariableisselectedandmodifiedto performthenextsetofexperiments,andsoonandforth.Intheend,everythingis repeatedforanothercolloidalbatchtoassessthevariabilitybetweencolloidal batches.This one-variable-at-a-time strategyhasbeenshowntobeinefficient,lackingtheabilitytoaddressinteractionsbetweentwoormorevariablesandoften needingmanyexperimentstoreachtheoptimum.Amorecomprehensiveresult canbeachievedbystudyingseveralvariablessimultaneouslyandsystematically bymeansofanappropriateexperimentaldesign.Byusing,forinstance,a factorial design with blocking,wheredifferentbatchesofsubstratescontainanequalnumber ofdesignedexperiments,itispossibletodetecttheinfluenceofallthevariablesand oftheirinteractionswithinonebatchandalsotheeffectofdifferentbatches.A coupleofdesignsusefulforSERSarebrieflydescribedin Table1.1.MoreinformationcanbefoundinRefs.[4 7].
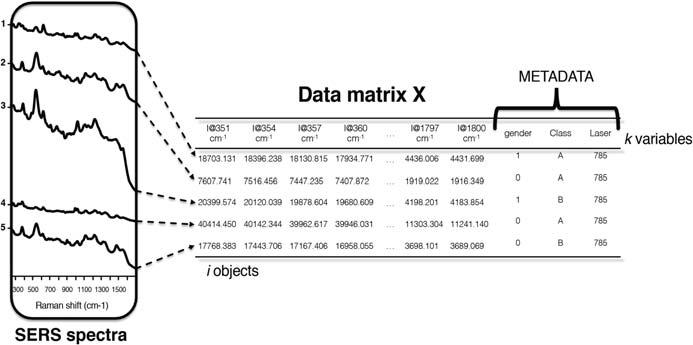
Datahandlingconsiderations(datastructures,organization) TypicalresearchgradecommercialRamanspectrometerswillhaveafullscanrange goingfrom50toabove3500.Withmoderninstruments,itiscommontomeasurethe entireindividualspectralrangewithasingleacquisition.Theusualintensityscale usedforspectralmeasurementsis“photoncounts”forSERSintensity,whichmay befurthernormalizedorscaled.Thesedataareusuallyarrangedintheformofamatrix X,havingasmanyrowsasthenumberofspectra(eachrowofthe X-matrixcorrespondstoawholespectrumofaparticularsample),andasmanycolumnsasthe numberofmeasuredvariables k (intensitiesataparticularwavenumber)overallthe samples.Accordingly,thenumberofsamples i maybeafewtentoafewhundred; k maybeafewhundredstoseveralthousands,dependingonthedensityofdatapoints. Fig.1.2 showsatypicaldatamatrixobtainedfromSERSmeasurement.
Table1.1 Examplesofdesignedexperiments. DesignDescription FullfactorialdesignTwoormorefactors,eachwithdiscretepossiblevalues(“levels”). Theexperimentsareperformedonallpossiblecombinationsof theselevelsacrossallsuchfactors.Inthisway,itispossibleto studytheeffectsofallthefactorsandoftheirinteractionsonthe responsevariables.Asanexample,consideringthree independenttwo-levelfactors(treated/untreated,male/female, young/old)willleadto23 ¼ 8experiments.Inthepresenceofa largenumberoffactorsorfactorlevels,fullfactorialdesignscan becomeprohibitivelyexpensive(intermsoftimeandcosts).In thesecases,itispossibletorelyonpartial/fractionaldesigns whereonlyasubsetofthedesignspaceisanalyzed.
BlockdesignAblockisagroupofsamples(treated)measuredinmoreorless homogeneousconditions(e.g.,onecolloidalbatch).Asan example,considermanybiologicalreplicatesforeachsample,and threecolloidalbatchestomeasurethemall.Thebestoptionisto assignthesamplesinsuchawaythateachbatchcorrespondsto theanalysisofonecompletesetofsamples.Often,however,this cannotbearrangedforseveralreasons.Insuchacase,an incompleteblockdesigncanbeused.
Split-plotdesignThistypeofdesignisusedtohandlenestedstructures,whereitis moredifficulttochangethelevelsofatreatmentduringthe experiment.Thefactorsdifficulttochangearerandomlyusedon eachblock,whiletheothersarefullyrepresentedwithineach block.
FIGURE1.2 Dataorganization.
Credit:StefanoFornasaro.
Metadataorganization Togetherwiththespectraldata,allthepossible metadata shouldalsobecollected andorganized.Theterm metadata referstothesetofinformationdescribingeach sampleandthewholeexperimentalpipeline.Theyhavetobeconsideredduring thedesignoftheanalyticalworkflow,inordertoensureproperrandomizationof thedatacollection.Moreover,theyareextremelyusefulforareliableinterpretation oftheresults:organizingandstoringadetailedrecordofthemetadataiscrucialto evaluatethepresence(andtheimpact)ofpossibleconfoundingfactors,anomalies, ortrendspresentinthedataset.Samplemetadataoftenmatchthefactorsincludedin theDoE(e.g.,typeoftreatment,referencediagnoses,gender,age,replicas),butthey alsoincludekeycharacteristicsofthesamples.Someexamplesare
•date/timeofsamplecollection
•date/timeofsampleanalysis
•instrumentonwhichtheanalysishasbeenperformed
•typeandpoweroflaser
•laserpowerdensityonthesample
•numberofaccumulations
•operatorwhohasperformedtheanalysis
•typeofSERSsubstrate
Recommendations •Adoptgoodlaboratorypracticeguidelines,possiblywiththehelpof spreadsheetsandlaboratorynotebooks,torecordmeasurementparameters andresults.
•organizethemetadatainmatrixform,insuchawaythattheimportcanbe performedinasinglestep.If,bycontrast,thedataarespreadinseveral filesorsheets(e.g.,onefileforeachsampleorforeachvariable),thenthe importprocedurewouldbemuchlongerandmoredifficult.
Abitofstatistics TointerpretthecomplexinformationhiddenintheSERSspectraandconnectitto specificbiologicalphenomena,severaldataanalysesandpattern-recognitionalgorithmsareused.Foranykindofanalysistobesuccessful,itisessentialtohave anoverallpictureofthespectraldataset.LookingatalltheSERSspectraatthe sametime(eitherstackedorsuperimposed)isnotfeasibleforalargedataset,so onehastofindotherwaystorepresentitsgeneralcharacteristics.Descriptivedata summarizationtechniquesarecommonlyappliedtovisualizethetypicalproperties ofSERSspectra.
Formany(pre)processingmethods,itisessentialtoevaluateinadvancethedistributionofthedatatoensureifthemethods’assumptionswillbemet. Descriptive statistics forcentraltendencyanddispersionareofgreathelpforthispurpose. Typicalmeasuresofcentraltendencyinclude mean and median,whilemeasures
ofdatadispersionincludequartiles,interquartilerange(IQR),andvariance.However,themeanisnotalwaysthebestwayofmeasuringthecenterofthedata,especiallyfornonnormaldistributions.Moreover,amajorproblemwiththemeanis itssensitivitytoextremevalues(e.g., outliers).Usually,theintensitydistributions ateachwavenumberarenot-normalforaSERSdataset,andthusrepresentingthe datasetwiththemeanspectrummightbenotentirelyappropriate.Fornon-normal datadistributions,abettermeasureofthecenterofdataisthe median,beingthemiddlevalueofthevariablewhenthedataarealignedeitherinincreasingordecreasing order.Thus,the medianspectrum isabetteroptiontorepresentthedatasetinterms of“spectralprofile,”havingtheadditionaladvantageofbeinglesssensitiveto outliers.Often,forSERSdatasetsthemedianandthemeanspectrumareverysimilar, withonlysmalldifferences,butifthedatasethasmanyoutliers,thesetwospectra canbequitedifferent,sothatusingthemedianspectrumtorepresentthedataisalwaysasaferchoice.
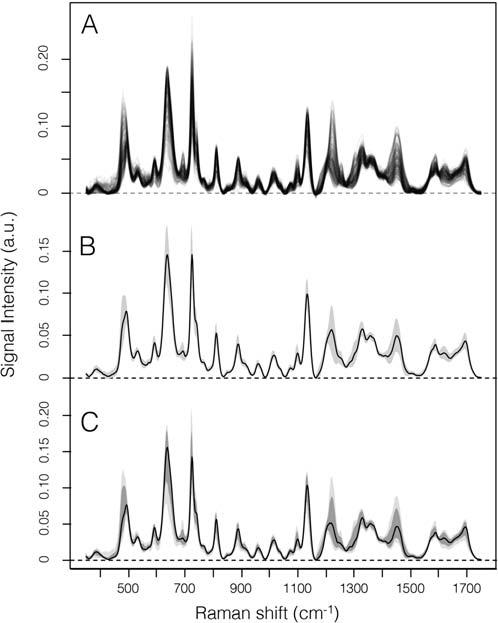
ThespectrainaSERSdatasetcanbequitedifferentfromeachother,thatisthe datasetpresentacertain spectralvariability.Thedegreetowhichintensitiesateach variabletendtospreadiscalledthe dispersion ofthedata.Apopularmeasureofthe dispersionisthe standarddeviation (s)definedasthesquarerootofthe variance (s2 ).Notethat s measuresthespreadaboutthemeanandshouldonlybeonly whenthemeanischosenasthemeasureofcenter.Sincewecannotassumeour datatohaveasymmetricdistribution,otheroptionstodescribetheintensitydispersion(i.e.,spectralvariability)shouldbeconsidered.The kth percentileofadatasetis thevalue xi havingthepropertythat k percentofthedatapointslieatorbelow xi The median is,bydefinition,the 50thpercentile.Themostcommonlyusedpercentilesotherthanthemedianare quartiles.The firstquartile (Q1)isthe 25thpercentile,whereasthe thirdquartile (Q3)isthe 75thpercentile.Thequartiles,including themedian,givesomeindicationofthecenter,spread,andshapeofadistribution. Thedistancebetweenthefirstandthirdquartilesisasimplemeasureofspreadthat givestherangecoveredbythe50%ofthedata.ItiscalledIQRanditisacommon measureofdispersionusedfornonnormaldistributions.Thus,the median andIQR oftheintensitiesforeachwavenumberofthedatasetgivesomeindicationofthe center,spread,andshapeofadistributionofintensitiesinthedataset.Mean standarddeviationormedian,1stand3rdquartilescoveronlyabout2/3and1/2ofthe data,respectively.Plottingthe5thto95thpercentileofintensitiesgivesabettervisualimpressionoftheactualrangecoveredbythedata.Box-and-whiskersplotsare averypopularwayofvisualizingadistribution,usingaboxwhoselengthistheIQR andwhosewidthisarbitrary.Alineinsidetheboxshowsthemedian,whereaswhiskersareconventionallyextendedtothemostextremedatapointthatisnomorethan 1.5 IQRfromtheedgeofthebox(Tukeystyle)orallthewaytominimumand maximumofthedatavalues(Spearstyle)[8].Thisgraphicalvisualizationcanbe used,forinstance,toprobethedistributionoftheintegratedareasofparticularbands thatarebiochemicallymeaningfulforeachspectrumofaspecificcondition/batch/ treatment.MeasuressuchasmedianandIQRaremorerobustthanmeanandstandarddeviation,andthustheycanbesafelyusedtosummarizethedatasetinafigure (Fig.1.3).Suchafigurewouldimmediatelyconveytheoverallspectralfeaturesof
FIGURE1.3
(A)Datasetcomposedof200SERSspectraofdeproteinizedserum;(B)mean spectrum 1standarddeviation;(C)5th,16th,50th,84thand95thpercentile.
Credit:AloisBonifacio.
thedataset(asthemedian),atthesametimegivinganideaofhowisthespectral variabilityforeachRamanshift.Alternatively,a“functionalboxplot”[9]approach couldbeused,withtheimmediateadvantageofbeingabletofindandvisualizethe spectrainthedatasetwhicharesignificantlydifferentfromtheothers.
Surveyofsoftwareavailable OneofthedifficultiesoftenencounteredbyresearchersapproachingSERSdata analysisistodecidethesoftwaretobeemployedinimporting,handling,andprocessingthedata.Theseinterconnectedtasksrequireflexibleandinteractivetools. Varioussoftwareprogramsandpackagesareavailable,rangingfromthosefor general-purposeusetothosetargetingspecificdataanalysistasks.Manyspectrometermanufacturersoffersoftwarecapableofdataanalysis,besidesoperatingtheir
instrumentandcollectingdata.However,thesesoftwareproductsareoftenlimited withrespecttodataanalysis,offeringbutfewoptions.Inspiteoftheselimitations, andevenifmanycommercialandnoncommercialalternativesdevotedtospectrum processingexist(seebelow),inthemajorityofthecasesfoundinliteraturethefirst steps(ormoreoftenthewholesequence)ofpreprocessingareperformedwithinthe proprietarysoftwareprovidedbytheinstrumentmanufacturer.Then,thepreprocesseddataareimportedinvariousstatisticalsoftware(e.g.,SPSS,Prism,Origin Pro,PLSToolbox,andothersavailableonthemarket)forfurtheranalysis.From ananalysisofliterature,alistofthemorefrequentlyusedsoftwareinSERSstudies wascompiledandisgivenin Table1.2.Mostoftheseapplicationshaveagraphical userinterface(GUI),resultinginarelativelyshallowlearningcurveforthenewuser. TheseGUI-basedsoftwareallowexperimentalscientistswithlittleornochemometricbackgroundtoperformdataprocessingandanalysisofthemeasurements.However,theseapplicationsarefrequentlyratherexpensive.Moreover,theuseof powerfulmultivariateanalysismethodsas“black-boxes”canleadtoincorrectconclusions,andingeneralwediscouragethereadertousethemwithoutaproper training.Inaddition,theuseofa“point-and-click”approachinGUIsandthe closed-sourcenatureofthesesoftwareusuallylimitthechoiceofalgorithms,the user’sabilitytoautomatetasksandtodevelopnewprocedureswithinthesoftware. Ontheotherhand,thereisagrowingcommunityofspectroscopiststhatpreferto programtheirownmethodsusingcommandlinetoolsandshortscriptswrittenin differentprogramminglanguagessuchasMatlab,R,orPython.Thelearningcurve tomanagetheselanguagesissteeperthanthoseofmostcommercialsoftware.The tradeoffforthistimeinvestmentisamuchtightercontrolandfinetuningonallthe
Table1.2 Resultsofsearchstring“surfaceenhancedRaman” AND“diagnostics”AND(softwarename)fromGoogleScholar: mostreportedsoftwaretogetherwiththenumberofpapersusing thekeyword.
differentstepsofdatahandling,processing,andvalidation.Thesestepscanbe developedindependently(alsowithdifferentprogramminglanguages),orconstitute the“nodes”ofauniquepipeline,leadingtoseveraladvantages.Sinceitisnoteasyto compareresultsfromdifferentsoftware,havingauniformpipelineaddressingallthe tasksisadefiniteadvantage.Moreover,awell-writtenworkflowscriptisalreadyan ordereddescriptionofthedataanalysisprocess,contributingtomaketheresults transparentandreproducible,asrequestedbytheFAIRprinciplesofscientific datamanagement[10].Alistofrecommendedsoftware/packagesisreportedin Table1.3
ThedataformatadoptedisalsorelevantinthelightoftheFAIRprinciples.Raw dataareusuallystoredinvendorproprietaryformatsthatcanusuallybereadonlyby proprietarysoftware.Aninstrumentmanufacturermayhavemorethan10different dataformats(includingolderformatversions).Acloseddataformatobstructsdata comparisonandsharing,whichisinsteadmadepossibleadopting“open”dataformatswithpubliclyavailablespecifications.Forthisreason,allthevendorsoftware allowsdataexportinopenfileformat(usuallyasASCIIfiles).Itisimportantto mentionthatnotalltheanalyticalmetadataareusuallyincludedintheexported opensourceversionofthedata.Itisthereforesuggestedtoalwaysstoreacopy oftheoriginal“closed”files,whichcanbeinspectedonlywiththevendorsoftware, toavoidimportantinformationloss.
Dataintegrity Thefirststepafterspectracollectionisalwaysthevisualassessmentofthequality andintegrityoftherecordedrawdata.Thisisusuallyaccomplishedbyexamination ofindividualspectra,checkingforanomaliesthatoccurredduringthemeasurement process(e.g.,spikes/cosmicrays;presenceofsaturatedchannels,verypoorsignalto-noiseratio),andforspectralcontaminationfromunknown/unwantedchemicals.
Afirstwaytocheckdata,asalreadymentioned,istoplotthemediantogether withthe16thand84thpercentileforeachRamanshift(Fig.1.3).Notethatfornormallydistributeddata,16th,50th,and84thpercentileareequaltomean onestandarddeviation(see“Abitofstatistics”section).
Sinceerrorscanalsooccurinthemetadatavalues(e.g.,concentrations,classmembership,etc.),itisimportanttocarefullycheckalsothecorrectassociationbetween spectraandthetablecontainingthissetof“numbers”beforestartingthepreprocessing step.Thisdata/metadatacheckingstepiscriticaltomaximizethepossibilityofperformingarobustandsuccessfulanalysis:manyalgorithmsusedfortheanalysisof SERSspectraaresensitivetothepresenceof“corrupted”spectraaffectedbytheissueslistedabove,ortoamismatchbetweendataandmetadata.Whethertheaimisto developamultivariatecalibrationmodeltoquantifyaspecificanalyteinacomplex matrix,ortryingtocorrectlyclassifyapatientfromtheanalysisofabiofluid,in manycasesjustfewcorruptedspectraormismatchedmetadatacandegradetheperformanceofthemodel.Forinstance,awronglyassignedconcentrationlabeltoa SERSspectrumcanseriouslyaffecttheperformanceofaregressionmodel.
Table1.3 Recommendedsoftware/packages.
SoftwareLicense
Ra O Generalpurposestatisticsplatformusing packagestoprovidespecificfunctionality (https://www.r-project.org )
OhyperSpec þ (þ) þb Infrastructureforhandlingofspectra;works togetherwithpreprocessingandmodeling packages(https://github.com/r-hyperspec/ hyperSpec)
OChemoSpec þþþ High-levelconveniencefunctionswrappers forwidelyuseddataanalysissuchasPCA orHCA(https://github.com/bryanhanson/ ChemoSpec)
Oprospectr- þ -Preprocessing,andsampleselection calibrationsampling(https://github.com/ antoinestevens/prospectr )
Omdatools- þþ
Projection-basedmethodsfor preprocessing,exploring,andanalysis (https://github.com/svkucheryavski/ mdatools)
Obaseline- þ -Variousbaselinecorrectionalgorithms (https://github.com/khliland/baseline )
OEMSC- þ -PackageforEMSC(https://github.com/ khliland/EMSC)
Opls,MASS, chemometrics, unmixR, . þ Rhasawidevarietyofpackagesthat providestatisticalmodels
Table1.3 Recommendedsoftware/packages. cont’d
SoftwareLicense
Pythona O
OQuasar/Orange
OScikit-(
OKeras--
MATLABP
PPLS_Toolbox
OMCR_ALS--
OBiodatatoolbox
UnscramblerP
Originfor spectroscopy P
General-purposeprogramminglanguage usingpackages(modules)toprovide specificfunctionality(https://www.python. org)
Collectionofdataanalysistoolboxesfor spectroscopy(https://quasar.codes ) expandingtheorangedataminingand machinelearningsoftwaresuite(https:// orange.biolab.si)
þ Frameworkforstatisticaldataanalysisand machinelearning(https://github.com/scikitlearn/scikit-learn)
þ Deeplearninglibrary(https://github.com/ keras-team/keras)
Multiparadigmprogramminglanguageand numericcomputingenvironment(https:// www.mathworks.com)
Suiteofmultivariateandmachinelearning tools(https://eigenvector.com/software/ pls-toolbox/)
GUIforMultivariateCurveResolutionAlternatingLeastSquares(MCR-ALS) algorithm(http://www.mcrals.info )
Frameworkforhandlingofspectra;works togetherwithpreprocessingandmodeling packages(https://www.mathworks.com/ matlabcentral/fileexchange/22068-biodatatoolbox)
Stand-alonec programforchemometrics (https://www.aspentech.com/en/products/ msc/aspen-unscrambler )
Stand-aloneprogramforchemistry-related data(https://www.originlab.com/index. aspx?go¼Solutions/Applications/ Spectroscopy)
þ recommended; (þ) usewithcaution; (-) notrecommended/notavailable/impossible; O,opensource; P,proprietary.
a Randpythonallowmutualinteraction,e.g.,apythonmoduleprovidingfileimportforaspecificfileformatcanbeusedfromRandtheresultingdatasetthen analyzedinR(orbothRandpython).
b Doesnotprovidedataanalysis,butseamlessuseofmodelingpackageslikePLS,MASS,etc.
c Currentversionoffersintegratedpythonscriptingsupport.
Outliers Insomecircumstances,visualinspectionofallthespectrainthedatasetcanhighlightclearoutliers,includingsubstantialspectralcontamination.Ideally,suchsamplesshouldbemeasuredagain,butifthatisnotfeasiblethentheyshouldberemoved fromthedatasetandthatexclusionreported.Otherspectracouldbeidentifiedasoutliersusingmorecomplexoutlierdetectionmethodsbasedonseveralpopulardistancemeasures,suchMahalanobisorEuclideandistances[11,12].
Despitethedifferencesbetweenalltheexistingmethods,theyhaveonefeaturein common:mostofthedetectionalgorithmsassumea(approximately)multivariate normaldistributionofthedata.Unfortunately,SERSdatafrequentlyfailtomeet thisassumption.
Othermethods,basedonso-calledrobustestimates,likeMinimumCovariance Determinant[13],orone-classclassificationapproacheslikeone-classsupportvectormachines(SVMs)havealsobeenproposed[14].Thesemethodsarebasedonthe assumptionthatthedatasetrepresentsasamplefromasingle“good”population, contaminatedbyoutliersfromdifferentpopulations.Thisassumptionisnotalways correct,especiallyinclinicalstudies,wherethesamplesdoactuallycomefrommultiplepopulations.The functionalbox-plots proposedbySunandGenton[15]could provideausefulstrategyforreliableandrobustmultipleoutlierdetection(even whendataarenotnormallydistributed),byimplementingtheideaofthe“depth ofacurve”(Fig.1.4).
Datapreprocessing Datapreprocessing,alsoknownasdatapretreatment,istobebroadlyintendedas thesetofproceduresperformedonrawdatatomakethemmoreadequateforthe analysisplanned.Mostoften,thissetofproceduresincludemethodstocompensate fortheeffectsoriginatedfrommeasurementnoise(randomvariation)andfromsystematicerrors(interferencesfromopticalandphysicaleffects)thatareuncorrelated tothebiochemicalfingerprintinthespectra.Thesemethodsareusuallyapplied beforetheactualanalysistoreducetheinfluenceoftherandomvariationand removeinsteadthesystematicvariation.However,itisnotalwayseasytoexactly definewhatthesetwovariationsareorhowtoquantifythem.Itmustbeclearthat althoughpreprocessingcanbeveryhelpfulin“cleaningup”aSERSsignal,itdoes not,byitself,improveitsquality.Ontheotherhand,theriskofremovingpertinent variationtospectraldataisalwayspresentwhenapplyingpreprocessing:anexcessivenoiseremovalorbackgroundcorrectioncansuppressimportantspectral information.
PreprocessingofSERSdatacouldbepursuedalongdifferentroutes,andawide rangeofmethodsexistintheliterature.Itisimportanttorememberthatthereisno oneandonlycorrectwaytochoosewhichpreprocessingtechniquetouse,ora sequenceofdifferentmethodsthatalwaysworkswithanydataset.Thechoiceof whichpreprocessingtechniquetousehighlydependsonthegoaloftheanalysis, thequalityofthespectra,theavailabilityofmethodsandsoftware,aswellason