https://ebookmass.com/product/pan-genomics-applicationschallenges-and-future-prospects-debmalya-barh/
Instant digital products (PDF, ePub, MOBI) ready for you
Download now and discover formats that fit your needs...
Advanced Biofuel Technologies: Present Status, Challenges and Future Prospects D.K. Tuli
https://ebookmass.com/product/advanced-biofuel-technologies-presentstatus-challenges-and-future-prospects-d-k-tuli/
ebookmass.com
Environmental Sustainability of Biofuels: Prospects and Challenges Khalid Hakeem
https://ebookmass.com/product/environmental-sustainability-ofbiofuels-prospects-and-challenges-khalid-hakeem/
ebookmass.com
Graphene: Preparations, Properties, Applications, and Prospects Kazuyuki Takai
https://ebookmass.com/product/graphene-preparations-propertiesapplications-and-prospects-kazuyuki-takai/
ebookmass.com
Truly Madly Plaid Eliza Knight
https://ebookmass.com/product/truly-madly-plaid-eliza-knight-2/
ebookmass.com
A Cowboy State of Mind (Creedence Horse Rescue 1) 1st Edition Jennie Marts
https://ebookmass.com/product/a-cowboy-state-of-mind-creedence-horserescue-1-1st-edition-jennie-marts/
ebookmass.com
The Shadow Princess: A Stone Veil Novel (Chronicles of the Stone Veil Book 6) Sawyer Bennett
https://ebookmass.com/product/the-shadow-princess-a-stone-veil-novelchronicles-of-the-stone-veil-book-6-sawyer-bennett/
ebookmass.com
The Holocaust Bystander in Polish Culture, 1942-2015: The Story of Innocence Maryla Hopfinger
https://ebookmass.com/product/the-holocaust-bystander-in-polishculture-1942-2015-the-story-of-innocence-maryla-hopfinger/
ebookmass.com
Data Structures & Algorithms in Python 1st Edition John Canning
https://ebookmass.com/product/data-structures-algorithms-inpython-1st-edition-john-canning/
ebookmass.com
Burnout For Dummies 1st Edition Eva Selhub
https://ebookmass.com/product/burnout-for-dummies-1st-edition-evaselhub/
ebookmass.com
Nurse Leader Certification Review, Second Edition (Ebook PDF)
https://ebookmass.com/product/clinical-nurse-leader-certificationreview-second-edition-ebook-pdf/
ebookmass.com
AcademicPressisanimprintofElsevier
125LondonWall,LondonEC2Y5AS,UnitedKingdom
525BStreet,Suite1650,SanDiego,CA92101,UnitedStates
50HampshireStreet,5thFloor,Cambridge,MA02139,UnitedStates
TheBoulevard,LangfordLane,Kidlington,OxfordOX51GB,UnitedKingdom
©2020ElsevierInc.Allrightsreserved.
Nopartofthispublicationmaybereproducedortransmittedinanyformorbyanymeans,electronicor mechanical,includingphotocopying,recording,oranyinformationstorageandretrievalsystem,without permissioninwritingfromthepublisher.Detailsonhowtoseekpermission,furtherinformationaboutthePublisher’ s permissionspoliciesandourarrangementswithorganizationssuchastheCopyrightClearanceCenterandthe CopyrightLicensingAgency,canbefoundatourwebsite: www.elsevier.com/permissions.
ThisbookandtheindividualcontributionscontainedinitareprotectedundercopyrightbythePublisher (otherthanasmaybenotedherein).
Notices
Knowledgeandbestpracticeinthisfieldareconstantlychanging.Asnewresearchandexperiencebroadenour understanding,changesinresearchmethods,professionalpractices,ormedicaltreatmentmaybecomenecessary.
Practitionersandresearchersmustalwaysrelyontheirownexperienceandknowledgeinevaluatingandusing anyinformation,methods,compounds,orexperimentsdescribedherein.Inusingsuchinformationormethods theyshouldbemindfuloftheirownsafetyandthesafetyofothers,includingpartiesforwhomtheyhavea professionalresponsibility.
Tothefullestextentofthelaw,neitherthePublishernortheauthors,contributors,oreditors,assumeanyliabilityfor anyinjuryand/ordamagetopersonsorpropertyasamatterofproductsliability,negligenceorotherwise,orfromany useoroperationofanymethods,products,instructions,orideascontainedinthematerialherein.
LibraryofCongressCataloging-in-PublicationData
AcatalogrecordforthisbookisavailablefromtheLibraryofCongress
BritishLibraryCataloguing-in-PublicationData
AcataloguerecordforthisbookisavailablefromtheBritishLibrary
ISBN978-0-12-817076-2
ForinformationonallAcademicPresspublicationsvisit ourwebsiteat https://www.elsevier.com/books-and-journals
Publisher: StacyMasucci
AcquisitionsEditor: RafaelE.Teixeira
EditorialProjectManager: SaraPianavilla
ProductionProjectManager: MariaBernard
Designer: GregHarris
TypesetbySPiGlobal,India
Editorsbiography SiomardeCastroSoares holdsanMScingenetics, PhDingeneticsandPhDinbioinformatics.Hewas aseniorbioinformaticsresearcherattheOfficialLaboratoryoftheMinistryofFisheries.Currently,heis workingasanassistantprofessoratFederalUniversity ofTri^anguloMineiro—UFTMandisanaffiliatememberoftheBrazilianAcademyofSciences.Hehas published79researchpublicationsand4bookchapters. Hisareasofexpertiseincludemoleculargenetics, genomicsequencing,andmicrobialcomparativegenomics,mainlyfocusedonpan-genomics,theroleof pathogenicityislandsandvirulencefactorsingenome plasticity,phylogenomics,molecularepidemiology, reversevaccinology,andsoftwaredevelopment.
SandipTiwari isapostdoctoralresearcheratthe LaboratoryofCellularandMolecularGenetics,Institute ofBiologicalSciencesInstitute,FederalUniversityof MinasGerais,Brazil.Heearnedabachelor’sdegreein microbiologyin2009fromDeenDayalUpadhyaya GorakhpurUniversity(India),amaster’sdegreein bioinformaticsin2011fromMadhyaPradeshStateUniversity(India),andadoctoratein2017inbioinformatics fromUFMG.Heworksintheareasofbioinformatics, genomics,transcriptomics,proteomics,anddrugtarget identificationagainstinfectiousdiseases.
DebmalyaBarh holdsanMScinappliedgenetics, MTechinbiotechnology,MPhilinbiotechnology, PhDinbiotechnology,PhDinbioinformatics,Postdoc inbioinformatics,andPGDMinpostgraduateinmanagement.HeisanhonoraryscientistattheInstituteof IntegrativeOmicsandAppliedBiotechnology(IIOAB), India.Dr.Barhisblendedwithbothacademicandindustrialresearchfordecadesandisanexpertinintegrative omics-basedbiomarkerdiscovery,moleculardiagnosis, andprecisionmedicineinvariouscomplexhumandiseasesandtraits.Heworkswithover400scientistsfrom morethan100organizationsacrossmorethan40countries.Dr.Barhhaspublishedover150researchpublications,morethan32bookchapters, andhaseditedmorethan20cutting-edge,omics-relatedreferencebookspublishedby Taylor&Francis,Elsevier,andSpringer.HefrequentlyreviewsarticlesforNaturepublications,Elsevier,AACRjournals,NAR,BMCjournals,PLOSONE,andFrontiers,to nameafew.Hehasbeenrecognizedby Who’sWhointheWorld and LimcaBookofRecords forhissignificantcontributionsinmanagingadvancescientificresearch.
VascoAzevedo isaseniorprofessorofgeneticsand deputyheadoftheDepartmentofGenetics,Ecology, andEvolutionatUniversidadeFederaldeMinasGerais, Brazil.HeisamemberoftheBrazilianAcademyofSciencesandisaknightoftheNationalOrderofScientific MeritoftheBrazilianMinistryofScience,Technology andInnovation.Heisalsoaresearcher1Aofthe NationalCouncilforScientificandTechnological Development(CNPq),whichisthehighestposition. ProfessorAzevedoisamoleculargeneticistwhograduatedfromveterinaryschool,FederalUniversityofBahia in1986.Heearnedhismaster’sin1989andPhDin1993 inmoleculargeneticsfromInstitutNationalAgronomiqueParis-Grignon(INAPG)and InstitutNationaldelaRechercheAgronomique(INRA),France.HeconductedapostdoctoralresearchatMicrobiologyDepartmentofMedicineSchoolfromUniversityof Pennsylvania,UnitedStates,in1994.In2017,heearnedanotherPhDinthefieldof bioinformatics.Histotalresearchpublicationsincludemorethan400researcharticles, 3books,andmorethan30bookchapters.ProfessorAzevedoisapioneerofgenetics oflacticacidbacteriaand Corynebacteriumpseudotuberculosis inBrazil.Hehasspecialized andcurrentlyresearchingonbacterialgenetics,genomes,transcriptomes,andproteomes fordevelopmentofnewvaccinesanddiagnosticagainstinfectiousdiseases.
Contributors TalitaEmileRibeiroAdelino
Laborato ´ riodeGeneticaCelulareMolecular,ICB,UniversidadeFederaldeMinasGerais; Fundac ¸aoEzequeilDias(Funed),BeloHorizonte,Brazil
JamilAhmad
ResearchCenterforModeling&Simulation(RCMS),NationalUniversityofSciencesand Technology(NUST),Islamabad,Pakistan
ShahbazAhmed
Atta-ur-RahmanSchoolofAppliedBiosciences,NationalUniversityofSciencesand Technology,Islamabad,Pakistan
LuizCarlosJuniorAlcantara
PGPrograminBioinformatics,InstituteofBiologicalSciences,FederalUniversityofMinas Gerais(UFMG),BeloHorizonte;Laborato ´ riodeFlavivı´rus,InstitutoOswaldoCruz, Fundac ¸ a ˜ oOswaldoCruz,RiodeJaneiro;Laborato ´ riodeGeneticaCelulareMolecular,ICB, UniversidadeFederaldeMinasGerais,BeloHorizonte,Brazil
AmjadAli
DepartmentofPlantBiotechnology,Atta-ur-RahmanSchoolofAppliedBiosciences(ASAB), NationalUniversityofSciencesandTechnology(NUST),Islamabad,Pakistan
RabiaAmir
DepartmentofPlantBiotechnology,Atta-ur-RahmanSchoolofAppliedBiosciences(ASAB), NationalUniversityofSciencesandTechnology(NUST),Islamabad,Pakistan
FabricioAraujo
InstituteofBiologicalSciences,FederalUniversityofPara ´ (UFPA),Belem,Brazil
MuneebaArveen
DepartmentofPlantBiotechnology,Atta-ur-RahmanSchoolofAppliedBiosciences(ASAB), NationalUniversityofSciencesandTechnology(NUST),Islamabad,Pakistan
VascoAzevedo
PGPrograminBioinformatics,InstituteofBiologicalSciences,FederalUniversityofMinas Gerais(UFMG),BeloHorizonte,Brazil
JahanzaibAzhar
Atta-ur-RahmanSchoolofAppliedBiosciences,NationalUniversityofSciencesand Technology,Islamabad,Pakistan
LucianaBalbo
StateUniversityofLondrina,Londrina,Brazil
LiBao
NationalClinicalResearchcenterforCancer,TianjinMedicalUniversityCancerInstituteand Hospital;KeyLaboratoryofCancerPreventionandTherapy,Tianjin,China
DebmalyaBarh
CentreforGenomicsandAppliedGeneTechnology,InstituteofIntegrativeOmicsandApplied Biotechnology(IIOAB),PurbaMedinipur,India
FernandaKhouriBarreto
Laborato ´ riodePatologiaExperimental,InstitutoGonc ¸aloMoniz,FiocruzBahia;Instituto MultidisciplinaremSau ´ de—IMS,UniversidadeFederaldaBahia(UFBA),Salvador,Brazil
AttyaBhatti
Atta-ur-RahmanSchoolofAppliedBiosciences,NationalUniversityofSciencesand Technology,Islamabad,Pakistan
AndreasBurkovski
Friedrich-Alexander-UniversitatErlangen-Nurnberg,Erlangen,Germany
RobertaTorresChideroli StateUniversityofLondrina,Londrina,Brazil
MauricioCorredor
GEBIOMICGroup,FCEN,UniversityofAntioquia,Medellin,Colombia
KennydaCostaPinheiro InstituteofBiologicalSciences,FederalUniversityofPara ´ (UFPA),Belem,Brazil
ArturLuizdaCostaSilva InstituteofBiologicalSciences,FederalUniversityofPara ´ (UFPA),Belem,Brazil
HamzaArshadDar
DepartmentofPlantBiotechnology,Atta-ur-RahmanSchoolofAppliedBiosciences(ASAB), NationalUniversityofSciencesandTechnology(NUST),Islamabad,Pakistan
Letı´ciadeCastroOliveira
DepartmentofImmunology,MicrobiologyandParasitology,InstituteofBiologicalScienceand NaturalSciences,FederalUniversityofTri^anguloMineiro(UFTM),Uberaba,Brazil
SiomardeCastroSoares
DepartmentofImmunology,MicrobiologyandParasitology,InstituteofBiologicalScienceand NaturalSciences,FederalUniversityofTri^anguloMineiro(UFTM),Uberaba,Brazil
JaquelineGoesdeJesus
Laborato ´ riodeFlavivı´rus,IOC,Fundac ¸ a ˜ oOswaldoCruz,RiodeJaneiro;Laborato ´ riode PatologiaExperimental,InstitutoGonc ¸aloMoniz,FiocruzBahia,Salvador,Brazil
ThiagodeJesusSousa
PGPrograminBioinformatics,InstituteofBiologicalSciences,FederalUniversityofMinas Gerais(UFMG),BeloHorizonte,Brazil
TuliodeOliveira
KwaZulu-NatalResearchInnovationandSequencingPlatform(KRISP),CollegeofHealth Sciences,UniversityofKwaZulu-Natal,Durban,SouthAfrica
StephaneFragadeOliveiraTosta
PGPrograminBioinformatics,InstituteofBiologicalSciences,FederalUniversityofMinas Gerais(UFMG);Laborato ´ riodeGeneticaCelulareMolecular,ICB,UniversidadeFederalde MinasGerais,BeloHorizonte,Brazil
UlissesdePa ´ duaPereira
StateUniversityofLondrina,Londrina,Brazil
VagnerdeSouzaFonseca
Laborato ´ riodeFlavivı´rus,IOC,Fundac ¸ a ˜ oOswaldoCruz,RiodeJaneiro;Laborato ´ riode GeneticaCelulareMolecular,ICB,UniversidadeFederaldeMinasGerais,BeloHorizonte, Brazil;KwaZulu-NatalResearchInnovationandSequencingPlatform(KRISP),Collegeof HealthSciences,UniversityofKwaZulu-Natal,Durban,SouthAfrica
DipaliDhawan
BaylorGenetics,Houston,TX,UnitedStates
CesarToshioFacimoto
StateUniversityofLondrina,Londrina,Brazil
NunoRodriguesFaria
DepartmentofZoology,UniversityofOxford,Oxford,UnitedKingdom
NosheenFatima
DepartmentofPlantBiotechnology,Atta-ur-RahmanSchoolofAppliedBiosciences(ASAB), NationalUniversityofSciencesandTechnology(NUST),Islamabad,Pakistan
HenriqueCesarPereiraFigueiredo
AQUACEN,NationalReferenceLaboratoryforAquaticAnimalDiseases,MinistryofFisheries andAquaculture,UniversidadeFederaldeMinasGerais,BeloHorizonte,Brazil
MartaGiovanetti
PGPrograminBioinformatics,InstituteofBiologicalSciences,FederalUniversityofMinas Gerais(UFMG),BeloHorizonte;Laborato ´ riodeFlavivı´rus,IOC,Fundac ¸ a ˜ oOswaldoCruz, RiodeJaneiro;Laborato ´ riodeGeneticaCelulareMolecular,ICB,UniversidadeFederalde MinasGerais,BeloHorizonte,Brazil
Aristo ´ telesGo ´ es-Neto
MolecularandComputationalBiologyofFungiLaboratory,DepartmentofMicrobiology, InstituteofBiologicalSciences(ICB),FederalUniversityofMinasGerais(UFMG),Belo Horizonte,Brazil
AnneCybellePintoGomide
PGPrograminBioinformatics,InstituteofBiologicalSciences,FederalUniversityofMinas Gerais(UFMG),BeloHorizonte,Brazil
LuisCarlosGuimara˜es
InstituteofBiologicalSciences,FederalUniversityofPara ´ (UFPA),Belem,Brazil
RaquelEnmaHurtado
PGPrograminBioinformatics,InstituteofBiologicalSciences,FederalUniversityofMinas Gerais(UFMG),BeloHorizonte,Brazil
FelipeCamposMeloIani
Laborato ´ riodeGeneticaCelulareMolecular,ICB,UniversidadeFederaldeMinasGerais; Fundac ¸ a ˜ oEzequeilDias(Funed),BeloHorizonte,Brazil
IzabelaCoimbraIbraim
PGPrograminBioinformatics,InstituteofBiologicalSciences,FederalUniversityofMinas Gerais(UFMG),BeloHorizonte,Brazil
MadangchanokImchen
DepartmentofGenomicScience,SchoolofBiologicalSciences,CentralUniversityofKerala, Kasaragod,India
ArunKumarJaiswal
DepartmentofImmunology,MicrobiologyandParasitology,InstituteofBiologicalScienceand NaturalSciences,FederalUniversityofTri^anguloMineiro(UFTM),Uberaba;PGProgramin Bioinformatics,InstituteofBiologicalSciences,FederalUniversityofMinasGerais(UFMG), BeloHorizonte,Brazil
SyedBabarJamal
DepartmentofBiologicalSciences,NationalUniversityofMedicalSciences,Rawalpindi, Pakistan
PeterJohn
Atta-ur-RahmanSchoolofAppliedBiosciences,NationalUniversityofSciencesand Technology,Islamabad,Pakistan
RodrigoBentesKato
MolecularandComputationalBiologyofFungiLaboratory,DepartmentofMicrobiology, InstituteofBiologicalSciences(ICB),FederalUniversityofMinasGerais(UFMG),Belo Horizonte,Brazil
JaspreetKaur
UniversityInstituteofEngineeringandTechnology(UIET),DepartmentofBiotechnology, PanjabUniversity,Chandigarh,India
BineypreetKaur
UniversityInstituteofengineeringandTechnology(UIET),DepartmentofBiotechnology, PanjabUniversity,Chandigarh,India
RanjithKumavath
DepartmentofGenomicScience,SchoolofBiologicalSciences,CentralUniversityofKerala, Kasaragod,India
XiaofengLiu
NationalClinicalResearchcenterforCancer,TianjinMedicalUniversityCancerInstituteand Hospital;KeyLaboratoryofCancerPreventionandTherapy,Tianjin,China
NguyenThanhLuan
DepartmentofVeterinaryMedicine,InstituteofAppliedScience,HoChiMinhCityUniversity ofTechnology—HUTECH,HoChiMinhCity,Vietnam
WajahatMaqsood
DepartmentofPlantBiotechnology,Atta-ur-RahmanSchoolofAppliedBiosciences(ASAB), NationalUniversityofSciencesandTechnology(NUST),Islamabad,Pakistan
WandersonMarquesdaSilva
InstituteofAgrobiotechnologyandMolecularBiology,INTA-CONICET,BuenosAires, Argentina
AnupriyaMinhas
UniversityInstituteofengineeringandTechnology(UIET),DepartmentofBiotechnology, PanjabUniversity,Chandigarh,India
FaizaMunir
DepartmentofPlantBiotechnology,Atta-ur-RahmanSchoolofAppliedBiosciences(ASAB), NationalUniversityofSciencesandTechnology(NUST),Islamabad,Pakistan
AmaliaMun ˜ oz-Go ´ mez
GEBIOMICGroup,FCEN,UniversityofAntioquia,Medellin,Colombia
KanwalNaz
DepartmentofPlantBiotechnology,Atta-ur-RahmanSchoolofAppliedBiosciences(ASAB), NationalUniversityofSciencesandTechnology(NUST),Islamabad,Pakistan
AnamNaz
DepartmentofPlantBiotechnology,Atta-ur-RahmanSchoolofAppliedBiosciences(ASAB), NationalUniversityofSciencesandTechnology(NUST),Islamabad,Pakistan
AyeshaObaid
DepartmentofPlantBiotechnology,Atta-ur-RahmanSchoolofAppliedBiosciences(ASAB), NationalUniversityofSciencesandTechnology(NUST),Islamabad,Pakistan
YanPantoja
InstituteofBiologicalSciences,FederalUniversityofPara ´ (UFPA),Belem,Brazil
RommelRamos
InstituteofBiologicalSciences,FederalUniversityofPara ´ (UFPA),Belem,Brazil
NoorUlSaba
DepartmentofPlantBiotechnology,Atta-ur-RahmanSchoolofAppliedBiosciences(ASAB), NationalUniversityofSciencesandTechnology(NUST),Islamabad,Pakistan
AlvaroSalgado
Laborato ´ riodeGeneticaCelulareMolecular,ICB,UniversidadeFederaldeMinasGerais,Belo Horizonte,Brazil
VartulSangal
FacultyofHealthandLifeSciences,NorthumbriaUniversity,NewcastleuponTyne,United Kingdom
Qurat-ul-AinSani
DepartmentofPlantBiotechnology,Atta-ur-RahmanSchoolofAppliedBiosciences(ASAB), NationalUniversityofSciencesandTechnology(NUST),Islamabad,Pakistan
NubiaSeyffert
BiologyInstitute,FederalUniversityofBahia,Salvador,Brazil
FaisalSherazShah
Atta-ur-RahmanSchoolofAppliedBiosciences,NationalUniversityofSciencesand Technology,Islamabad,Pakistan
FatimaShahid
DepartmentofPlantBiotechnology,Atta-ur-RahmanSchoolofAppliedBiosciences(ASAB), NationalUniversityofSciencesandTechnology(NUST),Islamabad,Pakistan
MuhammadShehroz
DepartmentofPlantBiotechnology,Atta-ur-RahmanSchoolofAppliedBiosciences(ASAB), NationalUniversityofSciencesandTechnology(NUST),Islamabad,Pakistan
AmnahSiddiqa
ResearchCenterforModeling&Simulation(RCMS),NationalUniversityofSciencesand Technology(NUST),Islamabad,Pakistan
GyanP.Srivastava
MolecularBiologyandGeneticEngineeringLaboratory,DepartmentofBotany,Universityof Allahabad,Allahabad,India
GuilhermeCamposTavares UniversidadeNiltonLins,Manaus,Brazil
HaiHaPhamThi
FacultyofBiotechnologyandEnvironmentalTechnology,NguyenTatThanhUniversity, HoChiMinhCity,Vietnam
SandeepTiwari
PGPrograminBioinformatics,InstituteofBiologicalSciences,FederalUniversityofMinas Gerais(UFMG),BeloHorizonte,Brazil
BasantK.Tiwary
CentreforBioinformatics,PondicherryUniversity,Pondicherry,India
NimatUllah
DepartmentofPlantBiotechnology,Atta-ur-RahmanSchoolofAppliedBiosciences(ASAB), NationalUniversityofSciencesandTechnology(NUST),Islamabad,Pakistan
RavaliKrishnaVennapu
DepartmentofGenomicScience,SchoolofBiologicalSciences,CentralUniversityofKerala, Kasaragod,India
JoilsonXavier
Laborato ´ riodeFlavivı´rus,IOC,Fundac ¸ a ˜ oOswaldoCruz,RiodeJaneiro;Laborato ´ riode
PatologiaExperimental,InstitutoGonc ¸aloMoniz,FiocruzBahia,Salvador,Brazil
NeelamYadav
MolecularBiologyandGeneticEngineeringLaboratory,DepartmentofBotany,Universityof Allahabad,Allahabad,India
BhupendraN.S.Yadav
MolecularBiologyandGeneticEngineeringLaboratory,DepartmentofBotany,Universityof Allahabad,Allahabad,India
RajivK.Yadav
MolecularBiologyandGeneticEngineeringLaboratory,DepartmentofBotany,Universityof Allahabad,Allahabad,India
DineshK.Yadav
MolecularBiologyandGeneticEngineeringLaboratory,DepartmentofBotany,Universityof Allahabad,Allahabad,India
TahreemZaheer
DepartmentofPlantBiotechnology,Atta-ur-RahmanSchoolofAppliedBiosciences(ASAB), NationalUniversityofSciencesandTechnology(NUST),Islamabad,Pakistan
PAN-GENOMICS: APPLICATIONS, CHALLENGES,AND FUTURE PROSPECTS PAN-GENOMICS: APPLICATIONS, CHALLENGES,AND FUTUREPROSPECTS Editedby
DEBMALYABARH,PhD Scientist,CentreforGenomicsandAppliedGeneTechnology,Instituteof IntegrativeOmicsandAppliedBiotechnology(IIOAB) Nonakuri,India
SIOMARSOARES,PhD AssistantProfessoratDepartmentofImmunology,Microbiology andParasitology,InstituteofBiologicalSciencesandNatural Sciences,FederalUniversityofTrianguloMineiro(UFTM) Uberaba,Brazil
SANDEEPTIWARI,PhD Post-DoctoralResearcher,LaboratoryofCellularandMolecularGenetics, FederalUniversityofMinasGerais(UFMG) BeloHorizonte,Brazil
VASCOAZEVEDO,PhD SeniorProfessor,InstituteofBiologicalSciences,FederalUniversityofMinas Gerais(UFMG)
BeloHorizonte,Brazil
Preface Sincethedevelopmentofthenext-generationsequencingtechnologies,manygenomes havebeendepositedinthedatabasesand,asaresult,thetermpan-genomewascoinedin 2005todescribeanewareaofgenomicsanalysesthatusedseveralstrainsofthesame speciestogaininsightsintothedevelopmentofbacterialgenomes.Thisareahasthen expanded,andnowotherapplicationshaveappearedtocomplementthepan-genomics, creatingthepan-omicsanalyses.Thisbookwasconceivedtobeacompendiumofpangenomicsandotherpan-omicsanalysesfromdifferentorganisms.
Thebook Pan-Genomics:Applications,Challenges,andFutureProspects beginswithan introductiononpan-omicsfocusedtoCrick’sCentralDogmaandabriefdescriptionof allthechaptersofthebook(Chapter1),inwhichsomebasicconceptsofpan-genomics areintroduced. Chapter2,ontheotherhand,discussestheuseofbioinformatics approachesappliedtopan-genomicsandtheirchallenges,withalistofsoftwarethat maybeusefulinthiscontext.In Chapter3,Dr.Tiwaryandcollaboratorsdiscussthe useofpan-genomicsinevolutionarystudiesbasedongenecontentandsinglenucleotide polymorphism.
Next,thechaptersexplorethepan-genomicsofmodelbacterialorganismsandits applicationsuchasinthediscoveryofvaccineanddrugtargetsagainstbacterialpathogens usingreversevaccinologyanddrugtargetanalyses(Chapter16). Chapter4 describesthe pan-genomicsanalysesof Corynebacteriumdiphtheriae and Corynebacteriumulcerans,the causativeagentsofdiphtheriaanddiphtheria-likediseases. Chapter5 describestheuse ofpan-genomicsinveterinarypathogens,focusingonthepan-genomeanalysisof Corynebacteriumpseudotuberculosis,thecausativeagentofCaseouslymphadenitisinsmallruminants.In Chapter6,Dr.Amirexploresthepan-genomesofplantpathogens,focusingon Pectobacteriumparmentieri, Pantoeaananatis, Erwiniaamylovora, Burkholderia, Xylellafastidiosa, Pucciniagraminis, and Zymoseptoriatritici.In Chapter7,Dr.Pereiraexploresthe pan-genomeoffoodpathogenssuchas Escherichiacoli, Salmonellaenterica, Clostridiumbotulinum, Clostridiumperfringens, Listeriamonocytogenes, and Staphylococcusaureus Chapter8 focusesonthepan-genomeofaquaticanimalssuchas Edwardsiella and Aeromonas.In Chapter9,Dr.Aliexploresthepan-genomesofmodelbacteriasuchas Streptococcusagalactiae, Neisseriameningitidis, Staphylococcusaureus, E.coli, Streptococcuspyogenes, Haemophilus influenzae, and Streptococcuspneumoniae.Finally,in Chapter10,thepan-genomeof multidrug-resistanthumanpathogenicbacteriaandtheirresistomearediscussed,focusingonbacteriasuchas Acinetobacterbaumannii and Pseudomonasaeruginosa
Otherchaptersfocusonvirus,plants,algae,fungi,andhumansinpan-canceranalyses. Chapter11 focusesonthepan-genomicsofvirusanditsapplicationstoprovideinsights
intothetransmission,biology,andepidemiologyofhealth-care-associatedviruspathogens,andalsoprovideadescriptionofsoftwareusedforthistask.In Chapter12, Go ´ es-Netoperformsanintensiveliteraturereviewandmetaanalysisofacustomized databasetoprovideinsightsintofunguspan-genomics,withdataonthemoststudied fungiofthe12moreexploredgenera.In Chapter13,Dr.Kaurdescribesthestateof theartinthegenomicsofalgaeorganisms,frommicrotomacroalgae. Chapter14 describesthepan-genomeofplantsanditsapplications,focusingon Brassicarapa, Brassica oleracea, Glycinesoja, Oryzasativa, and Brachypodiumdistachyon Chapter15 describesthe pan-cancerproject,whichmaybehelpfulincancerpreventionandinthedesignofnew cancertherapeutics.
Pan-omicsanalysesarefurtherdescribedinchaptersdedicatedtopan-proteomics, pan-metagenomics,pan-metabolomics,pan-interactomics,andpan-transcriptomics. Pan-metagenomicsisexploredin Chapter17 tobetterunderstandthemicrobiotaofa givenorganismorecosystemindifferentconditionsandalsotoexplorethecommonly sharedmicroorganismsintheseconditions.Theauthorsalsodiscusstheimportanceof pan-metagenomicsinpharmacokinetics.Pan-transcriptomics(Chapter18)andpanproteomics(Chapter19)areintendedtoanalyzethedatasetofdifferentiallyexpressed andcommonlyexpressedgenesindifferentconditionsinordertogiveinsightsintoadaptationtotheseconditions. Chapters20and21 explorepan-metabolomicsandpaninteractomics,respectively,whicharerecentareasofresearchandmayhelpinelucidating differentiallyregulatedmetabolicpathwaysandprotein-proteininteractionsindifferent conditions.
Atotalof65expertsfrom14countrieshavecontributedtothisbooktocoverwide areasofpan-genomics.Webelievethisbookwillprovidethereaderswiththemainstrategiesandtheirapplicationsutilizedsofarinpan-genomics.
Editors
DebmalyaBarh SiomarSoares
SandeepTiwari
VascoAzevedo
Pan-omicsfocusedtoCrick’scentral dogma ArunKumarJaiswal*ab,SandeepTiwari*a,GuilhermeCamposTavaresh, WandersonMarquesdaSilvad,LetíciadeCastroOliveirab,IzabelaCoimbra Ibraima,LuisCarlosGuimarãese,AnneCybellePintoGomidea,SyedBabarJamalc, YanPantojae,BasantK.Tiwaryi,AndreasBurkovskij,FaizaMunirk,HaiHaPhamThil, NimatUllahk,AmjadAlik,MartaGiovanettia,m,LuizCarlosJuniorAlcantaraa,m, JaspreetKaurn,DipaliDhawano,MadangchanokImchenp, RavaliKrishnaVennapup,RanjithKumavathp,MauricioCorredorq,HenriqueCesar PereiraFigueiredog,DebmalyaBarhf,VascoAzevedoa,SiomardeCastroSoaresb aPGPrograminBioinformatics,InstituteofBiologicalSciences,FederalUniversityofMinasGerais(UFMG),BeloHorizonte,Brazil bDepartmentofImmunology,MicrobiologyandParasitology,InstituteofBiologicalScienceandNaturalSciences, FederalUniversityofTri^anguloMineiro(UFTM),Uberaba,Brazil
cDepartmentofBiologicalSciences,NationalUniversityofMedicalSciences,Rawalpindi,Pakistan dInstituteofAgrobiotechnologyandMolecularBiology,INTA-CONICET,BuenosAires,Argentina eInstituteofBiologicalSciences,FederalUniversityofPara ´ (UFPA),Belem,Brazil fCentreforGenomicsandAppliedGeneTechnology,InstituteofIntegrativeOmicsandAppliedBiotechnology(IIOAB), PurbaMedinipur,India
gAQUACEN,NationalReferenceLaboratoryforAquaticAnimalDiseases,MinistryofFisheriesandAquaculture, UniversidadeFederaldeMinasGerais,BeloHorizonte,Brazil hUniversidadeNiltonLins,Manaus,Brazil iCentreforBioinformatics,PondicherryUniversity,Pondicherry,India jFriedrich-Alexander-UniversitatErlangen-Nurnberg,Erlangen,Germany kDepartmentofPlantBiotechnology,Atta-ur-RahmanSchoolofAppliedBiosciences(ASAB),NationalUniversityofSciences andTechnology(NUST),Islamabad,Pakistan lFacultyofBiotechnologyandEnvironmentalTechnology,NguyenTatThanhUniversity,HoChiMinhCity,Vietnam mLaborato ´ riodeFlavivı´rus,IOC,Fundac ¸aoOswaldoCruz,RiodeJaneiro,Brazil, nUniversityInstituteofEngineeringandTechnology(UIET),DepartmentofBiotechnology,PanjabUniversity, Chandigarh,India oBaylorGenetics,Houston,TX,UnitedStates pDepartmentofGenomicScience,SchoolofBiologicalSciences,CentralUniversityofKerala,Kasaragod,India qGEBIOMICGroup,FCEN,UniversityofAntioquia,Medellin,Colombia
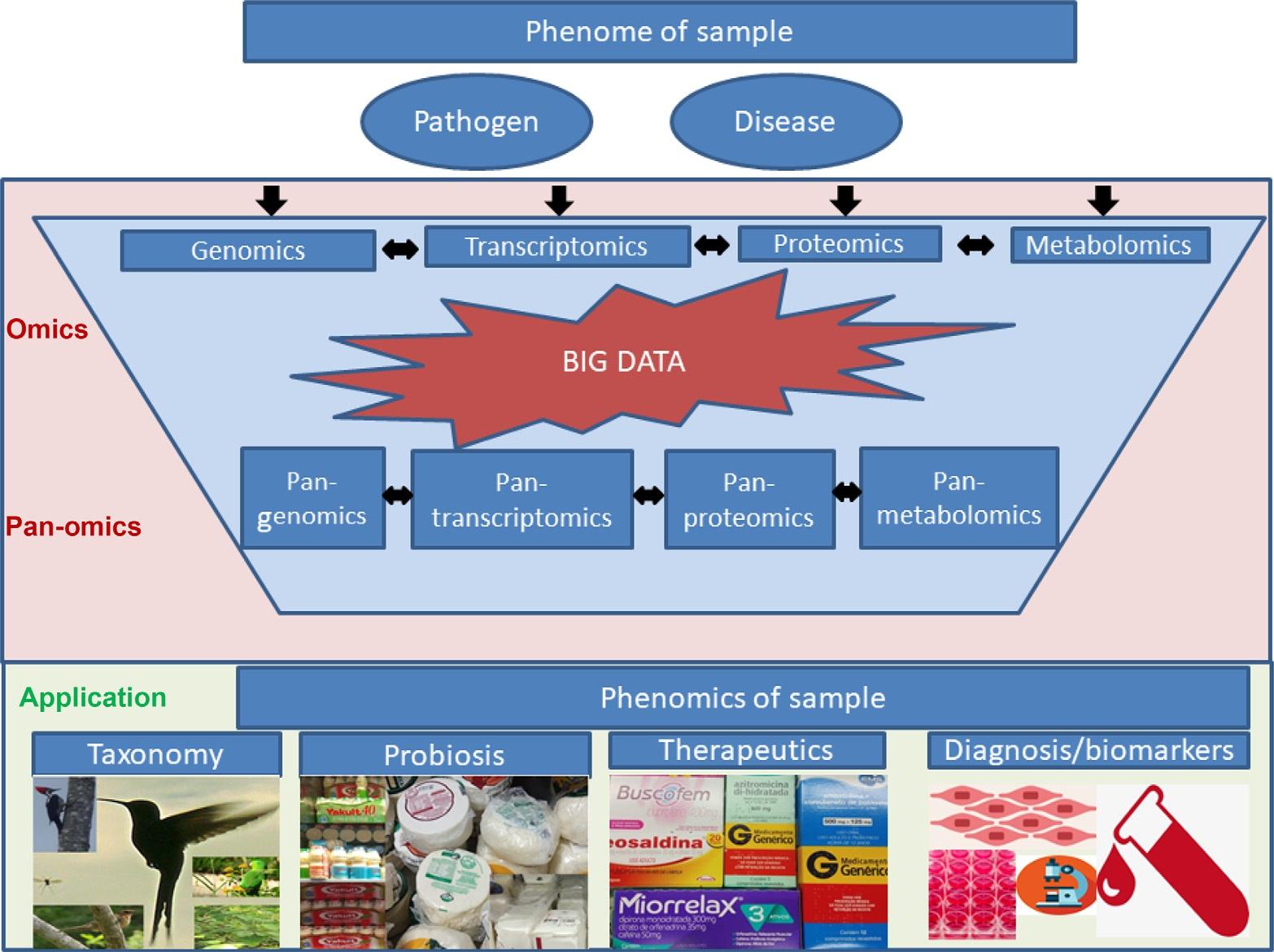
1Introduction SincethedevelopmentofthefirstDNAsequencingtechnologies,manyorganismshad theircompleteDNArepertoiresequencedbySangerandnext-generationsequencing (NGS)technologies,creatingtheareaofgenomics,whichwasoriginatedbythefusion ofthewordsgeneandchromosome [1].Inthisscenario,agenomeisthecompletedataset ofgenesofagivenorganism.Nowadays,therearemorethan200,000genomeprojects registeredattheGenomeOnlineDatabase(GOLD),whereasmorethan120,000are
* Theseauthorscontributedequallytothiswork.
Pan-genomics:Applications,Challenges,andFutureProspects
2020ElsevierInc. https://doi.org/10.1016/B978-0-12-817076-2.00001-9
2 Pan-genomics:Applications,challenges,andfutureprospects
genomesisolatedfrombacteria(https://gold.jgi.doe.gov/statistics).Bacteriaarewidely distributedallovertheworldandhaveimplicationsinhealth,agriculture,industry,and others.Besides,theirgenomesaresmall,highlycompact,anddonotpresentmanyrepetitions,makingthemgoodtargetsforgenomesequencing,oncetheirgenomesareeasier tosequencethantheonesfromotherorganisms.Also,fromthegenomesequenceof bacteria,itispossibletofindvirulencefactors,antibioticresistancegenes,newtherapeutictargetsforvaccineanddrugdevelopment,andindustriallyimportantgenes [2,3].
AnotherimportantpointofthedevelopmentofNGStechnologieswasthegenome sequencingprocessthathasbecomecheaperandfaster,makingitpossibleforsmalllaboratoriestousethetechnologyindailyroutine.NGSmadepossiblethecomparisonofseveralgenomesinamultiprongedstrategy,wherephylogenomics,genomeplasticity,and wholegenomesyntenyanalysesareeasiertoperformnowadays(Fig.1).Also,RNA sequencing(RNA-seq)bytheseplatformsandthedevelopmentofnewtechnologies forsequencingthecompletedatasetofproteinsofanorganismcreatedtheareasoftranscriptomicsandproteomics,respectively [4,5].Altogether,genomicsisresponsibleforthe identificationofthecompletedatasetofgenesofagivenorganism,whereastranscriptomics andproteomicsareimportantfortheidentificationofgenesthataredifferentiallyexpressed
Fig.1 Pan-omicsanditsapplications.
betweenstrainsorspecies.Finally,theeffortstocompareseveralgenomesatoncecreated theareaofpan-genomics,whichwillbefurtherdiscussedinthisbook.
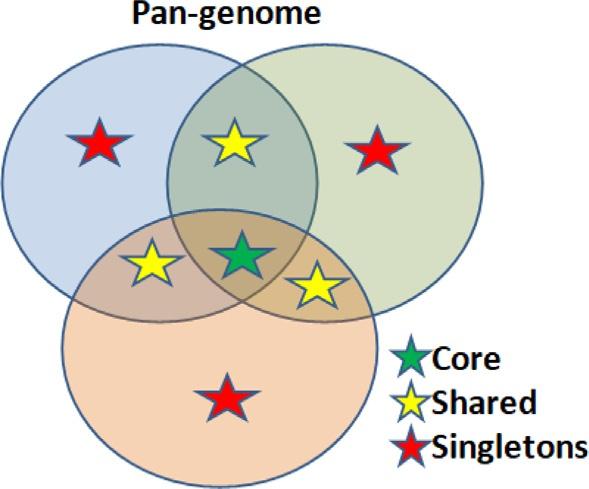
1.1Briefoverviewofpan-genomics Thetermpan-genomicswascreatedbyTettelinandcollaborators,in2005 [6],to describethecompletedatasetofgenesofagivenspeciesthroughthesequencingofseveralstrainsofthisspecies.Thepan-genomeiscomposedofthecoregenome,shared genome,andsingletonssubsets,whereasthecoregenomeiscomposedofallthecommonlysharedgenesbyallstrainsofthespecies;thesharedgenomecontainsgenesthatare presentintwoormore,butnotallstrainsfromaspecies;andthesingletonsarestrainspecificgenes(Fig.2).Fromthesesubsets,onecanextrapolatethedatatofindvaccines anddrugtargetsfromthecoregenome,whereasthesharedgenesandsingletonsare responsiblefordifferencesbetweenthestrainsthatarenormallyresponsiblefortheemergenceofnewpathogensandtheadaptationtonewtraits [6–10]
Normally,thecoregenomeiscomposedofhousekeepinggenesandothergenes importantformetabolismandotherimportantfunctionsoftheorganism,whereasthe sharedgenesandsingletonsaretheresultofgenomeplasticity.Genomeplasticityis thedynamicpropertyofDNAwhichinvolvesthegain,loss,andrearrangementofgenes throughplasmids,phages,andgenomicislands(GEIs).GEIsarehugeblocksofgenes acquiredthroughhorizontalgenetransfer(HGT)thatnormallyshareafunctionincommon.Theyareclassifiedaccordingtothefunctionsofthegenesinto:pathogenicity islands,harboringvirulencefactors;metabolicislands,composedofmetabolism-related genes;resistanceislands,withantibioticresistantgenes;andsymbioticislands,which shareincommonthepresenceofsymbiotic-relatedgenes [11,12]
Normally,thesubsetsofthepan-genomeareidentifiedbytheuseoforthologyanalyses,whichfirstidentifyallorthologousgenesfromthecompletedatasetusingall-vs-all blastsorotheralignmentsearchtools.Next,thedatasetsareclassifiedaccordingtotheir homologytogenesfromotherstrainsinthesubsets.Aftertheclassification,thedatais plottedinachartandmathematicalformulasareusedtofitthespecificcurves.Twosuch
Fig.2 Schematicrepresentationofthecoregenome,sharedgenome,andsingletonsubsetsofpangenomeanalysis.
formulasareHeaps’lawforthepan-genomedevelopmentandleast-squaresfitofthe exponentialregressiondecayforthecoregenomeandsingletonsubsets,whichare describedrespectivelyas: n ¼ k N α,where n isthenumberofgenes, N isthenumber ofgenomes,and k and α areconstantsdefinedbytheformula;and n ¼ k e x/τ + tgθ , where n isthenumberofgenes, x isthenumberofgenomes, e isEuler’snumber, and k, τ,and tgθ areconstantsdefinedbytheformula [6,9].
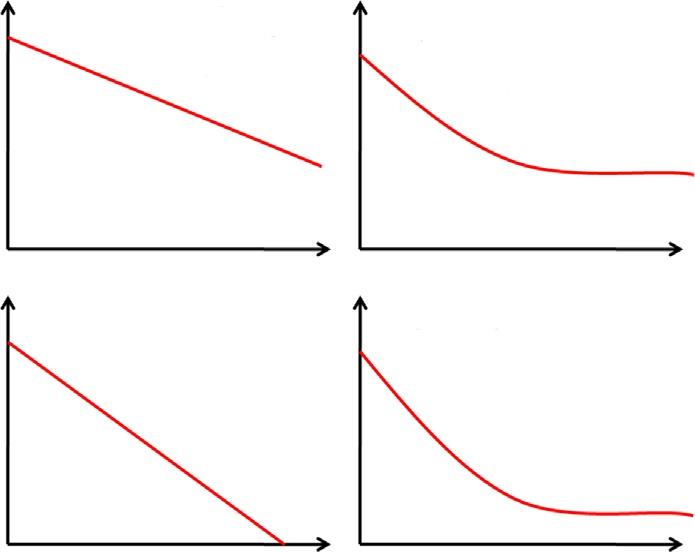
1.2Openandclosedpan-genomes AccordingtoHeap’slaw,the α valueisrepresentativeofthecurrentdynamicsofthepangenome,wherean α higherthan1isrepresentativeofaclosedpan-genomeandan α lowerthan1representsanopenpan-genome.Aclosedpan-genomehasallpossiblegenes representedandonlyfewgeneswillbeaddedtothepan-genomeifmoregenomesareto besequenced,whereasanopenpan-genomeisstillnotfullyrepresentedandthesequencingofnewgenomeswilladdmanygenestotheanalyses [6,9].Thisdefinitioniscontroversial,however,oncetheincorporationofGEIsmaychangethecompositionofthe pan-genomedrastically,evenforclosedpan-genomes,takingittobeopenagain.Most important,environmentalbacteriaandextracellularpathogensnormallyhaveopenpangenomes,oncetheystillneedtoadapttonewtraits,whereasobligateintracellularpathogenstendtohaveclosedpan-genomesoncetheyarenotinconstantcontactwithother bacteria.Also,intracellularpathogenshavelostmanygenesduringevolution,completely adaptingtothehostorganismand,thus,presentverycompactgenomeswithahighpercentageofessentialgenes [13]
Accordingtoleast-squaresfitoftheexponentialregressiondecay,the tgθ isrepresentative ofthenumberofgenespresentinthecoregenomeafterstabilizationofthecoregenome curveand,also,ofthenumberofgenesthatwillbeaddedtotheanalysesafteranewgenome issequencedfromthesingletondevelopmentcurve.Basedonthat,researchersmaychoose thespeciesthatneedmorestrainstobesequencedandwhichdonot.Finally,thehighestthe tgθ onthesingletondevelopment,thelowerthe α,onceahighnumberofgeneswillbeadded totheanalysestakingthepan-genometobemoreopenandthe α tobelower(Fig.3).The oppositeisalsotrue,thelowerthe tgθ ,thehigheristhe α value [6,7,10].
1.3Computationalmethodsusedinpan-genomics Computationalmethodstofindmoreefficientdatastructures,algorithms,andstatistical methodstoperformbioinformaticsanalysesofpan-genomeshavebeenstudiedbecauseit isknownthatinapan-genomeanalysisthegreaterthenumberofgenomestakentothe analysisthegreaterwillbethecomputationalcosts,thatis,thediscoveryofapan-genome contentisanNP-hardproblembecausecomparisonsbetweenallsetsofgenesarenecessarytosolvethetask.Furthermore,inanefforttocomputestandardizedpan-genome analysisandminimizecomputationalchallenges,severalonlinetoolsandsoftwaresuites
(q )=69±9
tg(q )=4±1.5
Fig.3 Theconceptofopenandclosepan-genome.
havebeendeveloped.Examplesofsuchapplicationsare:PGAP [14],oneofthemost completeprofileavailableforperformingfiveanalysismodules,buttheruntimeofthe analysisgrowapproximatelyquadraticallywiththesizeofinputdataandarecomputationallyinfeasiblewithlargedatasets.ThesoftwareRoary [15] andBPGA [16] was createdtoaddressthecomputationalissuesrelatedtoperformanceandexecutiontime. Roaryperformsarapidclusteringofhighlysimilarsequences,whichcanreducethe runtimeofBLAST.BPGAisanultrafastcomputationalpipelinewithsevenfunctional modulesforcomprehensivepan-genomestudiesanddownstreamanalyses.Pan-genome analysiscanbeappliedinmanydifferentapplicationdomains,suchasmicrobes,metagenomics,viruses,plants,cancer,andothers [17].Nowadays,theprocessesofsimilarity searchandpan-genomevisualizationaretwoofthewidevarietyofparticularcomputationalchallengesthatneedtobeconsidered.Forthis,noveldifferentcomputational methodsandparadigmsareneededovertheyears,makingthecomputationalpangenomicsasubareaofresearchinrapidextension.Furthermore,newtechnologiesthat areemerginginrapiddevelopmentallowtoinferthepan-genomewiththreedimensionalconformation,whichmeansthatpossiblyinthefuturethree-dimensional pan-genomeswillnotonlyrepresentallsequencevariationofthespeciesorgenus, butwillalsoencodetheirspatialorganization,aswellastheirmutualrelationshipsin thisregard.
1.4Applicationsofpan-genomicsinevolutionarystudies Themanifestationofrichgeneticdiversityintheformofapan-genomeinaspeciesisan evolutionarypuzzle.Thesethreedistinctpartsofapan-genome(core,shared,andsingletons)ofaparticularspeciesmayundergodifferentevolutionarytrajectoriesunderthe differentialinfluenceofevolutionaryforces.Anidealpan-genomeisexpectedtobevery complete,comprehensive,efficient,andstable [18].Thepan-genomeofaspecieshas someevolutionarysignaturesintheformofgenecontentandsinglenucleotide a <1 ® Open pan-genome a >1 ® Closed pan-genome Openpan-genome
polymorphism(SNP).Theseevolutionarysignaturesareusefulininferringthephylogeneticrelationshipamongdifferentstrainsofaspeciesbasedonthepan-genome.
Anevolutionarypan-genomicstudyofmicrobesprovidesaholisticpictureofallthe genomicvariationsofaspecies.Thesegenomicvariationsendowthebacteriawiththeir uniquepathogenicpropertiesandsubsequentdevelopmentofresistancetovariousantibiotics.Thus,acompletemechanisticdetailoftheprocessesinvolvedinthepathogenesis andfrequentantibioticresistanceinabacteriumwillfurtherpavethewayforbetter detectionmethodsandeffectivecontrolstrategiesforthepathogen.Inaddition,evolutionarypan-genomicsofausefulbacteriumwillhelpusinexploitingmaximallythefull potentialofthemicrobeinenhancingindustrialproductivity.Infact,itwillbeaboomfor theindustriesactivelyinvolvedintheproductionofpharmaceuticalsanddairyproducts usingmicrobialcultures.EukaryotesincludingcropplantsandfarmanimalshaveabundantgenomicvariationsintheformofSNP,copynumbervariants(CNVs),andpresence/absencevariants(PAVs).ThediscoveryofSNPsassociatedwithproductivityor diseaseresistanceinacroporafarmanimalwillbemuchmoreefficientwiththeavailabilityofacompletepan-genomeofthespecies [19].
Inarecentpast,aworkpublishedbyBenevidesetal. [20] utilized16SrRNAgene phylogeny,whole-genomemultilocussequencetyping(wgMLST),phylogenomics, genesynteny,averagenucleotideidentity(ANI),andpan-genometoexplainthephylogeneticrelationshipsinabetterwayamongstrainsof Faecalibacterium.Forthis,theyused 12newlysequenced,assembled,andcuratedgenomesof Faecalibacteriumprausnitzii, whichwereisolatedfromthefecesofhealthyvolunteersfromFranceandAustralia, andcombinedthesewithfivestrainsalreadypublished,whichweredownloadedfrom publicdatabases.Thephylogeneticanalysisofthe16SrRNAalongwiththewgMLST profileandthephylogenetictreebasedonthecomparisonofthesimilarityofgenome supportsthegroupingof Faecalibacterium strainsindifferentgenospecies [20].
InanotherworkpublishedbyChenetal. [21],thecomparisonofwholegenomeand coregenomemultilocussequencetyping(MLST)andSNPanalyseswerecarriedoutto showthemaximumbiasedpowerachievedbyusingmultipleanalyses.Itwasrequiredto differentiateisolatesassociatedwithoutbreakfromapulsed-fieldgelelectrophoresis (PFGE)-indistinguishableisolatecollectedin2012fromanonimplicatedfoodsource. Wholegenomesequencing(WGS)hasbeenprovenasapowerfulsubtypingtoolforbacterialike L.monocytogenes,afoodbornepathogen [21].Acompanyproducedanenvironmentalisolatethatwashighlysimilartoalloutbreakisolates.Thedifferenceobserved betweenunrelatedisolatesandoutbreakisolateswasonly7–14SNPs;consequently, theminimumspanningtreefromtheanalysesofwholegenome,phylogeneticalgorithm, andusualvariantcallingapproachforcoregenome-basedanalysescouldnotofferthe differencebetweenunrelatedisolates.ThisalsosuggestedthattheSNP/allelecounts shouldalwaysbepooledwithWGSclusteringanalysisproducedbyphylogenetically meaningfulalgorithmsonanadequatenumberofisolates,andtheSNP/allele
onsetalonedoesnotprovideenoughevidencetodemarcateanoutbreak [21].Hence,it wasproposedthatthecomparisonofpan-genomesubcategoriesandtheirrelated α value maybeutilizedasanalternateapproach,alongwithANI,intheinsilicocatalogingof newspecies [20,22].Wehopethattheever-expandingpan-genomeacrossdifferentspeciesandgenerawillgiveimpetustoabetterdatastructureofthepan-genomeandnovel computationalmethodsforarobustevolutionarypan-genomicanalysisinnearfuture.
2ApplicationsofPan-genomicsinBacteria 2.1Applicationsofpan-genomicsinmodelbacteria
Advancementinsequencingtechnologiesanddevelopmentinsophisticatedbioinformaticstoolscreatedanoverwhelmingnumberofmicrobialgenomicdataandallowed thescientificcommunitytoestimatethepan-genomeofaspecies.Identificationofnovel dispensablegeneshasapplicationsincharacterizingnovelmetabolicpathways,virulence determinants,andmolecularfingerprintingtargetsforepidemiologicalstudiesandcore genescanbeusedtopredicttheevolutionaryhistoryoftheorganism [9].Therefore,pangenomeanalysesarenowconsideredtheindispensableandgoldstandardforbacterial genomecomparisons,evolution,anddiversity.Itisalsousefultodevelopavaccine againstthepathogensofepidemicdiseasesbyfilteringdifferentfunctionalgenesinthe coregenomeusingreversevaccinologyapproaches [23]
Thereareanumberoffreelyaccessibletools,pipelines,andweb-serversavailableto estimatethemicrobialpan-genomeincludingRoary,BPGA,PGAP,PGAPx,Panseq, PanOCT,etc. [16].Anumberofmodelbacterialspeciespan-genomeisdetermined byresearchersandavastmajorityofthosehumanpathogensexhibitanopenpangenome,astheycolonizemultipleenvironmentsthatfacilitatethemtoexchangegenetic materials.Theseorganismsinclude Escherichiacoli, Meningococci, Streptococci, Salmonellae, Helicobacterpylori,etc. [24].Therefore,indealingwithsuchspeciesareasonablenumber ofgenomesisusuallyrequiredtodefinethecompletegenerepertoireofthesespecies.On theotherhand,specieslivinginisolated(close)habitatshavinglesspossibilitytoexchange geneticmaterialtendtohaveclosedpan-genome,forexample, Mycobacteriumtuberculosis, B.anthracis,and Chlamydiatrachomatis [25].Hence,pan-genomeanalysesserveasaframeworktodetermineandunderstandthegenomicdiversityinbacterialspecies.In Chapter17,wehavediscussedthebacterialpan-genomeanalysisperformedtilldatewith specificexamplesfrommodelorganismsalongwithstudyingapproaches,technical implementations,andtheiroutcome.
2.2Applicationsofpan-genomicsin Corynebacteriumdiphtheriae and Corynebacteriumulcerans
Thedevelopmentofdiphtheriatoxoidvaccinesinthe1920s,thestartofmassimmunizationinthe1940s,andtheglobalintroductionoftheExpandedProgramon
Immunization(EPI)bytheWorldHealthOrganization(WHO)in1974ledtoadramaticdecreaseofdiphtheriacases,bothinindustrializedanddevelopingcountries [26].However,despitethistremendoussuccessstory,diphtheriahasnotbeeneradicated yet.Thishasbeenillustrateddramaticallybyadiphtheriapandemicconnectedtothe breakdownoftheformerUnionofSocialistSovietRepublicswithmorethan 157,000casesandmorethan5000deathsreportedbetween1990and1998.Evenafter thepandemichasfinallystopped,localbreakoutshavebeenobservedconstantlyduring thelastyearsandthereportedglobalcasesincreasedfromabout7000in2016toalmost 9000in2017withafocusoncountrieswithlimitedorlackingpublichealthsystems,for exampleIndia,Indonesia,Nepal,Pakistan,Venezuela,andYemen.Consequently, Corynebacteriumdiphtheriae,theetiologicalagentofrespiratoryandcutaneousdiphtheria,isstill presentonthelistofthemostimportantglobalpathogens [27].Furthermore,thefrequencyofhumandiphtheria-likeinfectionsassociatedwith Corynebacteriumulcerans appearstobeincreasing [28].Thisspecies,whichwasrecognizedbeforeasacommensal ofalargenumberofanimalspecies,iscloselyrelatedto C.diphtheriae andrecognizedasan emergingpathogentoday [28,29].
Theneedoffastandunequivocalidentificationofespeciallypathogenic C.diphtheriae ledtotheearlydevelopmentofanumberofdifferentmethodssuchasbiovardiscriminationbasedondifferentbiochemicalreactions,Elek’stesttoimmunologicallydistinguishbetweentoxigenicandnontoxigenicstrains,restrictionfragmentlength polymorphism(RFLP),single-strandconformationpolymorphism(SSCP),phagetyping,spoligotyping,ribotyping,MLSTandothers.Thisplethoraofmethodswassignificantlyimprovedwhennext-generationsequencingwasintroduced.Thefirstgenome sequenceof C.diphtheriae waspublishedin2003andshowedthepresenceofthe tox gene onabacteriophageinadditiontoanumberofotherhorizontallyacquiredvirulenceassociatedgenes [30].Subsequentpan-genomestudiesallowedunravelingtheextent ofgenomicdiversitywithin C.diphtheriae andtheroleofHGTasasourceofvariation betweenstrains.Furthermore,pan-genomicsof C.ulcerans helpedtoestimatethevirulencepotentialofdifferentstrainsandtoverifyzoonotictransmissionfromanimalsto patients.Today,pan-genomicsof C.diphtheriae and C.ulcerans allowelucidatingglobal transmissiontraitsandlocaladaptationsofpathogeniccorynebacteriaand,hopefully,a betterunderstandingofpopulationdynamicsandstrainevolutionwillhelpcombatdiphtheriaandother Corynebacterium-associateddiseasesinfuture.
2.3Applicationsofpan-genomicsinmultidrug-resistanthuman pathogenicbacteriaandpan-resistome
Thepan-genomewillprobablybethelargestmolecularevolutionaryhistoryofthe organismeverwritten.Thiswillintegrateallthepan-phenotypesexistingonEarth,such asthepan-proteome,thepan-transcriptome,andespecially,aportionofpan-genome thathasmadetheorganismssuccessfulonEarth:thepan-resistome.Thepan-genome
representsthesetofallcurrentgenesinthegenomesofagroupoforganisms.Thebasic genomecommontoallbacteriacontainsabout250genefamiliesintheextended core,thespecificnicheadaptivegenomeofabout8000genefamiliesinthe charactergenepool,andthepan-genomicdiversity(accessorygenes)ofmorethan 139,000raregenefamiliesscatteredthroughoutthebacterialgenomes [31].Thepangenomeanalysis,wherebythesizeofthegenerepertoireaccessibletoanygivenspecies ischaracterizedalongwithanestimateofthenumberofwholegenomesequences requiredtoproperanalysis,andcurrentlyitisincreasing10yearsafterTettelinetal. [6] publication.Differentcurrentmodelsforthepan-genomeanalysis,accuracy,and applicabilitydependonthecaseathand [32].TheNCBI,EMBL,KEEG,PATRIC, MBGD,ENSEMBL,andJGI-IMG/Mdatabasesprovidecompletedownloadablegenomicsinformation,whichcanbeanalyzedforintraspeciesdiversity,anddeterminethe pan-genomeusingsoftwaretools,currentlydevelopedtoperformviaapersonalserver [32],orevenonlineresources.Thepan-genomicsisnowacuttingedgeofcomputational genomicsfield.Pan-genomicsisasubareaofcomputationalbiology [17].Therefore,the notionofcomputationalpan-genomicsintentionallypassesthroughmanyother bioinformatics-relateddisciplines.
Theresistome,atermcoinedbyWright [33],comprisesallthegenesandtheirproductsthatcontributetoresistwhateverenvironment,substance,orsomeextremegrow factor.Updateddatawillclosetothemetadataavailableforestablishingwhatpartofresistometraitsbelongbothtocore-genomeasaccessorygenomeinsideallbacterialspeciesas wellaswillofferabroaderperspectiveofbacterialantibioticresistance.TheWHOsummarizesantimicrobialresistance(AMR)astheresistanceofamicroorganismtoanantimicrobialdrugthatwasoriginallyeffectiveforthetreatmentofinfectionscausedby themselves.Anadequateapproachtosolvingmajorquestionsabouttheresistomeinside ofthebacterialgenome [34] istoperformapan-genomicsanalysis.Theupdatedpangenomedatawillbeclosetothemetadataavailableforestablishingthepartofresistome traitsthatbelongbothtocore-genomeasaccessorygenomeinbacterialspecies;aswellas abroaderperspectiveofantibioticresistanceinbacteria.Theemergentantibioticresistantpathogenicbacteriaareacurrentmenacingconcern. Pseudomonasaeruginosa, Acinetobacterbaumannii,andcoliformbacteriaarethenewemergentantibiotic-resistantbacteriaaccordingtotheWHO.Pan-genomicshastackledsomeimportantconcerns,which wouldbeimpossibletosolveusingclassicalmolecularbiologyordescriptivegenomics:it isveryimportanttodefinethecoreandaccessorygenomeforestablishingtheplasticityof resistome.Thousandsofunknownbacteriaandmicroorganismsareexposedtomanufacturedantibiotics,leadingustoassumethattherearenomeanstopreventthiscatastrophe. Inopposition,pan-genomicsisapowerfulapproachtopreventsuchdisaster.Wemust movetowardsequencingofknownandunknownspecies,classifythem,andestablishing itsantibiotic-resistantstatus,theirpan-genome,andcomeoutwithnewalternativesfor reducingantibioticconsumptionnowadays.
2.4Applicationsofpan-genomicsinveterinarypathogens
FollowingthedevelopmentofNGS,thenumberofsequencedgenomesfiledexponentially [35].Thus,projectsaimedatstudyinggroupsoforganismsbecameviable,andthus, severalstudiesappearedthatarecalledOmicsstudies.Thestudiesinvolvingpan-genomes areexposingimportantinformationonthedifferencesandsimilaritybetweenorganisms ofthesameorbetweenspecies.Forconceptpurposes,wehavethePan-genomeasaset ofgenesinagivengroupofindividuals [10].Thisinformationisbeingexploredand appliedbyseveralscientificfronts,forexample,inbacteriathatinfectanimalsand humans.Themainapplicationsofthesestudiesareinthedevelopmentofprophylactic anddiagnosticmethodsinlesstimeandwithlesscost,moreprecisetaxonomicstudies, studiesongeneticvariations,andpathogenesis [17].Inthischapter,wedescribemore recentresearchinvolvingpan-genomicsofthepathogenicbacteriathatcauseveterinary diseases,includingsomeresponsibleforzoonoses,theyare: Corynebacteriumpseudotuberculosis; Corynebacteriumulcerans; Streptococcussuis; Brachyspirahyodysenteriae; Moraxellabovoculi; Pasteurellamultocida; Mannheimiahaemolytica; Clostridiumbotulinum; Campylobacter; Streptococcusagalactiae; Francisellatularensis; Corynebacteriumdiphtheriae; Brucella spp.Finally, itisworthhighlightingthattheinfluenceoftheapproacheswithbigdataandartificial intelligenceareincreasingandtheinfluencesoftheseinPan-genomicstudieswillbringa neweraofstudiesanddiscoveries.
2.5Applicationsofpan-genomicsinaquaticpathogenicbacteria
Thesustainabilityofaquacult ureindustryiscriticalbothforglobalfoodsecurityand economicwelfare.However, themassivewealthofpathogenicbacteriaposesakey challengetothedevelopmentofasustainablebiocontrolmethod.Recentadvances ingenomesequencingstudycombinedwithpan-genomeanalysiscanbeanefficacious managementappliedtonumerousaquaticpathogens [36].Thus,routinepangenome analysesofgenomic-derivedaquaticpathogenswilldeducethephylogenomicdiversity andpossibleevolutionarytrendsofaquaticbacterialpathogenstrains,elucidatethe mechanismsofpathogenesis,aswellasestim atepatternsofpathogentransmissionacross epidemiologicalscales.Thewholegenome sequencingdataisth eopportunitytorevolutionizethemolecularepidemiologyofaquaculturepathogensasithasforthose pathogensofrelevancetopublichealth [37].Challengesofaquaculturediseasemanagementarethebiologicaldiversityofpathogens,host-pathogeninteractions(e.g.,differentmodesofadaptationandtransmission ),andshiftingenvironmentalpressures,in particularclimatechange.Hence,analysisofpathogenicphenotypecombinedwith genotypederivedfromthefullpotentialof genomesequencingdataiscriticaltoreconstructpathogentransmissionroutesonlocalandglobalscales,aswellasmitigatedisease emergenceandspread.
Comparativepan-genomeanalysesareaneffectivetoolwhichcouldpossiblybe extendedtotheanalysisofaquaticmicroorganismsandtodynamiccharacteristicsand adaptationtoabroadrangeoftheirhostsandenvironmentalniches.Conspicuously, ourpreviouspan-genomeanalysis [38] showedthatstrainWFLU12isolatedfrommarine fishexhibitedniche-specificcharacteristicsofenergyproductionandconversion,and carbohydratetransportandmetabolismbyexploringgenesinthegenerepertoireof strains.Basedonthepan-genomecategories,thefunctionalannotationsofselectedgenes canbereanalyzedwiththeVirulenceFactorsDatabase(VFDB),ClustersofOrthologous Groups(COG),KyotoEncyclopediaofGenesandGenomes(KEGG),andAntibiotic ResistanceGenesDatabase(ARDB).Also,comparativepan-genomehasadvancedto thepointwhengenesarepredictedasbelongingtocellsurface-exposedproteins (SEPs)fromimportantpathogens,includingoutermembraneproteins,andextracellular proteins.Thesepredictedgenesareservingasvaccinecandidatesinananimalmodel calledReversedVaccinology(RV) [39].Inaquaculture,SEPsfrompathogensinclude severalimportantvirulencefactorsthatplaykeyrolesinbacterialpathogenesisandhost immuneresponses.Forexample,theexpressionof esa1 from Edwardsiellatarda,aD15likesurfaceantigen,intheJapanesefloundermodelinducedtheexpressionofabroad spectrumofgenespossiblyinvolvedinbothinnateandadaptiveimmunity,aswellas ahighleveloffishsurvivalandproducedspecificserumantibodies [40].Vaccination usingSEPsresultsinthedevelopmentofprotectiveeffectsagainst Aeromonashydrophila infection, Flavobacteriumcolumnare infection, Pseudomonasputida infection,andEdwardsiellosis[asinthereviewofAbdelgayed [41]].Arecentstudy [42] hassuccessfullyimplementedapan-genomeanalysistoscreenSEPsfrom17representative Leptospirainterrogans strainscoveringmultiepidemicserovarsfromaroundtheworld,and118newcandidate antigenswereidentifiedinadditiontoseveralknownoutermembraneproteinsandlipoproteins.Wehighlyconsiderthattherapidincreaseinthenumberofgenomesequencing ofaquaticpathogenswillallowustodeveloparapid-responseinfectioncontrolprotocols,butalsobeapotentialtrendforstudyingaquaticpathogenicbacteriatoimprovethe cross-serotypeefficacyofvaccinesinfarmedfishandstemthediseaseoutbreakwhen implementingpan-genomeanalysis(usingRVstrategy).Inthechapter“Pan-genomics ofaquaticanimalpathogensanditsapplications,”wereviewedcomparativepan-genome analysiswithaparticularfocusoncontrollingaquaticdiseasesandgivereal-worldexamplesbyanalyzinggenomesequencingdataderivedfromaquaticbacterialisolates.
2.6Pan-genomicsapplicationsfortherapeutics Theemergenceofbacterialresistanceisoccurring,threateningtheabilityofantibiotics thathavetransformedmedicineandsavedmillionsoflivesaroundtheglobe [43,44].The occurrenceofbacterialresistancehasbeenidentifiedsincethebeginningoftheantibiotic erabuttheemergenceofmostdangerousandeasilycommunicatedstrainshasbeen
reportedinpasttwodecades [45,46].Afterseveralyearsofthefirstpatienttreatedwith antibiotics,bacterialinfectionsbecameathreatforsocietyonceagain.Thissituationis mainlybecauseofthemisuseand/oroveruseofantibioticsaswellastheinefficiency ofpharmaceuticalcompaniesfornotproducingadvanceddrugs,onceeconomicinvestmentshavebeenreduced [44].TheCentersforDiseaseControlandPrevention(CDC) hascategorizedseveralbacterialstrainsasanalarmingthreatthatneedseriousconsiderationforpropertreatmentandarealreadyresponsibleforputtingsignificantburdenon thehealth-caresystemintheUnitedStates(US),ultimately,affectingpatientsandtheir families [43,47,48].Theinfectionscausedbyantibiotic-resistantstrainsofbacteriaare pervasiveworldwide [43,44].Anationalsurveyofinfectious-diseasespecialistsledbythe IDSAEmergingInfectionsNetworkin2011foundthatabouttwo-third(2/3)ofthe participantshadseenapan-resistantanddeadlybacterialinfectionwithinthepastfew years [49].Therapidemergenceofresistantbacteriahasbeendescribedasanightmare byseveralpublichealthorganizationsthatcouldhavedisastrousresults [50].TheWHO cautionedin2014thatthedisasterofantibioticresistanceisbecomingdreadful [51] AmongGram-positivepathogens,auniversalendemicofresistant S.aureus and Enterococcus speciesarepresentlythebiggestintimidation [48].Vancomycin-resistantenterococci(VRE)andadditionalemergentpathogensareevolvingresistancetonumerous antibioticsusedcommonly [43].Theworldwidedistributionofcommonrespiratory pathogensincludes Streptococcuspneumoniae and Mycobacteriumtuberculosis,whichare reportedasepidemic [48].Gram-negativepathogensareingeneralmoretroublesome becauseofthefactthattheyarebecomingmoreresistanttoalmostalltheavailabletherapeutics,makingtheconditionsevocativetothepreantibioticera [44].Theoccurrence ofmultidrugresistant(MDR)Gram-negativebacillihasoutdatedallthepracticeinfield ofmedicine [43].ThemostcommoninfectionscausedbyGram-negativebacteriain health-caresettingsareusuallyby Enterobacteriaceae (mostly Klebsiellapneumoniae), Acinetobacter,and Pseudomonasaeruginosa [43,44].Theevolutionofbacterialstrainsanddevelopmentofantibiotic-resistantgenesthroughHGTmakeitnecessarytolookfornovel andadvancedstrategiestocopewiththeinfections [52]
Theinsilicoapproacheslikepan-genome,pan-modelome,subtractivegenomics,and reversevaccinologyareplayingvitalrolesinrapididentificationofnewtherapeutictargetsinthepostgenomicera [53–55].Comparativemicrobialgenomicsapproachalong withstatisticalanalysisareusefultoolsfortheidentificationofessentialgeneticcontents commonlypresentinallpathogenicisolates,basedonsequencesimilarity.Inadditionto essentialgeneticcontents,italsohelpstoidentifysubsetofgenesencodingvirulenceand novelfunctionsasthevariablegenome [56].Apan-genomeisusuallydividedintothree parts,thatis,coregenes,accessorygenes,andstrain-specificgenes.Inthedrugand vaccinediscoveryprocess,theveryfirststepisalwaystheidentificationofasuitable target.Subtractivegenomicsisawidelyusedprocessinthisregard.Inrecentpast,workingwithpathogenicbacteria,usingcomputationalapproaches,alargenumberofnovel