NewandFuture DevelopmentsinMicrobial Biotechnologyand
Bioengineering
TrendsofMicrobialBiotechnologyfor SustainableAgricultureandBiomedicine Systems:DiversityandFunctionalPerspectives
Editedby
AliAsgharRastegari
DepartmentofMolecularandCellBiochemistry, FalavarjanBranch,IslamicAzadUniversity,Isfahan, IslamicRepublicofIran
AjarNathYadav DepartmentofBiotechnology, AkalCollegeofAgriculture, EternalUniversity, BaruSahib,HimachalPradesh,India
NeelamYadav
VeerBahadurSinghPurvanchalUniversity, Ghazipur,UttarPradesh,India
Elsevier Radarweg29,POBox211,1000AEAmsterdam,Netherlands TheBoulevard,LangfordLane,Kidlington,OxfordOX51GB,UnitedKingdom 50HampshireStreet,5thFloor,Cambridge,MA02139,UnitedStates ©2020ElsevierInc.Allrightsreserved.
Nopartofthispublicationmaybereproducedortransmittedinanyformorbyanymeans,electronicormechanical,includingphotocopying, recording,oranyinformationstorageandretrievalsystem,withoutpermissioninwritingfromthepublisher.Detailsonhowtoseekpermission, furtherinformationaboutthePublisher’spermissionspoliciesandourarrangementswithorganizationssuchastheCopyrightClearance CenterandtheCopyrightLicensingAgency,canbefoundatourwebsite: www.elsevier.com/permissions
ThisbookandtheindividualcontributionscontainedinitareprotectedundercopyrightbythePublisher(otherthanasmaybenotedherein).
Notices
Knowledgeandbestpracticeinthisfieldareconstantlychanging.Asnewresearchandexperiencebroadenourunderstanding,changesin researchmethods,professionalpractices,ormedicaltreatmentmaybecomenecessary.
Practitionersandresearchersmustalwaysrelyontheirownexperienceandknowledgeinevaluatingandusinganyinformation,methods, compounds,orexperimentsdescribedherein.Inusingsuchinformationormethodstheyshouldbemindfuloftheirownsafetyandthesafety ofothers,includingpartiesforwhomtheyhaveaprofessionalresponsibility.
Tothefullestextentofthelaw,neitherthePublishernortheauthors,contributors,oreditors,assumeanyliabilityforanyinjuryand/ordamage topersonsorpropertyasamatterofproductsliability,negligenceorotherwise,orfromanyuseoroperationofanymethods,products, instructions,orideascontainedinthematerialherein.
LibraryofCongressCataloging-in-PublicationData AcatalogrecordforthisbookisavailablefromtheLibraryofCongress
BritishLibraryCataloguing-in-PublicationData AcataloguerecordforthisbookisavailablefromtheBritishLibrary
ISBN:978-0-12-820526-6
ForinformationonallElsevierpublications visitourwebsiteat https://www.elsevier.com/books-and-journals
Publisher: SusanDennis
AcquisitionsEditor: KostasKIMarinakis
EditorialProjectManager: AleksandraPackowska
ProductionProjectManager: BharatwajVaratharajan
CoverDesigner: GregHarris
TypesetbySPiGlobal,India
Contributorsxi
1.Tinymicrobes,bigyields: Microorganismsforenhancing foodcropproductionfor sustainabledevelopment 1
ArchanaSingh,RekhaKumari,AjarNathYadav, ShashankMishra,AshishSachan,and ShashwatiGhoshSachan 1.1Introduction
1.4.1Disease-suppressivesoils8
1.4.2Developmentofdisease suppressiveness8
1.4.3Beneficialrhizospheremicrobes: Modulationofthehostimmune system8 1.5Technicalchallengesandemerging
2.Thecontributionofmicrobial biotechnologytosustainable developmentinagricultureand alliedsectors
P.T.Pratheesh,SunainaLal,RandoTuvikene, SivakumarManickam,andSuryaSudheer
2.4Roleofmicroorganismsinimproving cropproduction 19
2.4.1Biofertilizers20
2.4.2Biopesticides22
2.4.3Bioherbicides23
2.4.4Bioinsecticides23
2.4.5Geneticallymodifiedplantsfor sustainableagriculture24
3.Microbialtechnologiesto enhancecropproductionfor futureneeds
LilianaAguilar-Marcelino, LaithKhalilTawfeeqAl-Ani, GloriaSarahiCastaneda-Ramirez, VirginiaGarcia-Rubio,and JuanJoseOjeda-Carrasco
3.1Introduction
3.2Organicagriculture
3.3Agroecology 30
3.4Microbialconsortia 31
3.4.1Bacteria32
3.4.2Microalgaeandcyanobacteria32
3.4.3Protozoa32
3.4.4Yeast33
3.4.5Filamentousfungi34
3.4.6Ediblemushrooms34
3.5Biofertilizers
3.6Biostimulantsofnaturalorigin 35
3.7Useofagro-industrialwaste 36
3.7.1Agro-industrialwastesas substratesforthegenerationof bioenergies38
3.8Agricultureandclimate change 39
3.9Conclusionandfuture
4.Roleandpotentialapplications ofplantgrowth-promoting rhizobacteriaforsustainable agriculture
PankajK.Rai,ManaliSingh,KumarAnand, SatyajitSaurabh,TanvirKaur,DivjotKour, AjarNathYadav,andManishKumar
4.1Introduction
4.2Plantgrowth-promotingrhizobacterial communities
4.3Microbe-mediatedalleviationofabiotic stressinplants 50
4.3.1Nutrientavailabilityforplant uptake53
4.3.2PGPRproducingplantgrowth regulators53
4.3.3Productionofphytohormones54
4.3.4IronchelationbyPGPR54
4.3.5PGPRproducingVOCs(volatile organiccompounds)54
4.3.6PGPRproducingenzymes54
4.4Formsofplantgrowth-promoting rhizobacteria
4.5PGPRasbiofertilizers
4.6PGPRasbiopesticides
4.7Conclusionandfutureprospects
5.Mechanisticunderstandingofthe
MuraliMohanSharaff,Gangavarapu Subrahmanyam,AmitKumar,and AjarNathYadav
5.1Introduction
5.2Rootmicrobiome
5.3Roleofrootmicrobiomeinteractionsin alleviatinginorganicpollutants 63
5.3.1PGPBstrategiestoremediate metaltoxicity67
5.3.2Microbes-assisted phytoremediationstrategy72
5.3.3Microbialbiosorptionand oxidativestressenzymes73
5.3.4Rhizobium-legumeasamodel73
5.4Roleofrootmicrobiomeinteractionsin alleviatingorganicpollutants
5.5Conclusionsandfutureprospects
6.Plantroot-microberelationship forshapingrootmicrobiome modificationinbenefit agriculture 85
SurendraSarsaiya,ArchanaJain, JingshanShi,andJishuangChen
6.1Introduction 85
6.2Plantrootecologyandmicrobial interactions 86
6.2.1Rootcolonization87
6.2.2Hostreactiontowardorganisms andsoilnetwork88
6.2.3Chemistryandmechanismsof action88
6.2.4Rootreactionstobacterialmajority detectingquorumsignals89
6.2.5Plantrootmicrobialvarietyfor sustainableagriculture89
6.3Plantandsoil-deriveddeterminants affectingmicrobialrootcommunities 91
6.4Researchadvancementonplant root-microberelationships 91
7.Biodiversity,phylogeneticprofiling, andmechanismsofcolonizationof seedmicrobiomes 99
KusamLataRana,DivjotKour, TanvirKaur,RubeeDevi,NeelamYadav, AliAsgharRastegari,andAjarNathYadav
7.1Introduction 99
7.2Isolationandcharacterizationofseed endophyticmicrobes 100
7.2.1Isolationandenumerationofseed endophyticmicrobes100
7.2.2Molecularcharacterizationof seedmicrobiomes101
7.2.3Diversityanddistributionofseed microbiomes101
7.3Endophyticlifestyle:Mechanismsof interactionwithplants 112
7.3.1Relationshipofflower microbiomeswithseed microbiomes113
7.3.2Interactionsbetweenmicrobes androotzone114
7.3.3Interactionsbetweenmicrobes andendosphere114
7.3.4Interactionsbetweenmicrobes andsoil115
7.3.5Microbialcolonization116
7.4Thegenomesofendophyticmicrobiomes 117
7.5Conclusionandfutureprospects 117 Acknowledgments 117 References 117
8.Biotechnologicalapplicationsof seedmicrobiomesforsustainable agricultureandenvironment 127
KusamLataRana,DivjotKour,TanvirKaur, RubeeDevi,NeelamYadav,Gangavarapu Subrahmanyam,AmitKumar,and AjarNathYadav
8.1Introduction 127
8.2Functionalattributesofseed microbiomes 128
8.3Biotechnologicalapplications 129
8.3.1Agriculturalapplications129
8.3.2Bioremediation136
8.4Conclusionsandfutureprospect 137 Acknowledgments
9.Microbialbiofilms:Beneficial applicationsforsustainable agriculture
MozhganGhiasian
9.1Introduction
9.2Plantsurfaces:Complexanddynamic environments
9.3Biofilmsintherhizosphere 146
9.4Biofilmsonseedsandsprouts 147
9.5Biofilmsinepiphyticplantcolonization 147
9.6Agriculturallyimportantmicroorganisms 147
9.7Agriculturallyimportantmicrobial biofilms 148
9.7.1Importanceofbiofilmformationin plantgrowthpromotion148
9.7.2Importanceofbiofilmformationin biocontrol149
9.7.3Importanceofbiofilmas biofertilizers149
9.7.4Importanceofbiofilmformation inbioremediation151
9.7.5Importanceofbiofilmformation innutrientmobilization151
9.8Conclusionandfutureprospect 151 References 152
10.Phytasesfrommicrobesin phosphorusacquisitionforplant growthpromotionandsoilhealth 157
DivjotKour,TanvirKaur,NeelamYadav, AliAsgharRastegari,BijenderSingh, VinodKumar,andAjarNathYadav
10.1Introduction 157
10.2Biodiversityofphytases-producing microbes 158
10.3Microbialphytases 160
10.3.1Productionofphytases160
10.3.2Factorsaffectingphytase production160
10.3.3Purificationofphytases161
10.3.4Propertiesofphytases161
10.3.5Regulationofphytaseformation164
10.4Biotechnologicalapplications 165
10.4.1Phosphorusacquisitionand plantgrowthpromotion165
10.4.2Fishfeed165
10.4.3Poultrynutrition166
10.4.4Pigdiet166
10.4.5Foodindustry166
10.4.6Humannutrition166
10.5Molecularbiology 167
10.5.1Geneexpression167
10.5.2Transgenicswithbacterialand fungalphytases168
10.6Conclusionsandfutureprospects 168 References 169 Furtherreading 176
11.Potassiumsolubilizingand mobilizingmicrobes:Biodiversity, mechanismsofsolubilization,and biotechnologicalimplicationfor alleviationsofabioticstress 177
DivjotKour,KusamLataRana,TanvirKaur, NeelamYadav,SumanKumarHalder, AjarNathYadav,ShashwatiGhoshSachan,and AnilKumarSaxena
11.1Introduction 177
11.2Dynamicsandfunctionsofpotassium 178 11.3Biodiversityandabundanceof K-solubilizingmicrobes 179
11.4MechanismsofK-solubilizationand mobilization 180
11.5Plantgrowthpromotingattributesof K-solubilizers 180
11.5.1K-solubilizersasplantgrowth promoters186
11.5.2OtherPGPattributesof K-solubilizingmicrobes186
11.6Biotechnologicalimplicationfor alleviationsofabioticstress 193
11.6.1Coldstresstoleranceand mitigation193
11.6.2Saltstresstoleranceand mitigation193
11.6.3Waterstresstoleranceand mitigation194
11.7RoleofKSMSasbiofertilizers 194
11.8RoleofKSMSinsoilfertility andhealth 194
11.9Conclusionandfuturescope 195 Acknowledgments 195
References 195
12.Scientifichealthassessmentsin agricultureecosystems—Towards acommonresearchframework forplantsandhuman 203
GanapathyAshok,ChandranViswanathan, GuruvuNambirajan,andKrishnanBaskaran
12.1Introduction 203
12.2Ecosystemhealthandcriteriafor indicatorselection 203
12.3Ecosystemhealthassessment methods 204
12.3.1Holisticapproach204
12.3.2Networkanalysis205
12.3.3Multimetricapproach205
12.3.4Predictivemodelapproach205
12.4Methodsforhuman-coupled ecosystems 206
12.4.1Analyticalhierarchyprocess (AHP)model206
12.4.2Mathematicalmodels206
12.4.3Artificialneuralnetworks206
12.4.4Geneticalgorithms207
12.5Theagroecosystem 207
12.5.1Energyflow208
12.5.2Nutrientcycling208
12.5.3Populationregulating mechanisms208
12.5.4Dynamicequilibrium208
12.5.5Land209
12.5.6Water209
12.5.7Soil209
12.6Agroecosysteminhumanhealth 209
12.7Conclusionandfutureprospects 210 Acknowledgments
13.Cyanobacteria:Aperspective paradigmforagricultureand environment 215
SandeepK.Malyan,SwatiSingh, ArchanaBachheti,MadhviChahar, MitaliKumariSah,Narender,AmitKumar, AjarNathYadav,andSmitaS.Kumar
13.1Introduction 215
13.2Cyanobacteriaandtheirrolein agriculture 215
13.3Roleofcyanobacteriain bioremediation 217
13.4Roleofcyanobacteriainglobal warming 218
13.4.1Atmosphericcarbondioxide sequestrationbycyanobacteria218
13.4.2Methaneemissionsmitigation frompaddysoilsthrough cyanobacteria219
13.5Biofuelproduction 219
13.6Utilizationofcyanobacteriaasfood supplements 220
13.7Conclusionandfutureprospects 220 References 221 Furtherreading 224
14.Theroleofmicrobialsignalsin plantgrowthanddevelopment: Currentstatusandfuture prospects 225
LaithKhalilTawfeeqAl-Ani, TheophileFranzino,LilianaAguilar-Marcelino, FethelZaharHaichar,EdsonLuizFurtado, WaqasRaza,GhulamHussainJatoi,and MubasharRaza
14.1Introduction 225
14.2Signalsbeneficialtoplantmicrobiota 226 14.2.1Enhancementofthemicrobial signalsfortheplantgrowthand development226
14.2.2Enhancementofthemicrobial signalsforinducedofplant defenses228
14.3Theformofrelationshipbetween plantdefenseandplantgrowth 228
14.4Theeffectofsignalsofplant-microbial pathogenonthegrowthand developmentofplantcomparedwith beneficialmicrobes 229
14.4.1Virusesandviroids230 14.4.2Phytoplasma232
14.4.3Bacteria232
14.4.4Fungi233
14.5Conclusionandfutureprospect 234 References 235 Furtherreading 242
15.Microbialbiopesticides: Currentstatusandadvancement forsustainableagricultureand environment 243
NeelamThakur,SimranjeetKaur, PreetyTomar,SeemaThakur,and AjarNathYadav
15.1Introduction 243
15.2Biopesticides 244
15.2.1Botanicals245
15.2.2Biochemical246
15.2.3Microbial248
15.3Biopesticide:Statusofresearch anddevelopment 253
15.3.1StatusinIndia253
15.3.2Biopesticidesworldwide254
15.4Commercialaspectsand applications 254
15.4.1Modeofaction254
15.4.2Modeofactiontakenupby differentmicroorganism256
15.4.3Formulationandproduction257
15.5Limitationsandchallenges 260
15.5.1Useofconventional pesticides261
15.5.2Replacementofchemical pesticides261
15.5.3Efficacyofdifferenttypesof biopesticides263
15.5.4Qualitycontrolof biopesticides264
15.5.5Regulationofbiopesticides264
15.6Roleofbiopesticidesinsustainable developments 268
15.6.1Microbialpesticidesin integratedpestmanagement268
15.6.2Roleinnanotechnology271
15.6.3Roleinsustainable environments272
15.6.4Roleinsustainable agriculture273
15.7Conclusionandfutureaspects 273 References
16.Salinemicrobiome:Biodiversity, ecologicalsignificance,and potentialroleinamelioration ofsaltstress 283
AjarNathYadav,TanvirKaur,DivjotKour, KusamLataRana,NeelamYadav, AliAsgharRastegari,ManishKumar, DibyPaul,ShashwatiGhoshSachan,and AnilKumarSaxena
16.1Introduction 283
16.2Biodiversityandecological significance 284
16.2.1Archaea287
16.2.3Fungi295
16.3Roleofsalt-tolerantmicrobesin cropsimprovements 297
16.3.1BiologicalN2-fixation297
16.3.2Phosphorusandpotassium solubilization300
16.3.3Phytohormonesproduction300
16.3.4ACCdeaminase production300
16.3.5Indirectplantgrowth-promoting attributes301
16.4RoleofPGPmicrobesforthe ameliorationofsaltstress 301
16.5Conclusionsandfuture
17.Globalscenarioandfuture prospectsofthepotential microbiomesforsustainable agriculture 311 AjayKumarandJoginderSingh
17.1Introduction 311 17.2Plant-associatedmicrobes 312
17.3Mechanismofrhizosphere microbesinteraction 314
17.3.1Directmechanisms315
17.3.2Indirectmechanisms316
17.4Quorumsensing 316
17.4.1Mathematicalmodelingof quorumsensinginbacteria316 17.5Biofilm 318
17.5.1Biofilmformation318
17.5.2Factorsinfluencingbiofilm formation319
17.5.3Biofilmgrowthmodel319
17.6Applicationsoftherhizosphere microbiome
17.6.1Bioremediation320
17.6.2Biofuelproductions322
17.6.3Biofertilizers323
17.6.4Biopesticides323
17.6.5Biocontrol324
17.6.6Biodegradation324
17.7Conclusionandfuture
18.Microbialbiotechnologyfor sustainableagriculture:Current researchandfuturechallenges
AjarNathYadav,DivjotKour,TanvirKaur, RubeeDevi,GeetikaGuleria, KusamLataRana,NeelamYadav,and AliAsgharRastegari
Contributors
Numbersinparenthesisindicatethepagesonwhichtheauthors’ contributionsbegin.
LilianaAguilar-Marcelino (29,225),CentroNacionalde Investigacio ´ nDisciplinariaenSaludAnimale Inocuidad,INIFAP,Jiutepec,Morelos,Mexico
LaithKhalilTawfeeqAl-Ani (29,225),Departmentof PlantProtection,CollegeofAgriculture,Universityof Baghdad,Baghdad,Iraq;SchoolofBiologyScience, UniversitiSainsMalaysia,Minden,PulauPinang, Malaysia
KumarAnand (49),DepartmentofBiotechnology, VinobaBhaveUniversity,Hazaribag,India
GanapathyAshok (203),DepartmentofBiotechnology, SreeNarayanaGuruCollege,Coimbatore,TamilNadu, India
ArchanaBachheti (215),DepartmentofEnvironmental Science,GraphicEraUniversity,Dehradun, Uttarakhand,India
KrishnanBaskaran (203),DepartmentofBiochemistry, SreeNarayanaGuruCollege,Coimbatore,TamilNadu, India
GloriaSarahiCastan ˜ eda-Ramirez (29),CentroNacional deInvestigacio ´ nDisciplinariaenSaludAnimale Inocuidad,INIFAP,Jiutepec,Morelos,Mexico
MadhviChahar (215),InstituteofPost-HarvestandFood Science,TheVolcaniCenter,AgriculturalResearch Organization(ARO),RishonLeZion,Israel
JishuangChen (85),BioresourceInstituteforHealthy Utilization,ZunyiMedicalUniversity,Zunyi;College ofBiotechnologyandPharmaceuticalEngineering, NanjingTechUniversity,Nanjing,China
RubeeDevi (99,127,331),DepartmentofBiotechnology, Dr.KSGAkalCollegeofAgriculture,EternalUniversity,BaruSahib,HimachalPradesh,India
TheophileFranzino (225),UniversitedeLyon,UMR5557 LEM,UniversiteLyon1,CNRS,INRA1418,VilleurbanneCedex,France
EdsonLuizFurtado (225),DepartmentofPlantProduction,CollegeofAgronomicScienceFazendaExperimentalLageado,SaoPauloStateUniversity,Sao Paulo,Brazil
VirginiaGarcia-Rubio (29),CentroUniversitarioAmecameca.UniversidadAuto ´ nomadelEstadodeMexico, Amecameca,Mexico
MozhganGhiasian (145),DepartmentofMicrobiology, FalavarjanBranch,IslamicAzadUniversity,Isfahan, Iran
GeetikaGuleria (331),DepartmentofBiotechnology, Dr.KSGAkalCollegeofAgriculture,EternalUniversity,BaruSahib,HimachalPradesh,India
FethelZaharHaichar (225),UniversitedeLyon,UMR 5557LEM,UniversiteLyon1,CNRS,INRA1418, VilleurbanneCedex,France
SumanKumarHalder (177),DepartmentofMicrobiology,VidyasagarUniversity,Midnapore,West Bengal,India
ArchanaJain (85),KeyLaboratoryofBasicPharmacologyandJointInternationalResearchLaboratoryof EthnomedicineofMinistryofEducation,Zunyi MedicalUniversity,Zunyi,China
GhulamHussainJatoi (225),DepartmentofPlant Pathology,FacultyofCropProtection,SindhAgricultureUniversity,TandojamSindh,Pakistan
SimranjeetKaur (243),DepartmentofZoologyand Entomology,AkalCollegeofBasicSciences,Eternal University,Sirmour,India
TanvirKaur (49,99,127,157,177,283,331),Department ofBiotechnology,Dr.KSGAkalCollegeofAgriculture,EternalUniversity,BaruSahib,Himachal Pradesh,India
DivjotKour (49,99,127,157,177,283,331),Department ofBiotechnology,Dr.KSGAkalCollegeofAgriculture,EternalUniversity,BaruSahib,Himachal Pradesh,India
AjayKumar (311),SchoolofBioengineeringandBiosciences,LovelyProfessionalUniversity,Phagwara, Punjab,India
AmitKumar (61,127,215),CentralMugaEriResearch& TrainingInstitute,CentralSilkBoard,MinistryofTextiles,Govt.ofIndia,Jorhat,Assam,India
ManishKumar (49,283),AmityInstituteofBiotechnology,AmityUniversity,Gwalior,MadhyaPradesh, India
SmitaS.Kumar (215),CenterforRuralDevelopmentand Technology,IndianInstituteofTechnologyDelhi,New Delhi,India
VinodKumar (157),QualityAnalysisLab.,Forage Section,CollegeofAgriculture,CCSHaryanaAgriculturalUniversity,Hisar,India
RekhaKumari (1),DepartmentofBio-Engineering,Birla InstituteofTechnology,Ranchi,Jharkhand,India
SunainaLal (17),DepartmentofMolecularMedicine, StJohn’sNationalAcademyofHealthSciences, Bangalore,India
SandeepK.Malyan (215),InstituteofSoil,Water,and EnvironmentalSciences,TheVolcaniCenter,AgriculturalResearchOrganization(ARO),RishonLeZion, Israel
SivakumarManickam (17),DepartmentofChemicaland EnvironmentalEngineering,FacultyofScienceand Engineering,UniversityofNottinghamMalaysia, Semenyih,Malaysia
ShashankMishra (1),BiotechPark,Lucknow,India
GuruvuNambirajan (203),DepartmentofMicrobiology, SreeNarayanaGuruCollege,Coimbatore,TamilNadu, India
Narender (215),DepartmentofSoilScience,Chaudhary CharanSinghHaryanaAgriculturalUniversity,Hisar, Haryana,India
JuanJoseOjeda-Carrasco (29),CentroUniversitario Amecameca.UniversidadAuto ´ nomadelEstadode Mexico,Amecameca,Mexico
DibyPaul (283),DepartmentofEnvironmentalEngineering,KonkukUniversity,Seoul,RepublicofKorea
P.T.Pratheesh (17),NehruArtsandScienceCollege, Coimbatore,India
PankajK.Rai (49),DepartmentofBiotechnology,Invertis University,Bareilly,India
KusamLataRana (99,127,177,283,331),Departmentof Biotechnology,Dr.KSGAkalCollegeofAgriculture, EternalUniversity,BaruSahib,HimachalPradesh, India
AliAsgharRastegari (99,157,283,331),Departmentof MolecularandCellBiochemistry,FalavarjanBranch, IslamicAzadUniversity,Isfahan,Iran
MubasharRaza (225),StateKeyLaboratoryofMycology, InstituteofMicrobiology,ChineseAcademyof Sciences,Beijing,China
WaqasRaza (225),DepartmentofPlantPathology,College ofAgriculture,UniversityofSargodha,Sargodha, Pakistan
AshishSachan (1),DepartmentofLifeSciences,Central UniversityofJharkhand,Ranchi,India
ShashwatiGhoshSachan (1,177,283),Departmentof Bio-Engineering,BirlaInstituteofTechnology,Ranchi, Jharkhand,India
MitaliKumariSah (215),RegionalAgriculturalResearch Station,Parwanipur,Bara,Nepal
SurendraSarsaiya (85),KeyLaboratoryofBasicPharmacologyandJointInternationalResearchLaboratoryof EthnomedicineofMinistryofEducation;Bioresource InstituteforHealthyUtilization,ZunyiMedical University,Zunyi,China
SatyajitSaurabh (49),DepartmentofBioengineering, BirlaInstituteofTechnology,Ranchi,India
AnilKumarSaxena (177,283),ICAR-NationalBureau ofAgriculturallyImportantMicroorganisms,Mau, India
MuraliMohanSharaff (61),DepartmentofBiological Sciences,P.D.PatelInstituteofAppliedSciences, CharotarUniversityofScienceandTechnology, CHARUSATCampus,Changa,Gujarat,India
JingshanShi (85),KeyLaboratoryofBasicPharmacology andJointInternationalResearchLaboratoryofEthnomedicineofMinistryofEducation,ZunyiMedicalUniversity,Zunyi,China
ArchanaSingh (1),DepartmentofBio-Engineering,Birla InstituteofTechnology,Ranchi,Jharkhand,India
BijenderSingh (157),DepartmentofBiotechnology, SchoolofInterdisciplinaryandAppliedLife Sciences,CentralUniversityofHaryana,Mahendergarh, India
JoginderSingh (311),SchoolofBioengineeringandBiosciences,LovelyProfessionalUniversity,Phagwara, Punjab,India
ManaliSingh (49),DepartmentofBiotechnology,Invertis University,Bareilly,India
SwatiSingh (215),DepartmentofEnvironmental Science,GraphicEraUn iversity,Dehradun, Uttarakhand,India
GangavarapuSubrahmanyam (61,127),CentralMuga EriResearch&TrainingInstitute,CentralSilkBoard, MinistryofTextiles,Govt.ofIndia,Jorhat,Assam,India
SuryaSudheer (17),DepartmentofBotany,Instituteof EcologyandEarthSciences,UniversityofTartu,Tartu, Estonia
NeelamThakur (243),DepartmentofZoologyand Entomology,AkalCollegeofBasicSciences,Eternal University,Sirmour,India
SeemaThakur (243),DepartmentofBiotechnology,Dr. KSGAkalCollegeofAgriculture,EternalUniversity, BaruSahib,HimachalPradesh,India
PreetyTomar (243),DepartmentofZoologyandEntomology,AkalCollegeofBasicSciences,EternalUniversity,BaruSahib,HimachalPradesh,India
RandoTuvikene (17),SchoolofNaturalSciencesand Health,TallinnUniversity,Tallinn,Estonia
ChandranViswanathan (203),DepartmentofBiotechnology,SreeNarayanaGuruCollege,Coimbatore, TamilNadu,India
AjarNathYadav (1,49,61,99,127,157,177,215,243, 283,331),DepartmentofBiotechnology,Dr.KSGAkal CollegeofAgriculture,EternalUniversity,BaruSahib, HimachalPradesh;KrishiVigyanKendra,Kandaghat, Solan,India
NeelamYadav (99,127,157,177,283,331),GopiNath P.G.College,VeerBahadurSinghPurvanchalUniversity,Ghazipur,UttarPradesh,India
Tinymicrobes,bigyields:Microorganisms forenhancingfoodcropproductionfor sustainabledevelopment
ArchanaSingha,RekhaKumaria,AjarNathYadavb,ShashankMishrac,AshishSachand andShashwatiGhoshSachana
a DepartmentofBio-Engineering,BirlaInstituteofTechnology,Ranchi,Jharkhand,India, b DepartmentofBiotechnology,Dr.KSGAkalCollegeof Agriculture,EternalUniversity,BaruSahib,HimachalPradesh,India, c BiotechPark,Lucknow,India, d DepartmentofLifeSciences,CentralUniversity ofJharkhand,Ranchi,India
1.1Introduction
Duetotherisingdemandforfoodandfiberatthegloballevel,improvedandsustainableagriculturalpracticesarerequired tocombatadverseclimaticconditions.Theincorporationofsuchsustainablepracticeswouldbepreferableoverclassical practicesforimprovedyieldofagriculturalgoodsfromtheexistingarablelandunderdeterioratingsoilandwaterquality conditions.“Sustainableincreasesinagriculturalproductivityarecriticaltoaddressmultiplesustainabledevelopment goals(SDGs)includingzerohunger(SDG2),nopoverty(SDG1),andgoodhealthandwell-being(SDG3).”According tothesurvey,globalpopulationisexpectedtoexceed9billionby2050.Hence,tomeetthefoodrequirementofsuchlarge population,cropproductivityneedstobeincreasedby70%–100%.Additionally,theseadvancedpracticesactasasaviorof agriculturalproduceagainstnewdevelopingandprevalentpestsandpathogens.‘Phytomicrobiome’isasustainableand effectiveapproachfortheimprovementinbothfarmproductivityandfoodquality(Yadavetal.,2020b,2020c).Theharnessingofnaturalresourcesintheagriculturesectornotonlyimprovesfarmproductivitybutalsopromotesenvironmental andsocialoutcomesinapositivemanner.Inconventionalfarming,widespreaduseofchemical-basedfertilizersandpesticidesisrequiredtoincreaseagriculturalproductivitysubstantially(Kouretal.,2020d; Yadavetal.,2020d).Theapplicationsofsuchchemical-basedstrategyinthefarmingsectorhavecontributedenormouslytofulfillthefoodavailability andpovertymitigationgoals.However,thischemical-basedconventionalfarmingisconsideredenvironmentally unfriendlyduetoextremeandindiscriminateuseofchemicals,whichcausespotentialthreatstoenvironmentwhichin turnhaveanegativeimpactonhumanhealthandfoodsecurity.Theseconventionalchemical-basedapproachesnotonly increasedcropssusceptibilitytowardpests/pathogensbutalsoresultedinfoodcontaminationandcontributedsignificantly tosoildegradationandbiodiversityloss(Tilmanetal.,2002).
Agriculturesectorinthedevelopingcountriesarefacingmajorchallengesassociatedwithyieldenhancementinterms ofqualityandquantityinanenvironmentallyfriendlyandeconomical(noincreaseinfarmingcosts)manner.Itisevident fromearlierreportsthatuseofconventionalagriculturalpracticestomaintainabalancebetweendemandandsupplyis neithereconomicallynorenvironmentallyfeasible.Hence,globalurgeforcomplimentaryandsustainableapproaches tosustainablymeettheglobalfoodsecuritydemandshasledtothedevelopmentofamendedandinnovativesustainable cropproductionmethods.
1.2Microbiometechnology
Themicrobiometechnologyisonesuchwayinwhichtheaidofbeneficialplant-associatedmicrobiomehelpstosustainablyenhancethequalityandquantityoffarmproduceusingminimumresources(Kouretal.,2020a,2020b).Thistechnologyhasthepotentialtominimizetheenvironmentaldistress.Microbiomesarethehost-associatedmicrobial communitieswhichinhabitmultipletissuetypesonthehostsurfacesaswellascolonizebothinter-andintracellularhost habitats(MedinaandSachs,2010; Huttenhoweretal.,2012).Amongdifferentmicrobialcommunities,eubacteriaoften
dominatetheplant-associatedmicrobiomesalongwithotherimportantmicrobiomesincludingfungi,protozoa,archaea, andviruses(MedinaandSachs,2010; Huttenhoweretal.,2012; BonkowskiandClarholm,2012; Hoffmannetal.,2013).
Anenormousdiversityofmicrobialcommunityisassociatedwithplantroots.Thesemicrobescanbeutilizedasthe potentialtooltoalterhostdevelopment,physiology,andsystemicdefenses,whichinturncansignificantlyenhancethe cropgrowthandvigor,enhancetheplantefficiencytoutilizenutrient,andimprovethebiotic/abioticstresstolerance anddiseaseresistance(Kouretal.,2020c; Yadavetal.,2020a).Applicationsofthesemicrobialcommunitiescanbeharnessedinasustainableandeconomicmannertoimprovefarmproductivityandfoodquality.Mutualisticrelationshiphas evolvedbetweenplantsandassociatedmicrobiotawherebothpartnersdevelopedamechanismtobenefitfromtheassociation(Ranaetal.,2020a; Saxenaetal.,2014).
Accordingtotheearlierreportmicrobiomeplaysanimportantroleinshapinghostphenotypesasitactsasaflexible shieldbetweentheeffectsofhost-genotypeinteractionandtheenvironmentaleffects(Ranaetal.,2020b; Saxenaetal., 2015).However,thisassociationhasbeenunintentionallyaffectedbyplantbreedingprogramswhichharmsthekeybeneficialmembersofthecropmicrobiome.Accordingtotheavailableliterature,itisevidentthatplantmicrobiomeplaysan importantroleincropyieldsandfitness(MuellerandSachs,2015).
Plantmicrobiomecanbepotentiallyharnessedtorevolutionizeagricultureandfoodindustriesby(i)thecumulative integrationofcrophealthwithvariousclimate-specificmanagementpracticestoimproveproductivityandquality; (ii)theapplicationofenvironmental-friendlyapproachesforpropereradicationofpestsandpathogensandthusminimizing theunavoidableuseofchemicalpesticideshavingenvironmentalandhealthimplications;(iii)thewidespreadutilizationof naturalresourcesincludingsoilandwaterbyconvenientandefficientmethods(iv)theenhancementoffoodqualitywith minimuminterferenceofchemicalcontaminantsandallergens;and(v)theimprovementofcropfitnessaccordingtothe prevalentextremeweathertherebyminimizinglosses(Kumaretal.,2019; Yadavetal.,2019a). Fig.1.1 representsthe isolationofplantmicrobiomesandtheirbiotechnologicalapplicationforsustainableagriculture.
1.3Rhizosphereversusphytomicrobiomeapproaches
Soilmicrobialcommunitiesareconsideredthegreatestreservoirofbiologicaldiversity(Curtisetal.,2002; Gams,2007; Torsviketal.,2002; Bueeetal.,2009).Thenarrowzoneofsoilwhichismajorlyinfluencedbytheplantrootsecretionsare knownasrhizosphere.Approximately,1011 microbialcellspergramrootwerereportedinthissoilzone(Egamberdieva etal.,2008)andmorethan30,000diversespeciesofprokaryotesandcompriserhizospheremicrobiome(Mendesetal., 2011).Duetolargecollectivegenomeofthemicrobialcommunityascomparedtothatoftheplant,itisdenominatedasthe secondgenomeofplants(Berendsenetal.,2012).Thegroupofmicrobiotaassociatedwithdifferentpartsoftheplant(e.g., root,stem,leaf,flower,seeds)comprisesphytomicrobiome(epiphytic,endophytic,andrhizospheric)(Yadavetal.,2019b, 2019c,2019d,2019e).Microbesassociatedwithrhizosphereofanyplantareknownasrhizosphericmicrobes.Therhizosphereisthezoneofsoilinfluencedbyrootsthroughthereleaseofsubstratesthataffectmicrobialactivity.Anumberof microbialspecieshavebeenreportedtobeassociatedwithplantrhizospherebelongingtothegenera Azospirillum, Alcaligenes, Arthrobacter, Acinetobacter, Bacillus, Paenibacillus,Burkholderia, Enterobacter, Erwinia, Flavobacterium, Methylobacterium,Pseudomonas, Rhizobium,and Serratia (Biswasetal.,2018; Kouretal.,2019a; Saxenaetal., 2016; Yadav,2017,2019; YadavandYadav,2019)(Table1.1).
Theendophyticmicrobesarereferredtothosemicroorganismswhichcolonizeintheinterioroftheplantparts,viz,root, stem,orseedswithoutcausinganyharmfuleffectonhostplant.Endophyticmicrobesenterplantsmainlythroughwounds, naturallyoccurringasaresultofplantgrowthorthroughroothairsandatepidermalconjunctions.Besidesprovidingentry avenues,woundsalsocreatefavorableconditionsforthebacteriabyallowingleakageofplantexudatesthatserveasa nutrientsourceforthebacteria.Theyexistwithinthelivingtissuesofmostplantspeciesbyformingassociationsranging fromsymbiotictoslightlypathogenic.Thesebacteriahavebeenisolatedfromavarietyofplantsincludingwheat(Verma etal.,2014,2015a,2016a),rice(ManoandMorisaki,2007; Naiketal.,2009; Piromyouetal.,2015),maize(Yadav,2017; Yadavetal.,2018),soybean(HungandAnnapurna,2004; Mingmaetal.,2014),pea(Narulaetal.,2013; Tariqetal.,2014), commonbean(Suyaletal.,2015),chickpea(Sainietal.,2015),andpearlmillet(BeatrizSa ´ nchezetal.,2014).Alarge numberofendophyticbacterialspeciesbelongingtodifferentgeneraincluding Achromobacter, Azoarcus, Burkholderia, Enterobacter, Gluconoacetobacter, Herbaspirillum, Klebsiella, Microbiospora, Micromonospora, Nocardioides,Pantoea, Planomonospora, Pseudomonas, Serratia,Streptomyces and Thermomonospora,etc.,havebeensortoutfromdifferent hostplants(Kouretal.,2019b; Ranaetal.,2019; Sumanetal.,2016; Vermaetal.,2017)(Table1.1).Thephyllosphere iscommonnicheforsynergismbetweenbacteriaandplant.Microbesonleafsurfacearemostadaptedmicrobesasthey toleratehightemperature(40–55 °C)andUVradiation.Manybacteriasuchas Agrobacterium, Methylobacterium, Pantoea, and Pseudomonas havebeenreportedinthephyllosphere(Vermaetal.,2014,2015a,2016a,b; Yadav,2009)(Table1.1).
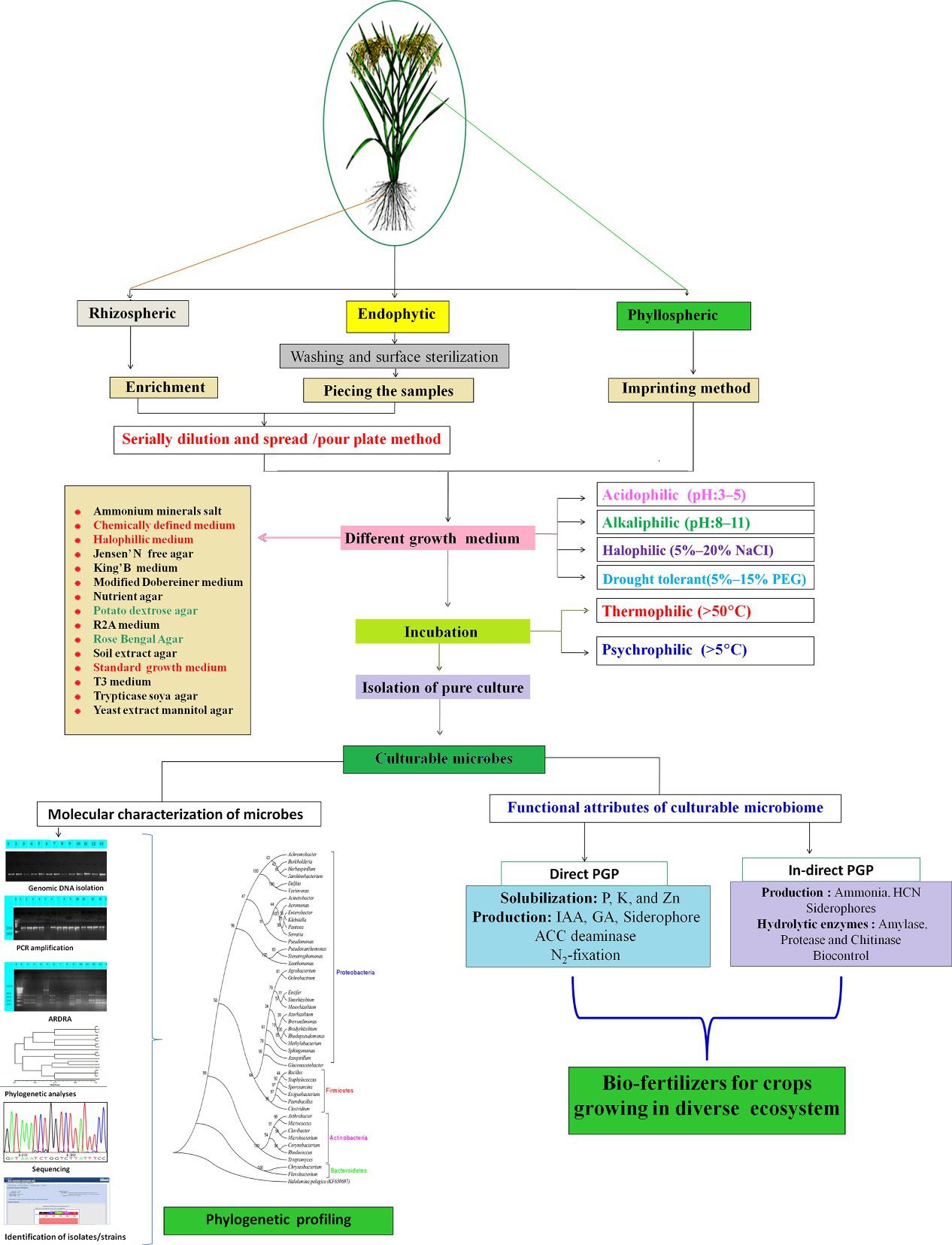
FIG.1.1 Aschematicrepresentationoftheisolation,characterization,identificationandpotentialapplicationofculturableandun-culturablemicrobiomesofcrops. (AdaptedwithpermissionfromVerma,P.,Yadav,A.N.,Kumar,V.,Singh,D.P.,Saxena,A.K.(2017)Beneficialplant-microbesinteractions:Biodiversityofmicrobesfromdiverseextremeenvironmentsanditsimpactforcropimprovement.In:SinghD.P.,SinghH.B.,PrabhaR.(Eds.) Plant-MicrobeInteractionsinAgro-EcologicalPerspectives:Volume2:MicrobialInteractionsandAgro-EcologicalImpacts.SpringerSingapore, Singapore,pp.543–580.https://doi.org/10.1007/978-981-10-6593-4_22.)
TABLE1.1 BeneficialmicrobiomeswithmultifariousPGPattributesunderthediverseabioticstress.
MicrobesStressPIAASidACCReferences
Achromobacterspanius Hightemp++ + Vermaetal.(2016b)
Acinetobacterrhizosphaerae BIHB723Cold++++ Gulatietal.(2009)
Acinetobacter sp.M05Drought+ + Zhangetal.(2017)
Aeromonashydrophila IARI-R-6Cold+++ Yadavetal.(2015a)
Alcaligenesfaecalis Hightemp++ + Vermaetal.(2016b)
Alcaligenes sp.Salinity + Baletal.(2013)
Arthrobactermethylotrophus IARI-HHS1–25Cold++++ Vermaetal.(2015b)
Arthrobacter sp.Salinity++ + Upadhyayetal.(2009)
Arthrobactersulfonivorans IARI-L-16Cold+++ Yadavetal.(2015b)
Azospirillumlipoferum B3Drought++ + Arzaneshetal.(2011)
B.sporothermodurances Salinity+ + Upadhyayetal.(2009)
B.subtilis,IARI-IIWP-2Drought++++ Vermaetal.(2014)
Bacillusaltitudinis Drought+++ Sunaretal.(2015)
Bacillusaltitudinis Hightemp++ + Vermaetal.(2016b)
Bacillusaltitudinis IARI-HHS2–2Cold++ Vermaetal.(2015b)
Bacillusamyloliquefaciens Drought++ + Vardharajulaetal.(2011)
Bacillusamyloliquefaciens Cold++ + Vermaetal.(2015b)
Bacillusamyloliquefaciens IARI-HHS2–30Cold++++ Vermaetal.(2015c)
Bacillusamyloliquefaciens IARI-R-25Cold+++ Yadavetal.(2015a)
Bacillusaquimaris Salinity+ + Upadhyayetal.(2009)
Bacillusaquimaris, IARI-IHD-17Drought+ Vermaetal.(2014)
Bacillusarsenicus Salinity++ + Upadhyayetal.(2009)
Bacillusaryabhattai IARI-HHS1–30Cold++ Vermaetal.(2015b)
Bacillusaryabhattai,IARI-IHD-34Drought+++ Vermaetal.(2014)
Bacilluscereus Cold + Vermaetal.(2015b)
Bacilluscereus Salinity++ + Upadhyayetal.(2009)
Bacillusfirmus IARI-L-21Cold++++ Yadavetal.(2015b)
Bacillusflexus Hightemp+ + Vermaetal.(2016b)
Bacillusflexus Cold+ Vermaetal.(2015b)
Bacillushalodenitrificans PU62Drought++++ Ramadossetal.(2013)
Bacilluslicheniformis Drought++ + Vardharajulaetal.(2011)
Bacilluslicheniformis Hightemp+ Vermaetal.(2016b)
Bacilluslicheniformis BGBA1Drought+++ PahariandMishra(2017)
Bacilluslicheniformis IARI-AL38Cold+++ Yadavetal.(2016)
Bacillusmegaterium Cold++ + Vermaetal.(2015b)
Bacillusmegaterium,IARI-IIWP-9Drought+++ Vermaetal.(2014)
Bacillusmojavensis Hightemp++ Vermaetal.(2016b)
Bacillusmuralis IARI-AR28Cold ++ Yadavetal.(2016)
TABLE1.1 BeneficialmicrobiomeswithmultifariousPGPattributesunderthediverseabioticstress—cont’d
MicrobesStressPIAASidACCReferences
Bacilluspumilus Salinity+ + Upadhyayetal.(2009)
Bacilluspumilus IARI-L-54Cold+++ Yadavetal.(2015b)
Bacillussiamensis Hightemp + Vermaetal.(2016b)
Bacillus sp.Salinity + Baletal.(2013)
Bacillus sp.(PS-12)Drought++ + Hussainetal.(2013)
Bacillus sp.,AW1Drought+ ++ Ranaetal.(2011)
Bacillussubtilis Drought++ + Vardharajulaetal.(2011)
Bacillussubtilis Hightemp++ + Vermaetal.(2016b)
Bacillussubtilis Salinity++ Upadhyayetal.(2009)
Bacillussubtilis IARI-L-69Cold++++ Yadavetal.(2015b)
Bacillusthuringenesis Drought++ + Vardharajulaetal.(2011)
Bacillusthuringiensiss Cold+ + Vermaetal.(2015b)
Bordetellabronchiseptica IARI-HHS2-29 Cold+++ Vermaetal.(2015b)
Brevundimonasdiminuta, AW7Drought+++ Ranaetal.(2011)
Cellulosimicrobiumcellulans IARI-ABL-30Cold++ + Yadavetal.(2015b)
Delftiaacidovorans Hightemp++ + Vermaetal.(2016b)
Delftialacustris Hightemp+ + Vermaetal.(2016b)
Delftia sp.,IARI-IIWP-31Drought+++ Vermaetal.(2014)
Desemziaincerta IARI-L-46Cold+++ Yadavetal.(2015b)
Duganellaviolaceusniger,IIWP-23Drought+++ Vermaetal.(2014)
Enterobacterhormaechei Drought + Niuetal.(2017)
Enterobacter sp.Halophilic++ HingoleandPathak(2016)
Enterobacter sp.Salinity++++ Sarkaretal.(2018)
Exiguobacteriumantarcticum HHS2–49Cold ++ Vermaetal.(2015b)
Flavobacteriumpsychrophilum HHS2–37Cold++++ Vermaetal.(2015b)
Flavobacterium sp.(PS-41)Drought++ + Hussainetal.(2013)
Kocuriakristinae IARI-HHS2–64Cold+++ Vermaetal.(2015b)
Lysinibacillussphaericus IARI-AR11Cold+ + Yadavetal.(2016)
Methylobacteriumextorquens, IIWP-43Drought+++ Vermaetal.(2014)
Methylobacteriummesophilicum Hightemp++ + Vermaetal.(2016b)
Methylobacteriummesophilicum, IIWP-45Drought+++ Vermaetal.(2014)
Methylobacteriumphyllosphaerae Cold+++ Vermaetal.(2015b)
Methylobacteriumradiotolerans, IHD-35Drought+++ Vermaetal.(2014)
Ochrobactrum sp.Salinity + Baletal.(2013)
Paenibacillusamylolyticus,IHD-24Drought+ Vermaetal.(2014)
Paenibacillusdendritiformis,IIWP-4Drought+++ Vermaetal.(2014)
Paenibacillusdurus,IARI-IIWP-40Drought+ + Vermaetal.(2014)
Paenibacillusfavisporus Drought++ Vardharajulaetal.(2011) Continued
TABLE1.1 BeneficialmicrobiomeswithmultifariousPGPattributesunderthediverseabioticstress—cont’d
MicrobesStressPIAASidACCReferences
Paenibacillus sp.,IARI-IHD-15Drought+++ Vermaetal.(2014)
Paenibacillustaichungensis M10Drought+++ Zhangetal.(2017)
Paenibacillustylopili IARI-AR36Cold+++ Yadavetal.(2016)
Pantoeaagglomerans IARI-R-87Cold+++ Yadavetal.(2015a)
Pantoeadispersa 1ACold+++ Selvakumaretal.(2008)
Planococcusrifietoensis Alkalinity+++ Rajputetal.(2013)
Providenciarustigianii IARI-R-91Cold++++ Yadavetal.(2015a)
Providencia sp.,AW5Drought+ ++ Ranaetal.(2011)
Pseudomonasaeruginosa Cold+++ Vermaetal.(2015b)
Pseudomonasbrassicacearum E85Drought+ ++ Aarabetal.(2015)
Pseudomonascedrina IARI-R-53Cold++++ Yadavetal.(2015a)
Pseudomonasentomophila Drought++ + Sandhyaetal.(2010)
Pseudomonasfluorescens Drought + Niuetal.(2017)
Pseudomonasfluorescens Cold++++ Vermaetal.(2015b)
Pseudomonasfluorescens Cold++ + Mishraetal.(2011)
Pseudomonasfluorescens Salinity++ Akhgaretal.(2014)
Pseudomonasfluorescens PPRs4Cold+++ Mishraetal.(2011)
Pseudomonasfluorescens, SorgP4Drought+ + Alietal.(2014)
Pseudomonasfluorescens,153Drought+ + Zabihietal.(2011)
Pseudomonasfragi IARI-R-57Cold++++ Yadavetal.(2015a)
Pseudomonasfuscovaginae,IIWP-29Drought+++ Vermaetal.(2014)
Pseudomonasgeniculata IARI-HHS1–19Cold+++ Vermaetal.(2015b)
Pseudomonasjaponica Hightemp+ Vermaetal.(2016b)
Pseudomonasjessani Cold++ + Mishraetal.(2011)
Pseudomonasjessani PGRs1Cold+++ Mishraetal.(2011)
Pseudomonaskoreensis Cold++ + Mishraetal.(2011)
Pseudomonaskoreensis PBRs7Cold+++ Mishraetal.(2011)
Pseudomonaslini,IARI-IIWP-33Drought+ ++ Vermaetal.(2014)
Pseudomonaslurida Cold++ + Mishraetal.(2011)
Pseudomonaslurida M2RH3Cold+++ Selvakumaretal.(2011)
Pseudomonaslurida NPRs3Cold+++ Mishraetal.(2011)
Pseudomonasmedicona Salinity+ + Upadhyayetal.(2009)
Pseudomonasmigulae Drought + Niuetal.(2017)
Pseudomonasmonteilii,IARI-IIWP-27Drought++++ Vermaetal.(2014)
Pseudomonasmoraviensis IARI-R-132Cold++++ Yadavetal.(2015a)
Pseudomonasplecoglossicida, S1 Drought+++ Rollietal.(2015)
Pseudomonaspoae Hightemp++ + Vermaetal.(2016b)
Pseudomonasputida Drought+ + Zabihietal.(2011)
TABLE1.1 BeneficialmicrobiomeswithmultifariousPGPattributesunderthediverseabioticstress—cont’d MicrobesStressPIAASidACCReferences
Pseudomonasputida Cold++ + Mishraetal.(2011)
Pseudomonasputida IARI-R-131Cold ++ Yadavetal.(2015a)
Pseudomonasputida PGRs4Cold+++ Mishraetal.(2011)
Pseudomonasreactans IARI-ABR-38Cold+++ Yadavetal.(2015a)
Pseudomonas sp.Drought++++ Poonguzhalietal.(2008)
Pseudomonas sp.Cold++ + Mishraetal.(2011)
Pseudomonas sp.Cold++ + Mishraetal.(2011)
Pseudomonas sp.NARs9Cold+++ Mishraetal.(2009)
Pseudomonas sp.PGERs17Cold+++ Mishraetal.(2008)
Pseudomonasstutzeri Drought++ + Sandhyaetal.(2010)
Pseudomonasstutzeri Hightemp++ Vermaetal.(2016b)
Pseudomonasthivervalensis, IHD-3Drought++++ Vermaetal.(2014)
Pseudomonastolaasii IEXbDrought+++ Virueletal.(2011)
Psychrobacterfozii,IIWP-12Drought++++ Vermaetal.(2014)
Psychrobacterfrigidicola IARI-R-127Cold++++ Yadavetal.(2015a)
Rahnella sp.BIHB783Cold++++ Vyasetal.(2010)
Rhodobactersphaeroides Hightemp + Vermaetal.(2016b)
Salmonellabongori Hightemp + + Vermaetal.(2016b)
Sanguibacterantarcticus IARI-L-33Cold++++ Yadavetal.(2015b)
Sanguibactersuarezii IARI-R-7Cold++++ Yadavetal.(2015a)
Serratiamarcescens,IIWP-32Drought+ Vermaetal.(2014)
Sporosarcinaglobispora IARI-AR111Cold + + Yadavetal.(2016)
Staphylococcusaureus 22FDrought+++ Toribio-Jimenezetal.(2017)
Staphylococcussuccinus Hightemp + Vermaetal.(2016b)
Stenotrophomonasmaltophilia HHS1–20Cold++++ Vermaetal.(2015b)
Stenotrophomonas sp.,IIWP-34Drought++ Vermaetal.(2014)
Streptoccoccusthoraltensis 5CR-FDrought++ Toribio-Jimenezetal.(2017)
P:PhosphorusSolubilization;IAA:Indoleaceticacids;Sid:Siderophores;ACC:1-aminocyclopropane-1-carboxylate.
However,phytomicrobiomehasbeengivenlessimportancecomparedtorootmicrobiomeduetosometechnicaldifficultiesassociatedwiththecharacterizationofotherpartsoftheplant(leaf,stem).Also,itischallengingtoamplifyplant tissuelocalizedbacterialmarkergenes(16SrDNA)duetosequencesimilarityofchloroplastandmitochondrialDNAwith Chlorobi/Chloroflexi/Cyanobacteriaphyla(SinghandTrivedi,2017).
However,majorityoftheresearchhasbeeninclinedtowardtherhizospheremicrobiome.Onthewhole,rhizosphereassociatedmicrobesandtheirgeneticelementsformrootmicrobiome.Plantrootsandsoilsareinvolvedininterfaceinteractionswhichlaidthebasisforbothplanthealthandresourceacquisition.Otherdemonstratedapplicationsofrhizosphere microbiomeincludediseaseresistance,nutrientacquisition,andabilitytofitinnovelandextremeenvironmentsbycombatingabioticstresses(SinghandTrivedi,2017).
1.4Effectsoftherootmicrobiomeonplanthealth
1.4.1Disease-suppressivesoils
Duetothestarvingconditionsofthesoilmicroflora,fiercebattleexistsintherhizospheremicrobesfortheacquisitionof plant-derivednutrients(Raaijmakersetal.,2009).Hence,toinfectthehosttissueaswellastoescapetherhizospherebattle zone,mostsoilbornepathogensneedtomaintaincloseassociationwiththehostbyachievingsufficientnumbersandalso needtogrowassaprophytesintherhizosphere.Hence,microbialcommunityassociatedwithinfectioussoilultimately decidesthesuccessofapathogen.Eachsoiltypehasthenaturalabilitytopreventthepathogeninvasionuptoacertain extent.Thiscanfurtherbeconfirmedfromthecommonoccurrenceofdiseaseseverityinthepathogeninoculatedpasteurizedsoilsincomparisontothatofnon-pasteurizedone.Thisphenomenonofdiseasesuppressionoccursduetothe presenceoftotalmicrobialactivityiscoinedasgeneraldiseasesuppression.Further,generaldiseasesuppressiveness canbeeffectivelyenhancedintheconducivesoilthroughorganicamendmentswhichstimulatestheactivityofmicrobial populations.
Anothermosteffectivemechanismofspecificdiseasesuppressivenessisunderdiscussion,inwhichtheabilityofspecificsuppressivenesscanbeintroducedtodiseaseconducivesoilbytheadditionofthesuppressivesoil(0.1%–10%).To studythebioticnatureofspecificdiseasesuppressiveness,pasteurizationofthesoilhasbeenperformedfortheremovalof diseasesuppressiveness(Welleretal.,2002; Garbevaetal.,2004; Raaijmakersetal.,2009; Mendesetal.,2011).
1.4.2Developmentofdiseasesuppressiveness
Somesoilscannaturallyretaintheirabilityofdiseasesuppressivenessforextendedperiodsandpersistsevenwhensoilsare leftbare.Insomecases,monocultureofacropforseveralyearsishelpfulinthedevelopmentofsoilssuppressiveness.In mostplantspecies,phenomenonofselfsuppressivenesspredominatesduetosuccessivemonoculturewhichinducesthe developmentofspecializedplantpathogens(Bennettetal.,2012).Nonetheless,therearesomecommoncropdiseases againstwhichsoilsuppressivenesshasbeenreportedincludingtake-alldiseaseofwheat(Triticumaestivum)causedby Gaeumannomycesgraminis var. tritici,damping-offdiseaseofsugarbeetcausedby Rhizoctonia,Fusariumwiltdisease ofnumerousplantspecies,and Streptomyces speciescausingpotatoscabdisease(Welleretal.,2002; Raaijmakers etal.,2009; Mendesetal.,2011).
Isolationofthemicroorganismsfrommanysuppressivesoilsthatcanconfersuppressivenessfollowedbytheirinoculationintheconductivesoilisahelpfulstrategytoboosttheconductivesoil.Earlierreportsevidencedaphenomenonof ‘take-alldecline’whichisadeclineinthesevereoutbreakof G.graminis var. tritici causingroot-associateddiseasein wheatduetomonocultureofwheatforseveralyears.Thiswidespreadphenomenonoccursduetothepresenceofantifungal compound2,4-diacetylphloroglucinol(DAPG)producedbytheantagonisticfluorescent Pseudomonas spp.(Welleretal., 2002).Additionallyothermicroorganismsincluding Proteobacteria and Firmicutes alongwithfungalmembersof Ascomycota divisionwerereportedtoconfersuppressiveness(Raaijmakersetal.,2009).Accordingtothestudy,rhizosphere microorganismsdevelopedamechanismtocounterasoilbornepathogenbytheproductionoflyticenzymesorantibiotic compounds,competingfor(micro)nutrientsandconsumepathogenstimulatorycompounds(Doornbosetal.,2012; LugtenbergandKamilova,2009).
1.4.3Beneficialrhizospheremicrobes:Modulationofthehostimmunesystem
Apartfromthedeleteriouseffectofpathogensassociatedwiththerhizosphere,evidencesexistforthepresenceofmany beneficialsoilbornemicroorganismsthatcanenhancethedefensivecapacityinabovegroundpartsoftheplant(Zamioudis andPieterse,2012).“Thisinducedsystemicresistance(ISR)isastateinwhichtheimmunesystemoftheplantisprimedfor acceleratedactivationofdefense”(Conrathetal.,2006; Bakkeretal.,2007; PozoandAzcon-Aguilar,2007; VanWees etal.,2008; DeVleesschauwerandHofte,2009).
Inearlierstudies,inductionmechanismofISRinArabidopsis(Arabidopsisthaliana)dueto Pseudomonasfluorescens WCS417(plantgrowth-promotingrhizobacterium)waswellstudied.Locally,thisroot-associated Rhizobacterium WCS417isinvolvedintheapoplasticsecretionofmultiplemoleculesoflowmolecularweightwhichinturnsuppress flagellin-triggeredimmuneresponses(Milletetal.,2010).Althoughplantimmunityisinthesuppressedstatelocally,systemicinitiationoftheirimmunesignalingcascadeisobservedtoprovideresistanceagainstabroadspectrumofpathogens andeveninsects(VanOostenetal.,2008; VanderEntetal.,2009; Pinedaetal.,2010).
1.5Technicalchallengesandemergingsolutions
Microbialproductscanactasthereliableandefficientalternativetoreplaceagriculturalchemicalsandfertilizersfor achievingenhancedcropyields.Manycompanieshaveshowninteresttowardtheuseofbiocontrolorbiofertilizerproducts whichcomprisetheformulationofindividualmicroorganismsaswellastowardthedevelopmentofcarrier-basedinoculantsofbeneficialstrains.Earlierstudieshavedemonstratedtheroleofeconomicallyimportantcropplantsinthe incrementofcropproductionupto10%–20%basedonthelarge-scalefieldtrials(Perez-Jaramilloetal.,2016).Though themicrobialtechnologieshavethehighestpotential,evidencesalsosuggestthefailureofgreenhousetrialsinthefield.
However,duetotheinconsistentandvarianteffectsofmicrobialproductsobtainedduringdifferentfieldtrialsunder differentclimatic/edaphicconditions,majorbottleneckisassociatedwiththelarge-scaleadoptionofthetechnology. Therefore,underfieldconditionsthereshouldbebetterandclearunderstandingoftheinteractionbetweeninoculated strainsandnativemicrobiomesfortheimprovementofselectionprocessandapplicationtechnique(SinghandYadav, 2020).Difficultypersistsinthisstudyduetothecomplexinteractionmechanisminvolvedamongmicrobes,plants,soil, andclimate.However,underthesimplifiedconditions,onlyfewoftheseinteractionshavebeenevidenced.
Theadventofmicrofluidic-basednewtechnologiessuchas‘MicrobiomeonaChip’ishelpfulinstudyingmultitrophic plant-microbiomeinteractionsthroughmultiplexedtreatmentsfollowedbytheincorporationofenvironmentalstimuli,host responses,andthecolonizationofmicrobes(StanleyandVanderHeijden,2017).Inrelevantfieldconditions,such knowledgeisdesirabletomonitorthebeneficialmicrobesbasedontheiractivitiesorassessmentofthenativemicrobiomes and/orthepotentialofinoculantstrainforcolonization.Combinedapplicationsofvarioustechnologiesandfacileformulationswithextendedshelflivesinthefieldconditionswillbeadvantageousintheimprovementofmicrobialproducts efficacy.Astrategyofmodifyingplantmicrobiomesandtraitsbytheintroductionofbeneficialbacteriaintotheprogeny seedsatfloweringstageishelpfulinachievingtheproductiongoal(Mitteretal.,2017).Thisstrategyofbacterialdelivery hasseveraladvantagesoverconventionalapplicationtechniques,forexample,apartfromintroducingbeneficialtraits withinonegeneration,italsoprovidesprotectionagainstcompetitivenativemicroflorafollowedbythesubstantialincrease intheabilityofinoculatedstrainforcolonizationandsurvival.
Survivalpotentialofconsortiaofmultiplecompatiblebeneficialmicrobesarecomparativelybetterthansingle-strain formulationsastheyformstructurednetworkandassociationwiththenaturalmicrobiomeoftherhizospheresoilswhichin turnbenefitsthehost(SinghandTrivedi,2017; Wallenstein,2017).Amongthenaturalmicrobialcommunitiesassociated withrhizosphereandphyllosphere,itispossibletoisolatemajorityofmicrobialspeciesusingvarioussystematicisolation approaches(Baietal.,2015).
1.6Insitumanipulationofmicrobiome
Insitumicrobiomeengineeringisthepowerfultooltoharnessthepotentialofbeneficialmicrobiomefordesiredoutcomes inagriculturesectorandfoodindustries(MuellerandSachs,2015).Incontrasttotheothercurrenttechnologiesinvolving theuseofselectiveantibioticsandprobiotics,thistechnologyprefersmicrobiomemanipulationwithoutculturing(Sheth etal.,2016).Syntheticbiologywillalsodisplayapplicationsintheformulationofcropprobioticsbytheengineeringof novelbutpredictablefunctionsinit.Theseengineeredcropprobioticshavethepotentialtomanipulatethemicrobiomeand/ oritsactivitiesinapredictedmanneruponadditiontoplantandsoils.Forexample,engineeredbacteriahavetheabilityto secretespecificchemicalandcanbeeffectivelyemployedforthemodulationofbothnativemicrobiomesandcropphysiology.Theseengineeredbacterialstrainsstimulatetheactivitiesofotherbeneficialmicrobiometherebyprovidingcrop resistanceagainstresourceandbioticstresses.Withthehelpoftheseoperationaltools,itispossibletobringrevolutionsin agriculturalproductivity.
Theselectionofthemicrobiomesisalsobasedonthegenotypicandphenotypicvariationsinplants.Thisselectionis supportedbythefactthatplanthasthenaturaltraittosupportabeneficialmicrobiome(Wallenstein,2017).Domestication isproposedfortheremovalof‘microbiome-mediatedtrait’therebynecessitatingtheapplicationofgrowthhormones, sprayingofinsecticides,andhighquantitiesofinorganicfertilizersforthemaintenanceofrequiredoutput(PerezJaramilloetal.,2016).Therehasbeenanemerginginteresttowardtheapplicationofoptimizedenvironmentalfriendly microbialbiofertilizersandbiopesticidesinvarioussoilsandcropvarieties.Duringcropbreedingprograms,theunderstandingofbeneficialplant-microbeinteractionswouldbebeneficialtobreed‘microbe-optimizedplants.’Theplant geneticisthebasisforbeneficialplant-microbiomeinteractions.Thoroughunderstandingofplantgeneticconstitution ishelpfultobreedandoptimize‘designerplants,’whichinturnattractandmaintainbeneficialmicrobes(Abhilash etal.,2012).Thesetraitshavebeenexploredbybreedingprograms;however,abetterandthoroughunderstandingof themechanismlyingbehindtheseattractionsofbeneficialmicrobestowardcropsisessential.Itispossibletomodel
microbe-optimizedplantsthroughapproachesofgeneticengineeringandplantbreeding.Thesemicrobe-optimizedplants generaterightexudatesandvolatilesthatmaintaintheattractionofbeneficialmicrobeseitherattherootorontheleafatthe righttime.
Asmentionedearlier,plantshavetheabilitytosecretespecificexudates,whichisnotonlyresponsiblefortheengineeringoftheirownrhizosphereenvironmentbutalsoimprovetheirinteractivepropertywithspecificbeneficialmicrobes aswellasnutrientavailability.Thisstrategyrequiredtheavailabilityofthetargetedmicrobesalongwiththecouplingof otherinoculatedcorrespondingmicrobes(Liuetal.,2016).PreliminaryworksuggestedthekeyroleofmiRNAinregulatingthestructureofrhizospheremicrobiome.Therefore,basicelucidationoftheinteractivemechanismbetweenmiRNA andthemicrobiomeactasavaluabletoolkittotransfermiRNAfromatargetsoiltorecipientsoilsaswellastoengineera definedbeneficialplantmicrobiomefordesirableoutcomes.
Techniqueofplantecologicalengineeringisrequiredforhostmicrobiomemanipulationaswellasforeffective enhancementofdiseasemanagementbymanagingtheintegrationofplantbreedingwithmicrobiomeselection.Inartificial ecosystemselection,differentcropsindifferentsoilsareinoculatedwithbothengineeredplant /soil-optimizedmicrobes. Itisalsoevidencedinearlierstudiesthatimprovedplant-microbeinteractionsaretheresultofdistinctiveabilityofsoil microbiomestoadapttotheircropsovertime(Berendsenetal.,2012).Atop-downapproachofartificialselectionon microbiomeshelpsintheimprovementofplantfitness,thusengineeringevolvedmicrobiomesispreferableforhostfitness (MuellerandSachs,2015).Thisapproachofhost-mediatedmicrobiomeengineeringisdesirablefortheselectionof microbialcommunitiesindirectlybasedontheirevolvedhosttraitsthatinfluencemicrobiomes.
Existingevidencesalsosuggestthecumulativeapplicationsofoptimizedmicrobiomesandphytohormonesfordisease resistanceasphytohormoneshavebeenreportedtoactivatemicrobiomedefenseresponses(Lebeisetal.,2015).Similarly, theroleofoptimizedmicrobiomesasinoculantsisnotonlyhelpfulforearlyorlatefloweringinplantsbutalsoprovides droughtresistancetoplants.Hence,itcanbeinferredthatduetotheapplicationsoftheseinoculants,plantsdevelopthe abilitytoresistseveralabioticstressesbyshowingalterationinfloweringtime(KazanandLyons,2016).
Host-mediatedartificialselectionofmicrobiomesisacheaperandmostpreferablewaytogeneratediseaseresistancein plantsascomparedtotheapplicationsofvariousgeneticallymodifiedorganisms,pesticides,andantibiotics.Furthermore, cross-compatibilityofmicrobiometransferwithphylogeneticallydistinctplantspeciescanbeconfirmedbythedetection ofoverlappingfunctionalcoremicrobiomesindifferentspeciesofplant. Shethetal.(2016) havealsohighlightedthe emergingroleofinsitugenomeengineeringasthehighlyspecificandeffectivetoolkitformicrobiomemanipulation uptoacertainlevel.
1.7Futureoutlook
Inrecentyears,theadventofhigh-throughputtechnologiesin“multiomics”andcomputationalintegrationprovidean insightintoplant-microbiomeinteractionsacrossscales.Thesetechnologiescanclearlybuilduptheconnectionwith thefunctionalgenenetworks/pathwayswiththehelpofindividualsignalmolecules,genesandgenecascades,andproteins. Technologicaladvancementsandadventofstrategieslikemutanttechnology,gene-editingsystems,genesilencing mediatedbyRNAinterference(RNAi),proteomics,andmetaboliteprofilinghavefacilitatedustorevealmechanism ofmicrobe-mediatedstrategiesadoptedbytheplantforitsgrowthpromotionandbiocontrol.Furtheradvancementin theautomationandlarge-scalebioinformaticstoolhaveaugmentedthegenomerepertoireofplant-associatedmicrobes. Thoroughinformationrelatedtothegenomeofplant-associatedmicrobesaswellasmechanisminvolvedintheirinteractionswithhost/environmentareresourcefulfortheresearcherstodiscovernovelandvaluablemicrobialgenesrelated toplantgrowthandproductivity.Theseadvancementslaidthebasisforpowerfulandconceptualframeworktoreveal complexplant-microbiomeinteractionsandalsohighlightedthepossibilitytoimproveplantproductivitythroughthe manipulationofbeneficialgenomiccircuitsofnovelpotentialplants.Accordingtotherecentsurvey,awidespreadcollectionofover250milliongenes(averageof5000genesperorganism)existsinthelibrary.Detailedprospectingofnovel genewouldbebeneficialforbiotechnologicalapplications.Approximately54,000completemicrobialgenomesequences areavailableinpublicdomain.Thedevelopmentofgeneticallyengineeredplantsthroughthecloningofnovelmicrobial geneshelpedtoconferresistanceagainstdiseases,byprovidingtolerancetowardherbicidesandotherstressconditions, whichinturnimprovestheplantyield(MacdonaldandSingh,2014).
However,theapplicationofinnovativegenecloningtechniqueswashelpfulinfewergeneinsertionsandmultiplexing (combinationofafewtargets).Futureresearchshouldfocusontheapplicationofmultigenicapproachinwhichmorethan onegenecanbeincorporatedtocreatetransgenicplants.Also,applicationofnewtoolsandresourcesforthebuildingof syntheticgenomeclustersfrommicrobiomescanalsobeexploredforthecomplexheterologouspathwayintroductioninto plants(Shihetal.,2016).Thisstrategywillbebeneficialfortransferoftraitslikediseaseresistanceandstresstolerance
betweencropplants.Further,therateofnovelgenediscoverycanbeintensifiedbyexploringnewcapabilitiesintrait discovery.
Accordingto BarakateandStephens(2016),aholisticapproachofCRISPR-Cas9-basedforwardgeneticscreenwillbe helpfulinthenearfuturetoelucidateplant-microbiomeinteractionsfollowedbythediscoveryofnovelgenesandtranscend ofindividualgenesforbiotechnologicalapplications.Genomeinformationneedstobeexploredtoexpandourunderstandingtowardnoveltrait-relatedmicrobialmetabolicpathways.Completeelucidationandinvestigationofnewmetabolicpathwayswillbehelpfulintheconstructionandengineeringofmicrobialcellfactorieswhichinturnwillleadto theproductionofdesiredcompoundsathigherlevelthatcanbecharacterizedfurther.Plantcultivationinasustainable mannerrequiresthecumulativecontributionofseveralbacterial-derivedmetabolitesfortheirhealthandgrowth improvement.
Metabolitesplayakeyroleinplanthealthinseveralwayssuchassomemetabolitestriggerthereleaseofsignalmoleculesdirectlyforenhancedplantgrowthwhereasotherscanindirectlycontributetoplantgrowthbyinhibitingplantpathogensorbybuildingupbeneficialmicrobiomearoundtheplant.Hence,toenhanceplant-microbeinteractions,theultimate futuregoalshouldbetheintegrationofsoilamendments,applicationofbiocontrolmicrobe-basedformulations,microbial biofertilizers,optimizedmicrobiomes,andsoil-specificmatchingmicrobe-optimizedcrops.Clearly,majorresearchefforts arerequiredforsustainableimprovementofcropyieldstoaddressfoodsecurityissues.Overall,newsustainablepractices involvingemergingmicrobiomesaswellastheexistingmicrobialtechnologiesaredesirabletoincreaseagriculturalproductivity.Increasingconcernofgrowersandconsumersforfoodsecurityinclinedthemtowardmicrobial-basedsolutions. However,successfulimplementationofthesesustainablepracticesfaceschallenges.Thesepracticesneedtobehighlighted followedbytheupgradationofregulatoryframework(e.g.,productsregistration,safetyrequirements).Transformationof thesustainableagricultureisbasedontheemergingmicrobial-basedsolutions.Successof Homosapiens iscertainlybased ontheagriculture;itispossibletoimplementmultipleSDGswhensuchanapproachisaddressedsystematically.
1.8Concludingannotationsandscenarios
Plant-microbeinteractionintherhizosphereandcomplexmechanismbehindthisneedsdetailedinvestigationasitisstillin itsinfancy.Earlierexperimentalevidencesrevealedimportantroleofrootmicrobiomeinplanthealthandalsosuggested thatitscompositioniscontrolledbytheplantitself.Themechanismbehindtheevolutionaryselectionofthemostfavored plantsmanagesorselectsthebeneficialmicrobiomefortheirreproductivesuccess.Itappearsthatplant-microbeinteraction istheresultofnaturalselectionpressureduetowhichplantscallformicrobialhelpwheneverneeded.Inthenearfuture manysuchselectiveforcesrequiredforshapingplantrootmicrobiomeandtheireffectsonplantshealthwillberevealed, becausetheadventofnext-generationsequencingtechniqueswillcertainlyopenupnovelopportunitiestowardthestudyof existinginterplaybetweenplantanditsassociatedmicroflora.
Hitherto,variousmetagenomicstudiesassociatedwiththerootmicrobiomehaverevealedtheirphylogeneticcompositionwhichinturnprovidesalimitedinsightintotheexistenceofspecificoperationaltaxonomicunits.Furtheradvancement inthefieldoffunctionalmetagenomicsandtranscriptomicswillprovideinformationrelatedtomicrobiome-associated activitiesandfunctions.Eventually,cumulativeapplicationsofalltheseadvancedtechnologiestoupliftcropquality andproductivitydependonthemechani smthroughwhichplantsandmicrobio mescontributetowardthehealthof eachother.
References
Aarab,S.,Ollero,J.,Megı´as,M.,Laglaoui,A.,Bakkali,M.,Arakrak,A.,2015.Isolationandscreeningofinorganicphosphatesolubilizing Pseudomonas strainsfromricerhizospheresoilfromNorthwesternMorocco.Am.J.Res.Commun.3,29–39.
Abhilash,P.C.,Powell,J.R.,Singh,H.B.,Singh,B.K.,2012.Plant–microbeinteractions:novelapplicationsforexploitationinmultipurposeremediation technologies.TrendsBiotechnol.30,416–420.
Akhgar,A.,Arzanlou,M.,Bakker,P.,Hamidpour,M.,2014.Characterizationof1-aminocyclopropane-1-carboxylate(ACC)deaminase-containing Pseudomonas spp.intherhizosphereofsalt-stressedcanola.Pedosphere24,461–468.
Ali,S.Z.,Sandhya,V.,Rao,L.V.,2014.Isolationandcharacterizationofdrought-tolerantACCdeaminaseandexopolysaccharide-producingfluorescent Pseudomonassp.Ann.Microbiol.64,493–502.
Arzanesh,M.H.,Alikhani,H.,Khavazi,K.,Rahimian,H.,Miransari,M.,2011.Wheat(Triticumaestivum L.)growthenhancementby Azospirillum sp. underdroughtstress.WorldJ.Microbiol.Biotechnol.27,197–205.
Bai,Y.,Muller,D.B.,Srinivas,G.,Garrido-Oter,R.,Potthoff,E.,Rott,M.,etal.,2015.FunctionaloverlapoftheArabidopsisleafandrootmicrobiota. Nature528,364–369.
Bakker,P.A.,Pieterse,C.M.,VanLoon,L.,2007.Inducedsystemicresistancebyfluorescent Pseudomonas spp.Phytopathology97,239–243.














