Gprotein-coupledreceptors:signaling, traffickingandregulationFirstEditionShukla
https://ebookmass.com/product/g-protein-coupled-receptorssignaling-trafficking-and-regulation-first-edition-shukla/
Instant digital products (PDF, ePub, MOBI) ready for you
Download now and discover formats that fit your needs...
The neuronal cytoskeleton, motor proteins, and organelle trafficking in the axon First Edition Pfister
https://ebookmass.com/product/the-neuronal-cytoskeleton-motorproteins-and-organelle-trafficking-in-the-axon-first-edition-pfister/ ebookmass.com
Advances in Protein Chemistry and Structural Biology: Protein Misfolding Donev
https://ebookmass.com/product/advances-in-protein-chemistry-andstructural-biology-protein-misfolding-donev/
ebookmass.com
Book of Love 1st Edition Aakash Shukla
https://ebookmass.com/product/book-of-love-1st-edition-aakash-shukla/
ebookmass.com
CompTIA Security+ Certification Practice Exams, Fourth Edition (Exam SY0-601) Lachance
https://ebookmass.com/product/comptia-security-certification-practiceexams-fourth-edition-exam-sy0-601-lachance/
ebookmass.com
Air and Gas Drilling Manual: Applications for Oil, Gas, Geothermal Fluid Recovery Wells, Specialized Construction Boreholes, and the History and Advent of the Directional DTH William C. Lyons
https://ebookmass.com/product/air-and-gas-drilling-manualapplications-for-oil-gas-geothermal-fluid-recovery-wells-specializedconstruction-boreholes-and-the-history-and-advent-of-the-directionaldth-william-c-lyons/ ebookmass.com
Charles III : New King. New Court. The Inside Story Robert Hardman
https://ebookmass.com/product/charles-iii-new-king-new-court-theinside-story-robert-hardman/
ebookmass.com
A Bird on Water Street Elizabeth O. Dulemba
https://ebookmass.com/product/a-bird-on-water-street-elizabeth-odulemba/
ebookmass.com
500 ACT Math Questions to Know by Test Day, 3rd Edition Inc. Anaxos
https://ebookmass.com/product/500-act-math-questions-to-know-by-testday-3rd-edition-inc-anaxos/
ebookmass.com
Pathways 1 answer key 2nd Edition Rebecca Tarver Chase
https://ebookmass.com/product/pathways-1-answer-key-2nd-editionrebecca-tarver-chase/
ebookmass.com
A Concise Introduction to Logic – Ebook PDF Version https://ebookmass.com/product/a-concise-introduction-to-logic-ebookpdf-version/
ebookmass.com
MethodsinCell Biology GProtein-CoupledReceptors: Signaling,Traffickingand Regulation Volume132 SeriesEditors LeslieWilson
DepartmentofMolecular,CellularandDevelopmentalBiology
UniversityofCalifornia
SantaBarbara,California
PhongTran
UniversityofPennsylvania
Philadelphia,USA&
InstitutCurie,Paris,France
MethodsinCell Biology GProtein-CoupledReceptors: Signaling,Traffickingand Regulation Volume132 Editedby
ArunK.Shukla
DepartmentofBiologicalSciencesandBioengineering, IndianInstituteofTechnology,Kanpur,India
AcademicPressisanimprintofElsevier
AcademicPressisanimprintofElsevier
50HampshireStreet,5thFloor,Cambridge,MA02139,USA 525BStreet,Suite1800,SanDiego,CA92101-4495,USA 125LondonWall,LondonEC2Y5AS,UK
TheBoulevard,LangfordLane,Kidlington,OxfordOX51GB,UK
Firstedition2016
Copyright © 2016ElsevierInc.AllRightsReserved.
Nopartofthispublicationmaybereproducedortransmittedinanyformorbyanymeans, electronicormechanical,includingphotocopying,recording,oranyinformationstorage andretrievalsystem,withoutpermissioninwritingfromthepublisher.Detailsonhowto seekpermission,furtherinformationaboutthePublisher’spermissionspoliciesandour arrangementswithorganizationssuchastheCopyrightClearanceCenterandthe CopyrightLicensingAgency,canbefoundatourwebsite: www.elsevier.com/permissions.
Thisbookandtheindividualcontributionscontainedinitareprotectedundercopyrightby thePublisher(otherthanasmaybenotedherein).
Notices
Knowledgeandbestpracticeinthisfieldareconstantlychanging.Asnewresearchand experiencebroadenourunderstanding,changesinresearchmethods,professional practices,ormedicaltreatmentmaybecomenecessary.
Practitionersandresearchersmustalwaysrelyontheirownexperienceandknowledgein evaluatingandusinganyinformation,methods,compounds,orexperimentsdescribed herein.Inusingsuchinformationormethodstheyshouldbemindfuloftheirownsafety andthesafetyofothers,includingpartiesforwhomtheyhaveaprofessionalresponsibility.
Tothefullestextentofthelaw,neitherthePublishernortheauthors,contributors,or editors,assumeanyliabilityforanyinjuryand/ordamagetopersonsorpropertyasa matterofproductsliability,negligenceorotherwise,orfromanyuseoroperationofany methods,products,instructions,orideascontainedinthematerialherein.
ISBN:978-0-12-803595-5
ISSN:0091-679X
ForinformationonallAcademicPresspublications visitourwebsiteat http://store.elsevier.com
Contributors
AgnesM.AcevedoCanabal
InstituteofNeurobiology,UniversityofPuertoRicoMedicalSciencesCampus, SanJuan,PR,USA;DepartmentofAnatomyandNeurobiology,Schoolof Medicine,UniversityofPuertoRico,SanJuan,PR,USA
D.Agranovich
SharettInstituteofOncology,Hadassah-HebrewUniversityMedicalCenter, Jerusalem,Israel
StefanAmisten
DiabetesResearchGroup,King’sCollegeLondon,London,UK
GabrielaAntunes
LaboratoryofNeuralSystems(SisNE),DepartmentofPhysics,Faculdadede FilosofiaCie ˆ nciaseLetrasdeRibeira ˜ oPreto,UniversidadedeSa ˜ oPaulo, Ribeira ˜ oPreto,Brazil
ChaitanyaA.Athale DivisionofBiology,IISERPune,Pune,India
NicolasAudet
DepartmentofPharmacologyandTherapeutics,McGillUniversity,Montreal,QC, Canada
MohammedAkliAyoub
BiologieetBioinformatiquedesSyste ` mesdeSignalisation,InstitutNationaldela RechercheAgronomique,UMR85,Unite ´ PhysiologiedelaReproductionetdes Comportements;CNRS,UMR7247,Nouzilly,France;LESTUDIUM LoireValley InstituteforAdvancedStudies,Orle ´ ans,France
R.Bar-Shavit
SharettInstituteofOncology,Hadassah-HebrewUniversityMedicalCenter, Jerusalem,Israel
DamianBartuzi
DepartmentofSynthesisandChemicalTechnologyofPharmaceutical SubstanceswithComputerModellingLab,FacultyofPharmacywithDivisionof MedicalAnalytics,MedicalUniversityofLublin,Lublin,Poland
MaikBehrens
DepartmentofMolecularGenetics,GermanInstituteofHumanNutrition Potsdam-Rehbruecke,Nuthetal,Germany
NicolasF.Berbari
DepartmentofBiology,IndianaUniversity-PurdueUniversityIndianapolis, Indianapolis,IN,USA
He ´ le ` neBonin
DepartmentofBiochemistryandMolecularMedicine,InstituteforResearchin ImmunologyandCancer,Universite ´ deMontre ´ al,Montreal,QC,Canada
MichelBouvier
DepartmentofBiochemistryandMolecularMedicine,InstituteforResearchin ImmunologyandCancer,Universite ´ deMontre ´ al,Montreal,QC,Canada
AmitabhaChattopadhyay
CSIR-CenterofCellularandMolecularBiology,Hyderabad,India
LinjieChen
InstituteofBiochemistry,CollegeofLifeSciences,ZijingangCampus,Zhejiang University,Hangzhou,Zhejiang,China
SantiagoCuevas
DivisionofRenalDiseases&Hypertension,DepartmentofMedicine,TheGeorge WashingtonUniversitySchoolofMedicineandHealthSciences,WA,USA
FrancheskaDelgado-Peraza
InstituteofNeurobiology,UniversityofPuertoRicoMedicalSciencesCampus, SanJuan,PR,USA;DepartmentofAnatomyandNeurobiology,Schoolof Medicine,UniversityofPuertoRico,SanJuan,PR,USA
DominicDevost
DepartmentofPharmacologyandTherapeutics,McGillUniversity,Montre ´ al,QC, Canada
AntonellaDiPizio
InstituteofBiochemistry,FoodScienceandNutrition,TheRobertH.Smith FacultyofAgriculture,FoodandEnvironment,TheHebrewUniversity,Rehovot, Israel
ShaliniDogra
DivisionofPharmacology,CSIR-CentralDrugResearchInstitute,Lucknow,Uttar Pradesh,India
ZyanyaP.Espinosa-Riquer
DepartamentodeFarmacobiologı´a,CentrodeInvestigacio ´ nydeEstudios
AvanzadosdelIPN,Me ´ xicoD.F.,Mexico
TimothyN.Feinstein
DepartmentofDevelopmentalBiology,UniversityofPittsburghSchoolof Medicine,Pittsburgh,PA,USA
ColleenA.Flanagan
SchoolofPhysiologyandMedicalResearchCouncilReceptorBiologyResearch Unit,FacultyofHealthSciences,UniversityoftheWitwatersrand,WitsParktown, Johannesburg,SouthAfrica
AlexandreGidon
MolecularMechanismsofMycobacterialInfection,CenterforMolecular InflammationResearch,NorwegianUniversityofScienceandTechnology, Trondheim,Norway
ClaudiaGonza ´ lez-Espinosa
DepartamentodeFarmacobiologı´a,CentrodeInvestigacio ´ nydeEstudios
AvanzadosdelIPN,Me ´ xicoD.F.,Mexico
S.Grisaru-Granovsky
DepartmentofObstetricsandGynecology,ShaareZedek,Jerusalem,Israel
AylinC.Hanyaloglu
InstituteofReproductiveandDevelopmentalBiology,ImperialCollegeLondon, London,UK
TerenceE.He ´ bert
DepartmentofPharmacologyandTherapeutics,McGillUniversity,Montre ´ al,QC, Canada
MellisaM.Hege
DepartmentofBiology,IndianaUniversity-PurdueUniversityIndianapolis, Indianapolis,IN,USA
IlpoHuhtaniemi
InstituteofReproductiveandDevelopmentalBiology,ImperialCollegeLondon, London,UK
M.Jaber
SharettInstituteofOncology,Hadassah-HebrewUniversityMedicalCenter, Jerusalem,Israel
KimC.Jonas
InstituteofReproductiveandDevelopmentalBiology,ImperialCollegeLondon, London,UK;InstituteofMedicalandBiomedicalEducation,StGeorge’s UniversityofLondon,London,UK
PedroA.Jose
DivisionofRenalDiseases&Hypertension,DepartmentofMedicine,TheGeorge WashingtonUniversitySchoolofMedicineandHealthSciences,WA,USA
ManaliJoshi
SavitribaiPhulePuneUniversity,Pune,India
AgnieszkaA.Kaczor
DepartmentofSynthesisandChemicalTechnologyofPharmaceutical SubstanceswithComputerModellingLab,FacultyofPharmacywithDivisionof MedicalAnalytics,MedicalUniversityofLublin,Lublin,Poland;Schoolof Pharmacy,UniversityofEasternFinland,Kuopio,Finland
A.Kancharla
SharettInstituteofOncology,Hadassah-HebrewUniversityMedicalCenter, Jerusalem,Israel
RafikKaraman
BioorganicChemistryDepartment,FacultyofPharmacy,Al-QudsUniversity, Jerusalem,Israel
HiroyukiKobayashi
DepartmentofBiochemistryandMolecularMedicine,InstituteforResearchin ImmunologyandCancer,Universite ´ deMontre ´ al,Montreal,QC,Canada
AjeetKumar
DivisionofPharmacology,CSIR-CentralDrugResearchInstitute,Lucknow,Uttar Pradesh,India
ChristianLeGouill
DepartmentofBiochemistryandMolecularMedicine,InstituteforResearchin ImmunologyandCancer,Universite ´ deMontre ´ al,Montreal,QC,Canada
AnatLevit
DepartmentofPharmaceuticalChemistry,UniversityofCalifornia San Francisco,SanFrancisco,CA,USA
BinLu
InstituteofBiochemistry,CollegeofLifeSciences,ZijingangCampus,Zhejiang University,Hangzhou,Zhejiang,China
ViktoryaLukashova
DepartmentofBiochemistryandMolecularMedicine,InstituteforResearchin ImmunologyandCancer,Universite ´ deMontre ´ al,Montreal,QC,Canada
MarinaMacı´as-Silva
DepartamentodeBiologı´aCelularyDesarrollo,InstitutodeFisiologı´aCelular, UniversidadNacionalAuto ´ nomadeMe ´ xico,Me ´ xicoD.F.,Mexico
M.Maoz
SharettInstituteofOncology,Hadassah-HebrewUniversityMedicalCenter, Jerusalem,Israel
DariuszMatosiuk
DepartmentofSynthesisandChemicalTechnologyofPharmaceutical SubstanceswithComputerModellingLab,FacultyofPharmacywithDivisionof MedicalAnalytics,MedicalUniversityofLublin,Lublin,Poland
JeremyC.McIntyre
DepartmentofNeuroscience,UniversityofFlorida,Gainesville,FL,USA;Center forSmellandTaste,UniversityofFlorida,Gainesville,FL,USA
MashaY.Niv
InstituteofBiochemistry,FoodScienceandNutrition,TheRobertH.Smith FacultyofAgriculture,FoodandEnvironment,TheHebrewUniversity,Rehovot, Israel;FritzHaberCenterforMolecularDynamics,TheHebrewUniversity, Jerusalem,Israel
CarlosNogueras-Ortiz
InstituteofNeurobiology,UniversityofPuertoRicoMedicalSciencesCampus, SanJuan,PR,USA
MelaniePhilipp
InstituteforBiochemistryandMolecularBiology,UlmUniversity,Ulm,Germany
CristinaRoman-Vendrell
InstituteofNeurobiology,UniversityofPuertoRicoMedicalSciencesCampus, SanJuan,PR,USA;DepartmentofPhysiology,SchoolofMedicine,Universityof PuertoRico,SanJuan,PR,USA
EwelinaRutkowska
DepartmentofBiopharmacy,FacultyofPharmacywithDivisionofMedical Analytics,MedicalUniversityofLublin,Lublin,Poland
JanaSelent
ResearchProgrammeonBiomedicalInformatics(GRIB),UniversitatPompeu Fabra,IMIM(HospitaldelMarMedicalResearchInstitute),Barcelona,Spain
DurbaSengupta
CSIR-NationalChemicalLaboratory,Pune,India
YingShi
InstituteofBiochemistry,CollegeofLifeSciences,ZijingangCampus,Zhejiang University,Hangzhou,Zhejiang,China
FabioMarquesSimoesdeSouza
CenterforMathematics,ComputationandCognition,FederalUniversityofABC, SaoBernardodoCampo,Brazil
MichalSlutzki
InstituteofBiochemistry,FoodScienceandNutrition,TheRobertH.Smith FacultyofAgriculture,FoodandEnvironment,TheHebrewUniversity,Rehovot, Israel
ChandanSona
DivisionofPharmacology,CSIR-CentralDrugResearchInstitute,Lucknow,Uttar Pradesh,India
KatarzynaM.Targowska-Duda
DepartmentofBiopharmacy,FacultyofPharmacywithDivisionofMedical Analytics,MedicalUniversityofLublin,Lublin,Poland
TeresaCasarTena
InstituteforBiochemistryandMolecularBiology,UlmUniversity,Ulm,Germany
B.Uziely
SharettInstituteofOncology,Hadassah-HebrewUniversityMedicalCenter, Jerusalem,Israel
GenaroVa ´ zquez-Victorio
DepartamentodeBiologı´aCelularyDesarrollo,InstitutodeFisiologı´aCelular, UniversidadNacionalAuto ´ nomadeMe ´ xico,Me ´ xicoD.F.,Mexico
Jean-PierreVilardaga
LaboratoryforGPCRBiology,DepartmentofPharmacology&ChemicalBiology, UniversityofPittsburghSchoolofMedicine,Pittsburgh,PA,USA
VanAnthonyM.Villar
DivisionofRenalDiseases&Hypertension,DepartmentofMedicine,TheGeorge WashingtonUniversitySchoolofMedicineandHealthSciences,WA,USA
RichardWargachuk
DepartmentofPharmacologyandTherapeutics,McGillUniversity,Montre ´ al,QC, Canada
KunhongXiao
LaboratoryforGPCRBiology,DepartmentofPharmacology&ChemicalBiology, UniversityofPittsburghSchoolofMedicine,Pittsburgh,PA,USA
PremN.Yadav
DivisionofPharmacology,CSIR-CentralDrugResearchInstitute,Lucknow,Uttar Pradesh,India
GuillermoA.Yudowski
InstituteofNeurobiology,UniversityofPuertoRicoMedicalSciencesCampus, SanJuan,PR,USA;DepartmentofAnatomyandNeurobiology,Schoolof Medicine,UniversityofPuertoRico,SanJuan,PR,USA
YapingZhang
InstituteofBiochemistry,CollegeofLifeSciences,ZijingangCampus,Zhejiang University,Hangzhou,Zhejiang,China
XiaoxuZheng
DivisionofRenalDiseases&Hypertension,DepartmentofMedicine,TheGeorge WashingtonUniversitySchoolofMedicineandHealthSciences,WA,USA
CynthiaZhou
DepartmentofPharmacologyandTherapeutics,McGillUniversity,Montre ´ al,QC, Canada
NaimingZhou
InstituteofBiochemistry,CollegeofLifeSciences,ZijingangCampus,Zhejiang University,Hangzhou,Zhejiang,China
Preface Gprotein coupledreceptors(GPCRs)alsoreferredasseventransmembrane receptors(7TMRs)lieattheheartofalmosteveryphysiologicalandpathophysiologicalprocessinourbody.Thesereceptorsbindtoandgetactivatedbyawide rangeofligandsrangingfromsmallmolecules,hormones,peptides,proteinsto lipids.TheoverallactivationandsignaltransductionmechanismsofGPCRsare highlyconservedwherebindingofanagonistresultsinaconformationalchange inthereceptorfollowedbyactivationofheterotrimericGproteinsandsubsequent generationofsecondmessengersanddownstreamsignaling.Downregulationof GPCRsisalsoprimarilyaconservedprocesswhereactivatedreceptorsare phosphorylatedbyGRKs(GPCRkinases)followedbybindingofbetaarrestins whichleadstoreceptordesensitizationandinternalization.GPCRsaretargetedby aboutone-thirdofthecurrentlyprescribeddrugswhichincludeangiotensinblockers forhypertension,beta-blockersforheartfailure,antihistaminesforallergymanagement,andopioidagonistsasanalgesicmedication.
InthisvolumeofMethodsinCellBiolo gy,wecovermultipleaspectsofGPCR signaling,trafficking,regulation,andcellularassaysinaformofeitheranovervieworasstep-by-stepprotocol.Thisisanefforttobringtogetherdifferent domainsofGPCRpharmacologyandsignalingontoacommonplatformandhighlighttheincrediblyversatilenatureanddiversefunctionalmanifestationof GPCRs.SectionIincludeschaptersonGPCRtraffickinginlipidraftsandcilia, imagingendogenousreceptorinneurons,singlemoleculeimagingofGPCRs, andacomprehensiveanalysisofGPCRsinadiposetissue.InSectionII,wecover topicsrangingfromGPCRsignalingfromendosomes,olfactoryreceptorsignal transduction,studiesofaspecializedGPCRsmoothenedinzebrafishmodel, andtheoutcomeofGPCRsignalingincytoskeletaldynamics.Inrecentyears,a keyfocusareainGPCRbiologyhasbeenthedevelopmentofnovelandmoresensitivecellularassaystoinvestigateGPCRexpression,signaling,anddownregulation.SectionIIIofthisvolumeisfocusedonGPCRassayswhichincludeclassical radioligandbinding,label-free,biosensorandfluorescence basedapproachesto studyGPCRtraffickingandsignaling,andTANGOassayformeasuringGPCRbeta-arrestininteraction.Finally,SectionIVconsistsofchaptersonstructural andcomputationalaspectsofprotease-activatedreceptors,bittertastereceptors, andGPCRdimerization.
Iwouldliketothankalltheauthorswhohavecontributedtothisfocusedvolume despitetheirbusyschedule.Ialsoexpressmysinceregratitudetothejournaleditorialstaffandproductionteamforawonderfuljobinputtingthisvolumetogetherina timelyfashion.Withthisbriefbackground,onbehalfoftheentireMethodsinCell
BiologyTeam,Ipresenttoyouthisvolumeentitled“GProtein CoupledReceptors: Signaling,Trafficking,andRegulation.”Isincerelyhopethatyouenjoythetopics coveredinthisissueandpleasefeelfreetoshareyourfeedbackwithus.
ArunK.Shukla IndianInstituteofTechnology,Kanpur,India
Localizationandsignaling ofGPCRsinlipidrafts 1 VanAnthonyM.Villar1,SantiagoCuevas,XiaoxuZheng,PedroA.Jose1
DivisionofRenalDiseases&Hypertension,DepartmentofMedicine,TheGeorgeWashington UniversitySchoolofMedicineandHealthSciences,WA,USA
1Correspondingauthors:E-mail:vvillar@gwu.edu;pjose@mfa.gwu.edu
CHAPTEROUTLINE 1.LocalizationofGPCRsinLipidRafts........................................................................6
1.1IsolationofLipidRafts............................................................................7
1.1.1Detergent-freemethod.......................................................................... 7
1.1.2Detergent-basedmethod...................................................................... 9
1.1.3Immunoblottinganddatainterpretation............................................... 10
1.2LocalizationofGPCRsinLipidRafts.......................................................11
1.2.1Cellsinsuspension.............................................................................
1.2.2Adherentcells....................................................................................
2.GPCRSignalinginLipidRafts...............................................................................15
2.1PerturbationofRaftStability..................................................................15
2.2ChangingtheCholesterolContent...........................................................16
2.3FluorescenceImaging............................................................................16
Abstract
Theunderstandingofhowbiologicalmembranesareorganizedandhowtheyfunction hasevolved.Insteadofjustservingasamediuminwhichcertainproteinsarefound, portionsofthelipidbilayerhavebeendemonstratedtoformspecializedplatformsthat fostertheassemblyofsignalingcomplexesbyprovidingamicroenvironmentthatis conduciveforeffectiveprotein proteininteractions.Gprotein-coupledreceptors (GPCRs)andrelevantsignalingmolecules,includingtheheterotrimericGproteins,key enzymessuchaskinasesandphosphatases,traffickingproteins,andsecondarymessengers,preferentiallypartitiontothesehighlyorganizedcellmembranemicrodomains, calledlipidrafts.Assuch,lipidraftsarecrucialforthetraffickingandsignalingof GPCRs.ThestudyofGPCRbiologyinthecontextoflipidraftsinvolvesthelocalization oftheGPCRofinterestinlipidrafts,atthebasalstateanduponreceptoragonism,and
MethodsinCellBiology,Volume132,ISSN0091-679X, http://dx.doi.org/10.1016/bs.mcb.2015.11.008
2016ElsevierInc.Allrightsreserved.
theevaluationofthebiologicalfunctionsoftheGPCRinappropriatecelllines.Thelack ofstandardizedmethodologytostudylipidrafts,ingeneral,andoftheworkingsof GPCRsinlipidrafts,inparticular,andtheinherentdrawbacksofcurrentmethodshave hamperedthecompleteunderstandingoftheunderlyingmolecularmechanisms.Newer methodologiesthatallowthestudyofGPCRsintheirnativeformareneeded.Theuseof complementaryapproachesthatproducemutuallysupportiveresultsappeartobethebest wayfordrawingconclusionswithregardstothedistributionandactivityofGPCRsin lipidrafts.
INTRODUCTION LipidRaftMicrodomains.Theplasmamembraneisasemipermeable,biological membranethatdemarcatestheintracellularmilieufromtheextracellularenvironment.Amphipathiclipids,suchasphospholipidsandsphingolipids,arethebuilding blocksofthesebilipidmembranesbecauseoftheiraggregativeproperties,i.e.,their hydrophobictailsassociatetogether,whiletheirhydrophilicheadsinteractwithboth extra-andintracellularaqueousenvironments(Sonnino&Prinetti,2013).The fluidityofthefattyacylgroupsofphospholipidsat37 Cenablesthemembranes toactasamediuminwhichdissolvedmembraneproteinsareaffordedamplelateral mobility,especiallyinresponsetoenvironmentalcues.Sincethefirstdescriptionof an“organizationofthelipidcomponentsofmembranesintodomains”(Karnovsky etal.,1982)andtheelaborationofthe“lipidrafthypothesis”bySimonsandvan Meer(vanMeer&Simons,1988;Simons&Ikonen,1997;Simons&vanMeer 1998),theexistenceoflipidraftsisnowestablished.
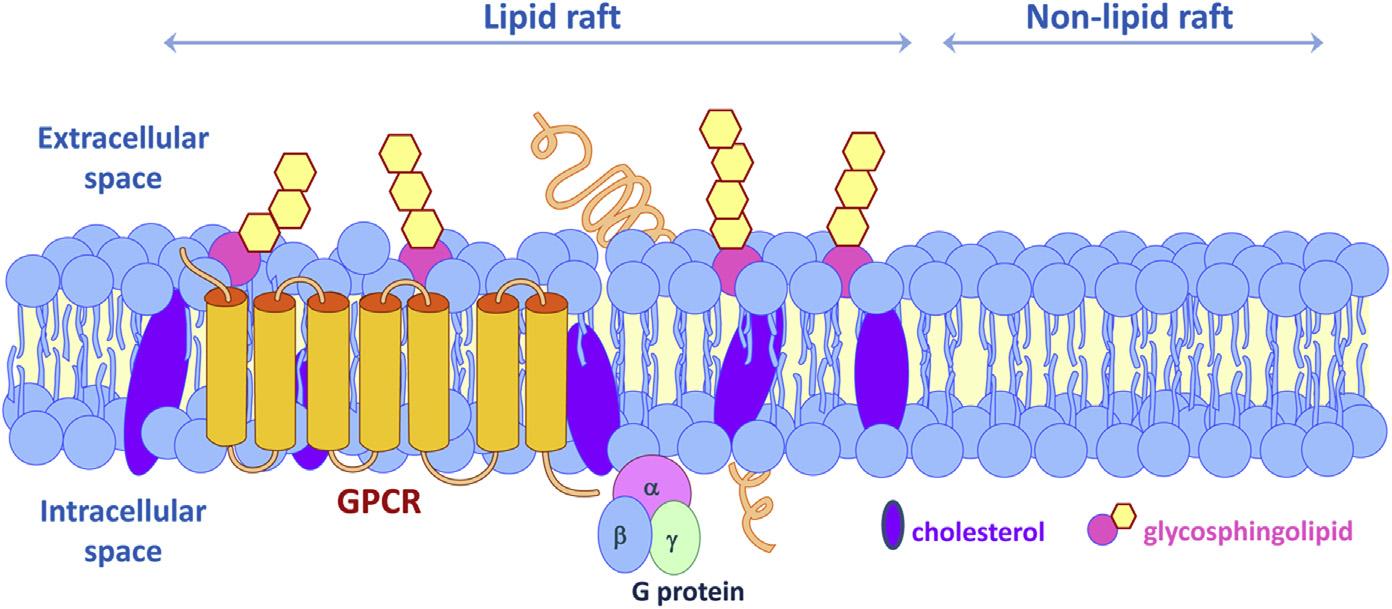
Lipidraftsaretightlypacked,highlyorganizedplasmamembranemicrodomains thatareenrichedinphospholipids,glycosphingolipids,andcholesterolandserveas aplatformfortheorganizationanddynamicinteractionofbiomoleculesinvolvedin variousbiologicalprocesses(Figure1).Thecholesterolbestowsasemblanceof rigidityandorderbyintertwiningintothehydrophobicgapsbetweenthephospholipidacylchains.Certainstructuralproteinsaboundinlipidraftstoserveasscaffold oranchorforotherproteins,includingcaveolins(Head,Patel,&Insel,2014;Quest, Leyton,&Pa ´ rraga,2004;Yu,Villar,&Jose,2013;Yuetal.,2004),flotillins (Rajendran,LeLay,&Illges,2007;Yuetal.,2004)andtetraspanins(Hemler, 2005),andglycosylphosphatidylinositol-linked(GPI-linked)proteins.Thespatial concentrationandorganizationofspecificsetsofmembraneproteinsallowgreater efficiencyandspecificityofsignaltransductionbyfacilitatingprotein protein interactionsandbypreventingcrosstalkbetweencompetingpathways.The nonhomogeneouslateraldistributionofmembranecomponentshelpsexplainthe differencesincompositionbetweenapicalandbasolateralmembranedomainsof polarizedepithelialcells(Sonnino&Prinetti,2013).
Thebestcharacterizedlipidraftmicrodomainsarethecaveolae,whichwerefirst describedbyPaladeandYamadainthe1950s(Palade,1953;Yamada,1955).These aresmall(60 80nm)invaginationsoftheplasmamembraneformedbythe polymerizationofcaveolinswithcholesterol(Parton&delPozo,2013).Caveolae
FIGURE1ALipidRaftMembraneMicrodomain.
Lipidraftsarehighlyorganizedplasmamembranemicrodomainsenrichedinphospholipids, glycosphingolipids,andcholesterol,andserveasmatrixforreceptors,suchasGproteincoupledreceptors(GPCRs),andothersignalingmolecules.(Seecolorplate) VanAnthonyM.Villar,MD,PhD.
havebeenimplicatedinavarietyofcellularprocesses,includingsignaltransduction,endocytosis,transcytosis,andcholesteroltrafficking(Barnett-Norris,Lynch,& Reggio,2005).Lipidraftsaccumulateintheapicalplasmamembraneinpolarized epithelialcellsandinaxonalmembranesinneurons.Basolateralanddendritic membranescontainlipidraftsbutinmorelimitedquantities(Simons&Ikonen, 1997).Interestingly,caveolaearefoundmostlyatthebasolateralmembranethat facesthebloodsupplyandismoreactiveduringsignaltransduction(Simons& Toomre,2000).Lipidraftsaremostlyfoundattheplasmamembrane;however, theymayalsobefoundinintracellularmembranesinvolvedinthebiosynthetic andendocyticpathways.Lipidraftmicrodomainsplayacrucialroleincellularprocessessuchasmembranesorting,receptortrafficking,signaltransduction,andcell adhesion.
GPCRSignalingandTrafficking.Gprotein-coupledreceptors(GPCRs) constitutethelargestsuperfamilyofseventransmembraneproteinsthatrespond toamyriadofenvironmentalstimulithataretransducedintracellularlyasmeaningfulsignalsthroughsecondarymessengers.AgoniststimulationofaGPCRleadstoa conformationalchangethatpromotestheexchangeofGDPforGTPontheGa subunitoftheGprotein,resultingintheuncouplingoftheGproteinfromtheGPCRand thedissociationofGa andGbg subunits.TheGa subuniteitheractivatesorinhibits intracellularsignalingpathwaysdependingonthereceptorsubtype,whiletheGbg subunitrecruitsGprotein-coupledreceptorkinaseswhichselectivelyphosphorylate serineandthreonineresidueslocalizedwithinthethirdintracellularloopand carboxyl-terminaltaildomainsofthereceptortopromotethebindingofcytosolic cofactorproteinscalledarrestins(Lefkowitz,1998).The b-arrestinsplayapivotal roleintheuncouplingprocessandinthesequestrationandinternalizationofGPCRs
throughadynamin-dependent,clathrin-mediatedendocytosis.Onceinternalized, theGPCRs,invesiclestermedasearlyendosomes,aresortedbysortingnexins andfollowdivergentpathways(Worby&Dixon,2002).Thereceptorsaresorted intorecyclingendosomesfortheirreturntothecellmembrane(recyclingand resensitization),accumulateinlateendosomeswhichtargetthelysosomesfortheir subsequentdegradation,ortransportedinitiallytotheperinuclearendosomes(transGolginetwork)andthentothelateendosomesforeventuallysosomaldegradation. Additionalproteolyticmechanisms,suchasproteasomesorcell-associatedendopeptidases,arealsoimplicatedinmediatingthedownregulationofcertainGPCRs (vonZastrow,2003).
ThesignaltransductionthatfollowsligandoccupationoftheGPCRishighly regulatedtoensurethespecificityofthecellularresponse,bothtemporallyand spatially.Thesignaltransductioncanbeattenuatedwithrelativelyfastkinetics throughaprocesscalleddesensitizationorthroughamuchslowerprocessofdownregulationfollowingprolongedorrepeatedexposuretoanagonist.Desensitization, orthewaningofareceptor’sresponsivenesstoagonistwithtime,isaninherent molecular“feedback”mechanismthatpreventsreceptoroverstimulationandhelps increatinganintegratedandmeaningfulsignalbyfilteringoutinformationfrom weakerGPCR-mediatedsignals(Ferguson,2001).
Itisaccomplishedthroughtwocomplementarymechanisms,i.e.,thefunctional uncouplingofGPCRsfromtheircognateGproteins,whichoccurswithoutany detectablechangeinthenumberofcellsurfacereceptors,andGPCRphosphorylation,sequestration,andinternalization/endocytosis.GPCRresensitizationprotects thecellsfromprolongeddesensitizationandiscarriedoutviadephosphorylation byphosphatasesastheGPCRtrafficsthroughtheendosomalpathway.GPCRactivityisthenetresultofacoordinatedbalancebetweenreceptordesensitizationand resensitization.
ItisnowestablishedthatlipidraftsserveasdynamicplatformsforGPCRsand pertinentsignalingmoleculessuchasGproteins,enzymes,andadaptors(BarnettNorrisetal.,2005;Lingwood&Simons,2010).However,understandingthe molecularmechanismsinvolvedhasbeenhamperedbythelackofstandardized methodologytostudylipidrafts,ingeneral,andoftheworkingsofGPCRsinlipid rafts,inparticular.Moreover,theminutesizeoflipidraftshasmadelipidrafts difficulttoresolvebystandardlightmicroscopy,unlessthelipidraftcomponents arecross-linkedwithantibodiesorlectins(Simons&Toomre,2000).Studying howGPCRworksinlipidraftsmaybeaccomplishedbydeterminingifthe GPCRofinterestlocalizestothelipidraftsandbyevaluatingifGPCRsignaling andactivityarelostwhenlipidraftsaredisrupted.
1. LOCALIZATIONOFGPCRsINLIPIDRAFTS SeveraltechniquesareavailableforthedetectionandlocalizationofGPCRsinlipid raftmicrodomainsincells.Themostcommonlyemployedapproachutilizescell
fractionationproceduresthatbreakthecellsapartanddestroycellmorphology beforeGPCRanalysisusingbiochemicalorimmunologicalassays.AcomplementarybiophysicalapproachinvolvesthevisualizationofGPCRsinintactcell membranes.
1.1 ISOLATIONOFLIPIDRAFTS Lipidraftsarecharacterizedbytheirrelativeinsolubilityinnonionicdetergentsat
4 Candlightbuoyantdensityonsucrosegradient(Schnitzer,McIntosh,Dvorak, Liu,&Oh,1995).Theisolationoflipidraftscanbeperformedusingeither detergent-basedordetergent-freemethods(Yuetal.,2013),withthelattergenerating agreaterfractionofinnerleafletmembraneraftsandproducingmorereplicable results(Pike,2004). Schnitzeretal.(1995) employedadetergent-freemethodto isolatelipidraftsusingcationiccolloidalsilicaparticles,whichisappropriatefor non-cellculturestudies.Lipidraftsmaybeextractedfromtotalcellmembranes (Songetal.,1996)orjustfromsurfaceplasmamembranes(Smart,Ying,Mineo,& Anderson,1995).Detergentinsolubilityresultsfromthesegregationofmembraneassociatedproteinsintothelipidrafts,whichareabundantincholesteroland glycosphingolipids.Nonionicdetergents,suchasTritonX-100, b-octylglucoside, CHAPS,deoxycholate,LubrolWX,LubrolPX,Brij58,Brij96,andBrij98,have beenusedtopreparelipidraftfractions(Macdonald&Pike,2005),resultingin varyingyieldsofproteins.Samplesobtainedbydetergent-basedmethodsaretermed detergent-resistantmembranesordetergent-insolublefractions.Differentdetergents mayyielddifferentlipidraftcomponentsbecauseofthevaryingdegreesofresistancebytheproteinstoextractionusingdifferentreagents.Themethodsdetailed belowarebasedon Yuetal.(2013)
1.1.1 Detergent-freemethod Materials
2-N-morpholinoethanesulfonicacid(Mes),250mM,pH ¼ 6.8
Mes-bufferedsolution(MBS),25mMMes þ 150mMNaCl
Sodiumcitrate,500mM,pH w 11(addproteaseinhibitors)
Sucrose,5%,35%,and80%inMBSsolution(addproteaseinhibitors)
Methyl-b-cyclodextrin(b-MCD),2%dissolvedincellculturemedia
Cholesterol þ b-MCD(Sigmacatalog#C4951),dissolvedincellculture media
1XPBS,forwashing
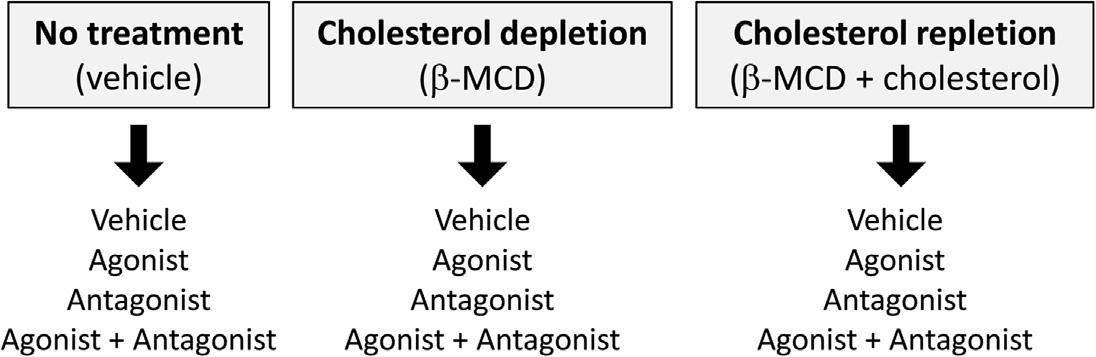
1. Cellcultureandcellpelletcollection.Toobtainsufficientamountsoflipidraft fraction,cellsshouldbegrownin150-mmdishesuntilalmostconfluentusing theappropriatemediaat37 Cwith95%airand5%CO2.Separatedishesof cellsshouldalsobetreatedforcholesteroldepletionandrepletionasexperimentalcontrols(Figure2).Cholesteroldepletiontodisruptthelipidraftsis commonlyperformedbypretreatmentwith b-MCDfor1hat37 C.
FIGURE2ComparisongroupsforGPCRlocalizationinlipidrafts.
Methyl-a-cyclodextrin(a-MCD)maybeusedascontrolfor b-MCD(Vial& Evans,2005).Cholesterolrepletionisperformedbypretreatingwithcholesterol/b-MCDsolutionfor1hat37 C.Cholestane-3,5,6-triol,aninactive analogofcholesterol,maybeusedascontrolfortheuseofexogenous cholesterol(Murtazina,Kovbasnjuk,Donowitz,&Li,2006).Todeterminethe effectofagonistorantagonisttreatment,cellsshouldbeserum-starvedforat least1hpriortotreatmenttoachieve“basal”conditionspriortotreatment. Additionalcontrols,suchastheuseofthedrugvehicle,shouldbeconcomitantlyperformed.
1.1 WashcellswithcoldPBSandscrapethecellsusingarubber-tippedcell scraper.
1.2 Transfercellsuspensioninto15-mLtubeandspinat2000 g for5min.
1.3 Decantthesupernatanttoobtainthecellpellet.
2. Cellhomogenatepreparation. Allstepsarecarriedoutat4 C.
2.1 Tothecellpellet,add1.5mL500mMsodiumcarbonateandvortex.
2.2 Homogenizethecellsuspensionbysonicationusingfive20-sburstsonice.
2.3 Add1.5mLof80%sucroseandmixbyvortexandsonication(three20-s bursts)onice.Proteinconcentrationmaybedeterminedatthistimeusinga BCAkit.
3. Sucrosegradientultracentrifugation.Prepare5%,35%,and80%sucrose solutionsinMBSsolution.TheuseofMBSsolutionwithpHcloseto7.0may beadvantageousformostproteins.
3.1 Place3mLofcellhomogenatesintothebottomofprecooled12-mL ultracentrifugetubes.
3.2 Overlaysequentially4.5mLof35%sucroseand4.5mLof5%sucroseto eachtube.
3.3 WiththetubessecurelybalancedinanSW-41bucket,spinat180,000 g (38,000rpm)for16hat4 CinaBeckmanSW-41centrifuge.
4. Lipidraftfractionpreparation.Alight-scatteringbandthatisenrichedwith caveolae/lipidraftscanbeobservedbetweenthe5%and35%sucrosegradients andcorrespondstothefourthfraction.
4.1 Carefullyaspirate121-mLfractionsfromthetopofthetubeandtransfer intoprelabeled1.5microcentrifugetubes.
4.2 Prepare0.5mLofeachfractionbyadding0.1mL6Xsamplebuffer,vortex, andboilfor5minbeforeuseforimmunoblotting.Thesesamplescanbe storedat 20 C,whiletherestofthefractionswithoutthe6Xsample buffercanbestoredat 80 C.
1.1.2 Detergent-basedmethod Materials
50%OptiprepStocksolution(45mLof60%Optiprep þ 9mLofOptiprep diluent)
MBSTSbuffer(MBS þ 0.5%TritonX-100 þ proteaseinhibitorsin10%sucrose)
Sucrosesolutions(Table1):
Table1 PreparationofOptiprepGradientSolutions
1. Cellcultureandcellpelletpreparation.Thesameaswiththedetergent-free method.
2. Cellextractpreparation.
2.1 Add0.3mLice-coldMBSTStocellpelletandpushthrougha25G needle10 .
2.2 Adjustcellextract(w0.4mL;cellpelletvolumeis w0.1mL)to40% Optiprepbyadding0.8mLofcold60%Optiprepandvortex.Determine proteinconcentrationusingaBCAkit.
3. Optiprepgradientultracentrifugation.
3.1 Place1mLofthecellextractintothebottomofprecooled5-mLultracentrifugetubes.
3.2 Overlaywith1mLeachof30%,25%,20%,and0%Optiprepsolutionsin MBSTSbuffer.
3.3 SecureeachtubeinaBeckmanSW50.1bucketandspinat175,000 g (42,000rpm)at4 Cfor4h.Otherrotorsmaybeused,suchastheSW55 (170,000 g for4h)orTLS55(250,000 g for2.5h).
4. Lipidraftfractionpreparation.
4.1 Carefullyaspirateten0.5-mLfractionsfromthetopofthetubeandtransfer intoprelabeled1.5microcentrifugetubes.
4.2 Prepare0.25mLofeachfractionbyadding0.5mL6Xsamplebuffer, vortex,andboilfor5minbeforeuseforimmunoblotting.Thesesamples canbestoredat 20 C,whiletherestofthefractionswithoutthe6X samplebuffercanbestoredat 80 C.
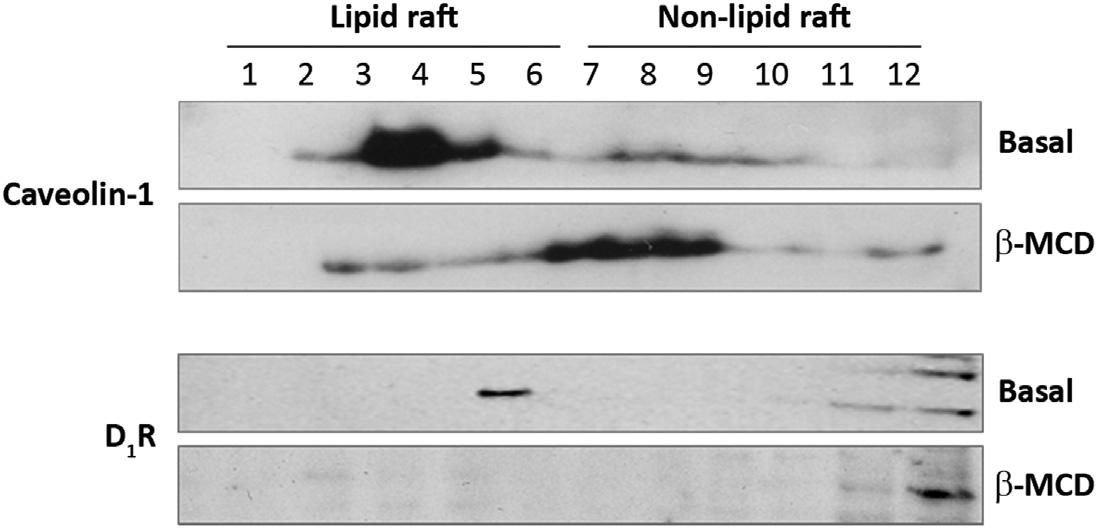
1.1.3 Immunoblottinganddatainterpretation Westernblotisthemostcommonlyusedmethodtodeterminethelipidraftdistributionofproteins,suchasGPCRs.Antibodyspecificityiscrucialfortheidentification oftheGPCRofinterest.Thelipidraftproteinsarefoundinthemorebuoyant fractions(top5 6fractions);however,theirdistributionamongthesefractionsis notuniform.Immunoblottingforlipidraftmarkersmayhelpindeterminingthe fractionswherethelipidraftsaremostabundant.Caveolin-1isthemostcommonly usedproteinmarkerforlipidrafts,specificallyforcaveolae(Inseletal.,2005; Lingwood&Simons,2010).Thereareseveralothermarkersforlipidrafts,such asflotillin-1,CD55,alkalinephosphatase,andpore-formingtoxins,suchascholera toxinsubunitB(CTxB),equinatoxinII,perfringolysin(Foster,DeHoog,&Mann, 2003;Salzer&Prohaska,2001;Skocajetal.,2013).Flotillin-1hasbeenusedasa lipidraftmarkerproteinincellsthatdonotcontaincaveolae,i.e.,bloodcells (Salzer&Prohaska,2001),neuralcells(Huangetal.,2007),andratrenalproximal tubulecells(Breton,Lisanti,Tyszkowski,McLaughlin,&Brown,1998;Riquier,Lee, &McDonough,2009)andhumanembryonickidney(HEK)-293cells(Yuetal., 2004).Thereisspeciesspecificitybecausehumanrenalproximaltubulecells expresscaveolin-1(Gildeaetal.,2009),whileHEK-293cellsexpresscaveolin-2. Thesemarkersmayalsobeusedtoindicatetheintegrityoflipidraftsincholesterol depletionorrepletionexperiments.Ingeneral,thesemarkersshouldbedistributedin themorebuoyantfractionsandshouldredistributeintothelessbuoyantfractions (fractions7 12)aftercholesteroldepletionwith b-MCD(Figure3).Cholesterol repletionreconstitutesthelipidraftsandthus,thesemarkersshouldbeobservedin themorebuoyantfractions.
FIGURE3LipidRaftDistributionofCaveolin-1andD1R.
Lipidraftandnon-lipidraftfractionsfromhumanrenalproximaltubulecellstreatedwith b-MCD,acholesterol-depletingandlipidraft-disruptingagent,werepreparedbydetergentfreemethodandsucrosegradientultracentrifugation.Thedistributionofcaveolin-1,alipid raftmarker,andthedopamineD1 receptor(D1R),aGPCR,isshownintheimmunoblots. ImagesarecourtesyofPeiyingYu,MD.
1.2 LOCALIZATIONOFGPCRsINLIPIDRAFTS AnotherwaytodemonstratethedistributionofGPCRsinlipidraftsisbyvisualizing theminintactcells,livingorfixed,andtissues.TherearenowcommerciallyavailablekitsthathavebeendevelopedforlabelingthelipidraftsusingtheCTxBthatis taggedwithfluorophores(Figure4).CTxBbindstothepentasaccharidechainof gangliosideGM1,whichselectivelypartitionsintolipidrafts.Forvisualizinglipid rafts,cellsarelabeledwithCTxBtaggedwithAlexaFluor 488,AlexaFluor
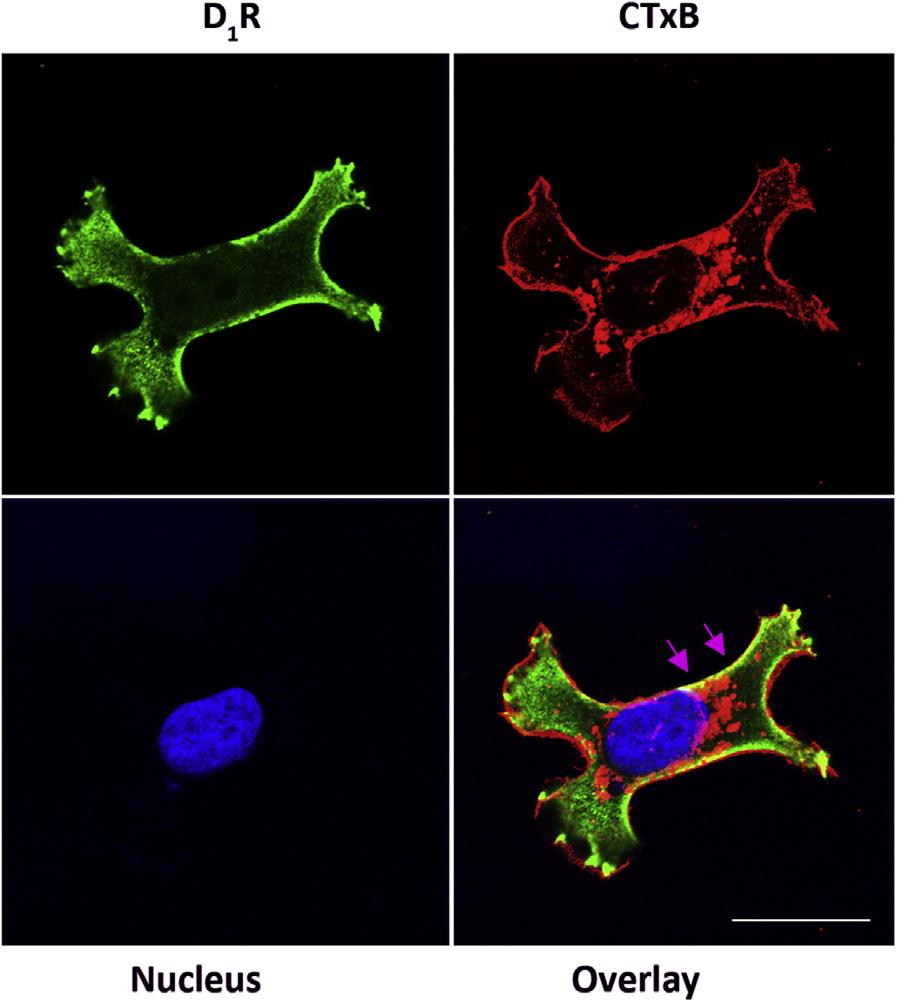
FIGURE4ColocalizationoftheD1 dopaminereceptor(D1R)inLipidRaftsofHumanRenal ProximalTubuleCells.
Humanrenalproximaltubulecellsweregrownonapoly-L-Lysine-coatedcoverslipto50% confluenceandserum-starvedfor1htodeterminethebasaldistributionofD1Rpriorto fixationwith4%paraformaldehydeandpermeabilizationwith0.5%TritonX-100.Thelipid raftswerelabeledusingcholeratoxinsubunitB(CTxB)taggedwithAlexaFluor 555 (MolecularProbes),whiletheendogenousD1Rwasimmunostainedusingaproprietary rabbit-anti-D1Rantibodyandadonkeyanti-rabbitsecondaryantibodytaggedwithAlexa Fluor 488(MolecularProbes).DAPIwasusedtovisualizethenucleus.Atthebasalstate, mostoftheD1Rwerefoundintracellularly,justbelowtheinnerleafletoftheplasma membrane,althoughsomecolocalizedwiththelipidrafts(yellowareaspointedatbyarrows). Therawimageswerecapturedvialaserscanningconfocalmicroscopeusingseparate channelsandthecompositeimagewasobtainedusingZen2011software.630X magnification,scalebar ¼ 10 mm.(Seecolorplate)
VanAnthonyM.Villar,MD,PhD.
555,orAlexaFluor 647beforecross-linkingwithananti-CTxBtomaintainthe insitu proteindistribution.TodemonstratethelipidraftdistributionofGPCRs,colocalizationexperimentsmaybeperformedvialaserscanningconfocalmicroscopyby labelingthelipidraftsusingCTxBandimmunostainingtheGPCRofinterestusing specificantibodiesonthesamecell.CTxBlabelingmayalsobeusedtodemonstrate lipidraftendocytosisuponagoniststimulationinlivecells(Qi,Mullen,Baker,& Holl,2010)andculturedexplants(Hansenetal.,2005).Thec-subunitofcytolethal distendingtoxin(cdt)mayalsobeutilizedforlipidraftcolocalizationexperiments (theprotocolisdetailedin Boesze-Battaglia,2006).Otherpore-formingtoxins,besidesCTxB,usedtovisualizelipidraftsincludeequinatoxinIIwhichbindsdispersed sphingomyelin,lyseninwhichbindsclusteredsphingomyelin,perfringolysinOwhich bindstocholesterol,andostreolysinwhichbindstothecombinationofsphingomyelin andcholesterol(Makinoetal.,2015;Skocajetal.,2013).
AnalternativetousingCTxB,cdt,andotherpore-formingtoxinsistouse antibodiesthatspecificallytargetthelipidraftproteinmarkers,suchas caveolin-1,caveolin-3,andflotillin-1.Conversely,transferrinreceptors,CD71, andgeranylatedproteinsarenon-lipidraftmarkers( Boesze-Battaglia,2006; Magee,Adler,&Parmryd,2005 ).ThegangliosideGM1 maybelabeledwithsingle quantumdotstomeasurethelateralmo bilityandextentofmovementofthelipid rafts(Chang&Rosenthal,2012 ).Recently,GPI-anchoredproteinsthatsegregate intolipidraftshavebeenvisualizedusin ganovelmethodcalledenzyme-mediated activationofradicalsources( Miyagawa-Yamaguchi,Kotani,&Honke,2015 ). Probesthattargetthelipidcontentoflipidraftshavealsobeenusedtovisualize thesemembranemicrodomains.Laurdan(6-dodecanoyl-2-(dimethylamino)naphthalene)andC-laurdan(6-dodecanoyl-2-[N-methyl-N-(carboxymethyl) amino]-naphthalene),whicharemembra neprobesthataresensitivetomembrane polarity,allowtheobservationoflipidraftsviatwo-photonmicroscopy(Gaus, Zech,&Harder,2006;Kimetal.,2007,2008 ).Afluorophore-taggeddomain D4ofperfringolysinO,acholesterol-bindingcytolysinproducedby Clostridium perfringens ,hasbeenusedasprobetostudymembranecholesterol(OhnoIwashitaetal.,2004).
Asidefromconfocalmicroscopy,othe rbiophysicalapproachesmayalsobe employedtostudylabeledGPCRsand/orli pidrafts.Singlefluorophoretracking microscopy( Schu ¨ tz,Kada,Pastushenko,&Schindler,2000 )andfluorescence recoveryafterphotobleaching(Kenworthy,2007 )maybeusedtomonitorlateral diffusionoflipidraft-anchoredGPCRs,w hilefluorescence lifetimeimaging microscopy fluorescenceresonanceenergytransfer(FLIM-FRET)(Kenworthy, Petranova&Edidin,2000;Thaa,Herrmann,&Veit,2010 )maybeusedtodeterminetheproximityofGPCRswithotherproteinsofinterest,oroflipidraftsizes dependingonmembranecomposition( deAlmeida,Loura,Fedorov,&Prieto, 2005).Atomicforcemicroscopymaybeused tovisualizetheeffectsofdetergent solubilizationofmembranesduringlipidraftstudies(Garner,Smith,&Hooper, 2008).Lipidraftscannowbevisualizedusingsuperresolutionimagingbelow the200nmlimitofconventionalmicroscopes,e.g.,includingstructured
illuminationmicroscopy,stimulatedemissiondepletion(STED)microscopy,nearfieldscanningopticalmicroscopy,pho toactivatedlocalizationmicroscopy (PALM),andstochasticopticalreconstructionmicroscopy(dSTORM)(Owen& Gaus,2013;Tobinetal.,2014;Wuetal.,2013 ).
Materials
Vybrant LipidRaftLabelingKits(Catalog#V-34403,V-34404,orV-34405) preparefreshworkingsolutionsaccordingtomanufacturer’sinstructions
PrimaryantibodyagainsttheGPCRofinterest
Secondaryantibodyagainstthehostoftheprimaryantibody
10%bovineserumalbumin(BSA)solution
4%ParaformaldehydeinPBS
Mountingmedium(EMScatalog#17985)without40 ,6-diamidino-2phenylindole(DAPI)
DAPI,anuclearstain,10mMstocksolution
TritonX-100,20%stocksolutionindeionizedwater 1XPBSforwashing
1.2.1 Cellsinsuspension ColocalizationofGPCRswithlipidraftscannowbeaccomplishedwiththeconcomitantuseofCTxBandanantibodyagainsttheGPCRofinterestoncells.Thecellscan belabeledinsuspensionandthenmountedonglassslidesforimaging,orthecellscan begrownandlabeledoncoverslipsorinTranswells cellcultureinsertswhencell polarityisimportanttodistinguishbetweenapical versus basolateralmembranes.
1. Fluorescentlabelingofcells.
1.1 Spincellsat2000 g for5minanddecantthemedium.
1.2 Resuspendthecellsincoldmedium,spin,anddecantthemedium.
1.3 Resuspendthecellsin2mLofCTxB AlexaFluor workingsolutionat 4 Cfor10min.TheprimaryantibodyagainsttheGPCRofinterestmay beaddedtothisworkingsolutionat1:100dilution.Theprimaryantibody againsttheGPCRshouldberaisedinmouse,goat,rat,orchickenbutnotin rabbitwhenusingtheVybrant LipidRaftLabelingKits.Alternatively, theprimaryantibodyagainsttheGPCR(especiallyifonlyarabbitantibodyisavailable)maybeprelabeledwithaFluorotherthantheoneused forCTxB.Directlylabelingtheprimaryantibodyprecludestheuseofa secondaryantibody(instep1.5).
1.4 Gentlywashcells3 withcoldPBS.Spincellsanddecantwashbuffer.
1.5 Resuspendin2mLoftherabbitCTxBantibodyworkingsolutionat4 C for30min.TherabbitCTxBantibodycross-linksittothelipidraftdomains.Thesecondaryantibodyagainsttheprimaryantibodymaybeadded tothisworkingsolutionat1:100dilution.Thesecondaryantibodyshould betaggedwithaFluorotherthantheoneusedtolabeltheCTxB. Ascounterstain,300nMDAPImayalsobeaddedtothisworkingsolution.
1.6 Gentlywashcells3 withcoldPBS.Spincellsanddecantwashbuffer.
2. Mountingandimaging.
2.1 (Optional)Fixcellswith4%paraformaldehydeatroomtemperaturefor 15min.Paraformaldehydeisacross-linkerfixativethatpreservesthe architectureofthecellbutmayreducetheantigenicityofsomecell componentsandthus,requiresanadditionalpermeabilizationstepif additionalintracellularproteinsareneededtobevisualized.Fixationmay alsobeachievedusingorganicsolvents,suchasalcoholsandacetone,but theseremovelipidsandprecipitatetheproteinsandoftendisruptthecell structure.
2.2 MountlivecellsincoldPBSorfixedcellsinmountingmediumonglass slideandcoverwithcoverslip.
2.3 Imagethecellsusingalaserscanningconfocalmicroscope.Theappropriate filtersshouldbeuseddependingontheAlexaFluor dyethatwasusedand whetherDAPIwasusedasanuclearstainornot(Table2).
1.2.2 Adherentcells
1. Cellcultureoncoverslips.
1.1 Growcellson12-mmcoverslipsplacedina24-welltissuecultureplateto w50%confluenceusingcompletecellculturemediumat37 Cin95%air and5%CO2.Coverslipscoatedwithlysine,laminin,orcollagenmay improvecellattachmentforcellsthateasilydetach,suchasHEK-293cells. Todeterminetheeffectofagonist/antagonisttreatmentonGPCR trafficking,cellsshouldbeserum-starvedforatleast1hpriortotreatment toachieve“basal”conditionspriortotreatment.Additionalcontrols,such asvehicletreatment,shouldbeperformed.
1.2 DrawoffthemediumandwashcellswithcoldPBS.Placethecellculture plateonicetostopfurtherreceptorendocytosisandendosomaltrafficking.
2. Fluorescentlabeling,fixation,andpermeabilization.
2.1 Add0.3mLofCTxB AlexaFluor workingsolutionat4 Cfor10min.
2.2 DrawoffthesolutionandwashcellswithcoldPBS.
2.3 Fixcellswith0.3mLof4%paraformaldehydeatroomtemperaturefor 15min.
2.4 WashcellswithPBS.Subsequentstepscanbeperformedatroom temperature.
Table2 FluorescenceSpectraofCTxBConjugates
AlexaFluor 488(V-34403)495/519 AlexaFluor 555(V-34404)555/565 AlexaFluor 594(V-34405)590/617
ThemaximumabsorptionandemissionforDAPIare358/461nm.
2.5 Permeabilizethecellswith0.3mLof0.5%TritonX-100indeionizedwater for10min.Permeabilizationprovidesaccesstointracellularantigens. TritonX-100caneffectivelysolvatecellularmembraneswithoutdisturbingprotein proteininteractions.Otherdetergentssuchassaponin, Tween-20,orsodiumdodecylsulfatemayalsobeused.
2.6 WashcellswithPBS.
3. Immunostaining.
3.1 Add0.3mLoftheprimaryantibodyagainsttheGPCRofinterestdissolved in10%BSA(1:100 200dilution)for30 60min.
3.2 Washcells3XwithPBS.
3.3 Add0.3mLofthesecondaryantibody(againstthehostoftheprimary antibodyusedinstep3.1)in10%BSA.Thesecondaryantibodyshouldbe taggedwithaFluorotherthantheoneusedtolabeltheCTxB.Ascounterstain,300nMDAPImayalsobeaddedtothisworkingsolution.
3.4 Wash2XwithPBSandoncewithdeionizedwater.Theuseofdeionized waterwashesawaytheresidualNaClcrystalsfromPBS.
3.5 Mountcoverslipsusingamountingmediumonglassslide.Gentlyremove excessmountingmediumbyaspiration.Allowthemountingmediumto hardencompletely.
3.6 Imagethecellsusingalaserscanningconfocalmicroscope.Theappropriate filtersshouldbeuseddependingontheAlexaFluor dyethatwasusedand whetherDAPIwasusedasanuclearstain.
2. GPCRSIGNALINGINLIPIDRAFTS TherearemanyestablishedprotocolsavailablethatallowthestudyofGPCRactivity perse usingcommerciallyavailablekitsor,lesscommonly,proprietarymaterials. StudyingtheactivityofGPCRsinthecontextoftheirresidencyinlipidraftsoften requiresadditionalstepsthatwoulddisrupttheintegrityofthelipidraftmicrodomainordissociatetheproteinofinterestfromtherafts.Mostofthecurrentstrategies todisruptlipidraftinvolveseitherperturbationoftheraftstabilityormodifyingthe cholesterolcontentofthelipidrafts.Mostofthesetreatmentsareperformedoncells priortoagonist/antagonisttreatmentandfunctionalassays,suchascAMPproduction,sodiumtransport,andNADPHoxidaseactivity(Gildeaetal.,2009;Hanetal., 2008;Yuetal.,2004,2014).
2.1 PERTURBATIONOFRAFTSTABILITY Lipidraftsaredynamicassembliesofphospholipidsandglycosphingolipidsthat containmostlysaturatedhydrocarbonchainswhichallowcholesteroltointercalate betweenthefattyacylchains.Thesurroundingmembranehasgreaterfluidity becauseofthepreponderanceofphospholipidswithunsaturatedacylgroups.The additionofexogenousgangliosides(Webb,Hermida-Matsumoto,&Resh,2000)