TranslationalEpigeneticsSeries
TrygveTollefsbol,Ph.D.,D.O.,DistinguishedProfessor-SeriesEditor
ProfessorofBiology,UniversityofAlabamaatBirmingham,andSeniorScientist,ComprehensiveCancerCenter, ComprehensiveCenterforHealthy,Birmingham,AL,UnitedStates Aging,ComprehensiveDiabetesCenterandNutritionObesityResearchCenter,Birmingham,AL,UnitedStates Director,CellSenescenceCultureFacility,Birmingham,AL,UnitedStates
TransgenerationalEpigenetics
EditedbyTrygveO.Tollefsbol,2014
PersonalizedEpigenetics
EditedbyTrygveO.Tollefsbol,2015
EpigeneticTechnologicalApplications
EditedbyY.GeorgeZheng,2015
EpigeneticCancerTherapy
EditedbyStevenG.Gray,2015
DNAMethylationandComplexHumanDisease
ByMichelNeidhart,2015
EpigenomicsinHealthandDisease
EditedbyMarioF.FragaandAgustinF.FFerna ´ ndez,2015
EpigeneticGeneExpressionandRegulation
EditedbySumingHuang,MichaelLittandC.AnnBlakey,2015
EpigeneticBiomarkersandDiagnostics
EditedbyJoseLuisGarcı´a-Gime ´ nez,2015
DrugDiscoveryinCancerEpigenetics
EditedbyGerdaEggerandPaolaBarbaraArimondo,2015
MedicalEpigenetics
EditedbyTrygveO.Tollefsbol,2016
ChromatinSignalingandDiseases
EditedbyOlivierBindaandMartinFernandez-Zapico,2016
GenomeStability
EditedbyIgorKovalchukandOlgaKovalchuk,2016
ChromatinRegulationandDynamics
EditedbyAnitaGondor,2016
NeuropsychiatricDisordersandEpigenetics
EditedbyDagH.Yasui,JacobPeedicayilandDennisR.Grayson, 2016
PolycombGroupProteins
EditedbyVincenzoPirrotta,2016
EpigeneticsandSystemsBiology
EditedbyLeonieRingrose,2017
CancerandNoncodingRNAs
EditedbyJayprokasChakrabartiandSangaMitra,2017
NuclearArchitectureandDynamics
EditedbyChristopheLavelleandJean-MarcVictor,2017
EpigeneticMechanismsinCancer
EditedbySabitaSaldanha,2017
EpigeneticsofAgingandLongevity
EditedbyAlexeyMoskalevandAlexanderM.Vaiserman,2017
TheEpigeneticsofAutoimmunity
EditedbyRongxinZhang,2018
EpigeneticsinHumanDisease,SecondEdition
EditedbyTrygveO.Tollefsbol,2018
EpigeneticsofChronicPain
EditedbyGuangBaiandKeRen,2018
EpigeneticsofCancerPrevention
EditedbyAnupamBishayeeandDeepakBhatia,2018
ComputationalEpigeneticsandDiseases
EditedbyLooKeatWei,2019
Pharmacoepigenetics
EditedbyRamo ´ nCacabelos,2019
EpigeneticsandRegeneration
EditedbyDanielaPalacios,2019
ChromatinSignalingandNeurologicalDisorders
EditedbyOlivierBinda,2019
TransgenerationalEpigenetics,SecondEdition
EditedbyTrygveTollefsbol,2019
NutritionalEpigenomics
EditedbyBradleyFerguson,2019
PrognosticEpigenetics
EditedbyShilpySharma,2019
EpigeneticsoftheImmuneSystem
EditedbyDieterKabelitz,2020
StemCellEpigenetics
EditedbyEranMeshorerandGiuseppeTesta,2020
EpigeneticsMethods
EditedbyTrygveTollefsbol,2020
HistoneModificationsinTherapy
EditedbyPedroCastelo-BrancoandCarmenJeronimo,2020
EnvironmentalEpigeneticsinToxicologyandPublicHealth
EditedbyRebeccaFry,2020
GenomeStability
EditedbyIgorKovalchukandOlgaKovalchuk,2021
TwinandFamilyStudiesofEpigenetics
EditedbyShuaiLiandJohnHopper,2021
EpigeneticsandMetabolomics
EditedbyPabanK.AgrawalaandPoonamRana,2021
MedicalEpigenetics,SecondEdition
EditedbyTrygveTollefsbol,2021
EpigeneticsinPrecisionMedicine
EditedbyJose ´ LuisGarcı´a-Gime ´ nez,2022
EpigeneticsofStressandStressDisorders
EditedbyNagyA.Youssef,2022
Post-TranscriptionalGeneRegulationinHumanDisease
EditedbyBuddhiPrakashJain,ShyamalK.Goswamiand TapanSharma,2022
Editedby
DepartmentofPharmacology,UniversidadeFederaldeSa ˜ oPaulo, Sa ˜ oPaulo,Brazil
LaboratoryofOntogenyandEpigenetics,EscolaPaulistadeMedicina, UniversidadeFederaldeSa ˜ oPaulo,Sa ˜ oPaulo,Brazil
MiriamGalvonasJasiulionis
AcademicPressisanimprintofElsevier 125LondonWall,LondonEC2Y5AS,UnitedKingdom 525BStreet,Suite1650,SanDiego,CA92101,UnitedStates 50HampshireStreet,5thFloor,Cambridge,MA02139,UnitedStates TheBoulevard,LangfordLane,Kidlington,OxfordOX51GB,UnitedKingdom
Copyright©2022ElsevierInc.Allrightsreserved.
Nopartofthispublicationmaybereproducedortransmittedinanyformorbyanymeans,electronicor mechanical,includingphotocopying,recording,oranyinformationstorageandretrievalsystem,without permissioninwritingfromthepublisher.Detailsonhowtoseekpermission,furtherinformationaboutthe Publisher’spermissionspoliciesandourarrangementswithorganizationssuchastheCopyrightClearance CenterandtheCopyrightLicensingAgency,canbefoundatourwebsite: www.elsevier.com/permissions.
ThisbookandtheindividualcontributionscontainedinitareprotectedundercopyrightbythePublisher (otherthanasmaybenotedherein).
Notices
Knowledgeandbestpracticeinthisfieldareconstantlychanging.Asnewresearchandexperiencebroadenour understanding,changesinresearchmethods,professionalpractices,ormedicaltreatmentmaybecome necessary.
Practitionersandresearchersmustalwaysrelyontheirownexperienceandknowledgeinevaluatingandusing anyinformation,methods,compounds,orexperimentsdescribedherein.Inusingsuchinformationormethods theyshouldbemindfuloftheirownsafetyandthesafetyofothers,includingpartiesforwhomtheyhavea professionalresponsibility.
Tothefullestextentofthelaw,neitherthePublishernortheauthors,contributors,oreditors,assumeanyliability foranyinjuryand/ordamagetopersonsorpropertyasamatterofproductsliability,negligenceorotherwise,or fromanyuseoroperationofanymethods,products,instructions,orideascontainedinthematerialherein.
ISBN978-0-323-91081-1
ForinformationonallAcademicPresspublications visitourwebsiteat https://www.elsevier.com/books-and-journals
Publisher: StacyMasucci
AcquisitionsEditor: PeterB.Linsley
EditorialProjectManager: SamW.Young
ProductionProjectManager: SreejithViswanathan
CoverDesigner: ChristianBilbow
TypesetbySTRAIVE,India
KhalilAzizian,MoeinShirzad,NegarGorjizadeh,andAnsarKarimian
NegarGorjizadeh,NassimGorjizadeh,KhalilAzizian,AnsarKarimian, andMoeinShirzad
Sirtuinsandepigeneticmodifications.......................................................................92
Histoneacetylationanddeacetylation..................................................................92
Histonemethylation..............................................................................................94
Sirtuinsandtelomeremaintenance...........................................................................95
Sirtuinsandgenomicstability...................................................................................96
Sirtuinsanddiseases..................................................................................................98
Concludingremarks...................................................................................................99
SECTION2DNAdamageandepigenetics—Theirimpactinthe onsetofdiseases
CHAPTER7DNAdamage,metabolism,andepigeneticregulation................
Introduction...........................................................................................................111
Theinterplaybetweenepigeneticsandmetabolism........................................111
MetabolicdysfunctionandDNArepair..........................................................112
Crosstalkbetweenepigeneticsandmetabolismincancer...................................114
DNAdamage....................................................................................................114
Roleofepigeneticalterationsincellularmetabolism.....................................114
Roleofmetabolicdysfunctioninthecontextofepigenetics..........................117
Metabolicdysfunction,epigeneticmodification,andgenomicinstability.........120
Epigeneticsandsteroidhormonemetabolisminendocrine-drivencancer.........123
Epigeneticalterationsinprostatecancerprogression.....................................124
Epigeneticalterationsinbreastcancerprogression........................................126
Conclusionsandperspectives...............................................................................127
CHAPTER8DNAdamage,epigenetics,andaging.......................................
Introduction...........................................................................................................139
EpigeneticmodificationsversusDNAdamage...................................................139
DNAdamagecausescellularsenescenceandaging...........................................140
EpigeneticmarksthatpreventDNAdamageandaging:Youth-associated genome-stabilizingepigeneticmarks...............................................................141
HowepigeneticmodificationspreventDNAdamageandtheaging process:DNAProtectionvs.DNArepair........................................................141
IRShypomethylationdrivesthemolecularpathogenesisofage-related noncommunicablediseases...............................................................................143
WhatareanIRS,thestructureofeachIRSelement,andthe distributionofmethylationmarksinthegenome?......................................143
IRSmethylationlevelsandpatternchangesinhumandiseases.....................145 MechanismsofIRShypomethylationandhypermethylation.........................146
CHAPTER9DNAdamagesignaling,cellreprogramming,
MikioShimadaandTomokoMiyake
CHAPTER10TheinterplaybetweenDNAdamageandepigenetics
DaynaChallisandKateH.Brettingham-Moore
CHAPTER11Oncometabolites,epigeneticmarks,andDNArepair.................. 191
JonathanDowandPeterM.Glazer
Oncometabolitebiology.......................................................................................191
Bridgingtwohallmarks:Alteredmetabolismand genomicinstability.......................................................................................191
Oncometabolites’identificationincancer.......................................................192
OncometabolitesinduceglobalhistoneandDNAhypermethylation via αKG-dependentdioxygenaseinhibition.................................................192
OncometabolitesandDNArepair........................................................................194
EarlyimplicationsofoncometabolitesinDNAdamage repair.............................................................................................................194
Identificationoftheoncometabolite-inducedHDRdeficiency......................194
OncometabolitesdisruptHDRsignalingviaH3K9 hypermethylation..........................................................................................196
Therapeuticstrategiestargetingtheoncometabolite-induced HDRdefect...................................................................................................198 References.............................................................................................................199
SECTION3TherapeuticstrategiesinducingDNAdamage andepigeneticalterations
DNAdamage-inducingchemotherapies..............................................................210
Alkylatingagents..............................................................................................211 Platinum-basedcompounds..............................................................................213
Cisplatinanalogs..............................................................................................215
CHAPTER13EpigenetictherapyandDNAdamageresponse..........................
MarinaBarettiandNiloferS.Azad
Introduction...........................................................................................................227
Commonlyalteredepigeneticregulatoryproteinsimplicated incancerandtheirimplicationfortherapy.....................................................228
TherapiesforaberrantDNAmethylation........................................................230
Inhibitorsofhistonemodifications..................................................................231
Targetingepigeneticreaders............................................................................234
TargetingtheepigeneticsoftheDNAdamageresponseincancer: Theconceptofsyntheticlethality....................................................................235
Epigeneticinhibitorsincombinationwithchemotherapy andradiotherapy:Syntheticlethalinteractionsandclinical applications...................................................................................................237
EpigeneticchangesofDNAdamagerepairgenesaspredictive markersofresponsetochemotherapy..........................................................238
EpigeneticinhibitorsincombinationwithdrugsthattargetDNA repaircomponentsforsyntheticlethaltherapy............................................240
Conclusion:Implicationsofepigeneticregulationforimproved cancertreatment................................................................................................244
GiovanadaSilvaLeandro,MarcelaTeatinLatancia, NathaliaQuintero-Ruiz,andCarlosFredericoMartinsMenck
Immunofluorescence(IF)todetectphosphorylatedhistone H2AX(Ser139).............................................................................................263 Immuno-slot-blotassayforpyrimidinedimers...............................................271
CHAPTER15Usefulmethodstostudyepigeneticmarks:DNAmethylation, histonemodifications,chromatinstructure, andnoncodingRNAs............................................................... 283
AnaLuisaPedrosoAyub,BrunadeOliveiraPerestrelo, GuilhermeCavalcantePessoa,andMiriamGalvonasJasiulionis
Introduction...........................................................................................................283
MethodsusedtostudyDNAmethylation............................................................284
Basedonbisulfitetreatment............................................................................285
BasedonCpGcapture.....................................................................................288
5mCcontent.....................................................................................................289
Methodsusedtostudyhistonemodifications......................................................290
Chromatinimmunoprecipitation(ChIP)..........................................................290
Immunodetectionwithspecificantibodies......................................................291
Proteomics........................................................................................................292
Methodsusedtostudychromatinaccessibility...................................................292
DeoxyribonucleasetreatmentfollowedbyDNAsequencing (DNase-seq)..................................................................................................293
Formaldehyde-assistedisolationofregulatoryelementsfollowed bysequencing(FAIRE-seq).........................................................................294
MicrococcalnucleasetreatmentfollowedbyDNAsequencing (MNase-seq)..................................................................................................294
Assayfortransposase-accessiblechromatinwithhigh-throughput sequencing(ATAC-seq)...............................................................................294
Methodsusedtoidentifychromatin-associatedproteins.....................................295
Chromatinenrichmentforproteomics(ChEP)................................................295
MethodsusedtostudynoncodingRNAs(ncRNA)............................................295
ncRNAgeneexpressionanalysis.....................................................................295
miRNA-mediatedposttranscriptionalregulationanalysis...............................297
NoncodingRNA-proteininteractions..............................................................298
LncRNAandchromatininteraction.................................................................299
ncRNAlocalization..........................................................................................300
ncRNAgeneexpressionmodulationmethods.................................................300
Conclusions...........................................................................................................303
References.............................................................................................................303
Contributors
DeboraKristinaAlves-Fernandes
DepartmentofPharmacology,EscolaPaulistadeMedicina,UniversidadeFederaldeSaoPaulo, Sa ˜ oPaulo,SP,Brazil
AnaLuisaPedrosoAyub
DepartmentofPharmacology,EscolaPaulistadeMedicina,UniversidadeFederaldeSa ˜ oPaulo, SaoPaulo,SP,Brazil
NiloferS.Azad
SidneyKimmelComprehensiveCancerCenteratJohnsHopkinsHospital,Baltimore,MD, UnitedStates
KhalilAzizian
DepartmentofMicrobiology,FacultyofMedicine,KurdistanUniversityofMedicalSciences, Sanandaj,Iran
MarinaBaretti
SidneyKimmelComprehensiveCancerCenteratJohnsHopkinsHospital,Baltimore,MD, UnitedStates
NikolaBowden
HunterMedicalResearchInstituteandUniversityofNewcastle,Callaghan,NSW,Australia
KateH.Brettingham-Moore
TasmanianSchoolofMedicine,UniversityofTasmania,Hobart,TAS,Australia
JessicaBuck
TelethonKidsInstitute,BrainTumourResearch,Perth;UniversityofWA,CentreforChildHealth Research,Crawley,WA,Australia
DaynaChallis
TasmanianSchoolofMedicine,UniversityofTasmania,Hobart,TAS,Australia
JonathanDow
YaleUniversitySchoolofMedicine,NewHaven,CT,UnitedStates
RaeleneEndersby
TelethonKidsInstitute,BrainTumourResearch,Perth;UniversityofWA,CentreforChildHealth Research,Crawley,WA,Australia
ShinjiniGanguly
ClevelandClinic,Cleveland,OH,UnitedStates
AnthonyGhanem
ClevelandClinic,Cleveland,OH,UnitedStates
SebastianoGiallongo
InternationalClinicalResearchCenter,St’AnneUniversityHospital;DepartmentofBiology,Faculty ofMedicine,MasarykUniversity,Brno,CzechRepublic
PeterM.Glazer
YaleUniversitySchoolofMedicine,NewHaven,CT,UnitedStates
NassimGorjizadeh
CellularandMolecularBiologyResearchCenter,HealthResearchInstitute,BabolUniversityof MedicalSciences,Babol,Iran
NegarGorjizadeh
DepartmentofCellandMolecularBiology,FacultyofBiologicalSciences,KharazmiUniversity, Tehran;CellularandMolecularBiologyResearchCenter,HealthResearchInstitute,Babol UniversityofMedicalSciences,Babol,Iran
MiriamGalvonasJasiulionis
DepartmentofPharmacology,EscolaPaulistadeMedicina,UniversidadeFederaldeSa ˜ oPaulo, SaoPaulo,SP,Brazil
PhilippeJohannToBerens
Institutdebiologiemoleculairedesplantes,Strasbourg,France
AnsarKarimian
CellularandMolecularBiologyResearchCenter,HealthResearchInstitute,BabolUniversityof MedicalSciences,Babol,Iran
MarcelaTeatinLatancia
DepartmentofMicrobiology,InstituteofBiomedicalSciences—UniversityofSa ˜ oPaulo(USP), SaoPaulo,Brazil
GiovanadaSilvaLeandro
DepartmentofMicrobiology,InstituteofBiomedicalSciences—UniversityofSa ˜ oPaulo(USP), SaoPaulo,Brazil
OrianaLoRe
InternationalClinicalResearchCenter,St’AnneUniversityHospital,Brno,CzechRepublic; DepartmentofStemCellBiologyandTransplantology,ResearchInstituteoftheMedicalUniversityVarna,Varna,Bulgaria
CarlosFredericoMartinsMenck
DepartmentofMicrobiology,InstituteofBiomedicalSciences—UniversityofSaoPaulo(USP), Sa ˜ oPaulo,Brazil
OmarY.Mian
ClevelandClinic,Cleveland,OH,UnitedStates
TomokoMiyake
LaboratoryforZero-CarbonEnergy,InstituteofInnovativeResearch,TokyoInstituteofTechnology, Tokyo,Japan
JeanMolinier
Institutdebiologiemoleculairedesplantes,Strasbourg,France
ApiwatMutirangura
DepartmentofAnatomy,FacultyofMedicine,CenterofExcellenceinMolecularGeneticsofCancer andHumanDisease,ChulalongkornUniversity,Bangkok,Thailand
BrunadeOliveiraPerestrelo
DepartmentofPharmacology,EscolaPaulistadeMedicina,UniversidadeFederaldeSa ˜ oPaulo, SaoPaulo,SP,Brazil
GuilhermeCavalcantePessoa
DepartmentofPharmacology,EscolaPaulistadeMedicina,UniversidadeFederaldeSaoPaulo, Sa ˜ oPaulo,SP,Brazil
NathaliaQuintero-Ruiz
DepartmentofMicrobiology,InstituteofBiomedicalSciences—UniversityofSaoPaulo(USP), Sa ˜ oPaulo,Brazil
SalimataOusmaneSall
Institutdebiologiemoleculairedesplantes,Strasbourg,France
MargaridaA.Santos
DepartmentofEpigeneticsandMolecularCarcinogenesis,TheUniversityofTexasMDAnderson CancerCenter;GraduatePrograminGenetics&Epigenetics,TheUniversityofTexasMDAnderson CancerCenterUTHealthGraduateSchoolofBiomedicalSciences,Houston,TX,UnitedStates
MikioShimada
LaboratoryforZero-CarbonEnergy,InstituteofInnovativeResearch,TokyoInstituteofTechnology, Tokyo,Japan
MoeinShirzad
CellularandMolecularBiologyResearchCenter,HealthResearchInstitute,BabolUniversityof MedicalSciences,Babol,Iran
HieuT.Van
DepartmentofEpigeneticsandMolecularCarcinogenesis,TheUniversityofTexasMDAnderson CancerCenter;GraduatePrograminGenetics&Epigenetics,TheUniversityofTexasMDAnderson CancerCenterUTHealthGraduateSchoolofBiomedicalSciences,Houston,TX,UnitedStates
ManlioVinciguerra
InternationalClinicalResearchCenter,St’AnneUniversityHospital,Brno,CzechRepublic; DepartmentofStemCellBiologyandTransplantology,ResearchInstituteoftheMedicalUniversityVarna,Varna,Bulgaria
Preface
Thegenomeisconstantlysubjectedtodamagecausedbybothendogenousandexogenousfactors.In additiontothewell-establishedroleinthegenetranscriptionregulation,epigeneticmechanismsalso playanimportantroleinDNAreplicationandrepair.Dynamicandorchestratedalterationsinthe chromatinconferredbyepigeneticmodificationsarefundamentalforDNAdamageresponse.After damage,epigeneticmechanismsinducechromatinremodelinginordertoaccessthedamageregion, recruitrepairproteins,andcontributetothedecisionofrepairpathway.Notonlytheentiremachinery involvedinDNArepairiscrucialformaintainingthegenomeintegrity,butalsothepreciseepigenetic regulationofthisprocess.Epigeneticderegulationduringthisprocessmaycontributetopremature agingandtheonsetofdiseases,includingcancer,neurodegenerativedisorders,andimmunedeficiencies.Recentadvancesinsequencingtechnologies,methodstoevaluateepigeneticmodifications,and chromatinstructurehavecontributedenormouslytoourknowledgeregardingtheepigenome,chromatinconfiguration,anddynamicassociationofproteinswiththechromatin.Althoughtheundeniable relevanceofepigeneticmechanismsintheDNAdamageresponse,thecomplexmechanisticinterplay betweenchromatinregulationandDNArepairisstillpoorlyunderstood.Comprehendinghowthese processesareconnectedintime,space,andcontextmaybringlighttonovelandmoreeffective therapeuticstrategiesfordiseasessuchascancer.
ThisbooksummarizestherecentadvancesinthisintriguingfieldofDNAdamageandepigenetics. Thisbookisdividedintofoursectionsand15chapters.Thefirstsectionincludeschaptersaddressing thebasicscienceofDNAdamageanditsrelationwiththedifferentepigeneticmechanisms,including DNAmethylation,posttranslationalhistonemodifications,histonevariants,chromatinremodeling, miRNAs,andlncRNAs.ThesecondsectiondescribestheimpactofDNAdamageandepigenetics onmetabolism,aging,differentiation,andcancer.Thethirdsectionbringsconceptsandupdatesabout therapiesinducingDNAdamageandepigenetictherapies.Thefinalsectioncompilesthemaincurrent methodologiesusedtoanalyzebothDNAdamageandepigeneticalterations.
Overthepastyears,theinterplaybetweenDNAdamageandepigeneticsincreasedinterestnotonly inbasicsciencebutalsoinmedicine.Forthisreason,thebookisdedicatedtobiologists,biochemists, geneticists,immunologists,andphysicians,andallstudentsofbiologyandmedicine.
DNAdamageandDNAmethylation 1
SalimataOusmaneSalla,PhilippeJohannToBerensa,andJeanMolinier
Institutdebiologiemoleculairedesplantes,Strasbourg,France
Introduction
Livingorganismshavetocopewithenvironmentalcuesandwithendogenouschemicalcompounds thataredeleteriousfortheirgeneticinformation.Indeed,genotoxicstresseschallengegenomesbyinducingchangesinthechemicalstructureofnucleotides.Thesealterationsinvolvingthefourcanonical bases(adenine(A),cytosine(C),guanine(G),andthymine(T))orbreakingtheDNAstrand(s)are definedasDNAdamage/lesionsandarespecificofparticulargenotoxicagents.1 Basemodification istheadditionofachemicalmoietyresultingeitherfromareactionwithagenotoxicagent(i.e.,alkylation)orfromanenzymaticreaction(i.e.,DNAmethylation).Thus,basescouldbeoxidized,deaminated,alkylated,ormethylatedleadingtodifferenttypesofmodifications.2 Importantly, methylationratherthanbeingexclusivelyconsideredasaDNAlesion perse isalsoanepigenetic mark.2 Indeed,onekeycomponentoftheepigenomeiscytosinemethylationleadingto5-methylcytosine(5-mC),whichisrequiredforthestablesilencingoftransposableelements(TE)aswellasforthe regulationofgeneactivity.3 WhileinmammalsDNAmethylationoccursintheCGcontext,inplants, 5-mCisfoundin2additionalsequencecontexts:CHGandCHH(whereH ¼ A,T,orC).4 Suchbase modificationlikelyaddsanotherlayerofcomplexityinthereactivityofgenomicregionssubjectedtoa genotoxicstress.GiventhatmostoftheDNAlesionsoccurataparticularbase,thenucleotidecompositionofthegenomeisanimportantfeaturetotakeintoaccount.Moreover,severalstudies highlightedthattheepigenomelandscapetendstoplayanimportantroleintheabilityoftheDNA/ genometobedamagedandtoefficientlyperformtheirrepair.5
Therefore,thischapterwillbedevotedtothepresentationofthedifferenttypesofbasemodificationsandtheirconsequencesongenomeintegrity.Aparticularemphasiswillbeplacedontheinfluence oftheDNAmethylationprofileondamageformationandonthemaintenanceofmethylomeintegrity uponrepair.Wewillfocusontheemergingnotionthatcomplexinterplaysexistbetweengenome, methylome,DNAdamage,andrepairandthatplantshavedevelopedsophisticatedstrategiestosimultaneouslymaintain(epi)genomeintegrity. aEqualcontribution.
EpigeneticsandDNADamage. https://doi.org/10.1016/B978-0-323-91081-1.00005-4 Copyright # 2022ElsevierInc.Allrightsreserved.
Basemodifications
Cellularprocessesaswellasenvironmentalfactorsinducegenotoxicstressleadingtotheformationof alargerepertoireofbasemodificationsthataffectgenomestability.ThesemodificationsalterDNA structureandaresensedeitherasDNAlesions(i.e.,baseoxidation)orasepigeneticmarks(i.e.,DNA methylation).2 Severalfeaturesofthegenomicregionssuchasnucleotidecompositionandspatialorganizationmayinfluencetheirabilitytoaccumulatebasemodificationsthatmustbeaccurately repaired(forDNAlesions)orfine-tuned(forDNAmethylation).Upondetection,thesemodifiedbases areactivelyremovedviaDNAsynthesis-dependentrepairprocessestomaintain(epi)genomeintegrity (Fig.1).
DNAmethylation
DNAmethylationistheadditionofamethylgrouponeitheradeninetoform6-methyladenine(6-mA) orcytosinetoform5-methylcytosine(5-mC).3 5-mCisthemoststudiedDNAmethylation/epigenetic mark.5-mCisdetectableatdifferentratesinplants,mammals,andbacteriaandisthoughttobelowin drosophilaorabsentinyeast.6 5-mCchangesDNAflexibilitybyenhancingstiffnessandmodulates DNAaccessibilitytodifferentfactors.7 Moreover,thepresenceof5-mCwithinalocusmayfavor thepreferentialformationofDNAdamage(i.e.,photolesions).8,9
Deamination
Inadditiontotheenzymatic-mediateddeamination,10–12 asignificantamountofhydrolyticdeaminationcanalsooccurespeciallyonsingle-strandedDNA.13–15 Thehydrolyticdeaminationofcytosine formsuracil(U)thatisrecognizedasaDNAlesion.Deaminationcanalsooccuron5-methylcytosine andproducesthymine(T)creatingaT:Gmismatch.ReplicationofthesedamagedsitesincludingC:G toT:AtransitionpreventsthemaintenanceofDNAmethylationatthisparticulargenomicsequence andthusleadstothealterationofbothgenomeandmethylomeintegrities.13 ToavoidsuchbasetransitionandpermanentlossofDNAmethylation,thedeaminationproductisremovedbyspecificglycosylase,viatheBERpathway.16,17
Alkylation
DNAalkylationincludesallmodificationsinducedbythebindingofanewalkylgrouponanoncanonicalpositionofDNAaftermonomolecularorbimolecularnucleophilicsubstitution(SN1/SN2)reactions.18–20 Alkylationcanoccurontheoxygenatomsinphosphodiesterbackbone,aswellason oxygenandnitrogenatomsofthe4nucleobases.18,21 ThesiteofDNAalkylationstronglydepends onthealkylatingagent,whichcanbeanendogenousmetabolitesuchasS-adenosylmethionine (SAM),22 oranexogenousagent,suchasalkylatingdrugscurrentlyusedaschemotherapeutics (i.e.,busulfan).18 Themostfrequentlytransferredalkylgroupisthemethylgroup,andthepredominant alkylationproductsareN7-methylguanine(N7-meG),N3-methyladenine(N3-meA),and O6-methylguanine(O6-meG).18–20 N7-meGandN3-meAarecleavedbyspontaneousdepurination orbyspecificDNAglycosylasesleavinginbothcasesanabasicsitewithcytotoxicandmutagenic potentialthatneedfurtherrepairbynucleotideexcisionrepair(NER)orbaseexcisionrepair(BER;
FIG.1
TypesofDNAdamages.DNAissubjectedtovarioustypesofalterationsthatleadtoeither(A)basemodifications, (B)mismatch,or(C)single /double-strandedbreaks(SSBs/DSBs).TheseDNAdamagesareprocessedand repairedbydifferentpathwaysthatrelyondenovoDNAsynthesis.WhentheoriginalDNAsequenceis methylated(M),theprocessingofthedamagedDNAleadstoatransientlossofDNAmethylationthatmustbe accuratelyre-establishedtomaintainmethylomeintegrity.
long-orshort-patchBER).23–25 Conversely,thehighlymutagenicO6-methylguanine,allowingapreferentialpairingwiththymine,26 canberepairedbydirectreversionleadingtothealkylationofa catalyticresidueofO6-methylguanine-DNAmethyltransferase(MGMT).27 Morerecently,N3methylcytosine(N3-meC),ausuallyraresideproductofclassicalalkylatingagents(i.e.,methylmethanesulfonate),20 wasshowntobeasignificantby-productofDNAmethyltransferaseenzyme(DNMT) activityresponsiblefor5-mCsynthesisinnematodes.28 Theseresultsmaynotablyexplaintheabsence ofDNAmethylationinthenematode Caenorhabditiselegans andtheobservedcoevolutionofmethylationandalkylationdamagerepairpathwaysallacrosseukaryotes.28
Oxidation
Asidedeaminationandalkylation,DNAoxidationisanothertypeofbasemodificationthataffectsgenomeintegrity.Environmentalfactors,suchasUVlight(UV-AandUV-B),aswellasendogenous cellularprocesses(i.e.,respiration)leadtotheformationofreactiveoxygenspecies(ROS):superoxide anion(O2 ),hydrogenperoxide(H2O2),andhydroxylradical(•OH).29–31 ROShaveagenotoxiceffect byreactingwithpurines,pyrimidines,andthedeoxyribosebackboneofDNA,toproducemorethan20 differenttypesofoxidativelyinducedDNAlesions.31 Theyieldofthesedifferentproductsishighly specifictothemolecularredoxcontext.Nevertheless,guanineisdescribedasthemajortargetof oxidation,especiallyatC8position,therebyformingthehighlymutagenic7,8-dihydro-8-oxoguanine (8-Oxo-G).31–34 Whenunrepaired,8-Oxo-GcanpairwiththeHoogsteenfaceofadenine,therebygeneratingaguaninetothyminetransversionmutationuponreplication,generatinganun-methylatable A:Tsite.35,36 Additionally,oxidationof8-Oxo-G/lysineformsbulkyproteins-DNAcross-linksthat aredeleterious.37 Asforalkylatingdamages,8-Oxo-GismainlyremovedbyspecificDNAglycosylases(i.e.,MutYandMutT)andfurtherprocessedbytheBERmachinery.38 Theoretically,5-mCcan alsoundergooxidativemodificationdespiteitsrelativelyhighreductionpotentialcomparedtoguanine.31,36,39 Inparticular,thedifferentoxidationproducts:5-hydroxymethylcytosine(5-hmC), 5-formylcytosine(5-fC),or5-carboxycytosine(5-caC)mainlygainedaninterestinthelastdecade. Inmammals,theactiveremovalof5-mCoccursuponsuccessiveenzymatic-mediatedoxidations.40,41 Theseserialoxidationsof5-mCproduce5-hmC,5-fC,and5-caCandarecatalyzedbytheten-eleven translocation(TET)proteinfamilymembers.42 Intheabsenceof bonafide 5-mCglycosylasesinmammals,thisalternativeprocessallowsefficientactiveDNAdemethylation.43 TheexistenceofaTETlikedemethylationpathwayinplants,asalternativestrategy,isasyetundetermined.44–46
Cross-links
DNA–DNAcross-link(CL)hasagreatpotentialtoaltergenomeintegrity.CLcanoccurwithinone DNAstrand(intra-strandcross-links)orinbetweenthe2DNAstrands(inter-strandcross-links).47 Intra-strandcross-linksareverycommonforlight-dependentlivingorganisms.Indeed,theUVspectrumofsunlightinducescross-linksbetweendi-pyrimidines.5,48,49 Twosuccessivepyrimidines(CC, TT,TC,orCT)canberaisedtotheirhighlyreactivesingletortripletstateswhenabsorbingUVradiation,especiallyintheUV-CandUV-Bwavelengthrangingfrom100to280nmand280to315nm, respectively.48,49 Oncereachingthereactivesingletortripletstates,fastphotochemicalreactionslead totheformationofthreemainDNAintra-strandcross-linkdamagesorphotoproducts:cyclobutane
pyrimidinedimers(CPDs),pyrimidine6–4pyrimidonephotoproducts(6-4PPs),andthe6-4PPsDewar isomer.49
Converselytointra-strandcross-links,inter-strandcross-links(ICLs)arerelativelyrareevents,oftenthoughttobeaconsequenceofdrugtreatments.50 Forexample,mitomycinCinducesinter-strand cross-linksbetweentheguaninesofbothstrandsandalsoshowsspecificityforCpGsequences.50,51 Recently,thebiologicalimpactoftheoxidationproductofadenine,7,8-dihydro-8-oxoadenine(8Oxo-A),wasshowntohavesignificantpotentialtoformICLwithadenineandguanineontheopposite strand.52 TherepairofICLiscomplexandtriggersdenovoDNAsynthesisofbothstrandsinthevicinityofthedamagedregions,endangeringthemaintenanceoftheDNAmethylationfootprints.47,53
DNAmethylationandDNAdamageability
EndogenousandexogenousstimuliformdirectlyorindirectlyDNAlesionsinasequence-specificcontext.20,32,49,54 RecentstudieshighlightedtheheterogenicityoftheformationofDNAlesionswithin genomecomplexity,hereaftercalled“damageability”.9,55–59 DNAdamageabilitycanbedefinedas thedegreeofsusceptibilityofalocus,withinagenome,tobedamagedbyaparticulargenotoxicagent. Thisdamageabilitydependsonthewholecomplexityofthelocalchemicalcontextandshouldbeconsideredashighlyvariableduringlifespanandonlypartiallypredictable.Theputativereverseinfluence oftheepigenomeontheDNAdamageabilitydrewtheattentionofmanyresearchgroupsinthelast decade.5,55–57,60,61
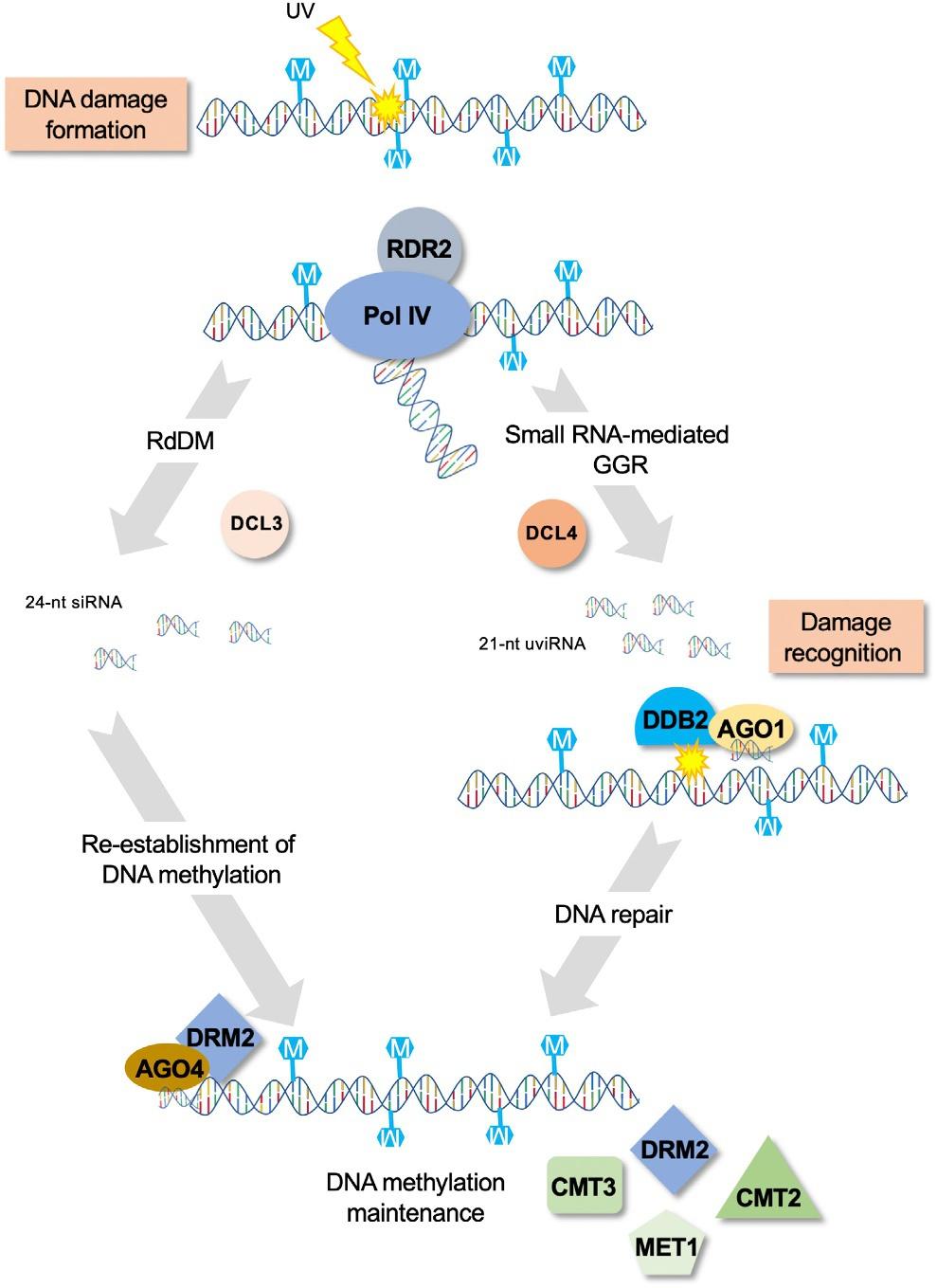
DNAmethylation(5-mC),asthemainepigeneticmark,hasthepotentialtoinfluencethedamageabilityofthegenome.Theinfluenceof5-mContheformationofspontaneoushydrolyticdeamination wasshown, invitro,tooccurtwicemoreoftencomparedtounmethylatedcytosine.15 Moreover,a 5-mCadjacenttoapyrimidineismorepronetoformphotoproductsthananunmethylatedcytosine incombinationwithanotherpyrimidine.49 Invitro ligation-mediatedPCR, invitro irradiationof genomicDNA,andinvivoimmunoprecipitationofUV-damagedDNA(IPOUD)experimentsall highlightedasignificantlyincreasedpotentialofmethylatedDNAtoformphotodamageuponUV exposure.9,55,62 Giventhat3/4ofthedi-pyrimidinecombinations(CC,TC,orCT)involveatleast onecytosineandthatinplants,DNAmethylationoccursalsoinCHGortheCHHcontexts (whereHisA,T,orC),thehighlymethylatedgenomicregionsmaybemorereactivetoformphotodamage.Inotherwords,thesequencecontextincombinationwiththemethylomelandscapemay influencetheUVdamageabilityandlikelytherepairmachinerythatactatparticularloci.9,55,62
Additionally,5-mCcanalsoindirectlyinfluenceDNAdamageabilitythroughitsroleintheestablishmentofhigherchromatinstructuressuchasnucleosomeoccupancy,histoneposttranslationalmodification(PTM),andgenomefolding.63,64 Nucleosomedisplacement,forexample,wasshownto influencetheformationofDNAstrandbreakuponZeocintreatment.56 8-Oxo-GdistributionwasidentifiedtoaccumulateinH3enrichedregionsoftheyeastgenome61 andwasconsistentlymorefrequently observedintherathercompactlamina-associateddomainsofchromatin,displacedatthenuclearperiphery,inrat.57
Finally,5-mCcanalsoindirectlyimpacttheDNAdamagedynamics,bycontrollingDNArepair. Forexample,whenpreventingtranscription,64 5-mCcanhinderthedamagerecognitionbythe transcription-coupledrepair(TCR)pathway.65