A Curse of Queens Amanda Bouchet
https://ebookmass.com/product/a-curse-of-queens-amanda-bouchet-8/
ebookmass.com
Editedby
DianaA
Elsevier
Radarweg29,POBox211,1000AEAmsterdam,Netherlands
TheBoulevard,LangfordLane,Kidlington,OxfordOX51GB,UnitedKingdom
50HampshireStreet,5thFloor,Cambridge,MA02139,UnitedStates
Copyright©2020ElsevierInc.Allrightsreserved.
Nopartofthispublicationmaybereproducedortransmittedinanyformorbyanymeans,electronicormechanical, includingphotocopying,recording,oranyinformationstorageandretrievalsystem,withoutpermissioninwritingfrom thepublisher.Detailsonhowtoseekpermission,furtherinformationaboutthePublisher ’spermissionspoliciesand ourarrangementswithorganizationssuchastheCopyrightClearanceCenterandtheCopyrightLicensingAgency,can befoundatourwebsite: www.elsevier.com/permissions
ThisbookandtheindividualcontributionscontainedinitareprotectedundercopyrightbythePublisher(otherthanas maybenotedherein).
Notices
Knowledgeandbestpracticeinthis fieldareconstantlychanging.Asnewresearchandexperiencebroadenour understanding,changesinresearchmethods,professionalpractices,ormedicaltreatmentmaybecomenecessary.
Practitionersandresearchersmustalwaysrelyontheirownexperienceandknowledgeinevaluatingandusinganyinformation,methods,compounds,orexperimentsdescribedherein.Inusingsuchinformationormethodstheyshouldbe mindfuloftheirownsafetyandthesafetyofothers,includingpartiesforwhomtheyhaveaprofessional responsibility.
Tothefullestextentofthelaw,neitherthePublishernortheauthors,contributors,oreditors,assumeanyliabilityfor anyinjuryand/ordamagetopersonsorpropertyasamatterofproductsliability,negligenceorotherwise,orfromany useoroperationofanymethods,products,instructions,orideascontainedinthematerialherein.
LibraryofCongressCataloging-in-PublicationData
AcatalogrecordforthisbookisavailablefromtheLibraryofCongress
BritishLibraryCataloguing-in-PublicationData
AcataloguerecordforthisbookisavailablefromtheBritishLibrary
ISBN:978-0-12-818196-6
ForinformationonallElsevierpublicationsvisitourwebsite at https://www.elsevier.com/books-and-journals
Publisher:JancoCandice
AcquisitionsEditor:LouisaMunro
EditorialProjectManager:SaraPianavilla
ProductionProjectManager:KumarAnbazhagan
CoverDesigner:MilesHitchen
TypesetbyTNQTechnologies
Contributors
KonstantinosA.Aliferis PesticideScienceLaboratory,AgricultureUniversityofAthens,Athens, Greece;DepartmentofPlantScience,McGillUniversity,Sainte-Anne-de-Bellevue,QC,Canada
DianaA ´ lvarez-Mun ˜ oz WaterandSoilQualityResearchGroup,DepartmentofEnvironmental Chemistry,IDAEA-CSIC,Barcelona,Spain
FranciscaArellano-Beltra ´ n DepartmentofChemistry,FacultyofExperimentalSciences, UniversityofHuelva,CampusElCarmen,Huelva,Spain;ResearchCenterofNaturalResources, HealthandtheEnvironment(RENSMA),UniversityofHuelva,CampusElCarmen,Huelva,Spain
AnaArias-Borrego DepartmentofChemistry,FacultyofExperimentalSciences,Universityof Huelva,CampusElCarmen,Huelva,Spain;ResearchCenterofNaturalResources,Healthandthe Environment(RENSMA),UniversityofHuelva,CampusElCarmen,Huelva,Spain
O ` scarAznar-Alemany WaterandSoilQualityResearchGroup,DepartmentofEnvironmental Chemistry,IDAEA-CSIC,Barcelona,Spain
Julia ´ nBlasco InstituteofMarineSciencesofAndalusia(CSIC),CampusRioSanPedro,Ca ´ diz,Spain
Be ´ nildeBonnefille HydroSciences,UnivMontpellier,CNRS,IRD,Montpellier,France
Bele ´ nCallejo ´ n-Leblic DepartmentofChemistry,FacultyofExperimentalSciences,Universityof Huelva,CampusElCarmen,Huelva,Spain;ResearchCenterofNaturalResources,Healthandthe Environment(RENSMA),UniversityofHuelva,CampusElCarmen,Huelva,Spain
PedroCarriquiriborde CentrodeInvestigacionesdelMedioambiente(CIM),Facultadde
CienciasExactas,UniversidadNacionaldelaPlata CONICET,LaPlata,BuenosAires,Argentina Chien-MinChen DepartmentofEnvironmentalResourcesManagement,ChiaNanUniversityof Pharmacy&Science,Tainan,Taiwan
Fre ´ de ´ riqueCourant HydroSciences,UnivMontpellier,CNRS,IRD,Montpellier,France
ArthurDavid UnivRennes,Inserm,EHESP,Irset(Institutderechercheensante ´ ,environnement ettravail),UMR_S1085,Rennes,France
XiaopingDiao StateKeyLaboratoryofMarineResourceUtilizationinSouthChinaSea,Hainan University,Haikou,HainanProvince,China;MinistryofEducationKeyLaboratoryofTropical IslandEcology,HainanNormalUniversity,Haikou,HainanProvince,China
ThibautDumas HydroSciences,UnivMontpellier,CNRS,IRD,Montpellier,France
MarinellaFarre ´ WaterandSoilQualityResearchGroup,DepartmentofEnvironmentalChemistry, IDAEA-CSIC,Barcelona,Spain
He ´ le ` neFenet HydroSciences,UnivMontpellier,CNRS,IRD,Montpellier,France
TamaraGarcı´a-Barrera DepartmentofChemistry,FacultyofExperimentalSciences,University ofHuelva,CampusElCarmen,Huelva,Spain;ResearchCenterofNaturalResources,Healthand theEnvironment(RENSMA),UniversityofHuelva,CampusElCarmen,Huelva,Spain
RubenGil-Solsona CatalanInstituteforWaterResearch(ICRA),ParcCientı´ficiTecnolo ` gicdela UniversitatdeGirona,Girona,Spain
ElenaGomez HydroSciences,UnivMontpellier,CNRS,IRD,Montpellier,France
Jose ´ LuisGo ´ mez-Ariza DepartmentofChemistry,FacultyofExperimentalSciences,University ofHuelva,CampusElCarmen,Huelva,Spain;ResearchCenterofNaturalResources,Healthand theEnvironment(RENSMA),UniversityofHuelva,CampusElCarmen,Huelva,Spain
AwadheshN.Jha UniversityofPlymouth,Plymouth,Devon,UnitedKingdom
VeraKovacevic DepartmentofChemistry,UniversityofToronto,Toronto,ON,Canada; EnvironmentalNMRCentre,DepartmentofPhysicalandEnvironmentalSciences,Universityof TorontoScarborough,Toronto,ON,Canada
MartaLlorca WaterandSoilQualityResearchGroup,DepartmentofEnvironmentalChemistry, IDAEA-CSIC,Barcelona,Spain
GemaRodrı´guez-Moro DepartmentofChemistry,FacultyofExperimentalSciences,University ofHuelva,CampusElCarmen,Huelva,Spain;ResearchCenterofNaturalResources,Healthand theEnvironment(RENSMA),UniversityofHuelva,CampusElCarmen,Huelva,Spain
SaraRodrı´guez-Mozaz CatalanInstituteforWaterResearch(ICRA),ParcCientı´ficiTecnolo ` gic
delaUniversitatdeGirona,Girona,Spain
PawelRostkowski NILU NorwegianInstituteforAirResearch,Kjeller,Norway
SaraRamı´rez-Acosta DepartmentofChemistry,FacultyofExperimentalSciences,Universityof Huelva,CampusElCarmen,Huelva,Spain;ResearchCenterofNaturalResources,Healthandthe Environment(RENSMA),UniversityofHuelva,CampusElCarmen,Huelva,Spain
AlbertSerra-Compte CatalanInstituteforWaterResearch(ICRA),ParcCientı´ficiTecnolo ` gic delaUniversitatdeGirona,Girona,Spain
MyrnaJ.Simpson DepartmentofChemistry,UniversityofToronto,Toronto,ON,Canada; EnvironmentalNMRCentre,DepartmentofPhysicalandEnvironmentalSciences,Universityof TorontoScarborough,Toronto,ON,Canada
HailongZhou StateKeyLaboratoryofMarineResourceUtilizationinSouthChinaSea,Hainan University,Haikou,HainanProvince,China;SchoolofLifeandPharmaceuticalSciences,Hainan University,Haikou,HainanProvince,China
Preface
Metabolomicsconsistsofthesimultaneouscharacterizationofthemetabolitespresentinan organismandoffersa“picture”ofthebiochemistryoftheorganismatanyonetime.Its applicationtotheenvironment,knownasEnvironmentalMetabolomics,allows characterizingtheinteractionthatoccursbetweenorganismsandthesurrounding environment.Concretely,thisbookisfocusedontheinteractionbetweenorganismsand contaminantsthatarepresentintheenvironmentduetohumanactivitiesandmayhave toxiceffects.
Theapplicationofmetabolomicsintheenvironmentalfieldforbiomarkersdiscoveryis relativelynew.Scientificpapersonthissubjectstartedbeingpublishedabout10yearsago, butithasbeeninthelast5yearswhentheapplicationofmetabolomicsforanalyzing biologicalsamplesinenvironmentalmonitoringhasstronglyattractedtheattentionof researches.Consequently,anincreasingnumberofpapersarecurrentlybeenpublished generatingahighamountofdatathatneedtobecompiledandharmonizedtogetrelevant information.
Thisbookgathersinformationonenvironmentalmetabolomicswhennaturalorganismsare exposedtometals,persistentorganicpollutants,andemergingpollutants.Itshowsthe readerdifferentexperimentalsetups,analyticaltechniques,dataprocessing,anddata analysis.Thisbookwillleadyouthroughthemetabolomicsworkflowandwillserveasa guideforimplementation.Besides,itallows,forthefirsttime,tohavegeneralbiomarkers snapshotveryusefulforriskassessment.Italsodiscussesthecurrentlimitationsandfuture perspectivesofenvironmentalmetabolomics.
Theaudienceofthisbookiswide-rangingfromundergraduatetograduatestudents interestedinenvironmentalresearch,researchersinthefieldofenvironmentaltoxicology andchemistry,legislators,andpolicy-makers.
We,theEditors,learnedalotfromtheauthorsandhopethatyoureadersalsodo.Weexpect thattheknowledgecontainedherewillhelptofurthergaininsightandadvanceon environmentalmetabolomicsscience.
Acknowledgments
Hugethankstoallthepeoplewhohavebeeninvolvedinthisproject,especiallytothe authors;withouttheirhardwork,thisbookwouldnothavebeenpossible.Thanksto Elsevier,particularlytotheacquisitionseditor,theeditorialprojectmanager,andthe productionprojectmanager.Finally,wearethankfultoourfamiliesfortheirsupport,for understandingourpassionforscience,andallthetimededicatedtothissubject.
CHAPTER1
Fundamentalsofenvironmental metabolomics
VeraKovacevic1, 2,MyrnaJ.Simpson1, 2
1DepartmentofChemistry,UniversityofToronto,Toronto,ON,Canada; 2EnvironmentalNMR Centre,DepartmentofPhysicalandEnvironmentalSciences,UniversityofTorontoScarborough, Toronto,ON,Canada
ChapterOutline
1.Environmentalstressors1
1.1Contaminantstressors2
2.Ecotoxicology4
3.Metabolomics5
4.Environmentalmetabolomics6
4.1Studydesign7
4.2Collectionoforganismsandexperimentalexposure8
4.3Samplecollection,samplepreparation,andmetaboliteextraction9
4.4Analyticaltechniquesfordatacollection11
4.5Rawdatapreprocessinganddataanalysis14
4.6Biologicalinterpretationoftheresults17
5.Exposuretocontaminantmixtures18
6.Summaryandenvironmentalbiomonitoringefforts19 References20
1.Environmentalstressors
Aquaticandterrestrialecosystemsareunderconstantthreatfromvariousenvironmental stressorsthatarisefromnaturaloranthropogenicactivities.Environmentalstressors includebioticstressorssuchaspathogensandabioticstressorssuchasdrought,flood, extremetemperatures,andsalinity(No ˜ gesetal.,2016).Thereleaseofanthropogenic contaminantsintotheenvironmentfromurbanization,transportation,andindustrial activitiesisalsocontributingtoenvironmentalstress(Schaefferetal.,2016).Increasesin theamountandvarietyofsyntheticchemicalsproducedhavecausedcontaminantstoenter theenvironmentataveryquickpaceonaglobalscale(Bernhardtetal.,2017).The
EnvironmentalMetabolomics. https://doi.org/10.1016/B978-0-12-818196-6.00001-7
Copyright © 2020ElsevierInc.Allrightsreserved. 1
transportandfateofcontaminantsisgovernedbytheirphysical chemicalproperties whichincludewatersolubility, n-octanol waterpartitioncoefficients(Kow),acid dissociationconstants(pKa)orbasedissociationconstants(pKb),andvaporpressure(De Laenderetal.,2015;Pereiraetal.,2016).EnvironmentalfactorssuchaspH,sunlight intensity,temperature,andorganicmattercontentalsoinfluencethetransportandfateof contaminantsintheenvironment(DeLaenderetal.,2015;Pereiraetal.,2016).Some contaminantsmaybetransportedlongdistancestoisolatedregionsandsomecontaminants maybioaccumulateinfoodwebs(Gaoetal.,2018;Xieetal.,2017).Forinstance, althoughmetalsarenaturallyoccurring,anthropogenicactivitieshavecausedincreased metalconcentrationsintheenvironmentandmetalsoftenaccumulateinorganismsasthey cannotbebiodegraded(Pengetal.,2018a;Wiseetal.,2018).Contaminantsarefrequently detectedinallthreeenvironmentalcompartmentsofair,water,andsoil(Gavrilescuetal., 2015;Netetal.,2015).Consequently,aquaticandterrestrialorganismsareexposedto variousclassesofcontaminantssuchasmetals,persistentorganicpollutants(POPs), pharmaceuticalsandpersonalcareproducts(PPCPs),industrialchemicals,plasticizers, flameretardantssuchasorganophosphateesters,andpesticides(Pengetal.,2018b;van denBrinketal.,2016;Wilkinsonetal.,2018).
1.1Contaminantstressors
Contaminantsofemergingconcern(CECs)arenotdefinitivelydefined,andthereisno comprehensivelistofCECs.Instead,CECsarethoughttobeanynaturallyoccurringor anthropogeniccompoundswhicharenowdetectedorsuspectedtooccurinsoil,air,or waterandwhosepersistenceortoxicitymaysignificantlyalterthemetabolismofan organism(Sauve ´ andDesrosiers,2014).TheUnitedStatesEnvironmentalProtection Agency’slistofCECsincludesPOPs,PPCPs,veterinarymedicines,endocrine-disrupting chemicals(EDCs),andnanomaterials(Ankleyetal.,2008, Fig.1.1).POPsarelegacy pollutantsthathaveexistedandpersistedintheenvironmentfordecadesandinclude polychlorinatedbiphenyls,dibenzo-p-dioxins,dibenzofurans,andorganochlorine pesticidessuchasdichlorodiphenyltrichloroethane(Nadaletal.,2015).TheStockholm ConventiononPOPsisaninternationalenvironmentaltreatythatwasinitiatedin2001to protecthumanhealthandtheenvironmentfromPOPs(Lallas,2001).Thescreening criteriaforPOPsincludepersistence,bioaccumulation,potentialforlongrangetransport intheenvironment,andadverseimpactstoorganisms(McLachlan,2018).Otherorganic contaminantssuchasPPCPs,veterinarymedicines,andEDCscanmoreeasilydegradein soilandwaterdependingontheirproperties,buttheirextensiveusehasresultedintheir frequentdetectionintheenvironment(Ba ´ rtı´kova ´ etal.,2016;Ebeleetal.,2017;Song etal.,2018).Pharmaceuticalsincludeover-the-countermedicationsandprescriptiondrugs, andpersonalcareproductsareineverydayproductssuchasshampoos,hairdyes, toothpaste,anddeodorants(Boxalletal.,2012).Veterinarymedicationsinclude
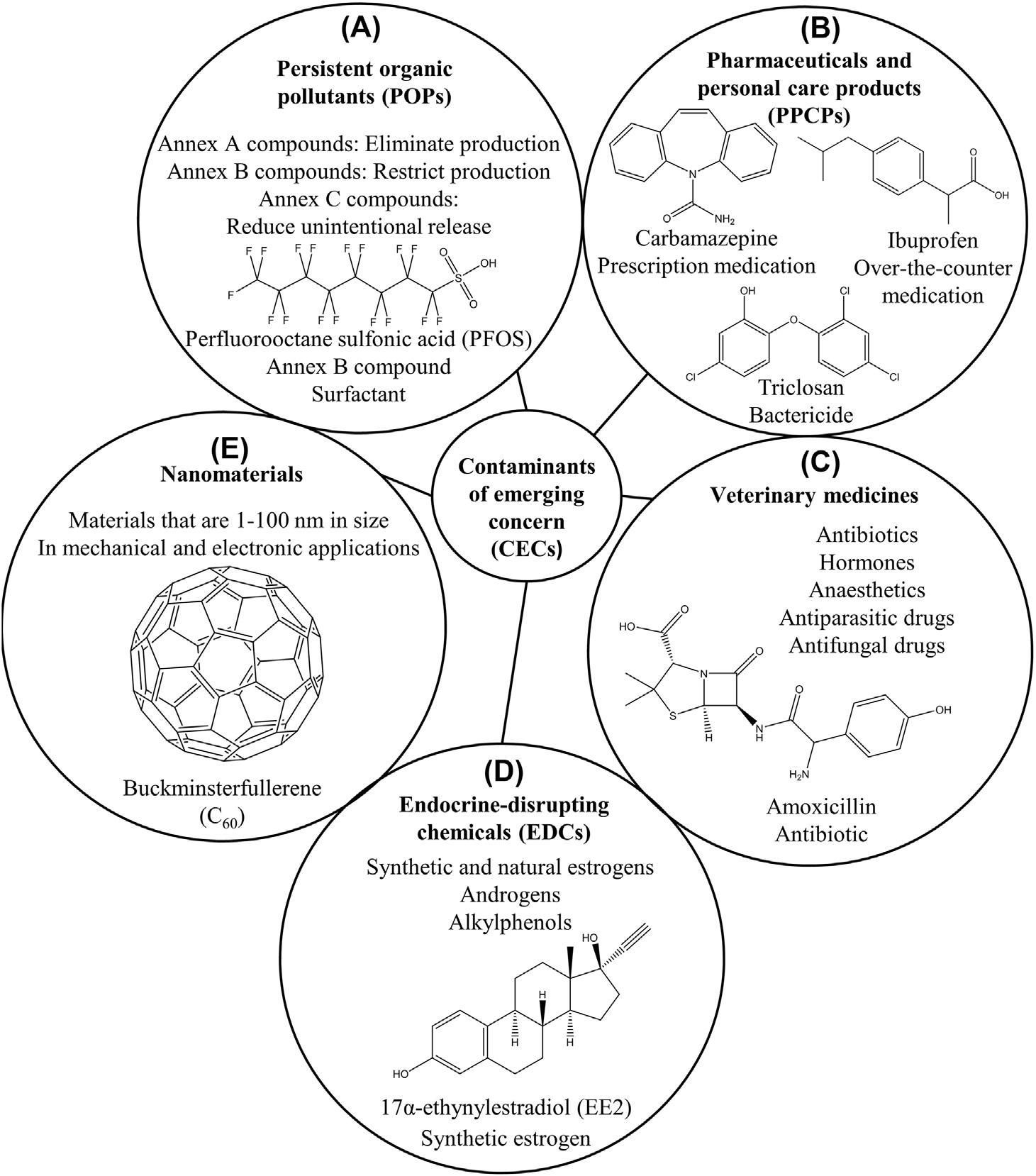
Figure1.1
Namesandchemicalstructuresofsomewell-knowncontaminantsofemergingconcern(CECs): (A)persistentorganicpollutants(POPs),(B)pharmaceuticalsandpersonalcareproducts (PPCPs),(C)veterinarymedicines,(D)endocrine-disruptingchemicals(EDCs),and (E)nanomaterials.
antibiotics,hormones,anesthetics,andantiparasiticandantifungaldrugs(Ba ´ rtı´kova ´ etal., 2016).Themainentryrouteofveterinarymedicationsintotheenvironmentisfrom treatmentoflivestock,aquaculture,andcompanionanimals(Ba ´ rtı´kova ´ etal.,2016).EDCs includealkylphenolcompounds,naturalestrogens,naturalandrogens,synthetichormones, andsomepharmaceuticalsandpesticides(Omaretal.,2016).Forinstance,oneofthe
mostpotentendocrinedisruptorsisthesyntheticestrogen17a-ethynylestradiolfoundin birthcontrolpills(Laurensonetal.,2014).Therearealsocontaminantsofindustrialorigin suchasperfluorinatedcompoundswhichcanbefoundincommonproductssuchas furniture,carpets,cookware,andfirefightingmaterials(vonderTrencketal.,2018). Terrestrialandaquaticecosystemsaretypicallyexposedtoamixtureofpesticidesfrom agriculturalapplications,andrunoffwatercanhavepesticideconcentrationsthatare substantiallyabovethelegallimit(Lefrancqetal.,2017).Nanomaterialshaveelectronic andmechanicalapplicationsandarebeingincreasinglydetectedintheenvironment,most likelyfromsewagetreatmentplantsludgeandsolidwaste(Sunetal.,2016).Thecommon detectionoforganiccontaminantsinsurfacewatersismainlyduetotheinabilityof conventionalwastewatertreatmentmethodstoefficientlyremovecompoundssuchas PPCPs,veterinarymedications,andEDCs(Herna ´ ndezetal.,2015;RiceandWesterhoff, 2017;Yangetal.,2017).Organiccontaminantsalsoentersoilandgroundwaterwhensoil isirrigatedwithwastewaterorwhensoilisfertilizedwithsewagesludge,andfrom municipalsoilwastelandfills(Healyetal.,2017;ProsserandSibley,2015).Thedetection ofevenverylowconcentrationsofCECsintheenvironmenthasraisedconcernsbecause organismsareconstantlyexposedtoinputsofCECswhichmaybioaccumulateto concentrationsthatcouldcausedeleteriousimpactsinbiota(Meadoretal.,2016).The potentialharmfulimpactsofCECsonecosystemandhumanhealthhaveresultedinthe developmentoftoxicitytestsandtheassessmentandmanagementofenvironmental contamination.
2.Ecotoxicology
Ecotoxicologyhastraditionallybeendescribedasthestudyofthetoxicimpactsof pollutantstoecosystemconstituents,includinganimals,plants,andmicroorganisms (Truhaut,1977).Assuch,environmentalqualitystandardshavebeeninitiatedtopreserve ecosystemsandhumanhealthbyplacingmaximumpermissibleconcentrationsof contaminantsthatmaybedetectedinwater,soil,orbiota(Lepper,2005).Ecologicalrisk assessmentisgenerallydonebycomparingmeasuredenvironmentalconcentrations (MECs)ofacontaminanttoitspredictednoeffectconcentrations(PNECs)from ecotoxicologicaldatawhichideallyrepresentthemostsensitivespeciesoverseveral trophiclevels(Papadakisetal.,2015).IftheriskquotientcalculatedfromtheMEC/PNEC ratioisgreaterthanone,thenthecontaminantisaconcernandactionshouldbe undertakentoconfirmtheenvironmentalrisk,identifythesourcesofcontamination,and reducethereleaseofcontamination(Papadakisetal.,2015;Thomaidietal.,2016).
Toxicologistshaveestablishedandusedacutetoxicitytestsonterrestrialandaquatic organismsbasedonmortalityorimmobilizationrateswithincreasingcontaminant concentrations(Bruce,1985;Buckleretal.,2005).Mostchronictoxicitytestsalsoassess
changesindevelopment,growth,andreproductionparameterssuchasamountof offspring,timetofirstbreeding,andnumberofbroods(Thomeetal.,2017;Toumietal., 2013;Wangetal.,2006).Othertestshavestudiedbehavioralchanges,mainlywithfish, andtheseincludechangesinmotoractivityduringlightanddarkphotoperiodsaswellas theabilityandlengthoftimeneededtocatchprey(GaworeckiandKlaine,2008; Kristofcoetal.,2016).Sincetheseapproachesarenotsensitiveforverylowsublethal concentrationsofcontaminants,newtechniquesweredevelopedusingbiomarkers (Coppolaetal.,2018;Valavanidisetal.,2006).Abiomarkerisdefinedasabiological responsewhichincludesanybiochemical,physiological,histological,andmorphological changesmeasuredinsideanorganismthatarisefromcontaminantexposure(VanGestel andVanBrummelen,1996).Substantiveprogresshasbeenmadeinmeasuring biochemicalimpactsfromcontaminantexposure,andthisincludesmeasuringchangesin biomarkersofoxidativestress,endocrinedisruption,immunomodulation,xenobiotic detoxificationsystems,andDNAdamage(Jasinskaetal.,2015;Lougheryetal.,2018; Valavanidisetal.,2006).Althoughbiomarkersgiveimportantinformationaboutthe potentialdeleteriousimpactsofcontaminantsonabiochemicallevel,theyarenotcapable ofevaluatingthemolecularmechanismofactionofcontaminants(Camposetal.,2012). Thereisaneedtostudythemechanismormodeofactionofcontaminantstobetter understandthemolecularprocessesofhoworganismsrespondtocontaminantstressorsin theenvironment.Environmentalomicsresearchwasinitiatedwhichinvolvestheuseof omicstechnologiestoinvestigatethemolecular-levelresponsesoforganismstovarious environmentalstressors(MartyniukandSimmons,2016).Theomicstechniquesinclude genomicstostudythestructure,function,andexpressionofgenes,transcriptomicsto measuregeneexpression,proteomicstomeasureproteinsandpeptides,andmetabolomics tomeasuremetaboliteswhicharetheendproductsofcellularevents(Lougheryetal., 2018;Marjanetal.,2017;Reveletal.,2017).Thedatacollectedfromomicstechniques formasystemsbiologyapproach,andtheirintegrationintothefieldofecotoxicologyis referredtoasecotoxicogenomics(Snapeetal.,2004).Also,omicstechnologiesare sensitiveandcandetectchangesinorganismbiochemistryatlowercontaminant concentrationsandmorerapidlythanconventionalmoralitytests,reproductiontests,or cytotoxicevaluations(deFigueire ˆ doetal.,2019;Shinetal.,2018).
3.Metabolomics
Metabolomicsistheuseofadvancedanalyticaltechniquestoidentifyandmeasurelowmolecularweightmetabolitesthataregenerallylessthan1000Daincells,tissues, biofluids,organs,orwholeorganisms(Gaoetal.,2019;Linetal.,2006).Thisincludes primarymetaboliteswhichareinvolvedinthedevelopment,growth,andreproductionof anorganismaswellassecondarymetaboliteswhichareproducedbybacteria,fungi,and
plantsandhavevariousecologicalfunctions(Mazzeietal.,2016;PalazzottoandWeber, 2018).Metabolomicscanbeconsideredasthedownstreamprocessofgenomics, transcriptomics,andproteomics,andthechangesofmetabolitelevelsdirectlyrelateto biochemicalactivityandthephenotype(Johnsonetal.,2016).Fundamentalmetabolic pathwayssuchasthoseinvolvedinenergy,carbohydrate,aminoacid,andlipid metabolismareconservedfrombacteriatoeukaryotes(Peregrı´n-Alvarezetal.,2009). Metabolomicshasapplicationsinseveralfieldsincludingmedicine(Wishart,2016), pharmacology(Pangetal.,2018),toxicology(Ramirezetal.,2018),plantbiochemistry (Sklirosetal.,2018),andtheenvironmentalsciences(Zhangetal.,2018a).
Themainanalyticaltechniquesthatareusedtocollectmetabolomicsdatasetsarenuclear magneticresonance(NMR)spectroscopyandmassspectrometry(MS)becauseofthe abilityforsmallmoleculedetectionandtheuniqueassetsofeachanalyticalinstrument. Targetedmetabolomicsanalysisdetectsapredeterminedsetofmetabolites,usuallychosen withregardtothebiologicalsampletobeanalyzedorfrommetabolitelibrariesin softwaredatabases(Bingol,2018).Nontargetedmetabolomicsanalysisisthenonbiased analysisofasmanymetabolitesthatcanbereliablyidentifiedandassignedbythe analyticalinstrumentandmetabolomicsdatabases(Bingol,2018).Nontargeted metabolomicsanalysisfrequentlyusesNMRspectroscopyorhighresolutionMS,while targetedmetabolomicsanalysisoftenusesMSastheanalyticalmethodofchoice(Emwas, 2015;Mullardetal.,2015).Therearearound114,000metaboliteslistedintheHuman MetabolomeDatabase(Wishartetal.,2017),butonlyaround1500metabolitesare identifiedinnontargetedanalysis,200 500metabolitesareidentifiedintargetedanalysis, anditisestimatedthatlessthantwodozenmetabolitesareregularlyquantifiedinmost metabolomicsstudies(Markleyetal.,2017;Psychogiosetal.,2011).Oncemetabolitesare identifiedandquantified,onlinedatabasesandtoolsmaybeusedtoaidindata interpretationandmechanisticunderstandingbyrelatingthechangesinmetabolitelevels tometabolicpathwaysthatarelikelyimpacted(Chongetal.,2018;Kanehisaetal.,2016). Throughthisprocess,metabolomicsgivesdetailedinformationaboutthemodeof actionthatmaybeoccurringinthebiologicalsampleandmaybeusedforhighthroughputtestingofindividualcontaminantsandmixtures(Ahmedetal.,2019;Zampieri etal.,2018).
4.Environmentalmetabolomics
Environmentalmetabolomicsinvolvesapplyingmetabolomictechniquestoanalyzethe metabolicresponseoforganismsasaresultoftheirinteractionswiththeenvironment (Bundyetal.,2009).Metabolomicsisusedtostudyvariousenvironmentalstressors includingUVlight(Zhangetal.,2018b),elevatedatmosphericcarbondioxide concentration(Creydtetal.,2019),ambientfineparticulatematter(Xuetal.,2019),
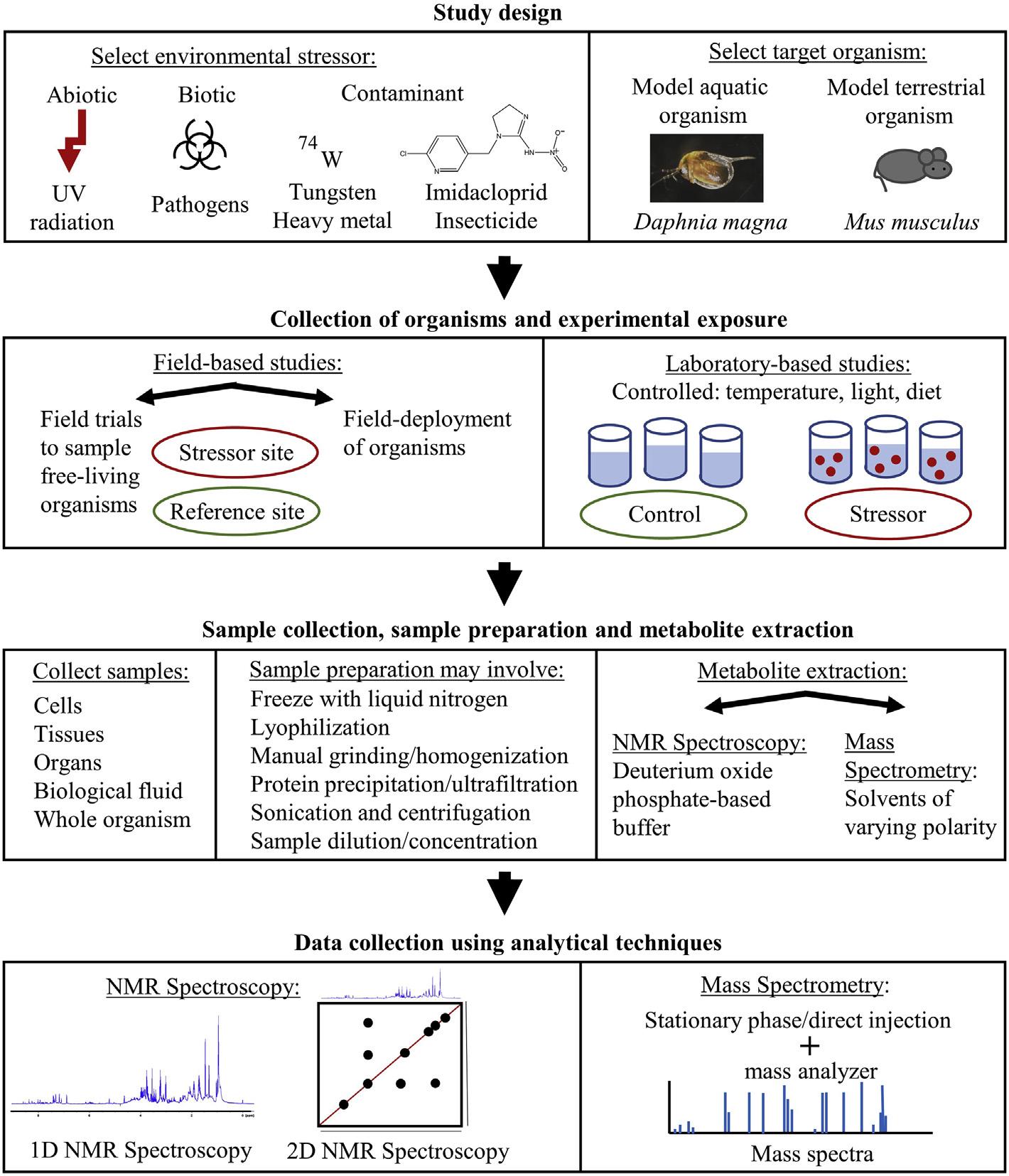
drought(Lietal.,2018b),extremetemperatures(Tomonagaetal.,2018),and contaminants(Roszkowskaetal.,2018).Controlledlaboratoryexposureswithtarget speciesareperformedtoacquireknowledgeofthemodeofactionofabioticstressors, bioticstressors,orcontaminants(Garreta-Laraetal.,2016;Sivarametal.,2019;Tang etal.,2017).Furthermore,field-basedstudiesmaybeconductedtounderstandhowthe metabolomeoforganismsisimpactedwhenexposedtoanecosystemunderenvironmental stress(Gauthieretal.,2018).Oneofthegoalsofenvironmentalmetabolomicsresearchis forutilizationinenvironmentalbiomonitoringandriskassessmentbyapplying metabolomicstechniquestokeystoneorganismsthatplayimportantrolesintrophiclevels andfoodwebs(Bahamondeetal.,2016).Toachievetheseaims,environmental metabolomicsstudieshaveaworkflowthatincludesstudydesign,exposure,sample preparation,metaboliteextraction,datacollection,dataanalysis,andfinallyabiological interpretationoftheanalyzeddata.
4.1Studydesign
Theexperimentaldesignofenvironmentalmetabolomicsprojectsinvolvestheselectionof anenvironmentalstressorandthetargetorganism.Environmentalmetabolomicsstudies aredoneonavarietyoforganismswhichspanfrommicroorganismstoplantsandanimals (Sivarametal.,2019;Tianetal.,2017;Tomonagaetal.,2018).Microorganismshave beenexposedtoenvironmentalcontaminants,forinstance,theyeast Saccharomyces cerevisiae hasbeenexposedtocopper(Farre ´ setal.,2016)andtetrachlorobisphenolA (Tianetal.,2017).Plantmetabolomicshasinvestigatedtheexposuretonanoparticles (Zhangetal.,2018a),howplantsrespondtopolycyclicaromatichydrocarbonsormetals fromremediationefforts(Pidatalaetal.,2016;Sivarametal.,2019),theimpactofmineral deficiency(Sungetal.,2015),UV-Bradiation(Zhangetal.,2018b),anddrought(Khan etal.,2019).Commonterrestrialorganismsusedinenvironmentalmetabolomicsstudies includenematodes(Ratnasekharetal.,2015),earthworms(Tangetal.,2017),flies(Cox etal.,2017),andmice(Wangetal.,2018a).Thechoiceoforganismshouldreflectthe environmentalcompartmentunderconsideration,forinstance,earthwormsarecommonly usedtoassesssoilcontaminationinmetabolomicsstudiesduetotheiroccurrenceina varietyofsoilsworldwide(Heetal.,2018;Tangetal.,2017).Aquaticorganisms frequentlyusedinenvironmentalmetabolomicsstudiesincludecrustaceans(Garreta-Lara etal.,2016;Go ´ mez-Canelaetal.,2016)andfishsuchasmedaka,rainbowtrout,salmon, andfatheadminnow(Kanekoetal.,2019).Thestudiedaquaticorganismshouldalso reflecttheresearchquestion,forinstance,bivalveshaveasessilelifestyleandcan accumulatecontaminants,andthereforeananalysisofthemetabolicprofileofbivalves mayreflectthecontaminationatthesiteofcollection(Watanabeetal.,2015). Metabolomicsstudiesmayalsobeperformedonatargetedselectionoforganismsthat serveasresearchmodels.Modelorganismsmayberepresentedbytheratandmousefor
mammals,zebrafish Daniorerio foraquaticvertebrates,thewaterflea Daphniamagna for aquaticinvertebrates, Arabidopsisthaliana forplants,theyeast S.cerevisiae for eukaryotes,and Escherichiacoli forprokaryotes(Kimetal.,2015;Reedetal.,2017).
4.2Collectionoforganismsandexperimentalexposure
Environmentalmetabolomicsstudiesmaybefield-basedwhereorganismsaresampled directlyfromtheenvironmentorlaboratory-basedwhereorganismsareculturedinthe laboratoryandthenexposedundercontrolledconditions(Campilloetal.,2019;Davis etal.,2016).Bothlaboratoryandfieldresearchshouldhavecarefulplanningofthe numberandtypeofcontrolorreferencesiteexposuresandenvironmentalstressor exposures,aswellasthenumberofsamplestobetakenfromeachtreatmentgroup.This isimportantasthereisnaturalvariationinbiologicalsamplesandadequatereplicationis neededforproperstatisticalanalysis(Simmonsetal.,2015).Regardingfield-basedwork, thereistheoptionoffieldtrialswhichinvolvesthesamplingoffree-livingorganismsin theenvironment(Gauthieretal.,2018;Melvinetal.,2018,2019)orthereistheoptionof fielddeploymentwhichinvolvesdeployingorganismsintoenvironmentsthatareunder stress(Ekmanetal.,2018).Infield-basedstudies,theorganismsthatrepresentthe stressor-exposedgroupsmaybecollectedatcontaminatedlocationsandmaybecompared toorganismscollectedatmorepristinelocationswhichserveasthegroupfroma referencesite(Watanabeetal.,2015).Environmentalmetabolomicsstudiesthatsample free-livingorganismsatafieldsiteareastepforwardforvalidatingthistechniqueforuse inenvironmentalmonitoring(Melvinetal.,2018,2019).Also,usingfield-deployed organismshasagreatvalueforenvironmentalriskassessment.Forinstance,astudythat usedcage-deployedfatheadminnows(Pimephalespromelas)atsitesacrosstheGreat Lakesbasinnotedthattheprofilesofendogenouspolarmetaboliteshadcovariancewithat most49contaminants(Davisetal.,2016).Takingorganismsfromthefieldtakesinto considerationthenaturalvariationfromdifferentlocations,forexample,thereare correlationsbetweenthemetabolomeofthepinetree(Pinuspinaster)anditsoriginal geographicoriginevenwhengrowninacommongardenfor5years(Meijo ´ netal.,2016). Samplingorganismsfromthefieldalsoconsiderstheindividualvariability,whichmay stemfromgeneticdifferences,inthemetabolicprofileoftheseorganismsastheyarenot onlyonelaboratorystrain(Quinaetal.,2019).Understandingthemetabolicresponseof organismswhicharesampledfromthefieldmaybechallengingbecausefieldpopulations maybeexposedtomultiplestressorsatonce,includingbothabioticandbioticstressorsas wellascontaminantstressors(Garreta-Laraetal.,2018;Mishraetal.,2019).Thechoice oforganismsshouldbeascloseaspossibletoidenticalageandsizetominimizenatural variationinpopulations(Coppolaetal.,2018).However,whensamplingorganismsfrom thefielditmaybedifficulttodistinguishgender,differentlifestages,andspecies,for instance,DNAbarcodingisusedtodistinguishspeciesofthegenus Atlantoscia,a
terrestrialisopod(Zimmermannetal.,2018).Organismssampledfromtheenvironment havevariousfactorsthatmayimpactthemetabolomewhichshouldberecorded,suchas seasonandgeographiclocation(Weietal.,2018),climate(Gargallo-Garrigaetal.,2015), disease(Zacheretal.,2018),andhabitatsurroundings(Quinaetal.,2019).Also,itisa challengetobecertainofallthecurrentenvironmentalstressorspresentatthetimeof collectionandthehistoryoforganismexposuretoenvironmentalstressors(Olsviketal., 2018).Field-basedstudiesalsofacethedifficultlyoflocatinganidealreferencesitethat canactasacontrolwhichhasminimalabiotic,biotic,andcontaminantstressorsbut wherethenaturalsettingsareanalogoustotheimpactedsite(Martyniuk,2018).However, toaidinthemetabolomicsanalysisandinterpretationoffield-basedstudies,thereare standardreportingrequirementsformetabolomicsexperimentsofbiologicalsamplestaken fromtheenvironment(Morrisonetal.,2007).
Laboratorypopulationsarestrictlycontrolled,andtemperature,light,anddietare maintainedconstantlytolimitanyperturbationstothemetabolome.Laboratory-based studieshavethebenefitofstandardprotocolsthatareavailable,forinstance,the OrganisationforEconomicCo-operationandDevelopmentprovidesguidancedocuments foraquatictoxicitytestingandsoiltoxicitytestingthatshouldbefollowedforlaboratory work(OECD,2002,2004).Controlledlaboratoryconditionsareoptimalfordetermining themodeofactionofcontaminantsasthemetabolicperturbationscanbedirectly accreditedtothestressor(Go ´ mez-Canelaetal.,2016).Forthisreasontherearestandard reportingrequirementsformetabolomicsexperimentsofmammalian/invivo work(Griffin etal.,2007)andformetabolomicsexperimentsofmicrobialand invitro work(vander Werfetal.,2007).Therouteofexposure,suchasdosing,air-borneinhalation,aqueous exposure,andothers,shouldbecarefullyselectedanddocumented(vanderWerfetal., 2007).Althoughthemetabolicdisturbancesfromexposuretostressorsinlaboratory conditionscanbeextrapolatedtopredictthehazardinenvironmentalsettings,caremust betakenwhendoingsotoavoidanoverestimationoranunderestimationofrisk(Murphy etal.,2018).
4.3Samplecollection,samplepreparation,andmetaboliteextraction
Thedecisionofwhichbiologicalsampletoanalyzeisimportantsincedifferentmetabolic responsescanbefoundacrosscells,tissues,organs,biologicalfluids,andthewhole organism(Tavassolyetal.,2018).Thereareadvantagesofchoosingeachbiological compartment.Forinstance,usingcellculturesprovidesspecificinformationonthe metabolicpathwaysinvolvingthechangesofendogenousmetabolitesincells,anddifferent celllinesmayhavedifferentmetabolicalterations(Rodriguesetal.,2019).The metabolomicsanalysisofacertaintissuecangiveinformationaboutthestateofanorgan anddifferenttissuesofanorganismcontaindifferentbaselinemetabolicprofiles(Cappello
etal.,2018).Thecollectionofabiofluidsuchasbloodorurineisminimallyinvasiveand canprovideinformationabouttheoverallbiologicalstateofanorganism(Liaoetal.,2018).
ComparedtoDNAsequenceswhichcanbeextractedfromsamplesover100,000yearsold (Rohlandetal.,2018),metabolitesamplesarehighlyunstableandtheremaybeadditional changesinmetabolitelevelsfromresidualenzymaticactivity(Berninietal.,2011). Therefore,propersamplepreparationisimportanttoavoidanydisturbancesinthe metabolomethatmayoccurfrompreparingsamples.Samplepreparationproceduresmay needtoquicklyhaltenzymaticactivitywithfreezinginliquidnitrogenandsamplesmust bekeptcoldthroughoutanystorage(Berninietal.,2011).Somesampletypesneedto havewaterremovedforanalysisbyNMRspectroscopy,forinstance,tissuesamplesneed tobelyophilizedsoonaftercollectionandthesolventextractsofcellculturesneedtobe driedandreconstitutedinanNMRbuffer(Beckonertetal.,2007;Carrolaetal.,2018).
NMR-basedmetabolomicsmostfrequentlyusesadeuteriumoxidephosphate-basedbuffer whichextractspolarmetabolites(Beckonertetal.,2007).Thephosphatebuffermaintains aconstantpHtominimizevariationsinchemicalshiftandadeuteratedsolventallowsfor alocksignalinNMRspectroscopy(Schripsema,2010).Themetaboliteextraction procedureforMSanalysistypicallyusessolventsofvaryingpolaritytoextractpolarand nonpolarmetabolitesandseveralextractionmethodsarebasedonthewidelyusedBligh andDyerextraction(BlighandDyer,1959;Wuetal.,2008).Themetaboliteextraction methodmustbedevelopedandverifiedbecauseoneextractionprotocolisusuallynotable toextractallthemetabolitesfromthesample.Also,themetaboliteextractionmethod dependsonthebiologicalmatrixchosentobeanalyzed,andtherearewell-developed protocolsformetaboliteextractionsfrommammalianandbacterialcellcultures(Dietmair etal.,2010;Winderetal.,2008),biologicalfluidssuchasbloodandurine(Beckonert etal.,2007;Bruceetal.,2009),andplant-andanimal-derivedtissues(Valledoretal., 2014;Wuetal.,2008).Toextractmetabolitesfromcells,coldsolventssuchasaqueous methanoloraqueousacetonitrileareappliedtocellculturestoquenchthemetabolismof cells(Dietmairetal.,2010;Rodriguesetal.,2019).Forbiologicalfluidssuchasblood, thereisusuallyaproteinprecipitationsteptoisolateproteinsfromthemetabolitesample bytreatmentwithasolventmixturesuchasmethanol/ethanolormethanol/acetonitrile/ acetone(Bruceetal.,2009).Urinesamplestypicallyhavesimplesamplepreparationand involvesampledilutionwithappropriatesolventsorultrafiltration(Khamisetal.,2017).
Fortissuesamplesorwholeorganismhomogenates,thereismanualgrindingor homogenizationwithprecooledextractionsolventssuchasmethanol(Ro ¨ misch-Margl etal.,2012).Tissuesamplesorsmallwholeorganismhomogenatesmaybefurther sonicatedtoenhancetheextractionefficiency(Wangetal.,2018b).Finally,centrifugation canseparatethesupernatant-containingmetabolitesfromlargemoleculessuchas precipitatedproteinsandcellularandtissuedebriswhicharepelletedfromcentrifugation
(Hamerlyetal.,2015).Besides,theorderofmetaboliteextractionanddatacollectionfrom samplesalsoshouldberandomizedtoavoidanybiasorbatchimpacts.
4.4Analyticaltechniquesfordatacollection
ThegeneralanalyticalmethodsusedinenvironmentalmetabolomicsareMSandNMR spectroscopy. Fig.1.2 illustratestheworkflowinanenvironmentalmetabolomicsstudy
Figure1.2
WorkflowIofagenericenvironmentalmetabolomicsstudy:fromstudydesigntodatacollection ontheanalyticalinstrumentofchoice.
beginningfromstudydesigntodatacollectionontheanalyticalinstrumentofchoice. BothNMRspectroscopyandMSanalyticaltechniquesencountersomechallenges,and thereisnosingleanalyticaltechniquethatcanidentifyandquantifythewhole metabolome.Severalstudiesproposedirectinfusionmassspectrometry(DIMS)coupledto highresolutionmassanalyzerssuchasOrbitrapmassanalyzersorFouriertransform ion cyclotronresonance(FT-ICR)massanalyzersbecauseoftherapidandnontargeted analysisofawiderangeofmetabolites(Ghasteetal.,2016;Southametal.,2017;Taylor etal.,2016).However,ionsuppressionandthecomplexityofthemassspectrawithout sampleseparationwithastationaryphasearedisadvantagesofDIMS(Ghasteetal., 2016).Therefore,MSisusuallycoupledtoliquidchromatography,gaschromatography,or capillaryelectrophoresisforanalyteseparationpriortomassdetection(Bealeetal.,2018; Gorrochateguietal.,2016;Sasakietal.,2018).Thestationaryphasecanaidinmetabolite identificationusingtheretentiontimeoftheanalyteofinterest(Chekmenevaetal.,2018).
Gaschromatography massspectrometry(GC-MS)ofpolarmetabolitestypicallyrequires chemicalderivatizationofmetaboliteextractsamplestoformvolatileandthermallystable analytesandtoalsoenhancetheseparationanddetectionofthesemetabolites(Miyagawa andBamba,2019).However,thequantitationofderivatizedmetabolitesbyGC-MS analysismaybedifficultasthechemicalderivatizationefficiencymayvaryandthe stabilityofthederivatizedmetabolitesmaynotbeconstant(MiyagawaandBamba,2019). MetabolomicsstudiesfrequentlyuseGC-MSwithelectronimpactorchemicalionization (Lietal.,2015).Capillaryelectrophoresis massspectrometry(CE-MS)involvesthe separationofhighlypolarandionicmetabolitesduetodifferentmigrationspeedsofthe metabolites(Ramautaretal.,2019).CE-MS basedmetabolomicsstudiesoftencouple capillaryelectrophoresistoatime-of-flight(TOF)massanalyzer(Ramautaretal.,2019). Liquidchromatography massspectrometry(LC-MS)analysisincludestheselectionofan appropriateliquidchromatographiccolumn,solventsforthemobilephases,potential additivestothemobilephase,andmobile-phasegradientsforseparationbyliquid chromatography(Wangetal.,2018b).MostMS-basedmetabolomicsstudiesuseliquid chromatographywithelectrosprayionization(ESI)thatissuitablefortheionizationofa rangeofmetabolites(Aszyketal.,2018).Nontargetedmetabolomicsstudiesforglobal profilingfrequentlyusehighresolutionandhighaccuracymassanalyzerssuchasFT-ICR massanalyzersorquadrupole-time-of-flight(QTOF)massanalyzers(Aszyketal.,2018). MS-basedmetabolomicsstudiesoftenusetriplequadrupolemassspectrometersfor targetedanalysisduetothehighsensitivityandselectivity(NaganaGowdaandDjukovic, 2014).
ThemainadvantagesofusingMSforenvironmentalmetabolomicsapplicationsisthatMS candetectandquantifymetabolitesatverylowconcentrationsinthefemtomolarto attomolarrangeandMShasahighdynamicrangeandresolution(MarshallandPowers, 2017).AdisadvantageisthatMSonlydetectsmetabolitesthatcanpromptlyionize,for
instance,upto60%ofcompoundsfromareferencechemicallibraryweredetectableby ESI-MS(Copelandetal.,2012).Ionsuppressionorionenhancementmayoccurin metabolomicssampleswithcoelutingcompoundsandacomplexmatrixand,asaresult, MShasissuesinreproducibility(Engskogetal.,2016).AnotherdownsideofMS-based metabolomicsisthatitmaybedifficulttoknowinadvancethemostoptimalionization techniqueandionizationpolaritytoselectforifthecompositionofthesampleis unknown,whichmaybeacircumstanceduringnontargetedMSanalysis(Panuwetetal., 2016).Also,metabolitesmayhaveretentiontimedriftsduringseparationinthestationary phaseandtheremaybeinstabilityinMSdetectionduetocontaminationoftheionsource ofthemassanalyzer(Zelenaetal.,2009).
NMRspectroscopycanidentifyunknownmetabolitesunambiguously,candistinguish isomers,andisexcellentforstructureelucidationofunknowncompounds(Bingoland Bruschweiler,2015).ThemostcommonlyusedNMRmethodinenvironmental metabolomicsisone-dimensional(1D)proton(1H)NMRspectroscopy(Ambergetal., 2017).NMRspectroscopyishighlyquantitativeandreproducible,forexample,the technicalvariationwithinmetabolomicsdatasetsusing1D 1HNMRspectroscopycanbe aslowas1.6%medianspectralrelativestandarddeviation(BhartiandRoy,2012;Parsons etal.,2009).WatersuppressionisusedwithNMRspectroscopytoavoidbaseline distortionsandlargeerrorsinpeakareassincewaterisatasubstantiallyhigher concentrationthanmetabolitesinsampleextracts(Giraudeauetal.,2015).Thereare severalwatersuppressiontechniquessuchasWATERGATE(Piottoetal.,1992), excitationsculpting(HwangandShaka,1995),andpresaturationutilizingrelaxation gradientsandechoes(SimpsonandBrown,2005).NMRspectroscopyhasnoneedfor sampleseparationbychromatographyandoftenrequiresminimalsamplepreparationsuch asforurinesamples(Takisetal.,2019).Onlyasingleinternalreferenceisneededforthe absolutequantitationofmetabolitesusingNMRspectroscopy(NaganaGowdaetal., 2018).AnotheradvantageofNMRspectroscopyisthatithasnondestructivedata acquisition,forinstance,NMRmethodssuchas 1Hhighresolutionmagic-anglespinning NMR(1HHR-MASNMR)canbeperformedonintacttissues(Battinietal.,2017).
Invivo NMRspectroscopyusingflow-basedsystemswithsolution-stateNMR(Tabatabaei Anarakietal.,2018),HR-MASNMR(Bunescuetal.,2010),andcomprehensive multiphaseNMR(Mobarhanetal.,2016)isalsoemergingforenvironmental metabolomicsapplicationsoflivingorganisms.AdisadvantageofNMRspectroscopyis thatonlythemostabundantmetabolitesatconcentrationsgreaterthan1 mMaredetected (NaganaGowdaandRaftery,2015).Also,theresolutionin1D 1HNMRspectroscopyis lowandalthoughtwo-dimensional(2D)NMRmethodsincreaseresolution,theacquisition timeof2DNMRspectraislonger(NaganaGowdaandRaftery,2015).However,thereare developmentsthatcanincreasethesensitivityandresolutionofNMRspectroscopyfor metabolomicsanalysis.Theseincludetechnicalimprovementsinpulsesequencessuchas
para-H2-inducedhyperpolarization(Reileetal.,2016),cryoprobetechnology,and microprobetechnology(Saboranoetal.,2019).Samplevolumesareminimizedfrom 500 600 mLusingstandard5mmNMRprobesto25 45 mLusing1.7mmmicroprobes whichisbeneficialformass-limitedsamples,andconcentratingthesamplecanimprove thesignal-to-noiseratio(Martin,2005;Nagatoetal.,2015).Thesensitivitygaincanbeup to40-foldforsamplesofequalmasswhentransitioningfroma5mmroomtemperature probetoa1.7mmmicrocryoprobe(Saboranoetal.,2019).Inaddition,microcoilNMR hasbeenusedfortheanalysisofsingleanimaleggsofthenematode Heligmosomoides polygyrusbakeri andthetardigrade Richtersiuscoronifer (Grisietal.,2017)aswellasthe fish Cypseluruspoecilopterus andthewaterflea D.magna (Fugariuetal.,2017).
MicrocoilNMRhastheabilitytoanalyzebiologicalsampleswithavolumeof approximately0.1nL(Grisietal.,2017),andcollectinganNMRspectrumona50 mm coilgives3000timesthemasssensitivitycomparedtoa5mmNMRprobe(Fugariuetal., 2017).
4.5Rawdatapreprocessinganddataanalysis
RawdatapreprocessingofspectracollectedfromNMRspectroscopyorMSisdone beforestatisticalanalysisandpatterndetectionwithchemometricsmodels.Fourier transformationofthefreeinductiondecaysignalproducestheNMRspectrumand typicallyalinebroadeningof0.3Hzisusedfor 1HNMRspectra(Kostidisetal.,2017). EachNMRspectrumshouldbemanuallyphasedformetabolomicsstudiessincemany automaticphasingmethodsmaydistortsignalswithsmallpeakareas(Emwasetal., 2018).Next,baselinecorrectionisperformed,andchemicalshiftsarealignedtoan internalcalibrantsuchas4,4-dimethyl-4-silapentane-1-sulfonicacidor3-(trimethylsilyl)2,20 ,3,30 -tetradeuteropropionicacid(Donaetal.,2016).TheNMRspectraundergo binning,alsocalledbucketing,mostcommonlyintoequalwidthsof0.02or0.04ppmfor 1HNMRspectraandthesignalineachbinisintegrated(Vignolietal.,2018).Binningthe dataallowsmultivariatemethodstoidentifytheregionsoftheNMRspectrathataremost responsibleforthevariancewithinthedataset(Karaman,2017).With 1HNMRspectraof polarmetabolites,theregionbetween4.7and4.9ppmistypicallyexcludedfromanalysis becausethisregionmayaddvariancetothedataduetoresidualwatersignals,andthe regionsoutsideof0.5 10ppmareexcludedfromanalysisastheseregionsarecomprised ofmostlynoise(Kimetal.,2016).Datascalingtoreducesample-to-samplevariationis oftencarriedoutbynormalizationtototalNMRspectralarea,wheretheintegratedvalue ofonebinoftheNMRspectrumisdividedbythesumoftheintegratedvaluesofallthe spectralbinsinonespectrum(Zachariasetal.,2018).Next,metabolitessuchas carbohydrates,aminoacids,organicacids,alcohols,osmolytes,andnucleotidesare identifiedby1D 1Hand2DNMRspectroscopy(Everett,2015;Guetal.,2019;Watanabe etal.,2015).Therelativequantitationofmetabolitescanbecompletedasthesignal
intensityofan 1HNMRpeakisdirectlyproportionaltotheconcentrationandnumberof protonsthatcontributetothatparticularresonancesignal(Kumaretal.,2014).Absolute quantitationof 1HNMRpeakscanbeperformedbymanuallyaddingaspecific concentrationofoneinternalstandard(NaganaGowdaetal.,2018),byaddinganexternal standardinacoaxialNMRtube(Gardneretal.,2018),orusingtheElectronicREference ToaccessIn-vivoConcentrationmethodtoproduceanartificialreferenceNMRsignal (Akokaetal.,1999).MSanalysisalsohasrawdatapreprocessingstepstosimplifyand convertaspectrumintopeaksforfurtherprocessingwithmultivariatetools.Thisincludes centroidingtochangeeachpeaktoonedatapointwithamass-to-chargeandintensity value,aswellasfilteringmethodstoremovethebaselineandmeasurementnoise (KatajamaaandOresic,2007).Peakdetectionanddeconvolutionstepsareimplementedto assigndifferentionsthatbelongtothesamemetaboliteortoseparatepeaksfrom coelutingcompounds(Nietal.,2016).ApreprocessingstepforLC-MSorGC-MSdata includesaligningthespectraalongthechromatographicruntimeasshiftsinretention timemayoccurduetocolumnaging,thesamplematrix,orchangesintemperature, pressure,orthemobilephase(Watrousetal.,2017).Peakextractionandpeak identificationmustbecompletedtoquantifymetabolites(Lietal.,2016).MS-based metabolomicsalsohasdatanormalizationmethodstoreducesample-to-samplevariability suchasMStotalusefulsignalnormalizationorprobabilisticquotientnormalization (Gagnebinetal.,2017).
Themultivariatestatisticalanalysisthatisappliedinenvironmentalmetabolomicsstudies isperformedtosimplifyandfacilitatedataanalysisintheoftenlargedatasetsfromNMR spectroscopyorMS.Thecommonlyusedmethodofprincipalcomponentanalysis(PCA) isanunsupervisedstatisticalmethodthathasnobiasorknowledgeaboutthetreatment groupofasample(Leveretal.,2017).PCAisusedtodeterminegeneralpatternsand visualdifferencesinthemetabolicprofilesofdifferenttreatmentgroups(Guetal.,2019). TheoutcomeofaPCAmodelisascoresplotwhereeachpointinatwo-dimensionalor three-dimensionalspaceisthespectrumofasinglesample(Huetal.,2017).Thefirstfew principalcomponentsexplainmostofthevariationinthedatasetandmostaccurately reflectthecontentofthedata(Leveretal.,2017).TheclusteringofsamplesinthePCA scoresplotsuggeststhatthosesampleshavesimilarfeaturesintheirmetabolome(Hu etal.,2017).Toeasilyidentifygroupmembershipandvisualizethedata,anaveraged PCAscoresplotismadewherethescoresofallthespectraofonetreatmentgroupare averagedandplottedasaveragevalueswithstandarderror(Wangetal.,2016).PCA loadingsplotsindicatetheregionsofthespectrathatcontributetothevariationineach principalcomponentandhenceindicatethemetabolitesthatcontributetothemetabolic variationbetweentreatmentgroups(Leeetal.,2016).
Supervisedstatisticalmethodssuchaspartialleastsquares(PLS),includingPLSregression (PLS-r),PLSdiscriminantanalysis(PLS-DA),andorthogonal(O)-PLS-DAmodels,
usesampleclassasafactortogeneratepredictionsaboutthedata(Davisetal.,2017; Tryggetal.,2007).Thedatafromclassifiedsamplesarethetrainingset,andthesedataare analyzedandusedtopredictthedatafromunclassifiedsampleswhicharethetestset(de CarvalhoRochaetal.,2018).PLSmodelsrequirepropervalidationbecausethese supervisedtechniquesmayoverfitthedataandresultinthefalseappearanceofgroup separation(Saccentietal.,2014).Forthisreason,validationtechniquesarerequiredto determinetheoptimalnumberofPLScomponentstoavoidoverfittingthedata(Franitza etal.,2018;Lietal.,2018a).Theseincludecross-validationmethodssuchasleave-one-out crossvalidation,K-foldcrossvalidation,MonteCarlocrossvalidation,anddoublecross validation(Lietal.,2018a).Thereisalsoanindependentvalidationwhereadifferentdata setisusedtotestthepredictiveabilityofthePLSmodel(Franitzaetal.,2018).To determinethestatisticalsignificanceofthegeneratedPLSmodel,apermutationtestis frequentlyusedwhichgivesrandomgroupassignmentstoevaluatethepredictionaccuracy (Zhangetal.,2017b).Forexample,aPLS-DAmodelwascreatedfromdatacollectedwith 1HNMRspectroscopyandunderwentsevenfoldcrossvalidationand200permutationtests todistinguishthemetabolomesofcontrolratsfromratsexposedtofineparticulatematter (Zhangetal.,2017b).
Inunivariatestatisticalanalysis,theindividualmetabolitesareidentified,andtheir absoluteorrelativequantificationiscompleted.NMRspectroscopyhasmultiplemethods formetaboliteidentification.In 1HNMR-basedmetabolomics,manymetabolitescanbe assignedfromthechemicalshiftandpeakmultiplicity(Cappelloetal.,2018).The MadisonMetabolomicsConsortiumDatabaseisachemicalshiftdatabasethatisoften usedinmetabolomicsthathasboththeoreticalandexperimentaland1Dand2DNMR dataaswellasMSdata(Cuietal.,2008).Metaboliteidentitiescanbevalidatedwith2D NMRmethodsbyusingsoftwarepackagestocompareachemicalshiftdatabaseof2D NMRspectratoregionsof2DNMRspectraofbiologicalsamples(Nagatoetal.,2015; Yuketal.,2013).The2DNMRspectrausedformetaboliteidentificationaremainly 1H-1Htotalcorrelationspectroscopyand 1H-13Cheteronuclearsinglequantumcoherence spectroscopy(HSQC)(Xiaetal.,2008).IdentificationofunknownsusingMScanbe accomplishedbymatchinghighaccuratemassexperimentaltandemmassspectraofan unknownmetabolitetothebestcorrespondingaccuratemassandtandemmassspectral fragmentationpatternofalllikelyisomericstructuresfromametabolitedatabase(Boiteau etal.,2018;Duhrkopetal.,2015).
Metabolitedatacanbedisclosedbyreportingtheabsoluteconcentrationsof metabolitesacrossalltreatmentgroupsorbyreportingthefoldchangeofmetabolites inanexposedgrouprelativetoacontrolgroup(Go ´ mez-Canelaetal.,2016).To determineifametabolitelevel thatwasobtainedfromabsolutequantificationor relativequantificationhasastatisticallysignificantchangebetweentwoindependent treatmentgroups,themainunivariatestatisticaltestsaretheStudent’s t-testfordata
withanormalitydistribution(Mosleyetal.,2018)andtheWilcoxon Mann Whitney testfordatathatarenotnormallydistributed( FayandProschan,2010;Pradhanetal., 2016).Forexample,aStudent’s t-testwasappliedtoliquidchromatographytandem massspectrometry(LC-MS/MS)datatodeterminesignificantchangesinmetabolitesof theskinmucusoffatheadminnows P promelas aftera21dayexposuretotreated wastewatereffluent(Mosleyetal.,2018).Whenmorethantwogroupsarebeing comparedunderthenormalitydistribution,ananalysisofvariance(ANOVA)testwith posthocanalysiscanbeusedtodeterminesignificantlydifferentmetabolitesfor multiplegroupmeans(Fuetal.,2019;Kokushietal.,2017;Renetal.,2015).For instance,one-wayANOVAwithTukey’sandFisher’sleastsignificantdifferencetests wereusedtodeterminethestatisticaldifferencesofmetabolitechangesinzebrafish D.rerio embryosfromexposuretofiveconcentrationsoftriclosan( Fuetal.,2019).
Theresultofallthesestatisticaltestsisa P -value,andifthis P-valueisbelowa significancelevelthatisgenerallytakentobe0.05,thenthealternativehypothesisthat thereisadifferencebetweentwoormoregroupmeansisaccepted(Saccentietal., 2014).Likewise,ifthe P-valueisbelowtheselectedsignificancelevelof0.05,thenthe nullhypothesisthatthereisnosignificantdifferencebetweentwogroupsisrejected (Saccentietal.,2014).
4.6Biologicalinterpretationoftheresults
Understandingtheimplicationsofmetabolitechangesischallengingbecausemetabolites areinterconnectedandcanbepartofseveralmetabolicpathways(Caspietal.,2015). OnlinedatabasesofmetabolicdataincludetheKyotoEncyclopediaofGenesand Genomes(KEGG)PathwayDatabasewhichshowsmanymetabolicpathwaysandthe individualmetabolitesineachstepofapathway(Kanehisaetal.,2016).Theonline databaseMetaboAnalyst4.0hasseveraldatainterpretationtoolsforbothNMR spectroscopyandMSdata,suchasmetabolitepathwayanalysis(MetPA)wherethe changesoftheindividualmetabolitesarecorrelatedtometabolicpathwayinformation (Chongetal.,2018).Theuserinputsalistofmetabolitesandtheirabundancesfromtwo treatmentgroupsandselectsoneofthe21modelorganismswhichMetaboAnalyst4.0has metabolitepathwaylibraryinformationfromadatabasesuchastheKEGG(Xiaand Wishart,2010).Next,theparametersforpathwayanalysisarespecified,whichincludes thealgorithmforpathwayenrichmentanalysisandthealgorithmfortopologicalanalysis (XiaandWishart,2010).Theoutputispresentedasamap-stylenetworkvisualization systemandasalistofmetabolicpathwaysinorderoftheirstatisticalsignificancebetween thetwotreatmentgroups(XiaandWishart,2011).Forinstance,metabolicpathway analysisperformedusingMetPAdeterminedthatninemetabolicpathwayswere significantlyalteredinthesoilnematode Caenorhabditiselegans fromexposureto sublethalconcentrationsoftitaniumdioxidenanoparticles(Ratnasekharetal.,2015).
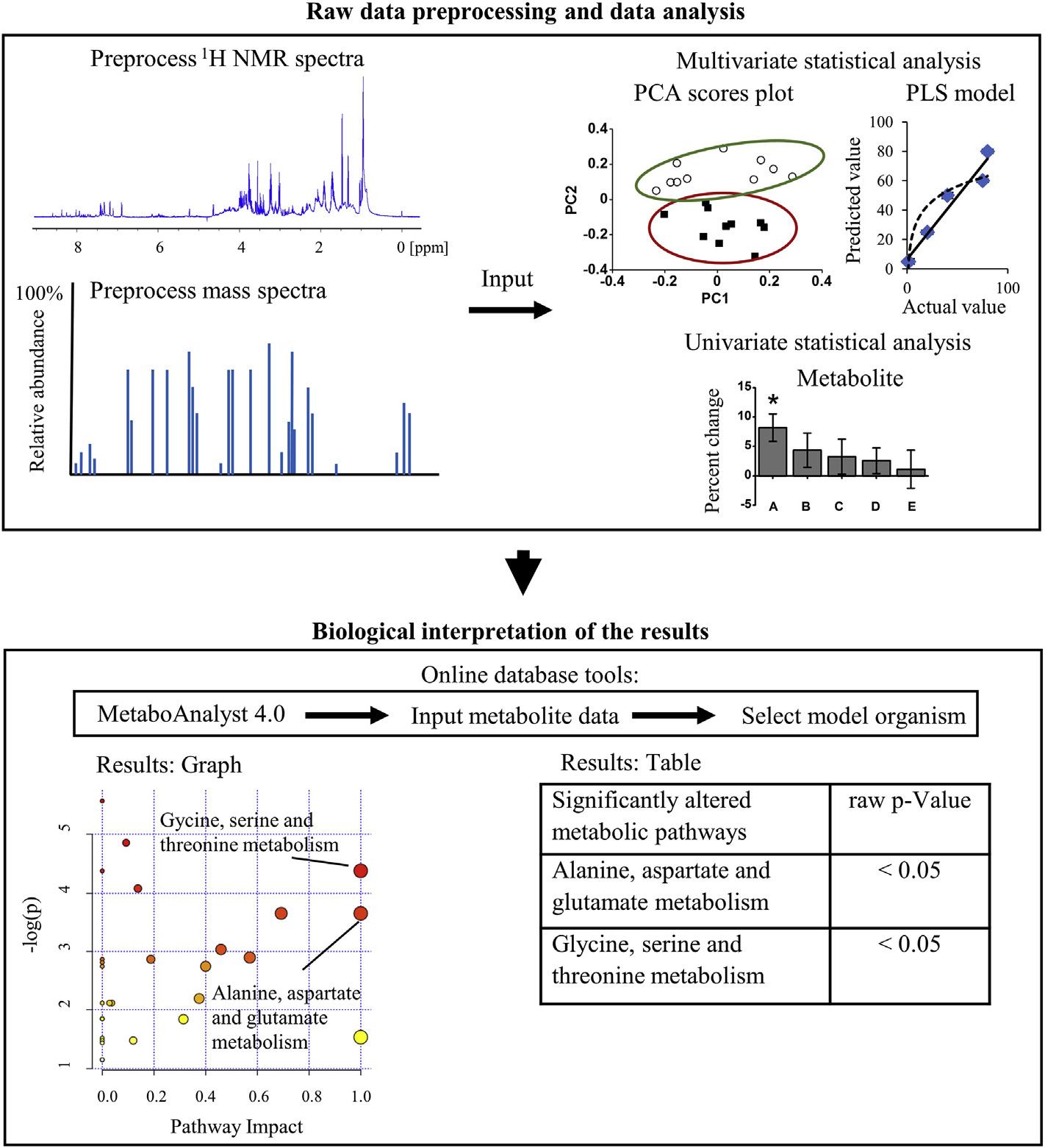
Figure1.3
WorkflowIIofagenericenvironmentalmetabolomicsstudy:fromdatapreprocessingto biologicalinterpretationwithmetabolitepathwayanalysis.
Fig.1.3 illustratestheworkflowinvolvedinanenvironmentalmetabolomicsstudy, beginningfromdatapreprocessingtobiologicalinterpretationofthedatausingmetabolite pathwayanalysistools.
5.Exposuretocontaminantmixtures
Contaminantsexistintheenvironmentascomplexmixtures,andthereforeefforts arebeingmadetoapplyenvironmentalmetabolomicstechniquestostudyexposures tocontaminantmixturesorenvironmentalsamplessuchaswastewatertreatment
Fundamentalsofenvironmentalmetabolomics19
planteffluent(VanMeteretal.,2018;Wagneretal.,2019;Zhenetal.,2018).
Environmentalmetabolomicsstudiesofcontaminantmixturesinvestigatethemetabolic responsefromthecombinedsublethalexposureofcontaminants(Ahmedetal.,2019; VanMeteretal.,2018).Itisbeneficialtoevaluatethemodeofactionoftheindividual contaminantsthatcomposeamixtureoftwoormorecontaminantstomoreeffectively assessanyinteractiveimpactsonthemetabolome(Altenburgeretal.,2018).For example,thejuvenilegreenfrogs Lithobatesclamitans wereexposedtofivepesticides assingle,double,ortriplepesticidemixtures,andmetabolomicsanalysiswithGC-MS revealeddifferentchangesinmetabolitesandmetabolicpathwaysbetweenthe treatments(VanMeteretal.,2018).Theriskassessmentofacomplexmixturethatmay haveunknownconstituentsorthathastensorhundredsofcontaminantsislikelybetter achievedbyevaluatingthetoxicityofthemixtureasawhole(Kienzleretal.,2016).
Forthisreasonenvironmentalmetabolomicsstudieshaveexaminedthetoxicityof environmentalsamplessuchaswastewatertreatmentplanteffluent,ambientfine particulatematter,andcontaminatedsediments,andthisisconsideredtobeawhole mixtureapproach(Berlioz-Barbieretal.,2018;Chiuetal.,2017;Mosleyetal.,2018; Songetal.,2019;Zhenetal.,2018 ).Forexample,nanoliquidchromatographycoupled toquadrupole-time-of-flight-massspectrometry(QTOF-MS)showedthatthe metabolomeofthebenthicinvertebrate Chironomusriparius wasimpactedwithmostly changesinlipidmetabolismfrombothfieldandlaboratoryexposuretowastewater treatmentplanteffluents(Berlioz-Barbieretal.,2018).Understandinganorganism’s metabolicresponseafterexposuretocomplexenvironmentalsamplesisachallenge becausethemolecular-levelresponsestotheindividualcontaminantsarenot alwaysknown.Also,thereareotherenvironmentalfactorsthatmayimpactthe metabolicresponseandthebioavailabilityofcontaminantssuchasdissolved oxygenconcentration,food,temperature,salinity,pH,andorganicmattercontent (Campilloetal.,2019;Garreta-Laraetal.,2018;Kovacevicetal.,2019;Mishra etal.,2019 ).
6.Summaryandenvironmentalbiomonitoringefforts
Environmentalmetabolomicshasmadegreatprogresssinceoneofitsfirst applicationsin1997using 1HNMRspectroscopytoanalyzechangesinendogenous metabolitesofearthwormexposedtocopper(Gibbetal.,1997).Sincethen,there havebeendetailedcharacterizationsofthemodeofactionofindividualcontaminants intargetspeciesusingadvancedanalyticaltechniquessuchas2Dgaschromatography coupledtotime-of-flight-massspectrometry(TOF-MS)(Liuetal.,2018)and2D 1H-13CHSQCNMR(Yuketal.,2013 ).Metabolomicstechniquesarenowbeingused todetectmolecular-levelchangesintargetorganismsfromexposureto environmentallyrelevantconcentrationsofcontaminantsintheng/Ltolow mg/L