CurrentDevelopments inBiotechnology andBioengineering
FilamentousFungiBiorefinery
Editedby
MohammadJ.Taherzadeh
SwedishCentreforResourceRecovery,UniversityofBora ˚ s,Bora ˚ s,Sweden
JorgeA.Ferreira
SwedishCentreforResourceRecovery,UniversityofBora ˚ s,Bora ˚ s,Sweden
AshokPandey
CentreforInnovationandTranslationalResearch,CSIR-IndianInstitute ofToxicologyResearch,Lucknow,India SustainabilityCluster,SchoolofEngineering,UniversityofPetroleumand EnergyStudies,Dehradun,India
Elsevier
Radarweg29,POBox211,1000AEAmsterdam,Netherlands
TheBoulevard,LangfordLane,Kidlington,OxfordOX51GB,UnitedKingdom 50HampshireStreet,5thFloor,Cambridge,MA02139,UnitedStates
Copyright©2023ElsevierInc.Allrightsreserved.
Nopartofthispublicationmaybereproducedortransmittedinanyformorbyanymeans,electronicor mechanical,includingphotocopying,recording,oranyinformationstorageandretrievalsystem,without permissioninwritingfromthepublisher.Detailsonhowtoseekpermission,furtherinformationaboutthe Publisher’spermissionspoliciesandourarrangementswithorganizationssuchastheCopyrightClearance CenterandtheCopyrightLicensingAgency,canbefoundatourwebsite: www.elsevier.com/permissions
ThisbookandtheindividualcontributionscontainedinitareprotectedundercopyrightbythePublisher (otherthanasmaybenotedherein).
Notices
Knowledgeandbestpracticeinthisfieldareconstantlychanging.Asnewresearchandexperiencebroadenour understanding,changesinresearchmethods,professionalpractices,ormedicaltreatmentmaybecome necessary.
Practitionersandresearchersmustalwaysrelyontheirownexperienceandknowledgeinevaluatingandusing anyinformation,methods,compounds,orexperimentsdescribedherein.Inusingsuchinformationormethods theyshouldbemindfuloftheirownsafetyandthesafetyofothers,includingpartiesforwhomtheyhavea professionalresponsibility.
Tothefullestextentofthelaw,neitherthePublishernortheauthors,contributors,oreditors,assumeanyliability foranyinjuryand/ordamagetopersonsorpropertyasamatterofproductsliability,negligenceorotherwise,or fromanyuseoroperationofanymethods,products,instructions,orideascontainedinthematerialherein.
ISBN:978-0-323-91872-5
ForinformationonallElsevierpublications visitourwebsiteat https://www.elsevier.com/books-and-journals
Publisher: SusanDennis
EditorialProjectManager: HelenaBeauchamp
ProductionProjectManager: KiruthikaGovindaraju
CoverDesigner: MatthewLimbert
TypesetbySTRAIVE,India
Contributors
RuchiAgrawal TheEnergyandResourcesInstitute,TERIGram,GwalPahari,Haryana, India
HamidAmiri DepartmentofBiotechnology,FacultyofBiologicalScienceand Technology;EnvironmentalResearchInstitute,UniversityofIsfahan,Isfahan,Iran
K.Amulya BioengineeringandEnvironmentalSciences,DepartmentofEnergyand EnvironmentalEngineering,CSIR-IndianInstituteofChemicalTechnology,Hyderabad, India
ElisabetAranda InstituteofWaterResearch;DepartmentofMicrobiology,Universityof Granada,Granada,Spain
MohammadtaghiAsadollahzadeh SwedishCentreforResourceRecovery,Universityof Bora ˚ s,Bora ˚ s,Sweden
AparnaBanerjee Centrodeinvestigacio ´ nenEstudiosAvanzadosdelMaule(CIEAM), Vicerrectorı´adeInvestigacio ´ nYPosgrado,UniversidadCato ´ licadelMaule,Talca, Chile
ParameswaranBinod MicrobialProcessesandTechnologyDivision,CSIR-National InstituteforInterdisciplinaryScienceandTechnology(CSIR-NIIST), Thiruvananthapuram,Kerala,India
KamalpreetKaurBrar DepartmentofCivilEngineering,LassondeSchoolofEngineering, YorkUniversity,Toronto,ON;IndustrialWasteTechnologyCenter,AbitibiTemiscamingue, QC,Canada
G € ulruBulkan SwedishCentreforResourceRecovery,UniversityofBora ˚ s,Bora ˚ s, Sweden
GustavoCabrera-Barjas UniversidaddeConcepcio ´ n,UnidaddeDesarrolloTecnolo ´ gico (UDT),Coronel,Chile
MartaCebria ´ n AZTI,FoodResearch,BasqueResearchandTechnologyAlliance(BRTA), ParqueTecnolo ´ gicodeBizkaia,Derio,Bizkaia,Spain
Chiu-WenChen DepartmentofMarineEnvironmentalEngineering,NationalKaohsiung UniversityofScienceandTechnology,KaohsiungCity,Taiwan
EduardoCoelho CEB—CentreofBiologicalEngineering,UniversityofMinho,Braga, Portugal
GabrielaA ´ ngelesdePaz InstituteofWaterResearch,UniversityofGranada,Granada, Spain
CedricDelattre UniversiteClermontAuvergne,ClermontAuvergneINP,CNRS,Institut Pascal,Clermont-Ferrand;InstitutUniversitairedeFrance(IUF),Paris,France
RatihDewanti-Hariyadi DepartmentofFoodScienceandTechnology,IPBUniversity, Bogor,Indonesia
Lucı´liaDomingues CEB—CentreofBiologicalEngineering,UniversityofMinho,Braga, Portugal
Cheng-DiDong DepartmentofMarineEnvironmentalEngineering,NationalKaohsiung UniversityofScienceandTechnology,KaohsiungCity,Taiwan
PascalDubessay UniversiteClermontAuvergne,ClermontAuvergneINP,CNRS,Institut Pascal,Clermont-Ferrand,France
LaurentDufosse ChemistryandBiotechnologyofNaturalProducts(CHEMBIOPRO), UniversityofReunionIsland,ESIROIFoodScience,Saint-DenisCedex9,Reunion Island,France
JorgeA.Ferreira SwedishCentreforResourceRecovery,UniversityofBora ˚ s,Bora ˚ s, Sweden
ShakuntalaGhorai DepartmentofMicrobiology,RaidighiCollege,Raidighi,India
DanielG.Gomes CEB—CentreofBiologicalEngineering,UniversityofMinho,Braga, Portugal
ShararehHarirchi SwedishCentreforResourceRecovery,UniversityofBora ˚ s,Bora ˚ s, Sweden;DepartmentofCellandMolecularBiology&Microbiology,Facultyof BiologicalScienceandTechnology,UniversityofIsfahan,Isfahan,Iran
JoneIbarruri AZTI,FoodResearch,BasqueResearchandTechnologyAlliance(BRTA), ParqueTecnolo ´ gicodeBizkaia,Derio,Bizkaia,Spain
SajjadKarimi SwedishCentreforResourceRecovery,UniversityofBora ˚ s,Bora ˚ s,Sweden
VinodKumar SchoolofWater,Energy,andEnvironment,CranfieldUniversity,Cranfield, UnitedKingdom
PatrikRolandLennartsson SwedishCentreforResourceRecovery,UniversityofBora ˚ s, Bora ˚ s,Sweden
HanifahNuryaniLioe DepartmentofFoodScienceandTechnology,IPBUniversity, Bogor,Indonesia
SaraMagdouli DepartmentofCivilEngineering,LassondeSchoolofEngineering,York University,Toronto,ON;IndustrialWasteTechnologyCenter,AbitibiTemiscamingue, QC,Canada
Manikharda DepartmentofFoodandAgriculturalProductTechnology,Universitas GadjahMada,Yogyakarta,Indonesia
PhilippeMichaud UniversiteClermontAuvergne,ClermontAuvergneINP,CNRS,Institut Pascal,Clermont-Ferrand,France
MarziehMohammadi SwedishCentreforResourceRecovery,UniversityofBora ˚ s,Bora ˚ s, Sweden
RafaelLeo ´ nMorcillo InstituteofWaterResearch,UniversityofGranada,Granada, Spain
SoumyaMukherjee UniversityofToledo,Toledo,OH,UnitedStates
VivekNarisetty SchoolofWater,Energy,andEnvironment,CranfieldUniversity, Cranfield,UnitedKingdom
SeyyedVahidNiknezhad BurnandWoundHealingResearchCenter;Pharmaceutical SciencesResearchCenter,ShirazUniversityofMedicalSciences,Shiraz,Iran
AshokPandey CentreforInnovationandTranslationalResearch,CSIR-IndianInstitute ofToxicologyResearch,Lucknow;SustainabilityCluster,SchoolofEngineering, UniversityofPetroleumandEnergyStudies,Dehradun,India
MohsenParchami SwedishCentreforResourceRecovery,UniversityofBora ˚ s,Bora ˚ s, Sweden
AnilKumarPatel DepartmentofMarineEnvironmentalEngineering;InstituteofAquatic ScienceandTechnology,NationalKaohsiungUniversityofScienceandTechnology, KaohsiungCity,Taiwan
GuillaumePierre UniversiteClermontAuvergne,ClermontAuvergneINP,CNRS,Institut Pascal,Clermont-Ferrand,France
EndangSutriswatiRahayu DepartmentofFoodandAgriculturalProductTechnology, UniversitasGadjahMada,Yogyakarta,Indonesia
G.Renuka DepartmentofMicrobiology,PingleGovernmentDegreeCollegeforWomen, Warangal,India
SaddysRodriguez-Llamazares CentrodeInvestigacio ´ ndePolı´merosAvanzados(CIPA), EdificioLaboratorioCIPA,Concepcio ´ n,Chile
NedaRousta SwedishCentreforResourceRecovery,UniversityofBora ˚ s,Bora ˚ s, Sweden
TanerSar SwedishCentreforResourceRecovery,UniversityofBora ˚ s,Bora ˚ s,Sweden
BehzadSatari DepartmentofFoodTechnology,CollegeofAburaihan,Universityof Tehran,Tehran,Iran
UlisesConejoSaucedo InstituteofWaterResearch,UniversityofGranada,Granada, Spain
ZohresadatShahryari SwedishCentreforResourceRecovery,UniversityofBora ˚ s,Bora ˚ s, Sweden;AvidzymeCompany,Shiraz,Iran
PoojaSharma EnvironmentalResearchInstitute,NationalUniversityofSingapore; EnergyandEnvironmentalSustainabilityforMegacities(E2S2)PhaseII,Campusfor ResearchExcellenceandTechnologicalEnterprise(CREATE),Singapore,Singapore
RuiSilva CEB—CentreofBiologicalEngineering,UniversityofMinho,Braga,Portugal
RaveendranSindhu MicrobialProcessesandTechnologyDivision,CSIR-National InstituteforInterdisciplinaryScienceandTechnology(CSIR-NIIST), Thiruvananthapuram,Kerala,India
ReetaRaniSinghania DepartmentofMarineEnvironmentalEngineering,National KaohsiungUniversityofScienceandTechnology,KaohsiungCity,Taiwan
MohammadJ.Taherzadeh SwedishCentreforResourceRecovery,UniversityofBora ˚ s, Bora ˚ s,Sweden
JoseA.Teixeira CEB—CentreofBiologicalEngineering,UniversityofMinho,Braga, Portugal
SunitaVarjani GujaratPollutionControlBoard,Gandhinagar,Gujarat,India
S.VenkataMohan BioengineeringandEnvironmentalSciences,DepartmentofEnergy andEnvironmentalEngineering,CSIR-IndianInstituteofChemicalTechnology, Hyderabad,India
RachmaWikandari DepartmentofFoodandAgriculturalProductTechnology, UniversitasGadjahMada,Yogyakarta,Indonesia
AkramZamani SwedishCentreforResourceRecovery,UniversityofBora ˚ s,Bora ˚ s,Sweden
Preface
AdvancesinFilamentousFungiBiorefinery isabookintheElsevierserieson Current DevelopmentsinBiotechnologyandBioengineering (Editor-in-Chief:AshokPandey).This bookexploresvariousfundamentalandindustrialaspectsoffilamentousfungiforthe manufactureofdifferentproductsusedinoursociety.
Fungiarepartoftheecosystem,andtheworldwouldlooktotallydifferentwithout them.Mostpeopleknowonlymushroomsandtheirfruitingbodiesasfungi.However, themainfungalbiomassistheirfilaments,whichcanhavemanyapplications.Filamentousfungicangrowonalargevarietyofmaterialsthatcontaincarbohydrates,proteins, fats,etc.,bydegradingthemacromoleculesandthenassimilatingthemonomerstogrow andproducevariousenzymesandmetabolites.Thismeansthattherearealargenumber ofsubstratesonwhichtogrowfungi,fromagriculturalandforestresidualstoindustrial residualsandproducts,tohouseholdwastesandwastewaters.Dependingontheecosystemandtheenvironmentalorcultivationconditions,fungicangrowinvariousmorphologies,andmanyfungalstrainsaredimorphic,meaningthattheycangrowbothlikeyeast andfilaments.Inaddition,theycangrowinvariousenvironmentalconditions,suchas aerobicoranaerobic.Theyadapttheirenzymemachineryasrequiredtotheseconditions, producingavarietyofmetabolitesthatarenecessaryforthefungitogrow.
Asfungigrowonalargenumberofsubstrates,theycanproducevariousextracellular enzymessuchashydrolyticenzymestodegradebiopolymers.Therefore,fungiarean industrialsourceofenzymeproduction.
Ultimately,weshouldnotforgetthattheonlygoaloffungiistogrow.However,incertainconditionstheycanproduce,forexample,enzymesand/orvariousmetabolites, whichcanbeusedasproducts.Bearinginmindthesinglegoaloffungi,oneoftheirmajor productsisalwaysfungalbiomassormycelium.Thisbiomassnormallycontainsprotein, fat,andotherbiopolymerssuchaschitosanorbeta-glucaninitscellwall,andavarietyof bioactivecompounds.Asaresult,thebiomassofmanyfilamentousfungicanbeagood sourceforfoodandfeed.Someofthesefungi,particularlyamongthezygomycetesand ascomycetes,areedibleandcanbeusedfordifferentfoodpreparationssuchastempeh, oncom,andkoji.However,thereisalsoaninterestnowadaysindevelopingnewfoodand feedsuchasfishfeedfromfungiasanenvironmentallyfriendlyandhealthyalternativeto meat,chicken,orevensoy-basedvegetarianproducts,forexample.However,assome fungiproducemycotoxins,thefungalstrainandtheprocessconditionsshouldbechosen carefullyinordertoavoidanyriskstohumansoranimals.
Theaimofthisbookistoexplorecomprehensivelytheadvancesinusingfilamentous fungiasthecoreofindustrialbiorefinery.Thebookprovidesathoroughoverviewand
understandingoffungalbiology,biotechnology,andecosystems,andcoversavarietyof industrialproductsthatcanbedevelopedfromfungi.
Thecontentsofthisbookareorganizedin18chapterstocover:(a)thefungalecosystem,biology,andbiotechnology;(b)thefungalgrowthandprocessinsolidstateandsubmergedfermentation,particularaspectsofbioreactors,andsampling,preservation,and processmonitoring;(c)mycotoxinsasanimportantaspecttochoosethefungalstrainsfor processes;(d)productsofbiorefinerythatthefungiincludingfood,feed,organicacids, alcohols,bioactivepigmentedcompounds,andantibiotics;and(e)fungiinnovel processes.
Wearegratefultotheauthorsforcompilingthepertinentinformationintheirchapters,whichwebelievewillbeavaluableresourceforboththescientificcommunityand readersingeneral.Wearealsogratefultotheexpertreviewersfortheirusefulcommentsandscientificinsights,whichhelpedshapethebook’sorganizationandwhich improvedthescientificdiscussionsandoverallqualityofthechapters.TheEditors (MohammadJ.TaherzadehandJorgeA.Ferreira)acknowledgesupportfromthe SwedishAgencyforEconomicandRegionalGrowth(Tillvaxtverket)throughaEuropean RegionalDevelopmentFund“Ways2Tastes.”Finally,oursincerethanksgotothestaff atElsevier,includingDr.KostasMarinakis(formerSeniorBookAcquisitionEditor), Dr.KatieHammon(SeniorBookAcquisitionEditor),andBernadineA.Miralles(Editorial ProjectManager),andtheentireElsevierproductionteamfortheirsupportinpublishing thisbook. Editors
MohammadJ.Taherzadeh
Worldoffungiandfungal ecosystems
GabrielaA ´ ngelesdePaza,UlisesConejoSaucedoa, RafaelLeo ´ nMorcilloa,#,andElisabetArandaa,b
a INSTITUTEOFWATERRESE ARCH,UNIVERSITYOFG RANADA,GRANADA,SPAIN
b DEPARTMENTOFMICROBIO LOGY,UNIVERSITYOFG RANADA,GRANADA,SPAIN
1.Introduction
Fungi,belongingtoEukarya,arehighlydiverseandlessexplored.Theyarecosmopolitan andplayimportantecologicalrolesassaprotrophs,mutualists,symbionts,parasites,or hyperparasites.Advancesinmolecularphylogenyhaveallowedtoclarifythecomplex relationshipsofanamorphicfungi(fungiimperfecti)andtoplacesomeofthemoutside thefungi.Exitingprogresshavebeenmadeindevelopingfungiformodernandpostmodernbiotechnology,suchasobtainingenzymes,alcohols,organicacids,pharmaceuticals, orrecombinantdeoxyribonucleicacid(DNA).Filamentousfungiandyeastsareextensivelyusedasefficientcellfactoriesintheproductionofbioactivesubstancesandmetabolitesorfornativeorheterologousproteinexpression.Thisisduetotheirmetabolic diversity,secretionefficiency,high-productioncapacity,andcapabilityofcarryingout post-translationalproteinmodifications.Thecommercialexploitationoffungihasbeen reportedformultipleindustrialsectors,suchasthoseinvolvedintheproductionofantibiotics,simpleorganiccompounds(citricacids),fungicidesorfoodandbeverages.
2.Fungalmorphology
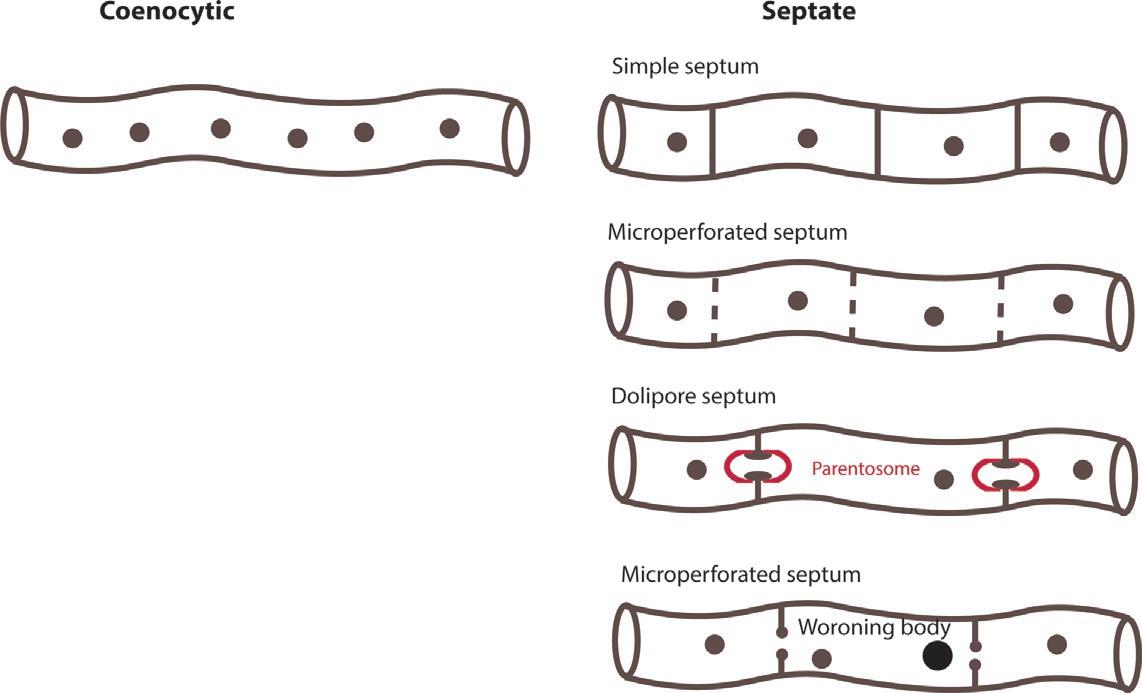
Fungicancolonizenewplacesbygrowingasasystemofbranchingtubes,knownas hyphae,whoseaggregatesformthe mycelium (filamentousfungi).Myceliumcanbefound inthesubstrateswherethefungigrowthorbelowgroundandplayanimportantrolein obtainingnutrientsforgrowthanddevelopment.Thehyphaearecharacterizedbythepresenceorabsenceofsepta,cross-wallsthataredistinctiveamongdifferenttaxonomicgroups. TheyareabsentinOomycotaandZygomycota,knownas coenocytichyphae (koinos ¼ shared,kytos ¼ ahollowvessel).ThepresenceofseptaisacommonfeatureofBasidiomycotaandAscomycota,inwhichtheexchangeofcytoplasmororganellesisensuredbyseptal
#Currentaffiliation:InstituteforMediterraneanandSubtropicalHorticulture“LaMayora”(IHSM),CSICUMA,CampusdeTeatinos,Ma ´ laga,Spain.
CurrentDevelopmentsinBiotechnologyandBioengineering. https://doi.org/10.1016/B978-0-323-91872-5.00010-7
Copyright © 2023ElsevierInc.Allrightsreserved.
pores.Theseporescanbesimpleordolipores,poreswithadistinctivemorphologythat haveabarrel-shapedswellingthatsurroundsthecentralpore(Fig.1).Undersenescence processes,differentiationorsimplyundermechanicalbreakingoff,differentorganelles actasseptalporeplugs,preventingthedetrimentaleffectoftraumasenescenceorpermittingdifferentiationprocesses.TheyincludeWoroninbodies,hexagonalcrystals,elongated crystallinebodies,nuclei,mitochondria,ordenovodepositionofpluggingmaterial (Markham,1994).Theseptumrepresentsaspecializedstructureforcelldivision.
Notallfungigrowashyphae;someoccurasyeasts(yeast-likefungi).Theyusually growonsurfaceswherepenetrationisnotrequired(suchasthedigestivetract).Suchfungi haveattractedtheattentionofbiotechnologistsbecausetheygrowrapidlyandcaneasily bemanipulated.Otherfungicanswitchbetweenyeast-likefungiandfilamentousfungi; theyareknownas dimorphicfungi andincludesomepathogenssuchastheplantpathogen Ustilagomaydis orthehumanpathogen Candidaalbicans.Thisattributeishighly importantasamodelofdifferentiationineukaryoticorganisms(Bosscheetal.,1993). Dimorphismisacommontreatinpathogenicfungi(animalsandplantspathogen)and usuallyisregulatedbydifferentfactorssuchastemperature,glucose,pH,nitrogensource, carbondioxidelevels,chelatingagents,transitionmetalsandinoculumsizeorinitialcell density(Romano,1966).However,anumberoffungiwithunknownpathogenicactivity haveimportantindustrialapplicationssuchastheproductionofchitosan,chitinfrom Saccharomyces,bioremediationprocessby Yarrowia,ethanolorenzymeproduction (Doiphodeetal.,2009).Thisfactisofspecialinterestinindustry,since(i)itispossible toovercometheoperationalproblemsgeneratedduringhyphalgrowthinbioreactors, (ii)morphologycouldbeanindicatorofbiotechnologicalprocess(enzymessecretion orproliferationvspenetration),or(iii)canbeanadvantageforbiocontrolformulations (Doiphodeetal.,2009).
Fromapointofviewofbiotechnology,themorphologyoffungihasanimportant implicationsincetheadaptationofthecultivationsystemmustbeoptimized.
FIG.1 Septumtypesinfungalhyphae.
Filamentousfungicangrowthasdispersemycelialorpellets,dependingonthemechanicalconditionsofthecultivation.Industrialcultivationprocesseswithfungihavebeen optimizedoverdecadestoincreaseproductivity( WalkerandWhite,2017).
Thefungal cellwall isadynamicandcomplexstructureandusuallybasedonglucans andchitin(Ruiz-Herrera,1991).However,thechemicalcompositionvariesamongdifferenttaxonomicgroups,withimplicationsforbiotechnologicalprocessessincedifferent enzymaticactivitiesoccurinthecellwall.Thebalancebetweenwallsynthesisandlysis couldinfluencehyphalmorphologyandcellgrowth,withimpactsontheretentionof chemicalcompoundsthroughbio-adsorptionprocesses,enablingthesuccessfulindustrialfermentationoffilamentousfungi.Inaddition,somecomponentsofthecellwall, suchaschitosanandchitinareconsideredhigh-valueproductsfortheiruseinbiomedicine,agriculture,papermaking,foodindustry,andtextileindustrythatcanbeeasily extractedusingdifferenttechnologies(Nweetal.,2011; Table1).
2.1Generalaspectsofreproduction
Fungicanreproducesexuallyorasexually(vegetativereproduction).Thesetworeproductionmodesdifferaccordingtothefungalmorphologyandtaxonomicgroup(yeast,filamentous,ordimorphicfungi)(Hawker,2016).Inyeasts,themostfrequentmodeof vegetativereproductionisbudding,whichhasbeenstudiedindetailfor Saccharomyces cerevisiae.Thismodecanbemultilateral,bipolar,unipolar,ormonopolarbudding.However,fissionbyformingaseptumcanalsooccurinyeastssuchas Schizosaccharomyces pombe (hostforheterologousexpression).Thisfissioncanbebinaryorabudfission, inwhichacross-wallatthebaseofthebudseparatesbothcells.Ballistoconidiogenesis isaspecificvegetativereproductioninspeciessuchas Bullera (β-galactosidase)or Sporobolomyces (carotenoidsandfattyacids).Pseudomyceliaaretypicalindimorphicspecies inwhichasinglefilamentisproducedwhencellsfailtoseparateafterbuddingorfission ( WalkerandWhite,2017).Infilamentousfungi,suchasbasidiomycetes,thisvegetative reproductioncanoccurbyfragmentationofthehyphae;insomeascomycetes,theformationofmitoticsporeshasbeenobserved.Sexualreproductionisacomplexmechanism anddiffersaccordingtothetaxonomicgroups;itinvolvestheformationofameioticspore byplanogameticcopulation,gametangialcontact,gametangialcopulation,spermatization,andsomatogamia.
Table1 Percentagesofdryweightofthetotalcellwallfractionofthemain components(chitin,cellulose,glucans,protein,andlipids)indifferentgroupsoffungi.
GroupChitinCelluloseGlucansProteinLipids
Oomycota0256542
Chytridiomycota5801610n/a Zygomycota904468
Ascomycota1–39029–607–136–8
Basidiomycota5–33050–812–10n/a n/a(Datanotavailable).
DataadaptedfromRuiz-Herrera,J.,Ortiz-Castellanos,L.,2019.Cellwallglucansoffungi.Areview.CellSurf.5,100022.
2.2Fungalnutrition
Filamentousfungiandyeastsarechemo-organotrophmicroorganismswithrelativelysimplenutritionalneeds.Fungiareheterotrophicorganismssincetheylackphotosynthetic pigments.Mostfungiareaerobes,butwecanfindrepresentativesofobligateanaerobes (Neocallimastix )orfacultativeanaerobes(Blastocladia).Inaerobicrespiration,theterminalelectronacceptorisoxygen;however,basedonoxygenavailability,wecanfindobligate fermentativeorfacultativefermentativefungi,includingCrabtree-positive(Saccharomyces cerevisiae),Crabtree-negative(Candidautilis),non-fermentative(Phycomyces,Rhodotorularubra)orobligateaerobes(mostfungi)( WalkerandWhite,2017).
2.2.1Nutrientuptake
Filamentousfungiandfewyeastspeciesobtaintheirnutrientsviaextracellularenzymes; theyabsorbsmallermoleculesproducedafterextracellulardigestion.Theseenzymescan bewall-bound-enzymesormaydiffuseexternallyintotheenvironment,dependingonthe lifestyleofthefungus(Section3).Nutrientdistributionthroughthehyphaemightoccur bypassive(diffusion-driven)oractivetranslocation(metabolicallydriven)throughthe protoplasm(OlssonandGray,1998; Perssonetal.,2000).Inbothcases,nutrienttranslocationallowsfilamentousfungitogrowthinhabitatswherethespatialdistributionof nutrientsandmineralsisirregularandvariable,includingenvironmentswithlownutrient concentrationsorpollutedareas,byexploitingtheresourcesavailableinotherpartsofthe mycelium(Boswelletal.,2002).
Theenzymaticsysteminfungidependsonthetaxonomicgroup.Someecophysiological artificialgroupshavebeenestablishedbasedonthecapabilitytoproduceenzymes.These groupsincludetheformerligninolyticfungibecauseoftheirabilitytosecreteasetofenzymes involvedinthedegradationoflignin.Theseenzymesplayasignificantroleinbiotechnology sincetheycanbeusedinbiorefineries.Theyincludelipasesproducedbysomeyeasts (Candida, Yarrowialipolytica),hydrolyticenzymessuchasglycosidehydrolases(GHs),polysaccharidelyases(PLs),glycosyltransferases(GTFs),carbohydrateesterases(CEs),lyticpolysaccharidemonooxygenases(LPMOs),non-catalyticcarbohydrate-bindingmodules (CBMs)andenzymeswith“auxiliaryactivities,”includingallenzymesinvolvedinlignocellulosicconversionsuchaslaccases,peroxidases,manganeseperoxidase,ligninperoxidases, versatileperoxidases,DyP-typeperoxidases(Levasseuretal.,2013).TheseenzymesareclassifiedintheCAZymesdatabase,whichdescribesthefamiliesofstructurallyrelatedcatalytic andCBMsofenzymesthatdegrade,modify,orcreateglycosidicbonds(http://www.cazy. org/).Theseenzymesareimportantinbiotechnology,particularlyinbiorefineries,forthe deconstructionofplantbiomassintosimplesugarstoobtainfermentation-basedproducts andfortherecoveryofvalue-addedcompounds(Contesinietal.,2021).
3.Lifestylesoffungi
Fungi,asheterotrophiceukaryoticmicroorganismsandefficientproducersofenzymes, canliveindifferenthabitatsandondifferentorganicsubstrates.Ingeneralterms,fungi
areeither saprophytic—theyfeedonnutrientsfromorganic,non-livingmatterinthesurroundingenvironment-, symbiotic—theyshareamutuallybeneficialrelationshipwith anotherorganism-, parasitic—theyfeedoffalivinghostthatmaysurvive(biotrophs) ordie(necrotrophs)-or hyperparasitic—theyliveattheexpenseofanotherparasites.
Saprotrophicfungi areimportantfortherecyclingofnutrients,especiallyphosphate mineralsandcarbonincorporatedinwoodandotherplanttissues.Theirroleasdecomposersoforganicmatterisfundamental,sincetogetherwithbacteria,theypreventthe accumulationoforganicmatter,ensurethedistributionofnutrientsandplayacrucialrole intheglobalcarboncycleinterrestrialandaquaticecosystems(KjøllerandStruwe,2002; Cebrian,2004; Mooreetal.,2004).Besides,filamentousfungiplayothersignificantrolesin naturalecosystems.Forinstance,interrestrialsystems,fungimaintainthesoilstructure duetotheirfilamentousbranchinggrowthandparticipateinthetransformationofrocks andminerals(Gadd,2008),incorporatingnewelementsintotheecosystemthatmaybe usedbyotherorganisms.
Inadditiontotheimportantroleinnaturalprocesses,andasstatedbefore,thedecompositionoforganicmatterbyfungirepresentsanimportanttraitforbiotechnologicalpurposesduetothepotentialuseofindividualmicrobialstrainsorenzymesfortheuseof renewableresources,suchasplantbiomass.Ingeneralterms,saprotrophicbasidiomycetescandegradeplantlitterandwoodmorerapidlythanotherfungibecauseoftheir highcapacitytodecomposeligninandotherplantpolymers,allowingthemtospreadrapidlyintheenvironment(OsonoandTakeda,2002; Martı´nezetal.,2005; Baldrian,2008). Nevertheless,litterandwooddecompositionisasuccessiveprocess,ofwhichbasidiomycetesandascomycetesgoverndifferentphases(Osono,2007; Vorı´s ˇ kova ´ andBaldrian, 2013).Thiscapacityhasbeenwidelyusedinatindustrialscaleinvariousapplications becauseoftheoxidationofphenolicandnon-phenoliclignin-derivedcompounds.Few examplesarefungallaccases,ligninolyticenzymeswhichdegradecomplexrecalcitrant ligninpolymersandarewidelyusedinthefoodindustry(Minussietal.,2002; MayoloDeloisaetal.,2020),incosmetic,pharmaceutical,andmedicalapplications(Golz-Berner etal.,2004; Niedermeyeretal.,2005; Huetal.,2011; Uedaetal.,2012; Sunetal.,2014),in thepaperandtextileindustry(Bourbonnaisetal.,1995;OzyurtandAtacag,2003; Rodrı´guezCoutoandTocaHerrera,2006;Virketal.,2012)orinthenano-biotechnology (Lietal.,2017; Kumarietal.,2018).Apartfromindustrialapplications,thepotentialcapacityofsaprophyticfungalintra-andextracellularenzymestodegrade/transformcomplex polymers,suchaslignin,isusedinthebiodegradationoforganicxenobioticpollutants. Amongthem,oxidoreductasesrepresentthemostimportantgroupofenzymesusedin xenobioticbioremediationtransformations,includingperoxidases,laccases,andoxygenases,andcancatalyzeoxidativecouplingreactionsusingoxidizingagentstosupport thereactions(Sharmaetal.,2018; Bakeretal.,2019).Theseenzymesareproducedbya widediversityoffungi,whicharesomeofthemostextensivelyfungiusedtodetoxify xenobioticcompounds;theybelongtothebasidiomycetesgroupcalled“whiterotfungi” andincludethegenera Trametes, Pleurotus,and Phanerochaete spp.(Aust,1995; Pointing, 2001; Baldrian,2003; Asifetal.,2017),butalsoascomycetesgenerasuchas Aspergillus or Penicillium (Aranda,2016; Arandaetal.,2017).
Pathogenicandparasiticfungi virtuallyattackallgroupsoforganisms,includingbacteria,otherfungi,plants,andanimals,includinghumans.Accordingtotheirnutritional relationshipwiththehost,parasiticfungicanbedividedintobiotrophicparasites,which obtaintheirsustenancedirectlyfromlivingcells,andnecrotrophicparasites,whichfirst destroytheparasitizedcellandthenabsorbitsnutrients.Besides,fungimightbefacultativeparasites,whicharecapableofgrowinganddevelopingondeadorganicmatter andartificialculturemedia,orobligateparasites,whichcanonlyobtainfoodfromliving protoplasmand,therefore,cannotbeculturedinnon-livingmedia(Brian,1967;Lewis, 1973)Fungipossessthebroadesthostrangespectrumofanygroupofpathogens.For instance,thefilamentousascomycetousfungus Fusariumoxysporum causesvascularwilt onmanydifferentplantspecies(Pietroetal.,2003),butitisalsoresponsibleforcausing life-threateningdisseminatedinfectionsinimmunocompromisedhumans(Boutatiand Anaissie,1997).Nonetheless,therearemanyexamplesoffungalpathogensthatinfect onlyonehost(ShivasandHyde,1997; ZhouandHyde,2001),highlightingthehighhost specificityofdiseasesproducedbycertainfungalinfections.Toexplainthisdualaspectof fungalinfectionspecificity,thepathogenicstrainsofparasiticfungiaredividedinto formaespeciales,definingtheexistenceofdifferentsubgroupswithinspeciesbasedontheir hostspecificity(ArmstrongandArmstrong,1981; Anikster,1984).
Intermsofagriculture,theestimatedcroplossesduetofungaldiseaseswouldbesufficienttofeedapproximately600millionpeopleayear(Fisheretal.,2012).Tocolonize plants,fungisecretehydrolyticenzymes,includingcutinases,cellulases,pectinasesand proteasesthatdegradethesepolymersandpermitfungalentrancethroughtheexternal plantstructuralbarriers.Theseenzymesarealsorequiredforthesaprophyticlifestyle offungi.Asmentionedabove,somefungiarefacultativeparasitesandmayattackplant rootsfromasaprophyticbaseinthesoilthroughthemycelium,progressivelycausingthe deathofthehostandthereafterlivingassaprophytes(ZhouandHyde,2001).Somefungi havedevelopedothermechanismstocolonizeplanthosts,suchasviaspecializedpenetrationorgans,calledappressoria,orviapenetratingthroughwoundsornaturalopenings, suchasstomata(Knogge,1996).
Fungalpathogensareresponsiblefornumerousdiseasesinhumansandfortheextinctionofamphibianandmammalpopulations(Brownetal.,2012; Fisheretal.,2012).For example, Batrachochytriumdendrobatidis,anaquaticchytridfungusthatattackstheskin ofover500speciesofamphibians,and Geomycesdestructans,aascomycetefungusthat attacksnumerousbatspecies,seriouslythreatenthesurvivaloftheseanimalsandmight leadtothedeclineinthepopulationsofotherspecies(Colo ´ n-Gaudetal.,2009; Fisher etal.,2009; Lorchetal.,2011).
Forhumans,fungalinfectionsarerarelylife-threatening;however,superficialfungal infectionsoftheskin,hairandnailsarecommonworldwideandaffectapproximately one-quarterofthehumanpopulation(Schwartz,2004).Airbornepathogenicfungican alsocausedifferentrespiratorydiseasesandalsobelethalinimmunocompromised patients(Mendelletal.,2011).Theinfectionprocessis,generally,similartothatinplants. However,andcontrarytoplantinfections,appressoriahavenotbeendescribedfor
animal-pathogenicfungi,exceptforafewsimilarlyshapedstructuresformedby Candida albicans (Krizniketal.,2005).Insteadtoappressoria,inanimals,fungalpathogensuse othermechanisms,suchasthebindingofspecificreceptorsthatfacilitatetheendocytosis ofhostcellsand,therefore,theirentranceintothelivingtissue( Woods,2003; N € urnberger etal.,2004).Nonetheless,andsimilartofungalinfectionsinplantsandotherfungallifestylessuchassaprophytes,thisprocessmayalsobemediatedbylyticenzymes,suchas proteases,thatdegradethesurfaceofthehostcellsandpermitfungalpenetrationintothe livinghost(N € urnbergeretal.,2004; Schalleretal.,2005).
Aspecialtypeofparasiticfungiisrepresentedby hyperparasites,fungithatliveatthe expenseofanotherparasite,whichishighlycommonamongfungi( JeffriesandYoung, 1994).Hyperparasitismisoftenusedinagricultureforplantprotectionasanalternative tochemicaltreatments(Broz ˇ ova ´ ,2004).Aclassicexampleis Trichodermaharzianum,a fungusextensivelyusedasbiologicalagentagainstawiderangeoffungalparasites.Nonetheless,itisestimatedthat90%ofallfungiusedinplantprotectionproductsbelongtothe genus Trichoderma (Benı´tezetal.,2004).
Fungimayalsoliveas mutualisticsymbionts, associatingwithotherorganismswith benefitsforbothparties.Remarkableexamplesofthesesymbiosesaremycorrhizaeand lichens.Mycorrhizaearethesymbioticassociationofsoilfungiwiththerootsofvascular plants.Generally,fungicolonizeplantrootsandprovidenutrientsandwater,whichare capturedfromthesoilthroughtheexternalhyphalnetwork,whereasplantssupply organicmoleculesderivedfromphotosynthesis,suchassugarsorfattyacids,totheobligatebiotrophicfungi(Harrison,1999; Keymeretal.,2017).Thisrepresentsauniversal symbiosis,notonlybecausealmostallplantspeciesaresusceptibletoformthesymbiosis, butalsobecausesuchsymbiosescanbeestablishedinthemajorityofterrestrialecosystems,evenunderhighlyadverseconditions(Mosseetal.,1981).Moreover,thissymbiosis contributestoglobalcarboncyclesasplanthostsdivertupto20%ofphotosynthatesto thehostfungi(SmithandRead,2010).Therearethreedifferenttypesofmycorrhizae: endomycorrhizae,ectomycorrhizae,andectendomycorrhizae.
Endomycorrhizaearecharacterizedbythepresenceofhyphaeinsidethecellsofthe rootcortex.Itisestimatedthatatleast90%ofallknownvascularplants,(about 300,000species),formthistypeofmycorrhizae.Ontheotherhand,intheectomycorrhizaesymbiosis,thehyphaeofthefungusdonotpenetratethecellsofthecortexoftheroots andformadensehyphalsheath,knownasthemantle,surroundingtherootsurface.Itis believedthatatleast3%ofvascularplantsdevelopthistypeofmycorrhizae,including almostallspeciesofthemostimportantforesttreegenera.Finally,theectendomycorrhizaepresentcharacteristicsofbothendo-andectomycorrhizae,namelyamantlesurroundingtheplantrootsandfungalhyphaethatpenetratetherootcells(Smithand Read,2010).
Mycorrhizalsymbiosisplaysanessentialroleintheestablishmentandfunctioningof terrestrialecosystems,beinginvolvedinnaturalprocessessuchasnutrientcyclingand,in part,inthestructureanddynamicsofpopulationsandplantcommunities(Newman, 1988; Klironomosetal.,2011).Intermsofagriculture,mycorrhizaeimprovethe
productivecapacityofpoorsoils,suchasthoseaffectedbydesertification,salinization, andwinderosion,becauseofthefungalcapacitytoobtainandtranslocatenutrients andwatertothehostplants(George,2000).Inaddition,thesymbiosisenhancessoil aggregationandstructure(MillerandJastrow,2000)andcontributestodefenceagainst diverseplantpathogens(Elsenetal.,2001; Azco ´ n-Aguilaretal.,2002; Garcı´a-Garrido andOcampo,2002)andabioticstresses,suchasdroughtorsalinity(Ruiz-Lozanoetal., 1996; Ruiz-Lozano,2003).
Anotherformofsymbioticassociationoffungiisrepresentedby lichens,whichare compositeorganismscomposedofalgaeorcyanobacteria,called photobionts,living amongfilamentsofafungus,theso-called mycobiont.Thealgae,asanautotrophicorganism,providesthefunguswithorganiccompoundsandoxygenderivedfromitsphotosyntheticactivity,whereasthefungus,asaheterotroph,suppliesthealgaewithcarbon dioxide,mineralsandwater,sincecontrarytoplants,lichenslackvascularorgansto directlycontroltheirwaterhomeostasis(ProctorandTuba,2002; LutzoniandMiadlikowska,2009).Mostofthelichenizedfungalspecies(98%approximately)belongtothe phylumAscomycota,whereasonlyfewordersareinthephylumBasidiomycotaand mitosporicfungi(Hawksworthetal.,1996).Althoughsomelichensinhabitpartially shadedareasandforests(NeitlichandMcCune,1997),mostlichensoftenliveinhighly exposedplacesunderintenselightintensities,suchasdesertsorarcticandalpineecosystems.Forthisreason,themycobiontnormallyproducessecondarycoloredcompounds, called lichencompounds,thatstronglyabsorbUV-Bradiationandpreventdamagetothe algae’sphotosyntheticapparatus(Fahselt,1994).Thesecompoundsareasourceofstructurallydiversegroupsofnaturalproducts,withawiderangeofbiologicalactivitiesincludingantibiotic,analgesic,andantipyreticactivities( Yousufetal.,2014)andhave traditionallybeenusedinthecosmeticanddyeindustryaswellasinfoodandnatural remedies(Oksanen,2006).
Innature,lichensareimportantasearly-stageprimarysuccessionorganisms.For instance,theyarethepioneersinthecolonizationofrockyhabitatsand,afterdying,their organicmattermightbeusedbyotherorganisms(LutzoniandMiadlikowska,2009; Muggia etal.,2016).Aspoikilohydricorganisms,theirwaterstatuspassivelyfollowstheatmospherichumidity(Nash,1996),andtheycantolerateirregularandextendedperiodsof severedesiccation.Thisallowsthemtocolonizehabitsthatcannotbecolonizedbymost plants.Despitethis,manylichensalsogrowasepiphytesonplants,mainlyonthetrunks andbranchesoftrees.However,theyarenotparasitesorpathogenssincetheydonotconsumeorinfecttheholdingplant(Ellis,2012).Inaddition,lichensadsorbandaresensitiveto heavymetalsandpollutants(Garty,2001),makingthemperfectenvironmentalindicators.
4.Taxonomyoffungi
Understandinghowfungihaveadaptedtosomanyecosystems,thewayinwhichthey haveevolved,butaboveall,taxonomicclassification,hasnotbeenaneasytask.Fungi, afterplantsandanimals,areoneofthemostdiverseanddominantgroupsinalmost
allecosystems.Itisestimatedthattherearebetween1.5and5.1millionspecies.However, anewestimationofthenumberoffungirangesbetween500,000andalmost10million (HawksworthandL € ucking,2017),althoughonlyalmost10%havebeenidentifiedsofar (Blackwell,2011;Hibbettetal.,2016)
Inthemiddleofthe18thcentury,thescientistCarlvonLinnaeusimplementedabinomialsystemtoclassifylivingbeings(Systemanaturae).Thissystemisbasedontheclassificationoforganismsaccordingtotheirmorphologicalcharacteristicsandphenotypic traits.Fungiwereconsideredaspartoftheplantkingdom(Linnaeus,1767).Someyears after,Whittakerclassifiedthefungiasanindependentgroup,whichhecalled“truefungi” (Eumycota)( Whittaker,1969).Then,differenttaxonomicclassificationscontinueduntil themiddleofthe19thcentury.Advancesintheclassificationoffungihavealwaysgone handinhandwiththedevelopmentofnewtechnologies,suchaselectronmicroscopy, newbiochemical,andphysiologicalanalysismethods,thestudyofsecondarymetabolites,cellwallcompositionandfattyacidcomposition,aswellasmoleculartechnologies, amongothers(Guarroetal.,1999).
Inthelasttwodecades,thedevelopmentofPCRtechniquesand,later,genome sequencing,hassignificantlycontributedtotheadvanceinfungaltaxonomy.Thispromotedrapidchanges,andtherefore,differentproposalsforthereclassificationoffungi havebeenmade,triplingthenumberofphylafrom4tomorethan12.However,lessthan 5%oftheidentifiedspecieshavebeentaxonomicallyclassified( Jamesetal.,2020).Nextgenerationsequencingtoolshaveallowedfungalgenomesequencing,transcriptomes, andmitochondrialgenomesthatprovidedrelevantinformationforphylogeneticstudies infungi.Additionally,specificregionsofribosomalRNA(rRNA),suchasinternaltranscribedspacers(ITSs),largesubunit(LSU),smallsubunit(SSU)andintergenicspacer ofrDNA,aswellasvariousmarkersincludingtranslationelongationfactor1(TEF1), glycerol-3-phosphatedehydrogenase(GAPDH),histones(H3,H4),calmodulingene, RNApolymeraseIIlargestsubunit(RPB1)genesandmitochondrialgenes(cytochrome c oxidaseIandATPasesubunit6),haveplayedanimportantroleinthedevelopment ofthefungaltaxonomy(Zhangetal.,2017).
Theorganizationofsuchinformation(genomes)bythescientificcommunityhas requireddifferentefforts.Ontheonehand,theY1000+projectaimstosequencethe genomesof1000yeastspecies(https://y1000plus.wei.wisc.edu),andontheotherhand, the1000FungalGenomesproject(http://1000.fungalgenomes.org/home)hastheobjectiveofsequencing1000fungalgenomes.ThedatabaseUNITEisarecentdatabasethat concentratesthesequencesoftheITSribosomalregionoffungiincludedintheInternationalNucleotideSequenceDatabase(http://www.insdc.org/).Thisdatabaseresulted fromthecollaborationofresearchersandtaxonomicspecialistswhocollectedand depositedfungalsequences,specificallywiththepurposeofbuildingadatabasethat registers,analyses,andsharesthisinformationwiththescientificcommunity(Koljalg etal.,2020).
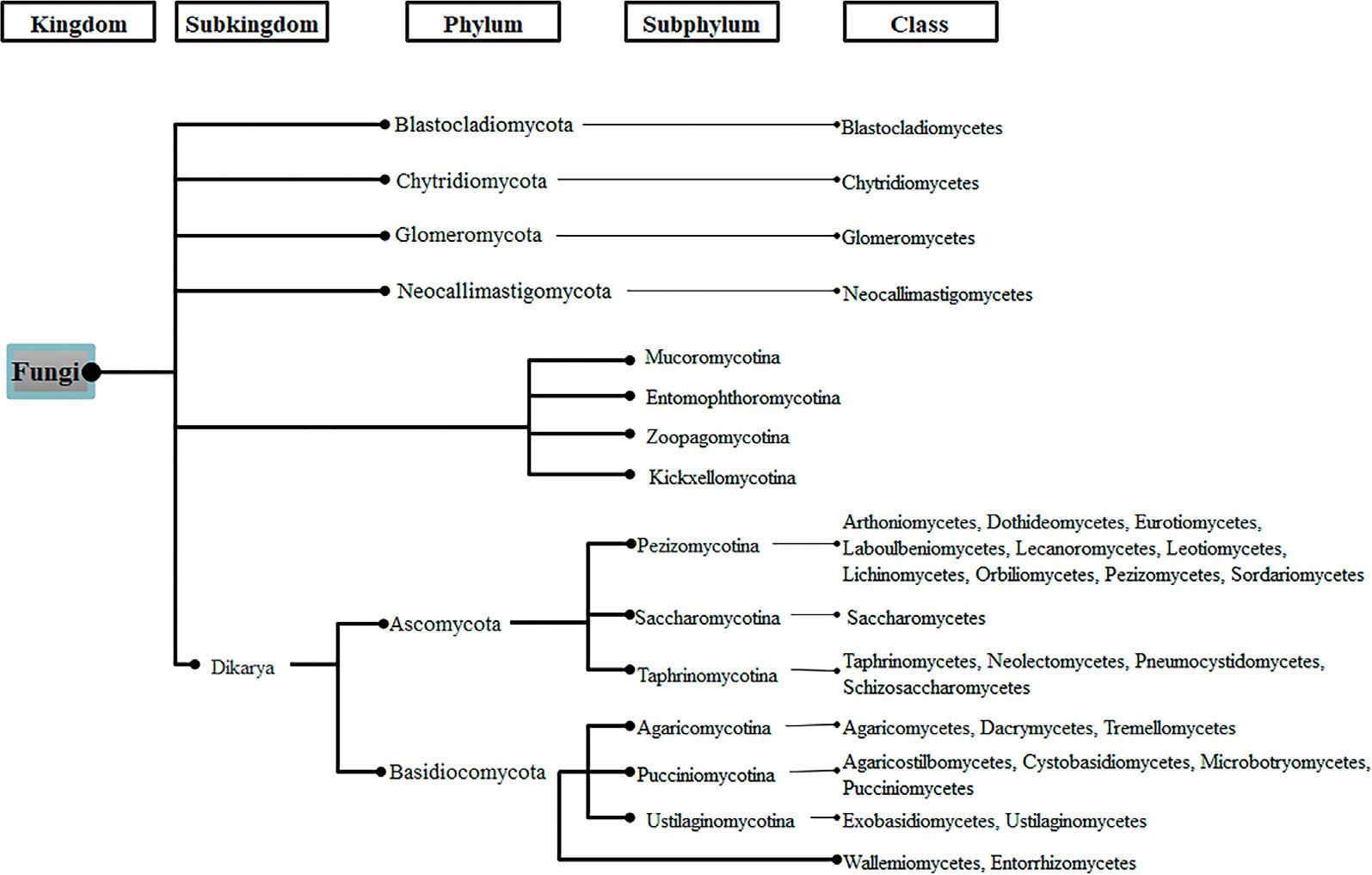
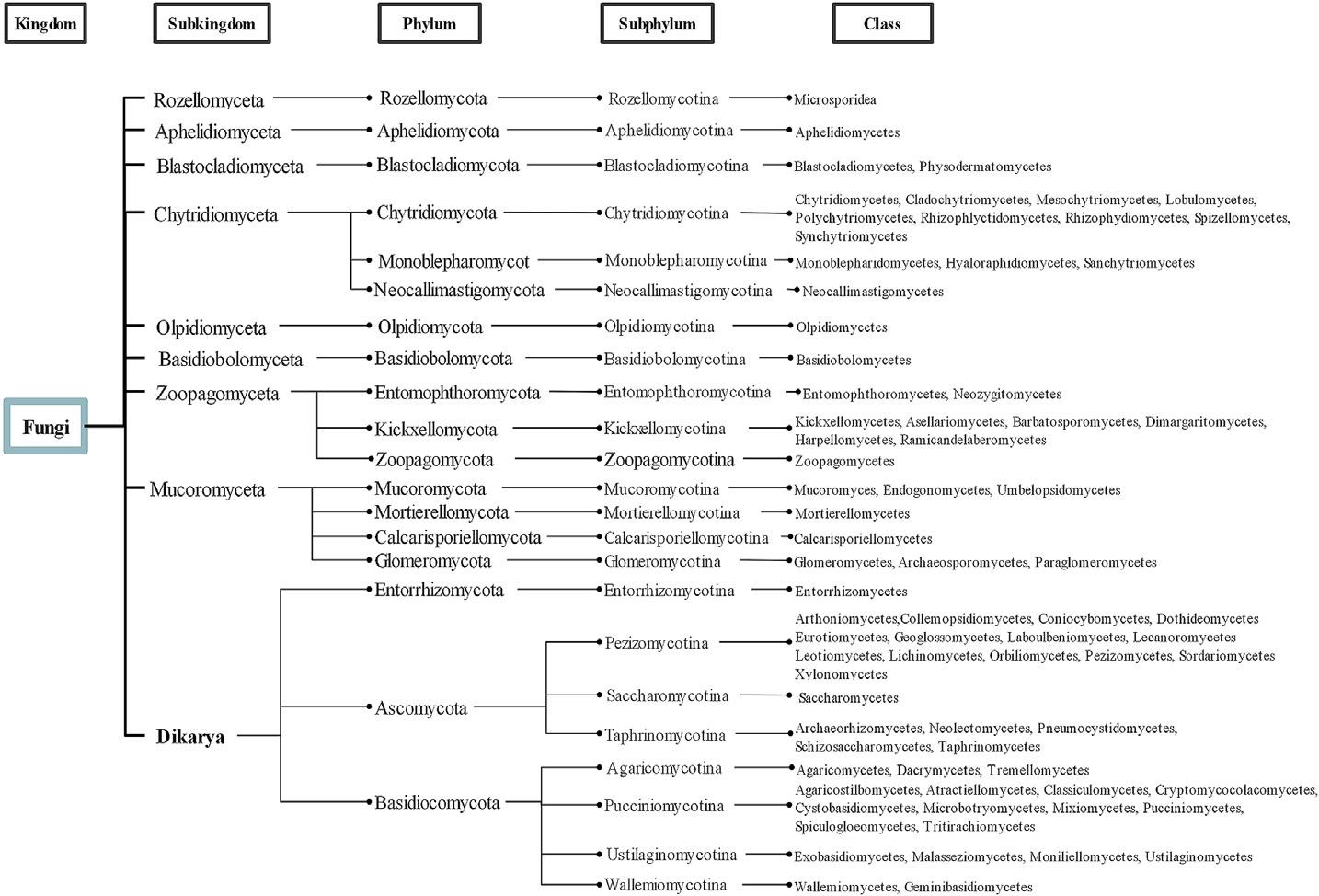
Inthelast14years,thetaxonomyoffungihasbeenundermajorchanges.Thekingdom offungi,proposedby Hibbettetal.(2007),includesonesubkingdom(Dikaria)andseven
Fungalclassificationproposedby Hibbettetal.(2007)
phyla:Blastocladiomycota,Glomeromycota,Chytridiomycota,Neocallimastigomycota, Microsporidia,Ascomycota,andBasidiomycota;foursubphyla,namelyEntomophthoromycotina,Kickxellomycotina,Mucoromycotina,Zoopagomycotina,andatotalof31classes(Hibbettetal.,2007)(Fig.2).Overthelastfewyears,differentapproaches, reclassifications,andupdatesonfungaltaxonomyhavebeenmade(Gryganskyietal., 2012; Hydeetal.,2013; Slippersetal.,2013; Phookamsaketal.,2014; Ariyawansaetal., 2015; Lietal.,2016; Spataforaetal.,2016; Marin-Felixetal.,2017,2019; Reblova ´ etal., 2018; Voglmayretal.,2019; Mitchelletal.,2021).
Tedersooetal.(2018) describedandproposedanupdatedclassificationforthefungal kingdombasedondivergencetimeandphylogeniesofparticulartaxa.Underthispointof view,ninesubkingdomshavebeenproposed(1Rozellomyceta, 2Aphelidiomyceta, 3Blastocladiomyceta, 4Chytridiomyceta, 5Olpidiomyceta, 6Basidiobolomyceta, 7Zoopagomyceta, 8Mucoromycetaand 9Dikarya);eachsubkingdomsdividesintooneormorephyla (18phyla-1Rozellomycota, 2Aphelidiomycota, 3Blastocladiomycota, 4Chytridiomycota, 4Monoblepharomycota, 4Neocallimastigomycota, 5Olpidiomycota, 6Basidiobolomycota, 7Entomophthoromycota, 7Kickxellomycota, 7Zoopagomycota, 8Mucoromycota, 8Mortierellomycota, 8Calcarisporiellomycota, 8Glomeromycota, 9Entorrhizomycota, 9Basidiomycota,and 9Ascomycota).Additionally,eachphylumdividesintooneormoresubphyla
FIG.2
FIG.3 Fungalclassificationproposedby Tedersooetal.(2018).
(20subphyla—1Rozellomycotina, 2Aphelidiomycotina, 3Blastocladiomycotina, 4Chytridiomycotina, 4Monoblepharomycotina, 4Neocallimastigomycotina, 5Olpidiomycotina, 6Basidiobolomycotina, 7Entomophthoromycotina, 7Kickxellomycotina, 7Zoopagomycotina, 8Mucoromycotina, 8Mortierellomycotina, 8Calcarisporiellomycotina, 8Glomeromycotina, 9Entorrhizomycotina, 9Agaricomycotina, 9Pucciniomycotina, 9Ustilaginomycotina, 9Wallemiomycotina, 9Pezizomycotina, 9Taphrinomycotina, 9Saccharomycotina),and76classes areincluded(Fig.3).
Analternativeclassificationwasproposedby Naranjo-OrtizandGabaldo ´ n(2019), basedonninemainlines:Opisthosporidia,Neocallimastigomycota,Blastocladiomycota, Chytridiomycota,Mucoromycota,Glomeromycota,Zoopagomycota,Ascomycota,and Basidiomycota.Themaindifferenceswithrespecttootherproposalscanbefoundin theincorporationofOpisthosporidia.Thisgroupincorporatesthreelineages:Rozellidea, Aphelidea,andMicrosporidia.Inturn,thegroupofChytridiomycotawasdividedinto threeclasses:Monoblepharidomycetes,Hyaloraphidiomycetes,andChytridiomycetes, whereasMucoromycotaincludesthetwosubphylaMucoromycotinaandMortierellomycotina.Additionally,BasidiomycotacomprisedPucciniomycotina,Ustilagomycotina, Agaricomycotina,Wallemiomycotina,andBartheletiomycetes.Finally,Ascomycotacontainsthreemainclades:Taphrinomycotin a,Saccharomycotina,andPezizomycotina. Mostrecently,theclassificationoffungiproposedby Wijayawardeneetal.(2020) hascoincided,formanyclades,withtheproposalof Tedersooetal.(2018).Thesubkingdomtaxonomicrankwasremoved,and16 phylawererecognized:Rozellomycota,
Blastocladiomycota,Aphelidiomycota,Monoblepharomycota,Neocallimastigomycota, Chytridiomycota,Caulochytriomycota,Basidiobolomycota,Olpidiomycota,Entomophthoromycota,Glomeromycota,Zoopagomycota,Mortierellomycota,Mucoromycota,Calcarisporiellomycota,andthreehigherfungi(Dikarya-Entorrhizomycota, Basidiomycota,Ascomycota).Inthisstudy,onlyfoursubphylawereproposed (MucoromycotaMortierellomycota,Entomo phthoromycota,andCalcarisporiellomycota);inthecaseofDycaria(AscomycotaandBasidiomycota),sevensubphylaare described(Fig.4).
Somechangesandadditionshavealsobeendescribed,includingthecreationofthe phylumRozellomycotawhichcontainstheclassesRudimicrosporeaandMicrosporidea. TheorderMetchnikovellidawasmovedtotheclassRudimicrosporea,andthemostsignificantchangesoccurredinthephylaAscomycotaandBasidiomycota;theclassBartheletiomycetes,whichwasincludedinthephylumBasidiomycota,waschangedtothe subphylumAgaricomycotina.Moreover,Agaricomycetes,Dacrymycetes,TremellomycetesandBartheletiomycetesweregroupedtogether.FromtheclassCollemopsidiomycetes,thesubphylumPezizomycotinawaseliminated,andnewclasseswereincluded (CandelariomycetesandXylobotryomycetes).TheorderCollemopsidialeswasmoved totheclassDothideomycetes.IntheclassGeminibasidiomycetes,thesubphylumWallemiomycotinawasexcluded,andonlytheclassWallemiomycetesremained.
FIG.4 Fungalclassificationproposedby Wijayawardeneetal.(2020).
Despitetheseefforts,therearelargenumbersofgenera,ordersandfamiliesthathave notyetbeenclassified.OnlyinthephylaAscomycotaandBasidiomycota,remainnot assignedfamiliesfor876genera( Wijayawardeneetal.,2018).Accordingtoacompilation fromtheRoyalBotanicGardens,inthelast10years,350newfamilieshavebeendescribed, includingPucciniaceaewith5000species,Mycosphaerellaceaewith6400species,CortinariaceaeandAgaricaceaeconsistingof3000species.Ontheotherhand,about30%ofthe newincorporationsarebasidiomycetes,whereas68%areascomycetes.Untilnow,the continuouscontributionofdifferentgroupsofresearchers,whichhasresultedinthe growthofdatabases,andthedevelopmentofnewtechnologiesandmoleculartoolshave helpedtoestablishauniversalclassificationforfungi.
Theorganizationofthisenormousamountofinformationthroughtaxonomyallows tocorrelatethedifferentstylesoflifeaswellasthestructural,genetic,andmetabolic characteristics,which,asmentionedhere,areusedtoclassifyfungi.Thistaxonomicpanoramaallowsustoshowthegreatdiversityofspeciesthatexistonearth,studytheirevolutionand,insomecases,takeadvantageofcertainmetabolicfunctionsthatcanbe appliedindifferentindustrialprocesses;mostofthefungiusedbelongtothephylaAscomycotaandBasidiomycota.Thesefungiplayanimportantroleintheproductionofvariousproductsorintermediatesinthegenerationofbioethanol(fungifromthesubphyla Agaricomycotina,Pezizomycotina,andMucoromycotina),biodiesel(fungifromthesubphylaPezizomycotina,Ustilaginomycotina,andSaccharomycotina),biogas(fungifrom thesubphylaMucoromycotina,Agaricomycotina,Pucciniomycotina,Pezizomycotina, Saccharomycotina),thepre-treatmentoflignocellulosicbiomass(fungifromthesubphylaBasidiomycotinaandclassAgaricomycetes),applicationsinthepulpandpaper industry(Agaricomycotina,Pezizomycotina),xylitolproduction(Saccharomycotina), andlacticacidproduction(Mucoromycotina,Saccharomycotina),amongothers (Kumarietal.,2018).
5.Fungaldiversity
Anincreasingnumberofstudiesareaddressingfungaldiversity.Thetotalrichnessand diversityoffungaltaxaacrossthestudiespublishedmainlyinvolveAscomycota(56.8% ofthetaxa)andBasidiomycota(36.7%ofthetaxa),withatotalfungaldiversityofaround 6.28milliontaxa.Thesestudiosrepresentaconservativeestimateofglobalfungalspecies richness(Baldrianetal.,2021).Inthelargeststudyoffungaldiversity,around45,000operationaltaxonomicunits(OTUs)wererecoveredfrom365sitesworldwide,using1.4millionITSsequences( Tedersooetal.,2014).One-thirdofthetotalOTUsshowed97%of similaritycomparedwithothersreportedinpublicdatabases;thereby, 30,000new anddifferentOTUshavebeendetected.Theseresultsgreatlycontributedtothediscovery ofnewfungalspecies.
ThesubkingdomDikarya(AscomycetesandBasidiomycetes)containsmostofthefungaldiversityonearthintermsofdescribedspecies,butissmallcomparedtothesizeofthe totalfungalkingdom.Recently,moretaxahavebeendescribed,suchasCryptomycota
( Jonesetal.,2011;Laraetal.,2010),achytridgroup,Archaeorhizomycetesandothersoil ascomycetegroups(Porteretal.,2008;Roslingetal.,2003;SchadtandRosling,2015).Furthermore,around150generahavebeenestimatedasbeingapartofthefungalgroup calledMicrosporidia,with1200–1300species(Leeetal.,2009);theactualfiguresarepresumablyhigherthanthefiguresforthehostdiversity.However,moleculardiversitystudieshavenotbeendetailedenoughtoelucidatethisinformation(Krebesetal.,2010; McClymont,etal.,2005).
ThoseapproximationshavebeenmadepossibleduetoDNAsequencingandthewidespreaduseoftheformalfungalbarcodeNuclearribosomalInternalTranscribedSpacer ITS1–2.Itisrecognizedastheofficialmolecularmarkerofchoicefortheexploration offungaldiversityinenvironmentalsamples(Ko ˜ ljalgetal.,2013)andcountswithavast andup-to-datedatabase,whichisnecessaryfordataanalysis(Schochetal.,2012).Nevertheless,itcannotdifferentiatebetweenallgroupsandcrypticspecies.Therefore,identificationanddiversityanalysisoffungiisstillgreatlychallenging(DeFilippisetal.,2017). Moreover,therelationshipsbetweenfungaldiversityandtheirenvironmentshavenot beencompletelydescribed,whichisalsothecasefortheprocessesandmechanisms involved(Branco,2019).
Asaconsequence,twodisciplineshavebeenrecognizedtounderstandtherelationship betweenfungaldiversityandtheirenvironment,communityecology,andpopulation genetics(Branco,2019).Communityecologystudiesfocusonthespecieslevel,addressing bothecologicalandbiologicalquestions,withahighlevelofaccuracyandreliability.In contrast,populationgeneticsstudieshavedeterminedspeciesassembliesandranges, comprehendingfungalintra-specificvariation,dispersion,andestablishmentandincludingtheidentificationofkeytraitsinfluencingfitness(MittelbachandSchemske,2015).
Crypticspeciesarebiologicalentitiesthathavealreadybeennamedanddescribed; however,theyaremorphologicallydifferent,andmolecularstudiesareneededtoelucidate,detectandenumeratethesedifferencesatthealphadiversitylevel(Bickfordetal., 2007; HortonandBruns,2001; RappeandGiovannoni,2003; Soginetal.,2006).These studieshighlighttheirdiversitypotential,proposingthemeasurementbygeneticdistancestoknowhowhyper-diversetheyareandtodeterminetheirspecies-leveldifferencesinmanymulticellulargroups.Althoughnovelmoleculartoolsandnewmethods ofidentificationhavebeenusedovertheyears,fungaldiversityisbarelyknown.Thisis mainlyduethespecieswithdifferentmorphologicalandecologicalfeatures (Hawksworth,2004).
6.Fungalecosystems
Asmentionedabove,fungiarehighlydiverseandconstituteamajorportionofvarious ecosystemsintermsofbiomass,geneticdiversification,andtotalbiosphereDNA (Bajpaietal.,2019).Theirdistributionisextraordinarilydiverseandshowsbiogeographicalpatternsdependinguponlocalandglobalfactors,suchasclimate,latitude,dispersal limitation,andevolutionaryrelationships(Bajpaietal.,2019).Intermsofdiversity,the
highestalphadiversityoffungihasbeenfoundinsoilsandterrestrialenvironments, mainlyinplantshoots,plantroots,anddeadwood(Baldrianetal.,2021).Theseassociationswithplantsleadustoinferthatfungiplayadominantroleinterrestrial environments.
6.1Terrestrialecosystems
Fungiinterrestrialhabitatsexhibitdifferentpreferencesrelatedtotheedaphiccondition, withahigherdiversityintropicalecosystems.Fungalendemicityisespeciallystrongin suchregions.Nevertheless,thisdistributiondependsonthegroupofthefungiandtheir features;forinstance,ectomycorrhizalfungiandotherclassesaremostdiverseintemperateorborealecosystems.Ingeneral,severaltaxashowacosmopolitandistribution throughouthabitats( Tedersooetal.,2014).
Sequencingstudieshavebeenperformedindifferentterrestrialenvironments,revealingseveralnumbersofnovelsequencesclusteredconservatively.Forexample,inforest soil,fungaldiversityshowedaround830OTUsthatwerenotmatchedtoanyfungaltaxon previouslydescribedwhenblastedagainstNCBI(http://www.ncbi.nlm.nih.gov/)or UNITE(http://unite.ut.ee/) Bueeetal.(2009).Thisanalysisresultedinanestimateddiversityof2240(71.5%)byusingChao1,anon-parametricrichnesstool.Theauthorsalso comparedthesequenceswithacurateddatabaseofrobustlyidentifiedsequencesand foundthat11%ofthetotalsequences,excludingall“unculturedfungi,”remainedunclassifiedandafurther20%belongedtotheunclassifiedDikarya(Bueeetal.,2009).
Fungaldiversityinforestsoilishighlyassociatedwithplants.Theirsymbiosisplaysan importantroleinvegetationdynamics.Moreover,strongrelationandsimilaritiesinfungal alphaandbetadiversitystudies(Hooperetal.,2000; Wardleetal.,2004; GilbertandWebb, 2007)havebeenreportedsince HawksworthandMound(1991) estimatedaround1.5Mof fungalspeciesonlyinthishabitat.Mycorrhizalandsaprotrophicfungiareusuallytheprimaryregulatorsofplant-soilfeedbacksacrossarangeoftemperategrasslandplantspecies;themostabundantfamiliesareParaglomeraceae,Glomeraceae,and Acaulosporaceae,whereasthemostabundantgeneraofsaprotrophicfungiare Mortierella and Clavaria (Semchenkoetal.,2018).
6.2Aquaticecosystems
Aquaticecosystemscomprisethelargestportionofthebiosphereandincludebothfreshwaterandmarineecosystems.Numerousstudiesindifferentaquatichabitatshaveindicatedthatfungiareabundanteukaryotesinaquaticecosystems(Grossartetal.,2019; Money,2016).Theycanreachrelativeabundancesof >50%infreshwaterandabout >1%insalinehabitats(Comeauetal.,2016; Monchyetal.,2011).However,theseresults canbeextremelyvariableanddependontherespectivehabitatanditsenvironmental settings.
Thepredominantfungiinaquatichabitatsaremostlydeterminedbycultivation methods.This,coupledwithtemporaldynamics,spatialconnectivity,andvectorssuch