ConceptsandExperimental ProtocolsofModellingand InformaticsinDrugDesign
OmSilakari
DepartmentofPharmaceuticalSciencesandDrugResearch, PunjabiUniversity,Patiala,India
PankajKumarSingh
DepartmentofChemistryandPharmacy,UniversityofSassari, Sassari,Italy
AcademicPressisanimprintofElsevier 125LondonWall,LondonEC2Y5AS,UnitedKingdom 525BStreet,Suite1650,SanDiego,CA92101,UnitedStates 50HampshireStreet,5thFloor,Cambridge,MA02139,UnitedStates TheBoulevard,LangfordLane,Kidlington,OxfordOX51GB,UnitedKingdom
Copyright©2021ElsevierInc.Allrightsreserved.
Nopartofthispublicationmaybereproducedortransmittedinanyformorbyanymeans,electronicormechanical, includingphotocopying,recording,oranyinformationstorageandretrievalsystem,withoutpermissioninwritingfromthe publisher.Detailsonhowtoseekpermission,furtherinformationaboutthePublisher’spermissionspoliciesandour arrangementswithorganizationssuchastheCopyrightClearanceCenterandtheCopyrightLicensingAgency,canbefound atourwebsite: www.elsevier.com/permissions.
ThisbookandtheindividualcontributionscontainedinitareprotectedundercopyrightbythePublisher(otherthanasmay benotedherein).
Notices
Knowledgeandbestpracticeinthisfieldareconstantlychanging.Asnewresearchandexperiencebroadenour understanding,changesinresearchmethods,professionalpractices,ormedicaltreatmentmaybecomenecessary.
Practitionersandresearchersmustalwaysrelyontheirownexperienceandknowledgeinevaluatingandusingany information,methods,compounds,orexperimentsdescribedherein.Inusingsuchinformationormethodstheyshouldbe mindfuloftheirownsafetyandthesafetyofothers,includingpartiesforwhomtheyhaveaprofessionalresponsibility. Tothefullestextentofthelaw,neitherthePublishernortheauthors,contributors,oreditors,assumeanyliabilityforany injuryand/ordamagetopersonsorpropertyasamatterofproductsliability,negligenceorotherwise,orfromanyuseor operationofanymethods,products,instructions,orideascontainedinthematerialherein.
BritishLibraryCataloguing-in-PublicationData AcataloguerecordforthisbookisavailablefromtheBritishLibrary LibraryofCongressCataloging-in-PublicationData AcatalogrecordforthisbookisavailablefromtheLibraryofCongress
ISBN:978-0-12-820546-4
ForInformationonallAcademicPresspublications visitourwebsiteat https://www.elsevier.com/books-and-journals
Publisher: StacyMasucci
SeniorAcquisitionsEditor: RafaelE.Teixeira
EditorialProjectManager: SamW.Young
ProductionProjectManager: NiranjanBhaskaran
SeniorCoverDesigner: MarkRogers
TypesetbyMPSLimited,Chennai,India
1.2.1Cartesiancoordinate
1.2.2Polarcoordinate
1.5.1VanderWaalssurface(VWS)
1.5.2Solventaccessiblesurface
1.5.3Solventexcludedsurface
1.5.4Chargedpartialsurfacearea(CPSA)
2.2FundamentalprincipleofQSAR.....................................................................31
2.3.1Datapreparation
2.4.1TypesofQSARdescriptors
2.5.1GeneralguidelinesforderivationofHanschQSARmodel
2.8.1Comparativemolecularfieldanalysis(CoMFA)
2.8.2Comparativemolecularsimilarityindicesanalysis,(CoMSIA)
Chapter4:Databaseexploration:Selectionandanalysisof targetproteinstructures .....................................................................89
4.1Introduction........................................................................................................89
4.2Proteindatabases................................................................................................89
4.2.1UniProt:theUniversalProteinknowledgebase .........................................89
4.2.2ResearchCollaboratoryforStructuralBioinformatics ProteinDataBank(RCSBPDB) ..............................................................95
4.2.3Bindingdatabase ....................................................................................100
4.2.4Therapeutictargetdatabase ....................................................................101
Chapter5:Homologymodeling:Developing3Dstructuresoftarget proteinsmissingindatabases
5.1Introduction......................................................................................................107
5.2Methodologyofhomologymodeling..............................................................110
5.2.1Templaterecognitionandinitialalignment
5.2.2Alignmentcorrection ..............................................................................113
5.2.3Step3:Backbonebuilding .....................................................................113
5.2.4Loopmodeling
5.2.5Side-chainmodeling
5.2.6Ligandmodeling
5.2.7Modeloptimization
5.2.8Modelvalidation ....................................................................................117
5.3Softwareforhomologymodeling....................................................................118
5.3.1Robetta
5.3.2Modeller
5.3.33D-JURY
5.3.4Swiss-model
5.4Conclusion........................................................................................................121
Chapter6:Moleculardockinganalysis:Basictechniquetopredict
6.1Introduction:whatismoleculardocking?.......................................................131
6.2Theoryofdocking............................................................................................132
6.2.1Samplingalgorithms
6.2.2Scoringfunctions
6.3Typesofmoleculardocking............................................................................137
6.3.1Rigiddocking:rigidligandandrigidreceptordocking
6.3.2Constraineddocking:flexibleligandandrigidreceptor
6.3.3Flexibledocking:flexibleligandandflexiblereceptordocking
6.4Standardmethodologyformoleculardocking................................................139
6.5Softwaresavailableformoleculardocking.....................................................143
6.6Conclusion........................................................................................................145
Chapter7:Moleculardynamicsimulations:Techniquetoanalyzereal-time interactionsofdrug-receptorcomplexes
7.1Introduction......................................................................................................157
7.2PrinciplesofMDsimulations..........................................................................158
7.2.1Definitions
7.2.2CalculatingaveragesfromaMDsimulation
7.2.3Classicalmechanics
7.2.4Algorithms
7.3StepsofMDsimulations..................................................................................165
7.3.1Initialization
7.3.2Energyminimization
7.3.3Heatingthesimulationsystem
7.3.4Equilibrationataconstanttemperature
7.3.5ProductionstageofMDtrajectory(NVEensemble)
7.4ApplicationsofMDsimulationsindrugdiscovery........................................168
7.4.1Identifyingcrypticandallostericbindingsites
7.4.2Improvingthecomputationalidentificationof small-moleculebinders ...........................................................................169
7.4.3Advancedfree-energycalculationsusingMDsimulations
8.1Introduction......................................................................................................179
8.3Predictinglocationandnatureofwatermolecule:Tobeor
8.4Strategiestoidentifycavity“waters”..............................................................187
8.4.1Moleculardocking..................................................................................187
8.4.2Moleculardynamics ...............................................................................189
8.4.3Freeenergycalculations .........................................................................190
8.5Loopholesandlimitations................................................................................192
Chapter9:Ligand-basedpharmacophoremodeling:Atechniqueutilizedfor virtualscreeningofcommercialdatabases
9.2Methodologyofpharmacophoremodelingormapping..................................205
9.2.1Input:Datasetpreparationandconformationalsearch
9.2.2Conformationalsearch
9.2.3Featureextraction
9.2.4Patternidentification
9.2.5Scoringofthemodel
9.2.6Validationofpharmacophore
9.2.7Applicationsofpharmacophoremodeling
9.3Inprocessdeterminantsforqualitypharmacophoremodeling.......................213
9.3.1Molecularalignments
9.3.2Handlingflexibility
9.3.3Alignmentalgorithms
9.3.4Keyaspectsofscoringandoptimization
9.4.1Geometry-andfeature-basedmethods ....................................................216
9.4.2Field-basedmethods ...............................................................................225
9.4.3Pharmacophorefingerprints ....................................................................226
9.4.4ChemX/ChemDiverse,PharmPrint,OSPPREYS, 3Dkeys,Tuplets ....................................................................................226
9.4.5Othermethods ........................................................................................227
Chapter10:Fragmentbaseddrugdesign:Connectingsmallsubstructures forabioactivelead
10.3.1Fragment-basedmolecularevolutionaryapproach ..............................242
10.3.2Constructionanddeconstructionapproach
10.3.3Computationalfunctionalgroupmapping
Chapter11:Scaffoldhopping:Anapproachtoimprovetheexisting pharmacologicalprofileofNCEs
11.2.1Pharmacophoresearching
Chapter12:Hotspotandbindingsiteprediction:Strategytotarget
12.6.1Hot-spotpredictionbasedonthesequence
12.6.2Hot-spotpredictionbasedonthestructure
13.4.1SIFT
13.4.5PhD-SNP
13.4.6SNPs&GO
13.5.1PolyPhen
13.5.2SNPs3D
13.5.7SAPRED
13.5.8MutPred
14.2.1Solubilityandsolubilization
14.2.2Permeabilityandactivetransporters
14.3.1Absorption
14.3.3Blood
14.3.4Transporters
14.3.5Dermalandocularpenetration
15.1Introduction....................................................................................................321
15.2Strategiesforcomputerassistedpredictionofsyntheticschemes................322
15.2.1Templatelibrary-based
15.2.2Template-free
15.2.3Focusedtemplateapplication
15.3Approachestovalidateselectedsyntheticroute...........................................324
15.3.1Classifyingreactionfeasibility
15.3.2Predictingmechanisticsteps
15.3.3Rankingtemplates
15.3.4Rankingproducts
15.3.5Generatingproducts
16.3.4Principalcomponentsregression(PCR)
16.3.5Partialleastsquareregressionanalysis
16.3.6Geneticfunctionapproximation(GFA)
16.3.7Geneticpartialleastsquares(G/PLS)
Preface
ThemainreasonforproposinganewbookinthefieldofCADDisincreasingfocusof educationalinstitutiontowardsmolecularmodelingasakeyandrationalapproachfordrug discovery.Researchscholars,undergradstudentsandsometimeseventeachersofdrug designandmedicinalchemistrydonotreadilyfindbookstoguidetheirresearchideasand CADD-basedexperiments.Studentsaswellasteachersusuallygaintheoreticalknowledge viavariousbooksavailableinthemarketbuttheylackproperexperimentalapplicationof thesetools.Mostofthebooksavailableinlibrariesfocusonthetheoreticalconceptsof computer-aideddrugdesignanduseofmolecularmodelingingeneral.Thorough examinationofsuchbooksdisappointscholarsandteachersastheydonotprovideany insightintotheactualapplicationofthesetheoreticalconceptsintosolvingresearch problems.Eventhebooksprovidingdiscussionaboutdifferentsoftwaresutilizedin in-silico analysisfailinprovidingtheclearguidelinestoa“lay-man”,toutilizethem.Therefore,we wantedtowriteabookthatreflectsontheissuesfacedbyresearchersinthepractical applicationofconceptsofCADD,ratherthanatheorybookondefinitionsandexplanations ofCADD.Thisbookwillbeahandbookforpracticalapplicationsofdifferentin-silico toolsavailabletodayalongwithvariousinformationabouthardwareaswellaslistof softwarescommonlyemployedinCAMM.
Ofallthecurrenttrendsinmedicinalchemistry,CADDbasedstudiesaremostcommon andrationaleapproach.Moreimportantlythecorrectutilizationofsuchtoolsisimportant toobtainreliableresultsandthereforetheuser,whichcouldbeanyonefromanundergrad studenttoadoctoralfellow,shouldbewell-versedabouttheapplicationofsuchtoolsand techniques.Additionally,experienceinsolvingtheproblemarisingduringperformingan analysisisnecessary.Weweregreatlyintriguedbythevastapplicationofferedbythisfield andbelievethatresearcherswouldbenefitmorefromabookthatprovidesampleexercises ineachchaptertoguideanenduserinpractisingaCAMMexerciseonanyandevery softwarepackage/onlineservers.Userscanre-performthegivenexerciseswhichwillhelp theminunderstandingthecorrectinterpretationincontextoftheirstudy.
SuchabookcanfacilitatetheactualapplicationofmultipleCAMMbasedapproaches employedintheprocessofdrugdesigning.EachuserwhoutilizeaCAMMbasedtoolsdo notactuallyhavesetstandardtocomparehis/herobtainedresults.Thisbookwillprovide userswithastandardprotocolandresultswhichcanbeutilizedtovalidatetheirownresults andwillaidinbuildingconfidenceovertheobtainedresults.
Wealsokeptinmindthatinformationregardingthegeneralprinciplesassociatedwiththe basicconceptsofmolecularmodelingisalsoimperativeforthebooktobecomea significantpieceofliterature.Weaimedatanapproachthatwouldmakesenseandappeal totoday’sresearchscholars.Thusweincorporatedasubsectionineachchapterthat specificallyunderstatedtheupdateinthecurrentknowledgeineachtoolsandtechniqueof thisfield.Toavoidcomplexityindiscussions,wehaveprovidedgraphicalrepresentations ofgeneralprotocolfollowedineach in-silico techniquefordrugdesigning.Wehave deliberatelyomitteddetaileddiscussionofobscuretheoreticalprinciplesandhaveonly focussedonsimpleexplanationsandinformationonhowtoutilizecomputersandartificial intelligenceintheprocessofdrugdesign.Thisbookwillalsohelpthereadersin understandingtheutilityoffreelyavailablesoftwaretoolsforthepurposeofunderstanding thecomplexprocessofidentificationofasuitabledrugforapathologicalconditionand theirdevelopment.
Eachchapterwilldiscussthebasicprotocolsutilizedintheprocessofleadidentificationto optimizationandfinallypredictionofitsmechanismofaction.Thebookwillprovideaset exercisewhichcouldbeutilizedbytheresearchertooptimizeandvalidatethetool employedbyhim.AdditionallythisbookwillprovidegoodnumberofexercisestoUG students(B.Pharm,B.Tech(bioinformatics),BSc(bioinformatics),BSc(biotechnology), BSc(biochemistry))andPGstudents(M.Pharm(allstreams),MSc(bioinformatics)), valuabletotrainthemfortheirpracticalapplications.Thus,thisbookmayservewelltothe beginnersofmolecularmodeling.Itmaybefollowedbythegraduatestudenttogainbasic knowledgeaboutthetoolsandrouteexercisesofmolecularmodeling.
Acknowledgment
Authorswouldliketothank Dr.BhawnaVyas,ResearchAssociate,Departmentof Chemistry,PunjabiUniversity,Patiala, Ms.ShalkiChoudhary,SeniorResearchFellow, DPSDR,PunjabiUniversity,Patialaand Ms.HimanshuVerma,SeniorResearchFellow, DPSDR,PunjabiUniversity,Patiala,fortheirprovidingassistanceandsuggestionswhile compilingthisbook.Authorsarealsoindebtedtothesupportive,andatthesametime critical,facultymembersofthedepartmentofPharmaceuticalSciencesandDrugResearch.
Authorswouldalsoliketoacknowledgethesupportandguidancefrom SamuelYoung, EditorialProjectManager,Elsevierand RafaelTeixeira,AcquisitionsEditor,Cancer Research/Oncology,MedicalInformatics/Bioinformatics,SystemsBiologyandBiostatistics, Elsevier,alongwithothermembersoftheeditorialteamatElsevier.Finally,thetimespent onthepreparationofthisbookwasmadeavailableonlywiththeforbearanceofour families,friends,andresearchgroups,andwethankallofthemfortheirpatienceand understanding.
Aspecialmentionto Prof.MarioSechi,DepartmentofChemistryandPharmacy, UniversityofSassari,Sassari,Italy,forhisconstantguidance,supportandforprovidingan excellentenvironmentduringtheexecutionofthiswork.
Fundamentalsofmolecularmodeling
1.1Molecularmodeling
Molecularmodelingdescribesthegeneration,representationand/ormanipulationof3-D structureofchemicalandbiologicalmolecules,alongwithdeterminationofphysicochemical propertiesthatcanhelptointerpretstructuralactivityrelationship(SAR)ofthebiological molecules.Molecularmodelingprovidesscientistwithfivemajortypesofinformation.
a.The3Dstructureofmolecules
b.Thechemicalandphysicalcharacteristicsofthemolecules
c.Comparisonofstructureofamoleculewithotherdifferentmolecules
d.Visualizationofcomplexesformedbetweendifferentmolecules/macromolecules
e.Predictionabouthownewrelatedmoleculesmightlook
Fortheanalysisofexponentiallyincreasingdataobtainedthroughtheintroductionofautomated wholegenomeandproteinsequencingtechniques,intheearly2000s,thefieldofbioinformatics emergedrapidly [1].Fromthepioneeringlaboriousmappingandcomparisonofproteinand genesequencesinmolecularbiology,viaanintensephase,whichtoalargeextentcanbe viewedas‘databasemining’andthedevelopmentofefficientcomputerbasedalgorithms,intoa scienceofitsown,whichtodayhasreachedahighlevelofmaturityandsophistication.Tools inbioinformaticsarenowadaysusedwithgreat successinstructuralbiology,computational chemistry,genetics,molecularbiology,pharmaceuticalindustry,pharmacologyandmore.In thischapter,abriefoutlineofthebasicsofmolecularmodelingisgiven,focusingonthe interfacesbetweenmedicinalchemistry,pharmacology,computationalchemistry,informatics, artificialintelligenceandmachinelearning.Thisincludesmolecularrepresentations,computer graphics,molecularsurfacesandtheirprinciplessuchasmolecularmechanics/quantum mechanics/moleculardynamics [2].Theaimistoprovideabriefintroductiontoavastand rapidlygrowingfield.Insubsequentchapters,morespecializeddrugdesigningtoolsand techniquesarepresented,thatbuilduponthefoundationsgivenherein.
1.2Molecularrepresentation
Oneofthemostbasicandusuallyignoredcomponentofmolecularmodelingstudyis representationofthemolecules.Sincethebeginningofthemolecularmodelingstudies
therehavebeenseveralrefinementinthemethodsutilizedtorepresentmoleculesin insilico studies.Torepresent3Dstructureofamoleculeandelectronicpropertiesassociated withit,certaincoordinatesystemsarerequired.Followingcoordinatesystemaregenerally usedformolecularrepresentation.
1.2.1Cartesiancoordinate
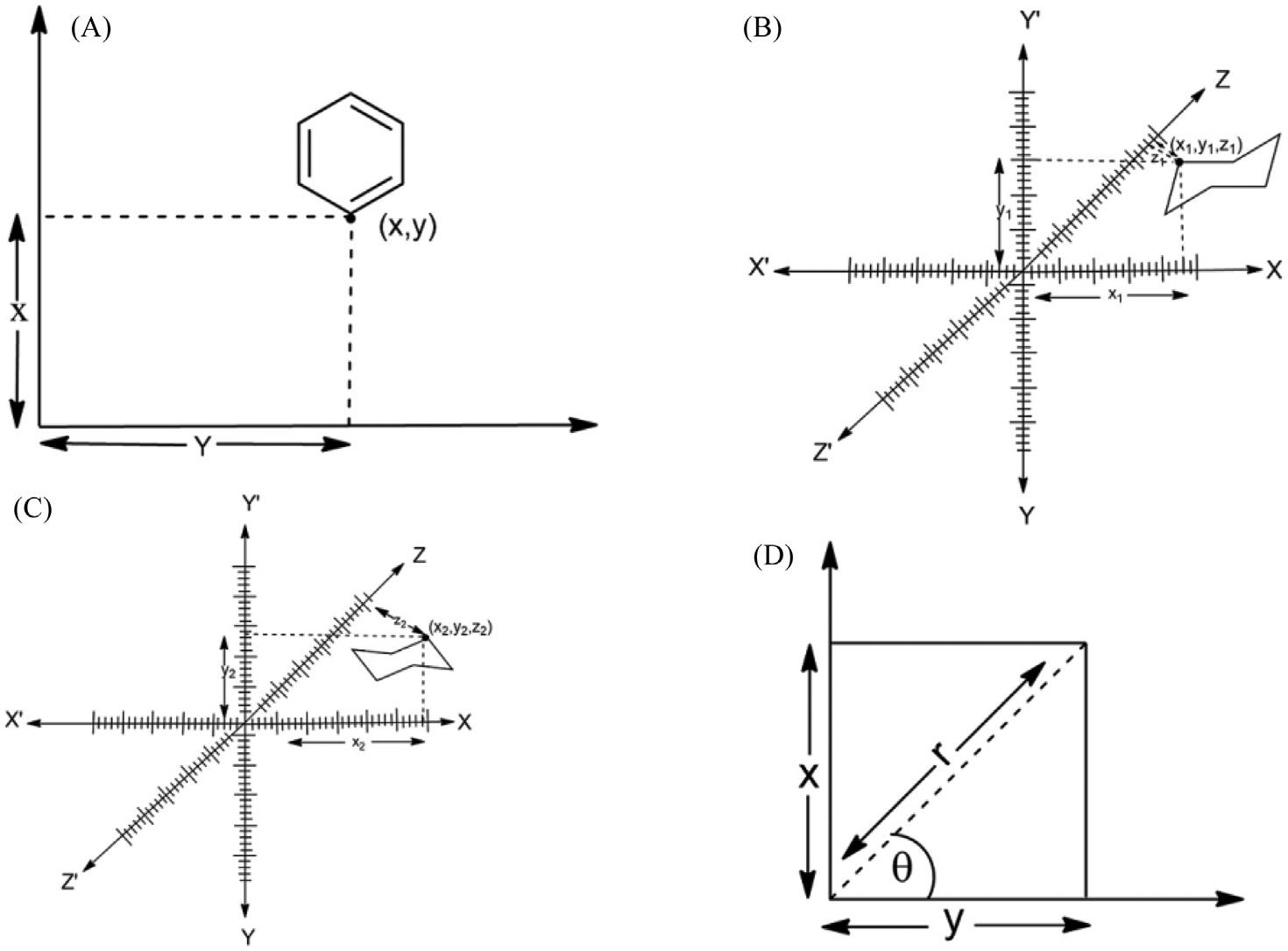
InCartesiancoordinatesystem,twoperpendicularlinesarechosenintheplaneandthe coordinatesofapointaretakentobeassignedasdistancestothelines(Fig.1.1A).
Similarlyfor3-Drepresentationofmolecules,threeperpendicularplanesarechosenand threecoordinatesofapointareassignedasdistancestoeachoftheplanes(Fig.1.1B). Dependingonthedirectionandorderofthecoordinateaxisthesystemmaybearighthand oralefthandsystem(Fig.1.1C) [3].Thiscoordinatesystemisimportanttounderstandthe orientationofthemoleculesinmolecularspaceoncomputerasin Fig.1.1B and 1.1C. orientationsofchairconformationsofcyclohexanearedifferentastheirCartesian coordinatearedifferent.
Figure1.1
Coordinatesystemforrepresentationofamolecule:Cartesian(A)2D,(B)&(C)3Dand(D) polarcoordinate.
1.2.2Polarcoordinate
Inthissystem,apointischosenasthepoleandarayfromthispointistakenas thepolaraxis.Foragivenangle θ,thereisasinglelinethroughthepolewhose anglewiththepolaraxisis θ.Thenthereisuniquepointonthislinewhosesigned distancefromtheoriginisrforgivennumberr.Foragivenpairofcoordinates (r, θ)thereisasinglepoint,butanypointisrepresentedbymanypairsof coordinates.Forexample,(r, θ),(r, θ 1 2 π)and(-r, θ 1 π)areallpolarcoordinates forthesamepoint( Fig.1.1D ) [1]
1.2.3Internalcoordinate
Amorechemicallyintuitivewayofwritingthecoordinatesistousetheinternal coordinatesofamolecule(i.e. bondlengths,bondanglesandtorsionangles).Internal coordinatesareusuallywrittenasaZ-matrix [1].HereisanexampleofaZ-matrix,for ethene(C2H4):
˚ )Bondangle( )Torsionangle( )
ThefirstlineoftheZ-matrixdefinesatomnumber1(carbon).Atom2isalsoa carbonatom,andisatadistanceof1.31A ˚ fromatom1(theapproximatelengthof acarbon-carbondoublebond).Thethirdcolumndefinestheatomtowhichthe distanceincolumn4refers,i.e.atom3(ahydrogen)is1.07A ˚ fromatom1(the lengthoftheC-Hbond).Similarlytheatomnumbersincolumns5and7define whichatomsareinvolvedinthebondangleandtorsionangle(valuesgivenin columns6and8respectively).So,forexample,atomnumber6isahydrogen.Itis 1.07A ˚ fromatom2,thebondangleinvolvesatoms6-2-1,andthetorsionangleis foratoms6-2-1 4.Allthetorsionanglesare180 ,showingthatthemoleculeis planar.Thissystemofcoordinateisrequiredtogenerateuniqueconformationofa molecularsystemo ncomputerscreen.
1.3Computergraphics
Computergraphicsdisplayareeithervectororraster.Rasterimages,alsoknownas bitmaps,arecomprisedofindividualpixelsofcolor.Eachcolorpixelcontributestothe
overallimage.Rasterimagesmightbecomparedtopointillistpaintings,whichare composedwithaseriesofindividually-coloreddotsofpaint.Eachpaintdotinapointillist paintingmightrepresentasinglepixelinarasterimage.Whenviewedasanindividualdot, it’sjustacolor;butwhenviewedasawhole,thecoloreddotsmakeupavividanddetailed painting.Thepixelsinarasterimageworkinthesamemanner,whichprovidesrichdetails andpixel-by-pixelediting.Unlikerastergraphics,whicharecomprisedofcoloredpixels arrangedtodisplayanimage,vectorgraphicsaremadeupofpaths,eachwitha mathematicalformula(vector)thattellsthepathhowitisshapedandwhatcoloritis borderedwithorfilledby.Sincemathematicalformulasdictatehowtheimageisrendered, vectorimagesretaintheirappearanceregardlessofsize.Theycanbescaledinfinitely. VectorimagescanbecreatedandeditedinprogramssuchasIllustrator,CorelDraw,and InkScape [4]
OnvectordisplaysthelinesmakinguptheimagearetracedonthefaceoftheCRT.The linesarecontinuousstrokesandappearverystraightandsmooth.Howeveronlylinesand dotscanbedrawnonvectorsystems.Filledareassuchasmolecularsurfaces,mustbe representedbymanycloselyspacedlinesordots,whichaddsgreatlytothecomplexityof theimage.Onrasterdisplays,theCRTisrepeatedlyhorizontallyscanned,asonatelevision screen.Theimageismadeupofdiscretepixels.Linescanappearjagged,dependingonthe resolutionoftheCRTbeingused.Becauseofthepixelsmethodusedinrastersystem,filled areasaremorereadilydrawnonthesesystemsthanonvectorsystems [5]
1.4Molecularmodels
ThesimplesttypesofmodelsareCPK,dreidingmodelsandcomputermodels,which provideabetterwaytorepresentmolecules.
1.4.1CPKmodels
CPKmodelsarephysicalmodelsinwhichacolorcodedmolecularmodelassemblykitis providedforrepresentingorganicmolecularstructures(Fig.1.2) [6].Thesemodelsconsist ofshapescomprisingtwobasicandcomplementaryconstructionunitscapableofbeing interlocked.Thebasicconstructionunitsarecolorcodedplastictubeswhichcanbecoupled tosecondbasicconstructionunits,representingthebondsbetweenadjacentatoms.The secondbasicconstructionunitsarecolorcodedcouplingspheres,accordingtothevalency oftheatomstobejoined,thecenterofsaidcouplingsphererepresentatomcenters.These couplingsphereshaveradialarmssubstantiallylocatedonthesurfaceofaspherewiththe centerofthecouplingunitbeingthecenterofthesphere.Theseunitsaremadeupof plasticandareoftwotypes,onetypeadaptedforplanar-trigonalcouplingofthreesaid tubesseparatedbyanglesofabout120 andtheothertypeadaptedfortetrahedralcoupling
CPKmodelrepresentationofquinazolinemolecule.
offoursaidtubesseparatedbyanglesofabout109 .Saidfirstandsecondconstruction unitsarecapableofbeingjoinedtogetherandheldimmobilebyfrictionbyhavingsaid radialarmsand/orthecavitiesofsaidtubestaperedsothatskeletalmodelsofcomplex organicmacromoleculesmaybeassembledsuchthatthedistancebetweenthecentersof twodirectlyconnectedcouplingmeansrepresentsthedistancebetweenjoinedatoms [7]. Thesemodelsgiveagoodrepresentationoftheshapeofamolecule.Theycanbe manipulatedtoproducevariousconformationsofthemolecule.Buttheycannotbeusedto presentelectronicpropertiesofmoleculesandtheycannotbesuperimposeduponone anothertocomparemolecularconformationandshape.Thebondlengthsandanglescannot beadjustedinthesemodels.
1.4.2Dreidingmodels
Dreidingmodelsarephysicalmodelsthatusethinmetalorplasticrodstorepresent bonds [8].Bondlengthsandanglesarefixed,althoughrotationsaroundbondscanbeeasily done.Onecanalsodemonstratethereadyconversionofoneboatformintoanotherand thenstophalfwaybetweenthetwoandpreservethetwistform.Selectionofappropriate ballsanduseofrubbertubingconnectorstoformdoublebondsandC3-C4cycloalkanes permitsconstructionofnumerousinterestingcisandtransolefins,opticallyactiveallenes, andsmallring-compounds,insomeofwhichopticalisomerismissuperimposedon geometricalisomerism.However,theygiveapoorrepresentationofmolecularvolumeand cannotbeusedtoshowelectronicproperties.Dependingonthecomplexityofthemodel, theycouldpossiblybesuperimposedupononeanotherforcomparisonofmolecular conformation(Fig.1.3).
Figure1.2
Dreidingmodelrepresentationofquinazolinemolecule.
Figure1.4
Computermodelrepresentationofligandproteincomplex.
1.4.3Computermodels
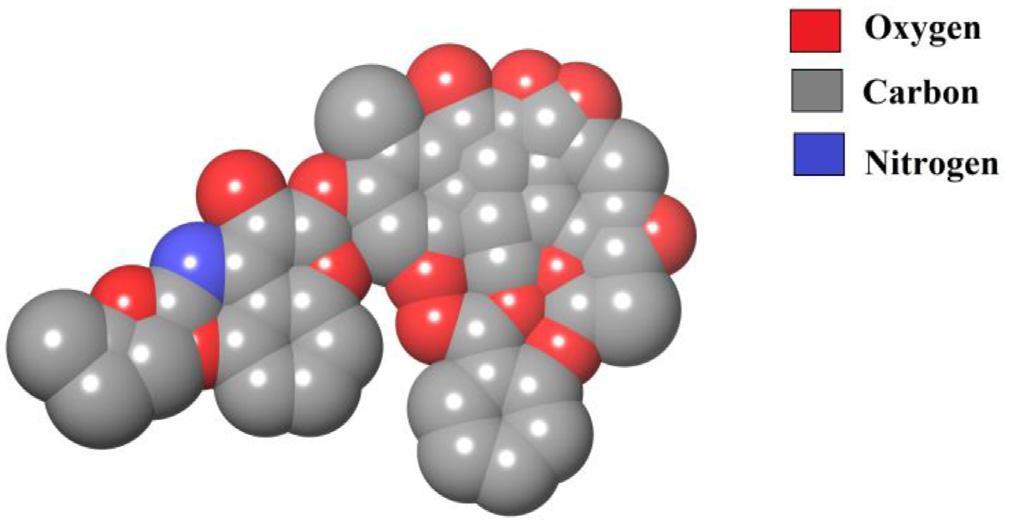
Computermodelscanbeusedtodrawavirtuallylimitlessvarietyofmolecular representationsfromstickfigurestomolecularsurfaces(Fig.1.4).Computergraphics modelscanalsoveryreadilybeusedtorepresentelectronicpropertiesofmolecules.These modelscanbeeasilysuperimposedandaccuratelyconstructedusinganybondlengths, anglesandtorsionangles.Elementsarecolorcodedandcaneasilyberecognizedonthe basisofcolorassignmenttothatparticularelement(Fig.1.5).
Figure1.3
Figure1.5
Filledcolorcodedcomputermodelrepresentationofligand.
Adisadvantageofcomputermodelsisthattheyarenotphysical,threedimensionalmodels. Thuscomputermolecularmodelingtoolsportrayimagesinawaythatseemsthree dimensional.Initiallythemodelsweredrawnoncathoderaytubes(CRTs)usingspecial purposecomputerhardware.InCRTstheimageswerelimitedtotwodimension.Thethird dimensionwasrealizedbyrapidlydisplayingslightlydifferenttwodimensionalimages.In thismethod,timewasusedasaparametertorepresentthirdCartesiandimension.This techniqueisreferredtoasreal-timegraphics.Anothertechniqueusedtogivegraphicsa threedimensionallookinvolveddrawing“infront”partsofimagemorebrightlythanparts whichare“inback”.MoreupdatedcomputergraphicsystemssuchasPS350andthesilicon graphicsIRISworkstationallowedtocombinethetechniquesofreal-timegraphics, stereographicsandintensitydepthcuingtoproducea3-Dimagewithmultiplecolors.
1.5Molecularsurfaces
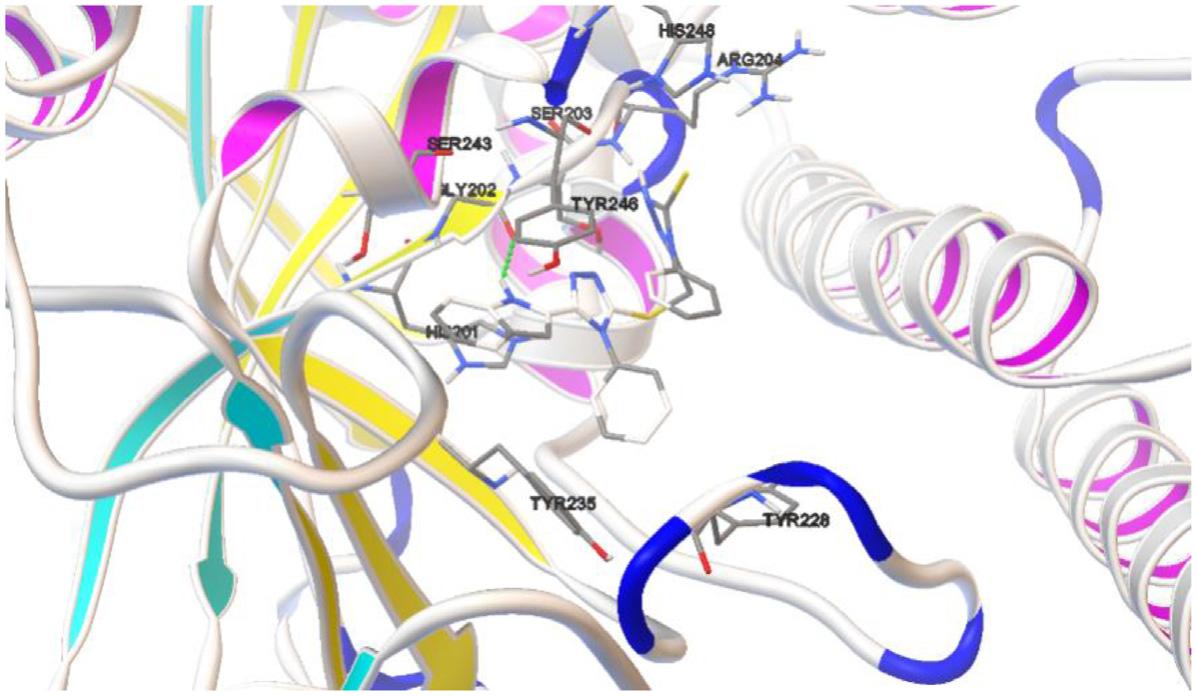
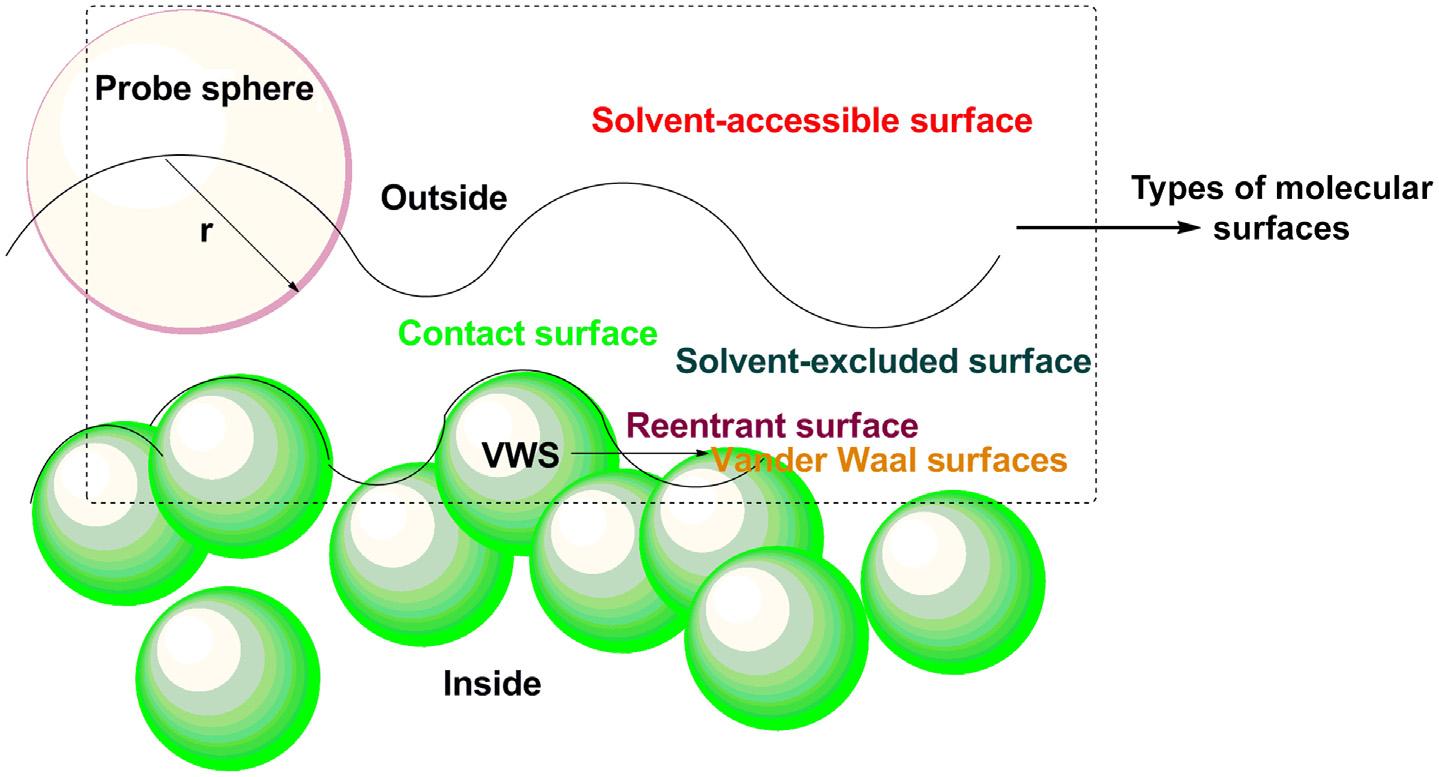
Molecularsurfaceisafundamentalaspectofastructureasitisthroughthecomplementarity ofshapeandchemistryofthesurfacethatmoleculesinteractwitheachother.Themolecular surfaceisdefinedas‘thesurfaceincontactwithaprobespherewhilethesphererollsover thesurfaceofthemolecule.Morerecentlytheincreasingpowerofarastergraphicssystem allowsmorecompleximagestobeviewedinteractivelyandthishasledtothedevelopment ofmanytechniquesforrepresentingsolidmolecularsurfaces(Fig.1.6).
Inmodelingofamacromoleculesuchasprotein,DNA,RNAetc.andsmallmoleculeswith biologicalsignificance,eachconstituentatomisconsideredasasimplespherewithitsVan derWaalsradius.Thevisualizationorsimulationsoftheseoverlappingballscanbedone bysurfacemeshgenerationtechnique,wheretheshapequalityoftheseoverlappingballs hasastronginfluenceonsimulationaccuracy [9].Therearemainlythreetypeofmolecular
Representationofdifferentmolecularsurfaces.
surfacesthatplaysimportantroleindrugdesigningprocesssuchasvanderwaalssurface (VWS),solvent-accessiblesurface(SAS)andsolventexcludedsurface(SES).
1.5.1VanderWaalssurface(VWS)
VWSsimplyrefertotheunionofallpossibleoverlappingballs.Theinteractionsofthese surfacesareimportantinvariouschemicalandbiologicalprocessessuchasformationofa tertiarystructureofbiopolymer,electrontunnelinginproteincrystalsetc.Theprobablerole ofweakVWSinteractionsonreactiondynamicsisanissueofgreatconcern [10].Usually, forthemacromoleculessuchasproteinsandnucleicacid,VWSmaybeburiedwithinthe interiorofthemolecule.vanderwaalSurfaceboundmoleculesareheldtogetherbyweak attractiveforceslikedispersive,electrostatic,chargetransferandhydrogenbondinteraction betweenclosedshellatomsormolecules,andmoleculesboundtoeachotherbythesetype ofattractiveforcespossesslowdissociationenergies.Theunderstandingofbothreactive andnon-reactivedynamicsinVWScomplexesrequiresdetailedinformationonpotential energies [11].TheVWradiiofeachspherevariesslightlywithitscovalentbonding environmentandtheseradiiareneededfortheevaluationofproteinvolume,interior packingandalsothepackingattheprotein-waterinterface [12].
1.5.2Solventaccessiblesurface
Thesolventaccessiblesurfacecanberecognizedasthesurfacecreatedbythecenterofthe solventthatisregardedasarigidsphere,whenitrollsaroundthevanderWaalssurface.
Figure1.6
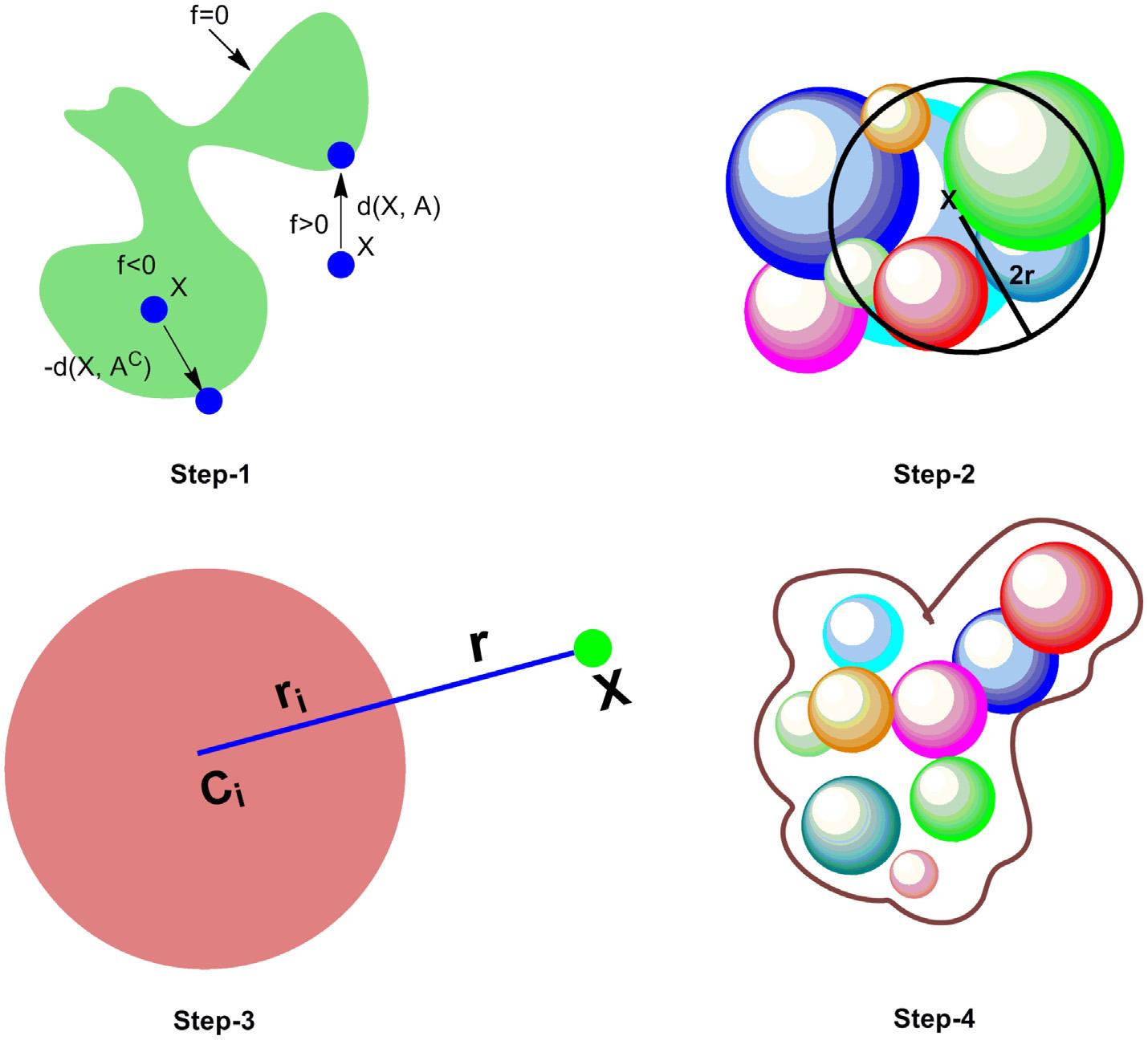
ThistermwasinitiallycoinedbyRichardandLee,whengoingthroughastudyon proteininteractions [13] .Theywereinterestedinanaly singtheinteractionofprotein withthesolventmoleculestodeterminethehydrophobicityandfoldingofproteins.In ordertoobtainmolecularsurfacethatasolventcouldaccess,aprobesphereismadeto rolloverthevanderWaalsurface,andthetracesofcenteroftheprobespherebest describetheSAS [14] .SAShasbecomeacommonthreadformostoftheresearchers especiallyinthecaseofthenon-polarmolecules,asthefreeenergyofaqueoussolvent isproportionaltoSAS,whichinturnproportionaltothenumberofsolventmolecules thatareincontacttothesolutemolecule.Thus,SASisacentralquantityinseveral solvationmodelsusedinmolecularmechanics(MM) [15] .Thissurfacecanbeusedto determinetheaminoacidenvironmentener gywhichdependsonanaccurateandrapid estimationofSAS.Itcanalsobeusedtocomp utepartitioncoefficient(logP),whichis animportantparameterextensi velyusedinstudyingthestructural-biologicalactivity. Further,inmolecularmodelingstudy,thei nteractionsofthecompoundwithnon-polar phasecanbedeterminedbyutilizingSASinformationboth invitro and invivo [16] . Theconstructivesolidoperationgeome tryoperationcanbeappliedforthe representationofSASandtheimplicitfunc tionsunderlyingthemolecularsurfacecan bedefinedthroughsomestepsasdisplayedin Fig.1.7 .Thefirststepissignchangeof thesefunctions,thenidentificationofato msanddefinitionoffunctionusedforthemap evaluationofmolecularsurfaceandeventuallytheclusteringofatoms.Function f SAS (.)canbecomputedbyaddingthecontributionofthoseatoms[(C i ,R i )] i εI ζ I thatbelong tothesphereofcenterxandradius2r,whichisaconstructivesolidgeometry operation [17] .
1.5.3Solventexcludedsurface
Solventexcludedsurface(SES)isoneofthemostpopularsurfacedefinitionsinthe fieldofbiophysicsandmolecularbiology.Itisdividedintotwopartsi.e.contact surfaceandthereentrantsurface.Thecontactsurface,asapartofthevanderWaals surfaceofeachatomisaccessibletoaprobesphereofagivenradius.Thereentrant surfaceisdescribedastheinward-facingpart oftheprobesphere,whileincontactwith morethanoneatom [13] .Later,Connollydevelopedthema thematicalrepresentationof theSESforarbitrarybiomoleculesintermsofconcavepatches,saddlepatches,and concexsphericalpatches [18] .Amongtheseregions,theconvexcontactsurface segmentofthevanderWaalssurfacepossessdirectcontacttothesolventsurrounding thesystem,andtheconcavere-entrantsurfacesegmentinwhichsolventspherehas contactwithtwoormoreatomsspheresofthestructure.CannollyrepresentationofSES isstandardtoolinmolecula rmodelingthatallowsquan titativeandqualitative comparisonofmolecules [14] .Theactualrepresentationofsolventexcludedsurfacesis displayedin Fig.1.8 .
Figure1.7
Stepsinvolveddefiningimplicitfunctionsunderlyingmolecularsurface.
1.5.4Chargedpartialsurfacearea(CPSA)
Thephysicalandchemicalpropertiesofchargedpartialsurfaces(CPSAs)playimportant roleinspecificityandselectiveinteractionsofligandwithprotein.CPSAsweredeveloped togettheinformationaboutthemolecularstructurethatinturnhelpindeterminingthe intermolecularinteractionsforQSAR.Practically,itisusefultoascertainthetoxicityand givedescriptionoflocalandglobalelectrophilicityinnon-covalentinteractions.CPSA descriptorshavebeenusedindistinguishingtheantagonistandagonistthatbindtoestrogen receptors.Thesedescriptorsalsohaveutilityindeterminingpartialchargeand conformationalchanges [19].Chargedpartialsurfaceslikehydrophilicandhydrophobic surfaceareahavetheirutilityinvariousphenomenonrelatedtoproteinadsorption.Usually, absorptionfromhydrophobicsurfacesismoreeffectivethanhydrophilicsurface.Notonly theadsorption,butalsosomeoftheproteinexchangeprocessesoccurswithmoreeaseover thehydrophobicsurfacesthanthatofhydrophilicsurfaces.Thereisastrongcorrelationof

targetselectivitywithphysicalandchemicalpropertiesofthesesurfaces [20].The modulationinhydrophobicsurfacecanbeachievedbysomeofthefactors,oneofthemis temperature.Temperature,asoneofthefactortoinduceexposureofhydrophobicsurface wasidentifiedbystudyingitseffectonthechaperoneactivityof α-crystallin [21].
1.6Workstations
Usuallyvectorsystemsarepreferredbymolecularmodelersbecauseoftheirspeedandhigh qualityoflinedrawing.Sincethesevectorsystemsusespecialpurposehardware,theyhave beenmoreexpensivethanrastersystemsandareusedasdisplaydevicesseparatefromthe hostcomputerwhichbeingusedtostoreandmodifymolecularcoordinates.However, nowadaysnewcomputergraphicworkstationshavebeenintroduced,whichincluderaster systemshavingacomputerwithfulloperatingsystemandmassstoragefacilityintegrated withthegraphicdisplay.
1.6.1GPUhardware
Thegeneralpurposegraphicsprocessingunit(GPU)computationalresourceisveryfastand hasalsobecomeverypopular.Itisstronglydependentonthehardware.Manycomputers haveGPUboardsthatareusedmainlyforgraphics.ToutilizetheGPUforanapplication program,wemustuninstalltheGPUgraphicsdriversoftwareandinstallCUDA,a computerlanguageforusewithGPU.MostoftheGPU-MDprogramsadoptCUDAfor
Figure1.8
Representationofmolecular/solvent-excludedsurface.
GPUcomputations.TheslotnumberofeachGPUboardinthecomputermustbeexplicitly indicatedinCUDA.Everyyear,newGPUhardwarehasappearinthemarketwithupdated CUDAversion.TheGPUprogramshouldbetunedupforeachGPU,sincetheperformance ofGPUprogramsdependsonthebalanceofthenumberofGPUcoresandmemory-band width.IncontrasttoCPUs,whichareusedforallapplicationprograms,theapplicationof GPUisquitelimited.ThismeansthattheGPUisusedonlywhentheGPUprogramsare available.
GPUcomputingisparticularlysuitabletorunmoleculardynamicssimulationprogramslike AMBER,Gromacs,NAMDandpsygene-G/myPresto.SomeoftheseGPUprogramsare freelyavailable.However,oneofthemostseriousproblemsforendusersishowtosetup theGPUmachinefortheseMDprograms.Theotherproblemisthatthesystemsizeforthe GPUcomputationmustbelargerthantheminimumsizethatisdeterminedbytheprogram. SincemostGPUprogramsadoptaspace-decompositionmethodforparallelcomputing,the systemmustbedecomposedintosubsystems.ThismeansthattheMDofasmallsystem (likeasinglemolecule)isnotsuitableforGPUcomputing.
1.7Principlesofmolecularmodeling
Modelingofmoleculesforunderstandingvarioustypesofmolecularphenomenonrelatedto chemistry,biochemistry,biophysics,molecularbiology,drugdiscoveryanddrugdesign, pharmacogenomics,pharmacologyetc.isbasedoncalculationofdifferentkindsofenergy associatedwithamolecularsystem.Amolecularsystemisassociatedwiththreetypesof energiesi.e.potentialenergy,kineticenergyandquantumenergy.Calculationand applicationoftheseenergiesdependuponthetypeofworktobeexecutedinproblems whichareunderconsideration.Differentfundamentalprincipleshavebeenemployedto calculatethesethreedifferentkindsofmolecularenergies.Potentialenergy,kineticenergy andquantumenergycanbecalculatedbyapplyingtheconceptsofmolecularmechanics (MM),MolecularDynamics(MD)andQuantumMechanics(QM)respectively.Nowbrief accountaboutthesethreeconceptsarediscussedinnextsections.
1.7.1Molecularmechanics
Molecularmechanicstreatsthemoleculeascollectionofatomsheldtogetherbyspring. ThisassumptionismadetoapplyNewtonian’smechanics(Huck’slawofmechanics)for calculatingpotentialenergyofmolecularsystembyconsideringatomsheldtogetherby elasticorharmonicforces.Theseforcescanbedescribedbypotentialenergyfunctionsof structuralfeatures(internalcoordinates)likebond-length,bond-anglestorsionalangleand non-bondedinteractionsofamolecule.Non-bondedatoms(greaterthantwobondsapart) interactthroughvanderWaalsattraction,stericrepulsion,andelectrostaticattraction/