ComputerAidedDrugDesign(CADD):FromLigandBasedMethodstoStructure-BasedApproachesMithun Rudrapal
https://ebookmass.com/product/computer-aided-drug-designcadd-from-ligand-based-methods-to-structure-basedapproaches-mithun-rudrapal/
Instant digital products (PDF, ePub, MOBI) ready for you
Download now and discover formats that fit your needs...
Research Design: Quantitative, Qualitative, Mixed Methods, Arts Based, and Community Based Participatory Research Approaches 1st Edition, (Ebook PDF)
https://ebookmass.com/product/research-design-quantitativequalitative-mixed-methods-arts-based-and-community-basedparticipatory-research-approaches-1st-edition-ebook-pdf/ ebookmass.com
Benzodiazepine-Based Drug Discovery Farzad Zamani
https://ebookmass.com/product/benzodiazepine-based-drug-discoveryfarzad-zamani/
ebookmass.com
Piperidine-Based Drug Discovery Ruben Vardanyan
https://ebookmass.com/product/piperidine-based-drug-discovery-rubenvardanyan/
ebookmass.com
Introduction to Computing Using Python: An Application Development Focus, 2nd Edition – Ebook PDF Version
https://ebookmass.com/product/introduction-to-computing-using-pythonan-application-development-focus-2nd-edition-ebook-pdf-version/
ebookmass.com
Cultivating Democracy: Politics and Citizenship in Agrarian India Mukulika Banerjee
https://ebookmass.com/product/cultivating-democracy-politics-andcitizenship-in-agrarian-india-mukulika-banerjee/
ebookmass.com
It Cannoli Be Murder Catherine Bruns [Bruns
https://ebookmass.com/product/it-cannoli-be-murder-catherine-brunsbruns/
ebookmass.com
Introduction to the Hebrew Bible: Third Edition
https://ebookmass.com/product/introduction-to-the-hebrew-bible-thirdedition/
ebookmass.com
Integration with Complex Numbers : A Primer on Complex Analysis Aisling Mccluskey
https://ebookmass.com/product/integration-with-complex-numbers-aprimer-on-complex-analysis-aisling-mccluskey/
ebookmass.com
Fire:
A Hutton Family Romance Brooks
https://ebookmass.com/product/fire-a-hutton-family-romance-brooks/
ebookmass.com
Arjan J. Weele
https://ebookmass.com/product/purchasing-and-supply-chain-managementseventh-edition-arjan-j-weele/
ebookmass.com
ComputerAidedDrug Design(CADD):From Ligand-BasedMethods toStructure-Based Approaches
Editedby
MithunRudrapal
DepartmentofPharmaceuticalChemistry,RasiklalM.DhariwalInstituteof PharmaceuticalEducationandResearch,Pune,Maharashtra,India
ChukwuebukaEgbuna
NutritionalBiochemistryUnit,AfricaCentreofExcellenceinPublicHealth andToxicologicalResearch(ACE-PUTOR),UniversityofPort-Harcourt, PortHarcourt,RiversState;DepartmentofBiochemistry,Chukwuemeka OdumegwuOjukwuUniversity,Uli,Anambra,Nigeria
Elsevier
Radarweg29,POBox211,1000AEAmsterdam,Netherlands
TheBoulevard,LangfordLane,Kidlington,OxfordOX51GB,UnitedKingdom 50HampshireStreet,5thFloor,Cambridge,MA02139,UnitedStates
Copyright©2022ElsevierInc.Allrightsreserved.
Nopartofthispublicationmaybereproducedortransmittedinanyformorbyanymeans, electronicormechanical,includingphotocopying,recording,oranyinformationstorageand retrievalsystem,withoutpermissioninwritingfromthepublisher.Detailsonhowtoseek permission,furtherinformationaboutthePublisher’spermissionspoliciesandourarrangements withorganizationssuchastheCopyrightClearanceCenterandtheCopyrightLicensingAgency,can befoundatourwebsite: www.elsevier.com/permissions
Thisbookandtheindividualcontributionscontainedinitareprotectedundercopyrightbythe Publisher(otherthanasmaybenotedherein).
Notices
Knowledgeandbestpracticeinthisfieldareconstantlychanging.Asnewresearchandexperience broadenourunderstanding,changesinresearchmethods,professionalpractices,ormedical treatmentmaybecomenecessary.
Practitionersandresearchersmustalwaysrelyontheirownexperienceandknowledgeinevaluating andusinganyinformation,methods,compounds,orexperimentsdescribedherein.Inusingsuch informationormethodstheyshouldbemindfuloftheirownsafetyandthesafetyofothers, includingpartiesforwhomtheyhaveaprofessionalresponsibility.
Tothefullestextentofthelaw,neitherthePublishernortheauthors,contributors,oreditors, assumeanyliabilityforanyinjuryand/ordamagetopersonsorpropertyasamatterofproducts liability,negligenceorotherwise,orfromanyuseoroperationofanymethods,products, instructions,orideascontainedinthematerialherein.
ISBN:978-0-323-90608-1
ForinformationonallElsevierpublications visitourwebsiteat https://www.elsevier.com/books-and-journals
Publisher: SusanDennis
EditorialProjectManager: KyleGravel
ProductionProjectManager: KumarAnbazhagan
CoverDesigner: GregHarris
TypesetbySTRAIVE,India
1.Introductiontodrugdesignanddiscovery
AndreM.deOliveiraandMithunRudrapal
1Definitionandconceptofdrugdesignanddiscovery 1
2Historicalperspectivesofdrugdiscovery 2
3Process,strategies,andstagesofdrugdiscoveryand development 4
3.1Discoveryphase5
3.2Preclinicalphase5 3.3Clinicalphase6
3.4Approvalandpostapprovalphases7
4Traditionalandmodernapproachestodrugdiscoveryand development 7
4.1Virtualscreening7
4.2High-throughputscreening8
4.3Phenotypicscreening8
4.4Structure-baseddrugdesign8
4.5Fragment-baseddrugdesign8
4.6Ligand-baseddrugdesign9
5Rationaldrugdesign(RDD)andCADD 9
5.1Structure-baseddrugdesign(SBDD)10
5.2Ligand-baseddrugdesign(LBDD)11
2.Fundamentalconsiderationsindrugdesign
ManojKumarMahapatraandMuthukumarKaruppasamy
1Fundamentalsofrationaldrugdesign(RDD) 17
1.1Rationaldrugdesign17
1.2Structure-baseddrugdesign(SBDD)18
1.3Ligand-baseddrugdesign(LBDD)19
2Conceptsofphysicochemicalproperties 19
2.1Structuralpropertiesandstereochemistry20 2.2Drugreceptorsandreceptortheories21
2.3Pharmacokineticsandpharmacodynamics22
2.4SARsandQSARs23
2.5Prodrugsanddrugmetabolism23
2.6Metaboliteantagonismandenzymeinhibition24
2.7Nucleicacid-baseddrugdesign25
2.8Leadcompounds25
2.9Peptidomimeticsandanalogdesign26
2.10Reversepharmacologyanddrugrepurposing strategies26
3Fundamentalsofcomputer-aideddrugdesign(CADD) 27
3.1Structure-baseddrugdesign(SBDD)28
3.2Ligand-baseddrugdesign(LBDD)36
3.3Virtualscreeningtechniques39
3.4ADMEanalysisandmeasuresofdrug-likeness45
3.Ligand-baseddrugdesign(LBDD)
VivekYadav,JurnalReang,Vinita,andRajivKumarTonk 1Introduction 58
2Randomandnonrandomscreening 58
2.1Drugmetabolismstudies59
2.2Serendipitymethod59
2.3Clinicalobservations60
3Drugdiscoveryprocess 60
3.1Ligand-baseddrugdesign(LBDD)61
3.2Structure-baseddrugdesign(SBDD)61
4Combinatorialchemistry 61
4.1Unbiasedlibrary62
4.2Biasedlibrary62
5Leadmodificationsandoptimizationapproaches 63
5.1Pharmacophore63
5.2Structure-activityrelationships(SARs)63
6Stereochemistryofdrugmolecules 64
6.1Importanceindrugaction65
6.2Stereoselectivityindrug-receptorinteraction66
6.3Stereospecificaspectsindrugdesign67
6.4Stereochemistryinbiologicalprocesses68
6.5Significanceofstereoselectivity69
7Bioisosterism 69
7.1Needanduseofbioisostericreplacements70
7.2Classificationofbioisosterism71
8Drugmetabolism 73
8.1Objectives74
8.2Prodrugs75
8.3Retrometabolism-baseddrugdesign(RMDD)80
9Virtualhigh-throughputscreening(vHTS) 85
9.1Toolsforvirtualhigh-throughputscreening(vHTS)85
9.2Techniquesforvirtualhigh-throughputscreening (vHTS)86
9.3Lipinski’srule88 9.4Veberrule88
9.5ADMETscreening89
9.6Toxicityprediction90
9.7Docking-basedvirtualscreening(DBVS)92
9.8Pharmacophore-basedvirtualscreening(PBVS)93
4.Quantitativestructure-activityrelationships(QSARs)
AndreM.deOliveira
6.1Regardingthevariableselection111
6.2Regardingthevariablevalidation111
6.3Regardingthemodelvalidation111
6.4Regardingtheamountofvariables114
6.5Regardingthebiologicalvalidation114
5.Fundamentalsofmolecularmodelingindrugdesign
ManishKumarTripathi,ShabanAhmad,RashmiTyagi, VandanaDahiya,andManojKumarYadav
1Fundamentalsofcomputationalchemistry 125 2Basicconceptsofquantummechanics 126
3Sketchapproach,conversionof2Dstructuresin3Dform,and generationof3Dcoordinates 128
4Moleculardynamicssimulationanditscomponents 129 4.1Forcefields133
4.2Geometryoptimization134
4.3Energyminimization135
4.4Conformationalsearch135
4.5Geneticalgorithms136
4.6MonteCarlosimulation137
4.7Artificialintelligencemethods138
4.8Pharmacophoreidentificationandmolecularmodeling138
5Molecularrecognitionindrugdesign 140
6Thermodynamicconsiderationofdrugdesigning 141
6.1Methodsofthermodynamicmeasurementfor bimolecularinteractions142
6.2Physicalbasisofintermolecularinteraction143
6.Pharmacophoremodelingindrugdesign
SiddharthaMaji,SubratKumarPattanayak,AnikSen,and VishnuNayakBadavath
3.1Ligand-basedpharmacophore161
3.2Structure-basedpharmacophore(SBP)163
4Pharmacophoremodel-basedvirtualscreening(VS)
5Pharmacophoreelementsandrepresentation 166
6Generationofpharmacophoremodelsfromreceptor-ligand complex 168
7ApplicationsofpharmacophoresinADME-Tox 169
7.1Pharmacophore-guideddrugtargetidentification170
7.2Multitargetsbypharmacophore170
7.3Possibleapplicationsofmultitargetligands171
7.Structure-baseddrugdesign(SBDD)
GouravRakshit,SheikhMurtuja,BanothKaranKumar, SankaranarayananMurugesan,and VenkatesanJayaprakash
1Computer-aideddrugdesign 181
2Structure-baseddrugdesign(SBDD) 183
2.1Overviewoftheprocessesinvolvedinstructure-baseddrug design(SBDD)190
2.2ExamplesofSBDD190
2.3CasestudyofSBDD191
3Moleculardocking 195
3.1Variousmodelspertainingtomoleculardocking196
3.2Classificationofmoleculardockingsystems198
3.3Dockingbasedscreening198
3.4Moleculardockingstepsandprocedure/docking protocol200
4Moleculardynamics 209
4.1ApplicationsofMD210
4.2BindingfreeenergycalculationswithMMGBSA/PBSA212
4.3MoleculardynamicssimulationusingDESMOND215
4.4Casestudy217
5Conclusion 222 References 223
8.RecentadvancesinCADD
TriptiSharma,SujataMohapatra,RasmitaDash, BiswabhusanRath,andChitaRanjanSahoo
1Introduction 232
2Roleofinformaticsindrugdiscovery 232
2.1Chemoinformaticsindrugdiscovery232
3Databasesusedindrugdiscovery 239
3.1RecenttrendsofADMET239
3.2Predictionofphysicochemicalproperties240
3.3PredictionofADMETproperties242
4Fragment-baseddrugdesign 244
4.1LibrarydesignforFBDD244
4.2Strategiesinfragment-baseddrugdesign246
4.3Generalconsiderationonfragment-baseddrug design248
5Receptor-baseddenovodesign 251
5.1Methodsinvolvedindenovodrugdesign251
6Nucleicacid-baseddrugdesign(NABDD) 253
7Advancesindrugdesigning 257
7.1Similaritysearching257
7.2Artificialintelligence(AI)257
7.3Machinelearning(ML)258
7.4Datamining258
7.5Networkanalysisandsystembiologytools258
7.6Dataanalysistools259
8Insilicoapproachesindrugrepurposing 259
8.1Knowledge-basedapproach266
8.2Target/structure-basedapproach266
8.3Ligand-basedapproach266
8.4Pathway/network-basedapproach266
8.5Signature-basedapproach266
8.6Targetedmechanism-basedapproach267
8.7Examplesofsuccessfuldrugrepurposing267
9Designofbiologicsandproteindrugdesign 270
9.1Spaceforcomputationalbiologicsdesign270
9.2Insilicoproteindesigning270
9.3Boundarybetweenusandproteindesigning271
9.Limitationsandfuturechallengesofcomputer-aided drugdesignmethods
AshishShahandManavJain
2Limitationsofcomputer-aideddrugdesignmethods (CADD)
2.1Limitationsofstructure-baseddrugdesign(SBDD)285
2.2Limitationsofligand-baseddrugdesign(LBDD) methods287
3Challengesincomputer-aideddrugdesignmethods 290
3.1ChallengesinSBDD291
3.2ChallengesinLBDD292
3.3Challengesassociatedwiththeforcefields292
3.4Challengesassociatedwiththescoringfunction292
3.5ChallengesinADME/Tprediction293
3.6Challengesinmoleculardynamics(MD)simulation293
4FuturedevelopmentinCADD 293
4.1Nature—Asourceoflearningindrugdiscovery293
4.2Precisionmedicine294
4.3CombiningCADDwithothertechniquesofdesign, synthesis,andtesting294
Contributors
Numbersinparenthesesindicatethepagesonwhichtheauthor’scontributionsbegin.
ShabanAhmad (125),DepartmentofBiomedicalEngineering,SRMUniversity,DelhiNCR,Sonepat,Haryana,India
VishnuNayakBadavath (157),ChitkaraCollegeofPharmacy,ChitkaraUniversity, Rajpura,Punjab,India
VandanaDahiya (125),DepartmentofBiomedicalEngineering,SRMUniversity, Delhi-NCR,Sonepat,Haryana,India
RasmitaDash (231),DepartmentofPharmaceutics,SchoolofPharmaceuticalSciences, Siksha‘O’AnusandhanDeemedtobeUniversity,Bhubaneswar,Odisha,India
AndreM.deOliveira (1,101),DepartmentofEnvironmentStudies,FederalCentreof TechnologicalEducationofMinasGerais(CEFET-MG),Contagem,MG,Brazil
ManavJain (283),DepartmentofPharmacology,PostgraduateInstituteofMedical EducationandResearch,Chandigarh,Punjab,India
VenkatesanJayaprakash (181),DepartmentofPharmaceuticalSciencesand Technology,BirlaInstituteofTechnology,Mesra,Ranchi,Jharkhand,India
MuthukumarKaruppasamy (17),YaAnPharmaceuticalandMedical Communications,Sivakasi,TamilNadu,India
BanothKaranKumar (181),MedicinalChemistryResearchLaboratory,Departmentof Pharmacy,BirlaInstituteofTechnologyandSciencePilani,PilaniCampus,Pilani, Rajasthan,India
ManojKumarMahapatra (17),KanakManjariInstituteofPharmaceuticalSciences, Rourkela,Odisha,India
SiddharthaMaji (157),InstituteofPharmacy,Ram-EeshInstituteofVocational& TechnicalEducation,GreaterNoida,UttarPradesh,India
SujataMohapatra (231),DepartmentofPharmaceutics,SchoolofPharmaceutical Sciences,Siksha‘O’AnusandhanDeemedtobeUniversity,Bhubaneswar,Odisha, India
SheikhMurtuja (181),DepartmentofPharmaceuticalSciencesandTechnology, BirlaInstituteofTechnology,Mesra,Ranchi,Jharkhand;KIETSchoolofPharmacy, KIETGroupofInstitutions,Ghaziabad,UttarPradesh,India
SankaranarayananMurugesan (181),MedicinalChemistryResearchLaboratory, DepartmentofPharmacy,BirlaInstituteofTechnologyandSciencePilani,Pilani Campus,Pilani,Rajasthan,India
SubratKumarPattanayak (157),DepartmentofChemistry,NationalInstituteof TechnologyRaipur,Raipur,Chhattisgarh,India
GouravRakshit (181),DepartmentofPharmaceuticalSciencesandTechnology,Birla InstituteofTechnology,Mesra,Ranchi,Jharkhand,India
BiswabhusanRath (231),AirisPharmaPrivateLimited,Hyderabad,Telengana,India
JurnalReang (57),DepartmentofPharmaceuticalChemistry,SchoolofPharmaceutical Sciences,DelhiPharmaceuticalSciencesandResearchUniversity,NewDelhi,India
MithunRudrapal (1),DepartmentofPharmaceuticalChemistry,RasiklalM.Dhariwal InstituteofPharmaceuticalEducationandResearch,Chinchwad,Pune,India
ChitaRanjanSahoo (231),CentralResearchLaboratory,InstituteofMedicalSciences andSUMHospital,Siksha‘O’AnusandhanDeemedtobeUniversity,Bhubaneswar, Odisha,India
AnikSen (157),DepartmentofChemistry,InstituteofScience,GITAM(Deemedtobe University),Visakhapatnam,AndhraPradesh,India
AshishShah (283),DepartmentofPharmacy,SumandeepVidyapeeth,Vadodara, Gujarat,India
TriptiSharma (231),DepartmentofPharmaceuticalChemistry,SchoolofPharmaceutical Sciences,Siksha‘O’AnusandhanDeemedtobeUniversity,Bhubaneswar,Odisha,India
RajivKumarTonk (57),DepartmentofPharmaceuticalChemistry,Schoolof PharmaceuticalSciences,DelhiPharmaceuticalSciencesandResearchUniversity, NewDelhi,India
ManishKumarTripathi (125),DepartmentofPharmaceuticalEngineeringand Technology,IndianInstituteofTechnology,BanarasHinduUniversity,Varanasi, UttarPradesh;DepartmentofBiophysics,AllIndiaInstituteofMedicalSciences, NewDelhi,India
RashmiTyagi (125),DepartmentofBiomedicalEngineering,SRMUniversity,DelhiNCR,Sonepat,Haryana,India
Vinita (57),DepartmentofPharmaceuticalChemistry,SchoolofPharmaceutical Sciences,DelhiPharmaceuticalSciencesandResearchUniversity,NewDelhi,India
ManojKumarYadav (125),DepartmentofBiomedicalEngineering,SRMUniversity, Delhi-NCR,Sonepat,Haryana,India
VivekYadav (57),DepartmentofPharmaceuticalChemistry,SchoolofPharmaceutical Sciences,DelhiPharmaceuticalSciencesandResearchUniversity,NewDelhi,India
Chapter1
Introductiontodrugdesign anddiscovery
AndreM.deOliveiraa andMithunRudrapalb aDepartmentofEnvironmentStudies,FederalCentreofTechnologicalEducationofMinasGerais (CEFET-MG),Contagem,MG,Brazil, bDepartmentofPharmaceuticalChemistry,Rasiklal M.DhariwalInstituteofPharmaceuticalEducationandResearch,Chinchwad,Pune,India
Chapteroutline
1Definitionandconceptofdrugdesignanddiscovery
Daybyday,alongsidepopulationgrowthandenvironmentalproblems,newdiseasesariseandoldonesrespawn,anditdemandsnewandbettertreatments.The developmentofnewbioactivecompoundsisataskthathasbeenbenefitedby thevarioustechnologiescurrentlybeingdevelopedbythepharmaceutical industry,universities,andresearchcenters.Theimmediateeffectofthese advancesistheevidenceofanincreaseinlifeexpectancy,estimatedataround 2yearsmoreeachweek.1 Thebasisofmanynewdrugdevelopmentmethodsis thesearchforbiologicalreceptors,notablyproteins.Itisestimatedthatofthe approximately20,000typesofknownproteins,approximately500areeligible asbiologicaltargetsfordrugs.2 DatafromtheNationalCenterforHealthStatisticsindicatethat,consideringonlytheUnitedStates,themaincausesofdeath
Intentional self-harm (suicide)
Influenza and Pneumonia
Nephritis, nephrotic syndrome, and nephrosis
Alzheimer´s disease
Stroke (cerebrovascular diseases)
Chronic lower respiratory diseases
Accidents (unintentional injuries)
100200300400500600700
FIG.1.1 Numberofdeathsforleadingmortalitycauses. (https://www.cdc.gov/nchs/fastats/ deaths.htm (accessed15November2021).)
areassociatedwithcardiovascularandcerebrovasculardiseases,cancer, chroniclowerrespiratorydiseases,andkidneydiseases(Fig.1.1).
Theconceptsofdrugdiscoveryanddrugdesigncanoccasionallybeconfused.Drugdiscoveryinvolvesseveralpaths,rangingfromchancetosystematic search,usingexperimentalandcomputationalresources.Thedesignofdrugs,in turn,referstotheuseofasystematicmethod,usuallybasedonthestructureofthe biologicaltargetandonpreviousknowledgeaboutthebiochemistryofthedisease,tofindpromisingcompounds.3 Itisalsopossibletosearchfornewdrugs basedonthestructureofdrugsalreadyknownandtested,whichsupposedly inhibitcertainenzymes.Thisapproachisknownasligand-baseddrugdesign.
Anoffshootofthedrugdesignconceptiscomputer-aideddrugdesign(CADD), whichemployscomputationalmethodsasmolecularmodelingtostudydrugreceptorinteractionatamicroscopiclevel.Thetraditional(notmediatedbya computer)processofdevelopinganewdrugcantake,accordingtoestimates,about 10–14years,atanaveragecostofmorethan $1billion,withtheexclusionofabout 10,000candidates.TheCADDapproachcanreducethiscostbyupto50%.4
2Historicalperspectivesofdrugdiscovery
Althoughtheuseofmedicinalplantsisreportedevenintheoldestsources,the systematicdevelopmentofnewdrugsisarelativelyrecentscience.Between 1872and1874,theanatomistWilhelmWaldeyer,whoworkedinthelaboratory ofPaulEhrlich(whostudiedtheaffinityofdyesforbiologicaltissues),
proposedtheconceptof“chemoreceptors.”Ehrlichproposedthatthedifferencesinaffinityofthechemoreceptorsofparasites,microorganisms,andtumor cellsagainststructurallyanalogoussubstanceswerethebasisfortheemergence ofchemotherapy.5
Thefirststepstakenbymedicinalchemistryinthenineteenthcenturyowe itsprogresstothemethodsofanalyticalchemistry,whichenabledtheisolation ofnaturalproductssuchasmorphine(isolatedfromopiumbyF.W.Serturnerin 1815)andnumerousaromaticcompoundsobtainedfromcoaltar.6 ThefoundationsofpharmacologywerelaidbyOswaldSchmiedebergbetween1871and 1918,7 certainlysupportedbytheexperimentsinphysiologybyFrancois MagendieandClaudeBernard.Thedyeindustryalsocontributedtothedevelopmentofsulfadrugs,whichwerefundamentalinfightinginfectionsuntilthey weresupplantedbythediscoveryofpenicillinbyAlexanderFlemingin1928 (firstusedasanantibioticin1940).Thesuccessofpenicillinledtothesearch forotherantimicrobialcompounds,leadingtothediscoveryoflovastatin,ivermectin,cyclosporins,amongothers.
Thedevelopmentofbiochemistryledtothestudyofmetabolicpathways andmacromolecularreceptors,whichservedasthebasisfortheproposition ofenzymeinhibitors,agonists,andantagonists.Thediscoveryofthetwotypes ofadrenergicreceptorsbyR.P.Ahlquist8 wasanimportantstepintothis approach.
TheuseofthemostdiversemoleculartargetshasgrownduetotheinvaluablecontributionsofX-raydiffractiontechniquesandtheconstructionofcrystallographicdatabasessuchastheProteinDatabase(PDB).9 Asurvey conductedintheUSFDA’sOrangeBookbyOveringtonandcolleaguesin 20062 drawsascenarioofthenatureandnumberofmoleculartargetscurrently availablefornewdrugdevelopment(Table1.1).
Themostrecentphaseinthehistoricalprocessofdevelopingnewdrugs involvescomputer-aidedmoleculardesign(CAMD),whichusescomputational resourcestostudydrug-targetmolecularinteractions.Thistypeofstrategy accompaniesthedevelopmentofnewsoftwareandhardwareresources,which allowworkingwithincreasinglychallengingsystemsintermsofsizeandcomplexity.1 Computationalmethodsapplicabletopharmaceuticalplanning include:semiempiricalmethods,whichmakeitpossibletocalculateelectronic properties,energies,andmolecularconformations10–14;empiricalmethods, suchasmolecularmechanics,whichareusedtodealwithlargestructuressuch asproteins,ionchannels,membranereceptors,nucleicacids,amongothers, obtainingenthalpicinteractionenergies,interactionfreeenergies,andinhibitionconstants,15–18 andpursuingmoleculardynamicsanddockingstudies19–21;statisticalmethods,withquantitativestructure-activityrelationships,22–26 whichallowestablishingcorrelationsbetweenthepropertiesofcompounds andtheirbiologicalactivityandtoxicity.27,28
TABLE1.1 MoleculartargetsofFDA-approveddrugs.
ClassofdrugtargetSpecies
Numberofmolecular targets
TargetsofapproveddrugsPathogenand human 324
Humangenometargetsofapproved drugs Human266
Targetsofapprovedsmall-molecule drugs Pathogenand human 248
Targetsofapprovedsmall-molecule drugs Human207
Targetsofapprovedoralsmallmoleculedrugs Pathogenand human 227
Targetsofapprovedoralsmallmoleculedrugs Human186
Targetsofapprovedtherapeutic antibodies Human15
TargetsofapprovedbiologicalsPathogenand human 76
(AdaptedfromreferenceOveringtonJP,Al-LazikaniB,HopkinsAL.Howmanydrugtargetsarethere? NatRevDrugDiscov2006;5(12):993–996. https://doi.org/10.1038/nrd2199.)
3Process,strategies,andstagesofdrugdiscovery anddevelopment
Theprocessofgeneratinganewdrugislongandcostly.Itisestimatedthatit takes10to15yearsandfiguresashighasUSD985millionfromconceptionto commercializationofanewdrug.29 Aconsiderablenumberofclinicallytested candidatesareexcludedduetotheirtoxicityandlowbioavailability.Itisestimatedthatonly1outof10newdrugsthatpassclinicaltrialswillreachthemarket.Tounderstandthereasonsbehindthishighcost,wemustrememberthe stagesorphasesinvolvedindevelopinganewdrug.30 Thefollowingphases areconsideredindrugdevelopment:discoveryphase,preclinicalphase,Phase I,PhaseII,PhaseIII,andtheapprovalandpostapprovalphases(Fig.1.2). Despitethesewell-definedphasesonthepathtofindinganewdrug,thisdoes notmeanthatthesestepsdonotreverseorintermingleeventually,withcharacteristicstudiesofone-stepsupportingstudiesofanother.
FIG.1.2 Drugdiscoveryanddevelopmentphases. (Nopermissionrequired.)
3.1Discoveryphase
Themainsourceofbioactivecompoundsthatcanbecomedrugcandidatesis nature,throughitsnaturalproducts.31,32 Manyofthesecompoundsareisolated fromterrestrialplants33–35 andanimals,36 andthereisalsoanimportantandunexploredsourceofnewcompoundsinmarineorganisms.10,37,38 Thediscoveryofa newdrugcanoccurindifferentways.Therearedrugsthatweretheworkofserendipity,suchaspenicillin,discoveredbyAlexanderFleming.WorkingatSt. Mary’sHospital,London,in1928,hewasstudyingthebacteria Staphylococcus aureus,responsibleforabscessesinopenwoundscausedbyfirearms.Whenhe wentonvacation,heleftthelaboratory’sglasscontainers,withthebacteriaculturesexposedandcontaminatedwiththemoldfromtheatmosphereitself.Ashe wasabouttodiscardallthematerial,henoticedthatwheremoldhadformed, therewasnoactive Staphylococcus:Themold,originatingfromthe Penicillium fungus,actedbysecretingasubstancethatdestroyedthebacteria.In1940,penicillinwasused,inEngland,inthefirsthumanpatient,apoliceman,avictimofa seriousbloodinfection.Thediscoveryofthisantibioticchangedthecourseof WorldWarIIandthefateofrecenthistory.
Despitetheimportanceofserendipity,whichbequeathedusalsootherrugs, suchassulfadrugs,37 andnewusesforalreadyknowndrugs,suchassildenafil,39 moresystematicandmethodicalpathshavebeenfollowedinthesearchfor thesesubstances.Thesemethodshavebeentraditionallyclassifiedintothefollowingcategories:virtualscreening,high-throughputscreening,phenotypic screening,structure-baseddrugdesign,fragment-baseddrugdesign,and ligand-baseddrugdesign.Wewillfurtherdiscussthenatureandextentofthese methodsinthenextsection.
3.2Preclinicalphase
Onceasetofleadcompoundsisavailable,preclinicaltestinghelpstoguidethe studybyexcludingcompoundswithlittleornopharmacologicalpotential.This phaseincludes:invitrotestswithcellandtissuecultures,measurementsof interactionwithproteinsandotherbiologicaltargets,whichcanbeperformed bymeansofisothermaltitrationmicrocalorimetry40;invivotestswithanimals, chosenaccordingtotheirphysiologicalandbiochemicalcharacteristicsthatcan resemblehumans(Fig.1.3).Thepreclinicalphaseallowsforthepromotionof leadcompoundstocandidatestatus,whichwillbesubmittedtothesubsequent phasesofthesearchfornewdrugs. Discovery Pre-clinical Phase I
3.3Clinicalphase
PhaseI. PhaseIbeginswiththefirsttestsinhumans,healthyvolunteerswithout cytotoxicproblems(suchascancerpatients),toassesssafety,tolerance,pharmacodynamics,pharmacokinetics,andtoxiceffectsthatdonotdependon populationeffects.PhaseIstudiesrequireapprovalfromaresearchethicscommitteeandregulatoryagencies.Otherstudieslikedrug-druginteraction,the effectoffoodonabsorption,age,andgeneticinfluencesarealsoconducted.30
PhaseII. ThemainobjectiveofPhaseIIistodeterminethedrug’sefficacy andsafetyinatargetpopulation.Groupswithcommonandcontrolledpathologies(suchashypertension)arechosentotrythedrug,andtheresultsarecomparedwithcontrolgroups,whichweregivenaplacebo.PhaseIIisusually dividedintoPhaseIIaandPhaseIIb.InPhaseIIa(calledthe“proofofconcept”),thedrugistestedinasubgroupofpatients(namely12–100individuals); PhaseIIbisperformedwithseveraldoseleveltestsinthetargetpopulation (dose-rangingstudies).Thistaskaimstodefinetheminimallyeffectiveorineffectivedoseandtofindtheoptimaldose(Fig.1.4).
PhaseIII. Thelastphaseofthedevelopmentofanewdrugbeforeitscommercializationaimstodetermineitsclinicaldosage,prescription,andfrequency ofuse.Thisstudycouldinvolvethousandsofpatientsandshedsnewlightonthe drug’sefficacyandsafetyinlargepopulations.Thisphasecantakeseveral years,withmonitoringofmortalityandcomorbidityrates,anditsdetailsvary fromcountrytocountry.Inpopulations,geneticissuesareveryrelevantand accountfordifferencesineffectivenessbetweendifferentcommunities.Itis
Cow Monkey
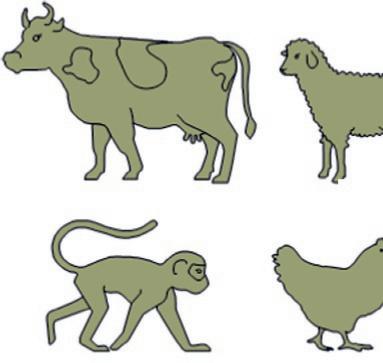
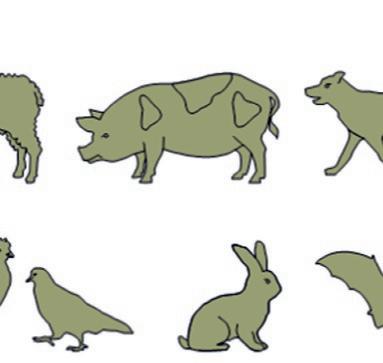
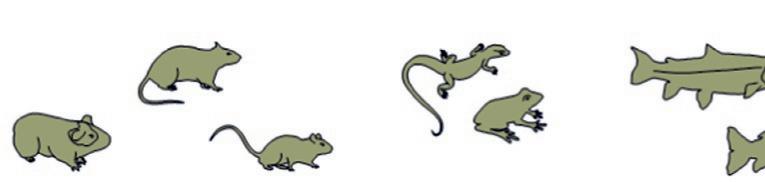
Guineapig Rat
BirdRabbit
SheepPigDog
Fish
Amphibian
Mouse Bat
FIG.1.3 Animalsusedinpreclinicaltests. (Nopermissionrequired.)
FIG.1.4 OptimaldosepursuitinPhaseIIdrugdevelopment. (Reproduced,bypermission,from OptimizingNutrition(2021).)
estimatedthattheoverallsuccessrateofPhaseIIIisaround70%andcancostup toUSD100million.
3.4Approvalandpostapprovalphases
Thelegalissuesrelatedtopublichealthineachregionoftheworldwillguide thisadditionalphaseofthebirthofanewdrug,anditinvolvesgovernment agenciesthatworkinareasasdiverseas:economy,sanitation,hospitaladministration,healthsurveillance,andmanufacturingrights.Patentissuesareraised andmanagedbyregularintra-andinternationalagencies.Dependingonthe performanceofthenewdrug,itsusemaybereconsidered,limitedorregulated. Fromthispointonwards,repositioningstudies,whichaimtofindnewapplicationsforit,canbeundertaken.
4Traditionalandmodernapproachestodrugdiscovery anddevelopment
Thediscoveryofanewdrugcanoccuralongverydifferentpaths.Verybriefly, themethodscanbeclassifiedintothefollowingcategories:
4.1Virtualscreening
Thevirtualscreeningmethodisbasedonacomparativeanalysisbetweendifferentleads(candidatesfornewdrugcandidates),usingcomputationalresources, generallybasedonthecorollarythatthedrug’sactionisdirectlyrelatedtoits affinityforabiologicaltarget.Inthiscontext,dockingisthemostusedtechnique, whichconsistsoftheinteractionbetweenamoleculeandabiologicaltarget,calculatingtheinteractionenergybetweenthem.Thecalculationsgenerallyemploy
moleculardynamics,makingitpossibletoobtaininteractionfreeenergiesand, fromthere,theinhibitionconstants(anexperimentalparameterthatcanbeverified).Thereareonlineresourcesthatenablevirtualscreening,oftencomparing interactionenergieswithbiologicalactivitydata.41 Theuseofartificialintelligencetechniqueshasbeenfundamentalinthesystematizationofthesestudies.42–44 Virtualtissueandorganmodelsareanelegantsolutiontothis approach,beingabletoanticipateevenphysiologicalandneuralphenomena.45
4.2High-throughputscreening
High-throughputscreening(HTS)isanunfoldingofvirtualscreening,withthe dockingofthousandsofmoleculescontainedinthestructuredatabase,using statisticalclusteringtechniquestochoosethebestcandidates.Fromanexperimentalpointofview,combinatorialchemistrymakesitpossibletotestnumerouscompoundsinanautomatedway.Anexampleofthisapproachisthe automaticsynthesisofoligopeptidesandthesubsequentbiologicaltestingof eachofthem.18
4.3Phenotypicscreening
Accordingtothisstrategy,compoundsarescreenedincellularoranimaldisease modelstoidentifycompoundsthatcauseadesirablechangeinphenotype.43 Whenanewtargetissearchedforsomepropertiesthatareimportanttothedisease,wecallittargetdeconvolution.Manyresearchstudiesusingphenotypic screeninghavebeendescribed,appliedtoinfectiousdiseases,46–48 contraceptivetreatments,49 childhooddiarrhea,50,51 amongothers.
4.4Structure-baseddrugdesign
TheavailabilityofnumerousbiologicaltargetbankssuchasPDB,ZINC,and TTD52 hasservedthestructure-basedstudyofnewdrugs.Themaindifference betweenthisapproachandphenotypicscreeningisthatthesearchfornewcompoundsisbasedonthecrystallographicstructuresoftargetsandcompoundsand theirinteractions,andnotnecessarilyonpharmacologicalproperties,although informationfrombothcanbesharedandcompared.
4.5Fragment-baseddrugdesign
Since1981,whenJencksintroducedtheideathatanentiremoleculecanbe interpretedasacombinationoffragmentsthatindividuallycontributetothe interaction,thefragmentscreeningstrategyhasbeendeveloped.53 GRIDprogramcreatedbyGoodford54 employedprobes(molecularfragments)tomap ontoaproteinstructureandyieldaninteractionsurfacearoundsomehotspots. Thisstrategywasfundamentalforthedevelopmentofcomparativemolecular
fieldanalysis(CoMFA)method55 thatappliesapartialleastsquare(PLS)multipleregressionwiththeinteractionenergiesofthehotspotandstericandelectrostaticfactors.Fragmentlibrariesbecamepopular56 andservedasabasisfor thediscoveryofnewcompoundswithoutanysimilarityorkinshipwithtraditionalnaturalproducts.Animportantadditiontothesemethodswastheinclusionofsyntheticpenalties,thatis,factorsthatpotentiallyhinderedthesynthesis oftheproposedcompounds(suchasthepresenceofchiralcenters,fusedrings, etc.)andthatmadeitpossibletolimittheproposalstocompoundswithbetter accessibility.
4.6Ligand-baseddrugdesign
Theproposalofnewdrugsbasedonasetofstructurallysimilarcompoundsof recognizedbiologicalactivityisthebasisofquantitativestructure-activityrelationships(QSARs).Sinceitispossibletobuildpharmacophoricmodelsofa receptorevenwithoutknowingitsstructure,basedsolelyonthestructureactivityrelationshipsofthecompoundswithwhichitinteracts,thisisa ligand-basedstrategy.Molecularmodelingtechniquescanalsobebasedon theligand,withthepredictionofelectronic,topological,andsoon,properties thatbytheirturnshallbeusedinQSARmodels.57,58
5Rationaldrugdesign(RDD)andCADD
Thecentralideaaroundmoderndrugdesignisrationalityandmethod.Despite thesuccessfulexperiencesofdrugdiscoverybychanceortheisolationofnaturalproducts,whosestructuralcharacteristicsfollowthepatternsdeterminedby thebiochemistryoftheorganismthatsynthesizedthem,thepharmaceutical industryislookingformorecontroltechniquestoproposenewstructures. Therationaldrugdesign(RDD)usesthetechniquesmentionedaboveona semanticbasisaimingtoimprovethepharmacologicalpropertiesanddecrease thetoxicityofthecandidates.
Mandalandcollaborators59,60 divideRDDintotwocategories:
1. Developmentofsmallmoleculeswithdesiredpropertiesfortargets,and biomolecules(proteinsornucleicacids),whosefunctionalrolesincellular processesand3Dstructuralinformationareknown.
2. Developmentofsmallmoleculeswithpredefinedpropertiesfortargets, whosecellularfunctionsandtheirstructuralinformationmaybeknown orunknown.Knowledgeofunknowntargets(genesandproteins)canbe obtainedbyanalyzingglobalgeneexpressiondataofsamplesuntreated andtreatedwithadrugusingadvancedcomputationaltools.
Thefirstcategoryisthemostusedbythepharmaceuticalindustry,andthesecondoneismoreusedforacademicpurposes,althoughtheconstantcooperation betweenthetwosegmentsallowsbothcategoriestodialoguewitheachother.
Computer-aideddrugdesign(CADD)isthenamegiventothesetofcomputationaltechniquesthatusethestateoftheartofRDDtobuildaninventoryof bioactivesubstances.Inthiscontext,bioinformaticstechniques(includingproteomics,metabolomics,andgenomics)arefundamental.Asurveyofthemain CADDtechniquesismadebyMacalinoandcoworkers,59 andalthoughthey havealreadybeendescribedinourshortessay,itisusefultomakeadigest:
5.1Structure-baseddrugdesign(SBDD)
Theknowledgeacquiredfromthebindingsiteofa3Dmacromoleculestructure isusedtodesignandevaluateligandsbasedontheirpredictedinteractionswith theproteinbindingsite,bymeansofstructuraltechniqueslikeX-raycrystallography,nuclearmagneticresonance(NMR),cryo-electronmicroscopy(EM), homologymodeling,andmoleculardynamic(MD)simulations.
5.1.1Moleculardocking
Itstartswithbindingsitepredictionoridentification.Thebindingsiteisusually aconcaveregionontheperipheryoftheproteinwhereinteractionwiththe ligandoccurs.Topologicalmethodsforidentifyingactivesites,suchas CASTp,60 allowustostudythesuitablesitesforthisinteraction.Dockinguses knowledgeofproteinbindingsitestotestforinteractionswithsmallmolecules. Numerouspositionsaretestedandthenclassifiedaccordingtoscoringcriteria. Thisscoringfollowsamathematicalfunctionthatvariesdependingontheprogramandthatdistinguishesthedifferentdockingmethodsintheirapplicability. Theinteractionenergyisobtainedthroughanempiricalexpressionthat involvesenthalpic,entropic,andhydrophobiccontributions,inadditiontosolvationeffects.Dockingcanberigid,whichisusefulwhenscanninglargedatabasesofstructures,orflexible,whenwewanttodrawamoredetailedprofileof theligand-receptorinteraction.Theeffectsofthesolventontheinteractioncan besimulatedindirectly,throughtheintroductionofacorrectionintheexpressionoftheinteractionenergy,orexplicitly,throughtheuseofsolventboxes.
5.1.2Moleculardynamics
Moleculardynamicscanbedefinedasthesetofexperimentalprotocolsthat simulatetheconformationalvariationsexperiencedbythemoleculebythe actionofforcesactingonthemedium.ThisprocedureappliesNewton’sequationstotheinternalcoordinatesofthemolecule,withinagiventimeframe,in ordertofollowthevariationsintheinternaldegreesoffreedomofthemolecule. ThedifferentialformofNewton’sclassicalequationFi ¼ mi ai.Anerrorinherenttothemethodwillbemorepronouncedthelongerthetimestepused,which istypicallyontheorderof0.5to1femtosecond(1fs ¼ 10 15 s).Ingeneral,we startwithaminimizedstructureandprocessthedynamics,recordingthe
resultingstructurefromtimetotimeandminimizingeachoftheseconformationsagain,sothat,intheend,weproceedtoselecttheconformationwhose energynolongervaries.
5.2Ligand-baseddrugdesign(LBDD)
Inthelackofthetargetstructureandtheavailabilityofasetofsimilarcompounds,onecanmaketheassumptionthatstructurallysimilarcompoundsdisplaysimilarbiologicalresponseandinteractionwiththetarget.
5.2.1Quantitativestructure-activityrelationship(QSAR)
Fromasetofsimilarmolecules,physicochemicaldescriptors(electronic, hydrophobic,steric),topological,amongothers,areobtained.Amultivariate regressionanalysisisperformedbetweenthesedescriptorsandbiologicalactivity,resultinginanequationthatrepresentsamodelofthesysteminvestigated. The3D-QSARapproachusesthethree-dimensionalstructuresofcompounds, properlyaligned,onwhichaprobe(forexample,apositivecarbon)isplaced aroundagridofpoints,calculatingthestericandelectrostaticenergy.Each valueoftheseenergiesbecomesaphysicochemicaldescriptorinthemodelthat usesbiologicalactivitydataasadependentvariable.Thepointsofinteraction favorableorunfavorabletobiologicalactivityareconvertedintoathreedimensionalmapshowingregionsfavoredbylargegroupsandregionsfavored bysmallgroups,andregionsfavoredbypositivegroupsandthosefavoredby thesegroups.
5.2.2Pharmacophoremodelingandsimilaritysearch
Throughpharmacophorescreening,itispossibletoidentifycompoundscontainingdifferentscaffolds,butwithasimilar3Darrangementofkeyinteractingfunctionalgroups,ontowhichbindingsiteinformationcanbeincorporated.61–63
5.2.3Absorption,distribution,metabolism,excretion,andtoxicity (ADMET)prediction
Thedeterminationofpharmacodynamic,pharmacokinetic,andtoxicological propertiesisessentialforobtainingrelevantbioactivecompounds.Lipinski’s rules64 (amoleculewithamolecularmasslessthan500Da,nomorethan5 hydrogenbonddonors,nomorethan10hydrogenbondacceptors,andan octanol-waterpartitioncoefficientlogPnotgreaterthan5)allowdetermining whetherthecompoundhaspotentialasadrugwithoutpresentingtoxicological characteristicsthatmakeitsuseunfeasible.ManytoolsforADMETareavailable,suchasQikProp.65
12
6Conclusion
Populationgrowthandenvironmentalproblemshaveledtotheemergenceor worseningofmanydiseases.Inthisscenario,thechallengeofdiscovering newdrugscapableofalleviatinghumansufferingandprovidingabetterquality oflifeisimposed.Universitiesandresearchcentershaveagreatopportunityto partnerwithgovernmentsandcompaniesinthesearchformoreaccessibleand efficienttherapeuticmethods.Strategiesfordiscoveringnewbioactiveagents willbeofgreatimportanceinthefuture,especiallyinaglobalizedeconomy, whichhasproventruewiththerecentSARS-CoV-2pandemic.Remediation techniques,combinedwithimmunology,weredecisiveincombatingitsworldwideconsequences.Drugrepositioningandmolecularmodelingtechniques haveonceagainprovedtheirworthinacontextofmanyuncertainties.
References
1.SeddonG,LounnasV,McGuireR,etal.Drugdesignforever,fromhypetohope.JComput AidedMolDes2012;26(1):137–50. https://doi.org/10.1007/s10822-011-9519-9.
2.OveringtonJP,Al-LazikaniB,HopkinsAL.Howmanydrugtargetsarethere?NatRevDrug Discov2006;5(12):993–6. https://doi.org/10.1038/nrd2199.
3.AndersonAC.Theprocessofstructure-baseddrugdesign.ChemBiol2003;10(9):787–97. https://doi.org/10.1016/j.chembiol.2003.09.002
4.SurabhiS,SinghB.Computeraideddrugdesign:anoverview.JDrugDelivTher2018;8 (5):504–9. https://doi.org/10.22270/jddt.v8i5.1894
5.DrewsJ.Drugdiscovery:ahistoricalperspective.Science2000;287(5460):1960–4. https://doi. org/10.1126/science.287.5460.1960
6. FarberE.Theevolutionofchemistry:ahistoryofitsideas.In:Methods,andmaterials.New York:RolandPress;1952.
7.Koch-WeserJ,SchechterPJ.SchmiedeberginStrassburg1872-1918:themakingofmodern pharmacology.LifeSci1978;22(13–15):1361–71. https://doi.org/10.1016/0024-3205(78) 90099-1.
8.AhlquistRP.Astudyoftheadrenotropicreceptors.AmJPhysiol1948;153(3):586–600. https:// doi.org/10.1152/ajplegacy.1948.153.3.586
9.BermanHM,WestbrookJ,FengZ,etal.Theproteindatabank.NucleicAcidsRes2000;28 (1):235–42. https://doi.org/10.1093/nar/28.1.235
10.KaramanR,FattashB,QtaitA.Thefutureofprodrugs-designbyquantummechanicsmethods. ExpertOpinDrugDeliv2013;10(5):713–29. https://doi.org/10.1517/17425247.2013.786699
11.KromannJC,LarsenF,MoustafaH,JensenJH.PredictionofpKavaluesusingthePM6semiempiricalmethod.PeerJ2016;2016(8). https://doi.org/10.7717/peerj.2335.
12.Krı ´ z ˇ K, Řeza ´ cJ.Benchmarkingofsemiempiricalquantum-mechanicalmethodsonsystemsrelevanttocomputer-aideddrugdesign.JChemInfModel2020;60(3):1453–60. https://doi.org/ 10.1021/acs.jcim.9b01171.
13.Leps ˇ ı ´kM, Řeza ´ cJ,Kola ´ rM,PecinaA,HobzaP,Fanfrlı´kJ.Thesemiempiricalquantum mechanicalscoringfunctionforinsilicodrugdesign.ChemPlusChem2013;78(9):921–31. https://doi.org/10.1002/cplu.201300199
14.ShivakumarD,WilliamsJ,WuY,DammW,ShelleyJ,ShermanW.Predictionofabsolute solvationfreeenergiesusingmoleculardynamicsfreeenergyperturbationandtheoplsforce field.JChemTheoryComput2010;6(5):1509–19. https://doi.org/10.1021/ct900587b.
15.BekonoBD,SonaAN,EniDB,OwonoLCO,MegnassanE,Ntie-KangF.Molecularmechanics approachesforrationaldrugdesign:forcefieldsandsolvationmodels.PhysSciRev2021. https://doi.org/10.1515/psr-2019-0128
16.ErenD,Yalc ¸ in _ I.Theaimofimplementationofthemolecularmechanicandthemolecular dynamicmethodsinrationaldrugdesign.AnkaraUnivEczacilikFakDerg2020;44(2): 334–55. https://doi.org/10.33483/jfpau.688351
17.PissurlenkarRRS,ShaikhMS,IyerRP,CoutinhoEC.Molecularmechanicsforcefieldsand theirapplicationsindrugdesign.Anti-InfectAgentsMedChem2009;8(2):128–50. https:// doi.org/10.2174/187152109787846088
18.WangJ,LiH,PanJ,etal.Oligopeptidetargetingsortaseaaspotentialanti-infectivetherapyfor Staphylococcusaureus.FrontMicrobiol2018;9. https://doi.org/10.3389/fmicb.2018.00245.
19.FerreiraLG,DosSantosRN,OlivaG,AndricopuloAD.Moleculardockingandstructurebaseddrugdesignstrategies.Molecules2015;20(7):13384–421. https://doi.org/10.3390/ molecules200713384
20.TorresPHM,SoderoACR,JofilyP,Silva-JrFP.Keytopicsinmoleculardockingfordrug design.IntJMolSci2019;20(18). https://doi.org/10.3390/ijms20184574
21.ZoeteV,GrosdidierA,MichielinO.Docking,virtualhighthroughputscreeningandinsilico fragment-baseddrugdesign.JCellMolMed2009;13(2):238–48. https://doi.org/10.1111/ j.1582-4934.2008.00665.x
22.BradshawJ.3DQSARindrugdesign.TrendsPharmacolSci1994;15(12):469–70. https://doi. org/10.1016/0165-6147(94)90062-0.
23.DanishuddinKAU.DescriptorsandtheirselectionmethodsinQSARanalysis:paradigmfor drugdesign.DrugDiscovToday2016;21(8):1291–302. https://doi.org/10.1016/j.drudis.2016. 06.013.
24.TandonH,ChakrabortyT,SuhagV.AconcisereviewonthesignificanceofQSARindrug design.ChemBiomolEng2019;4(4):45. https://doi.org/10.11648/j.cbe.20190404.11
25.VermaJ,KhedkarVM,CoutinhoEC.3D-QSARindrugdesign—areview.CurrTopMed Chem2010;10(1):95–115. https://doi.org/10.2174/156802610790232260
26.S ´ ledz´P,CaflischA.Proteinstructure-baseddrugdesign:fromdockingtomoleculardynamics. CurrOpinStructBiol2018;48:93–102. https://doi.org/10.1016/j.sbi.2017.10.010
27.DeardenJC.Predictionofenvironmentaltoxicityandfateusingquantitativestructure-activity relationships(QSARs).JBrazChemSoc2002;13(6):754–62. https://doi.org/10.1590/S010350532002000600005.
28.SchultzTW,HewittM,NetzevaTI,CroninMTD.AssessingapplicabilitydomainsoftoxicologicalQSARs:definition,confidenceinpredictedvalues,andtheroleofmechanismsof action.QSARCombSci2007;26(2):238–54. https://doi.org/10.1002/qsar.200630020
29.WoutersOJ,McKeeM,LuytenJ.Estimatedresearchanddevelopmentinvestmentneededto bringanewmedicinetomarket,2009-2018.JAMA2020;323(9):844–53. https://doi.org/ 10.1001/jama.2020.1166
30.TamimiNAM,EllisP.Drugdevelopment:fromconcepttomarketing!NephronClinPract 2009;113(3):c125–31. https://doi.org/10.1159/000232592
31.AtanasovAG,ZotchevSB,DirschVM,etal.Naturalproductsindrugdiscovery:advancesand opportunities.NatRevDrugDiscov2021;20(3):200–16. https://doi.org/10.1038/s41573-02000114-z.