https://ebookmass.com/product/biophysical-characterizationof-proteins-in-developing-biopharmaceuticals-second-editionedition-damian-j-houde-editor/
Instant digital products (PDF, ePub, MOBI) ready for you
Download now and discover formats that fit your needs...
Biophysical Characterization of Proteins in Developing Biopharmaceuticals 2nd Edition Damian J. Houde (Editor)
https://ebookmass.com/product/biophysical-characterization-ofproteins-in-developing-biopharmaceuticals-2nd-edition-damian-j-houdeeditor/
ebookmass.com
Proteins Galanakis
https://ebookmass.com/product/proteins-galanakis/
ebookmass.com
Total Chemical Synthesis of Proteins 1st Edition Ashraf Brik
https://ebookmass.com/product/total-chemical-synthesis-ofproteins-1st-edition-ashraf-brik/
ebookmass.com
Fluid Mechanics (2nd ed.) Global Edition Russell C. Hibbeler
https://ebookmass.com/product/fluid-mechanics-2nd-ed-global-editionrussell-c-hibbeler/
ebookmass.com
The Innkeeper of Ivy Hill Julie Klassen
https://ebookmass.com/product/the-innkeeper-of-ivy-hill-julie-klassen/
ebookmass.com
Nutrition and Functional Foods in Boosting Digestion, Metabolism and Immune Health Debasis Bagchi
https://ebookmass.com/product/nutrition-and-functional-foods-inboosting-digestion-metabolism-and-immune-health-debasis-bagchi/
ebookmass.com
Les fantômes du vieux pays Hill
https://ebookmass.com/product/les-fantomes-du-vieux-pays-hill/
ebookmass.com
Conditions of Visibility Richard Neer
https://ebookmass.com/product/conditions-of-visibility-richard-neer/
ebookmass.com
Circuit Theory and Networks Analysis and Synthesis 2nd Edition Ravish R. Singh
https://ebookmass.com/product/circuit-theory-and-networks-analysisand-synthesis-2nd-edition-ravish-r-singh/
ebookmass.com
Teaching in the Online Classroom: Surviving and Thriving in the New Normal Doug Lemov [Lemov
https://ebookmass.com/product/teaching-in-the-online-classroomsurviving-and-thriving-in-the-new-normal-doug-lemov-lemov/
ebookmass.com
BIOPHYSICALCHARACTERIZATIONOF PROTEINSINDEVELOPING BIOPHARMACEUTICALS SECONDEDITION Editedby
DAMIAN J.HOUDE
BiomolecularDiscovery,RelayTherapeutics, Cambridge,MA,UnitedStates
STEVEN A.BERKOWITZ Consultant,Sudbury,MA,UnitedStates
Elsevier
Radarweg29,POBox211,1000AEAmsterdam,Netherlands
TheBoulevard,LangfordLane,Kidlington,OxfordOX51GB,UnitedKingdom 50HampshireStreet,5thFloor,Cambridge,MA02139,UnitedStates
Copyright©2020ElsevierB.V.Allrightsreserved.
Nopartofthispublicationmaybereproducedortransmittedinanyformorbyanymeans,electronicor mechanical,includingphotocopying,recording,oranyinformationstorageandretrievalsystem,without permissioninwritingfromthepublisher.Detailsonhowtoseekpermission,furtherinformationaboutthe Publisher’spermissionspoliciesandourarrangementswithorganizationssuchastheCopyrightClearanceCenter andtheCopyrightLicensingAgency,canbefoundatourwebsite: www.elsevier.com/permissions
ThisbookandtheindividualcontributionscontainedinitareprotectedundercopyrightbythePublisher(other thanasmaybenotedherein).
Notices Knowledgeandbestpracticeinthis fieldareconstantlychanging.Asnewresearchandexperiencebroadenour understanding,changesinresearchmethods,professionalpractices,ormedicaltreatmentmaybecomenecessary.
Practitionersandresearchersmustalwaysrelyontheirownexperienceandknowledgeinevaluatingandusingany information,methods,compounds,orexperimentsdescribedherein.Inusingsuchinformationormethodsthey shouldbemindfuloftheirownsafetyandthesafetyofothers,includingpartiesforwhomtheyhaveaprofessional responsibility.
Tothefullestextentofthelaw,neitherthePublishernortheauthors,contributors,oreditors,assumeanyliability foranyinjuryand/ordamagetopersonsorpropertyasamatterofproductsliability,negligenceorotherwise,or fromanyuseoroperationofanymethods,products,instructions,orideascontainedinthematerialherein.
LibraryofCongressCataloging-in-PublicationData
AcatalogrecordforthisbookisavailablefromtheLibraryofCongress
BritishLibraryCataloguing-in-PublicationData
AcataloguerecordforthisbookisavailablefromtheBritishLibrary
ISBN:978-0-444-64173-1
ForinformationonallElsevierpublicationsvisitour websiteat https://www.elsevier.com/books-and-journals
Publisher: SusanDennis
AcquisitionEditor: AnitaKoch
EditorialProjectManager: SaraPianavilla
ProductionProjectManager: SuryaNarayananJayachandran
CoverDesigner: MatthewLimbert
TypesetbyTNQTechnologies
Contributors YvesAubin CentreforBiologics,Regulatory ResearchDivision,Evaluation,Biologicsand GeneticTherapiesDirectorate,HealthProducts andFoodBranch,HealthCanada,Ottawa,ON, Canada
StevenA.Berkowitz Consultant,Sudbury,MA, UnitedStates
GeorgeM.Bou-Assaf AnalyticalDevelopment, Biogen,Cambridge,MA,UnitedStates
MarkL.Brader DrugProductAnalytical Development,Moderna,Cambridge,MA, UnitedStates
RichardK.Burdick BurdickStatisticalConsulting,LLC,ColoradoSprings,CO,United States
JohnF.Carpenter DepartmentofPharmaceuticalSciences,CenterforPharmaceuticalBiotechnology,UniversityofColoradoAnschutz MedicalCampus,Aurora,CO,UnitedStates
StephenJ.Demarest EliLillyBiotechnology Center,SanDiego,CA,UnitedStates
ErtanEryilmaz Takeda,Cambridge,Massachusetts,UnitedStates
VernaFrasca FieldApplicationsManager,MalvernPanalytical,Northampton,MA,United States
DarronL.Freedberg StructuralBiologySection, LaboratoryofBacterialPolysaccharides,Silver Spring,MD,UnitedStates
JohnP.Gabrielson ElionLabs,ADivisionof KBIBiopharma,Inc.,Louisville,CO,United States
AndreaHawe CoriolisPharma,Munich, Germany
DamianJ.Houde BiomolecularDiscovery, RelayTherapeutics,Cambridge,MA,United States
DavidA.Keire FoodandDrugAdministration, St.Louis,MO,UnitedStates
FrancisKinderman Amgen,ThousandOaks, California,UnitedStates
LeeMakowski DepartmentofBioengineering andDepartmentofChemistryandChemical Biology,NortheasternUniversity,Boston,MA, UnitedStates
JohnP.Marino NationalInstituteofStandards andTechnology,InstituteforBioscienceand BiotechnologyResearch,Rockville,MD,United States
AlanG.Marshall IonCyclotronResonance Program,NationalHighMagneticField Laboratory,FloridaStateUniversity,Tallahassee,FL,UnitedStates;Departmentof Chemistry & Biochemistry,FloridaState University,Tallahassee,FL,UnitedStates
A.J.Miles InstituteofStructuralandMolecular Biology,BirkbeckCollege,UniversityofLondon,London,UnitedKingdom
JohnS.Philo AllianceProteinLaboratories,San Diego,CA,UnitedStates
AngelikaReichel CoriolisPharma,Munich, Germany
DenizB.Temel Bristol-MyersSquibb,Devens, Massachusetts,UnitedStates
B.A.Wallace InstituteofStructuralandMolecularBiology,BirkbeckCollege,Universityof London,London,UnitedKingdom
DanielWeinbuch CoriolisPharma,Munich, Germany
WilliamF.WeissIV BioproductResearchand Development,LillyResearchLaboratories,Eli LillyandCompany,Indianapolis,IN,United States
SarahZölls CoriolisPharma,Munich,Germany
Prefacesforthesecondedition Criticaltothedevelopmentofany successfultherapeuticdrugisourabilityto identifyandmanufacturethedrugsuch thatitsbene fi cialtherapeuticeffectcanbe safelydeliveredtothepatient.Inthecaseof proteinbiopharmaceuticals,theselarge, heterogeneous(complex)andmarginally stablemoleculesareoftenverysensitiveto theirmicro-environment.Thismakesthe processofdevelopingandmanufacturinga proteintherapeuticextremelychallenging. Throughoutthisentireprocessaproteinbiopharmaceuticalmustmaintainitscomplex anddelicatestructure(orconformation)to realizeitsbeneficialtherapeuticattributes, whileavoidingthepotentialharmfuleffects infailingtoachievethisgoal.
Whenwesetouttowritethe firstedition ofthisbook,ourgoalwastoprovidea generalresourcethatspeci ficallydealtwith themanychallengesassociatedwiththe testingandcharacterizationofthehigher orderstructureandbiophysicalpropertiesof proteinbiopharmaceuticalsfromapractical pointofviewtosupportitssafetyand bene ficialtherapeuticactivity.Asstatedin thebook’ s firstprefacewewantedtokeep thereaderfocusedonobtainingapragmatic understandingandknowledgeoftheutility ofbiophysicaltoolsandhowtheyareusedto meetthesechallengesbyunderstanding whatinformationcanrealisticallybeextractedfromthesetools.Whilewefeltwehad initiallyachievedourgoal,theprogressionof timeinevitablyledtobetterandimproved
scienti ficdevelopmentsandtotherealizationthattherewasroomforimprovements.
Asaresult,inwritingthissecondedition wehaveundertakenthejobofupdatingold information,correctingmistakes,improving clarityandtheintroductionofnewtopics thatwerenotcoveredinthe fi rstedition. Therefore,wegatheredourco-authorsonce again,invitedafewnewones,andtasked ourselveswiththegoaltoachievethese objectives.Insodoing,alloriginalchapters havebeenupdated,correctedand enhanced,whilenewchaptershavebeen added.
Globally,theformatofthebookhas remainedthesame,consistingofthreesections.SectionI,whichdealswiththe complexityofproteinsandtherelevanceof biophysicalmethodsinthebiopharmaceuticalindustry.Ithasforthemostpartbeen alteredtoremoveerrorsandachieveclarity. SectionII,whichdiscussesthebiophysical toolsandtechniquesmostcommonlyusedin thebiopharmaceuticalindustrytocharacterizeproteintherapeuticmoleculeshas similarlybeenaltered,buthasalsobeen enhancedbytheadditionofanewchapter (Chapter14)dedicatedtotheareaofchromatographyandelectrophoresis.Thetoolsin thischapter,whichwedidnotcoverinthe firsteditionofthebook(withtheexceptionof size-exclusionchromatography),aretypically notthoughtoforclassifiedasbiophysical tools.Nevertheless,animportantobjectivein addingthischapteristobringmoreattention
totheirunrealizedlinkageaseffectivebiophysicalcharacterizationtoolswithoutgettingtoodeepintothedetailsoftheirinner workings(whichareextensivelycoveredin manyexcellentbooksandreviewarticlesthat aresolelydedicatedtothesetwoenormously importanttechniques).
Overall,however,SectionIIIofthebook hasexperiencedthemostsigni ficantchange andexpansionviatheadditionoffournew chaptersthatcoverthefollowing:
• Chapter15,whichdealswiththebiophysical characterizationofcomplexbiopharmaceuticals;
• Chapter16,whichdealswiththerigorof statisticalanalysis;
• Chapter17,whichdealswithbiopharmaceuticaldevelopability;
• Chapter18,whichdealswithtechnicaldecisionmaking.
Finally,wewouldliketopointoutthat inwritingthissecondeditionwehave madeaparticulareffort,whereverpossible, tobetterlinkandcross-referenceinformationineachchaptertobringmorecohesion tothebookasapposetojustproviding thereaderwithacollectionofisolated chapters.Wethinkhiscohesionisinparticularmadeapparentbythefouradditional chaptersinSectionIII(describedabove).
Intheend,weandourcoauthorshopewe havefurtherenhancedtheinitialobjectiveof the firsteditionofthebook,ofenlightening thereadertothechallenges,toolsandinner workingsofthetaskassociatedwiththe biophysicalcharacterizationofproteinbiopharmaceuticals.Anintegralpartoftoday’ s modernandchallengingworldofdevelopinglifesavingdrugs.
Listofabbreviationsandsymbols (T)RPS (Tunable)resistivepulsesensing
3D Threedimensional
A22 orB2
Secondviralcoefficient
AAV Adeno-associatedvirus
AC Alternatingcurrent
AC-SINS Affinity-captureself-interactionnanoparticlespectroscopy
AFFF-MALLS
Asymmetric field flowfractionationwithmulti-anglelaserlightscattering
ACN Acetonitrile
ACS Ammoniumcamphorsulfonate
ADCorADCs AnalogtodigitalconverterorAntibodydrugconjugate(s)
ADCC Antibodydependentcell-mediatedcytotoxicity
AF4 Asymmetric flow field flowfractionation
AFM Atomicforcemicroscopy
API Activepharmaceuticalingredient
APR Aggregationproneregions
AQL Acceptablequalitylevel
ASTM Americansocietyfortestingandmaterials
ATP Analyticaltargetprofile
ATR Attenuatedtotalreflectance
AUC Analyticalultracentrifugation
BiAborbsAb Bispecificantibody
BLA Biologicallicenseapplication
BMI Backgroundedmembraneimaging
BSA Bovineserumalbumin
CA Capsidprotein
CAD Collision-activateddissociation
CCD Charge-coupleddevice
CD Circulardichroism
CDER Centerfordrugevaluationandresearch
CDR Complementarity-determiningregion
CEX Cation-exchangechromatography
cGMP Currentgoodmanufacturingpractices
CH Immunoglobulingammaheavychainconstantdomain
CH1orCH1
Immunoglobulingammaheavychainconstantdomain1
CH2orCH2 Immunoglobulingammaheavychainconstantdomain2
CH3orCH3 Immunoglobulingammaheavychainconstantdomain3
CHO Chinesehamsterovary
CIC Cross-interactionchromatography
CID Collisioninduceddissociation xv
cIEF
Capillaryisoelectricfocusing
CIU Collision-inducedunfolding
CL Immunoglobulingammalightchainconstantdomain
CMC ManufacturingandControl
COSY Correlationspectroscopy
cP Centipose
CPL Circularlypolarizedlight
CQAorCQAs Criticalqualityattribute(s)
CSA Camphorsulfonicacid
CZE Capillary(free)zoneelectrophoresis
D Deuteriumortranslationaldiffusioncoefficientorelectricdipole
DAC Deutscherarzneimittel-codex
DC Directcurrent
DI Developabilityindex
DLS Dynamiclightscattering
DoE Designofexperiment
DNA Deoxyribonucleicacid
DOSY Diffusionorderedspectroscopy
DP Drugproduct
dPLIMSTEX
DilutionPLIMSTEX
DRI Differentialrefractiveindexdetector
DS Drugsubstance
DSA 4,4-dimethyl-4-silapentane-1-ammoniumtrifluoroacetate
DSC Differentialscanningcalorimetry
DSF Differentialscanning fluorimetry
DSLS Differentialstaticlightscattering
DSS 4,4-dimethyl-4-silpentane-1-sulfonicacid
DTT Dithiothreitol
ECD Electroncapturedissociationorequivalentcirculardiameter
ECHOS Easycomparabilityofhigherorderstructure
EDTA Ethylenediaminetetra-aceticacid
EM Electromagneticradiationorelectronmicroscopy
EMEA EuropeanMedicinesAgency
ESD Equivalentspherediameter
ESI Electrosprayionization
ESZ Electricalsensingzone
ET Electrontomography
ETD Electrontransferdissociation
EX1
H/Dexchangemechanisminwhichtherateconstantforproteinfolding/unfolding ismuchslowerthantherateconstantforH/Dexchange
EX2 H/Dexchangemechanisminwhichtherateconstantforproteinfolding/unfolding ismuchfasterthantherateconstantforH/Dexchange
Fab Immunoglobulingammafragmentantigenbinding
Fc Immunoglobulingammafragmentcrystallizable(constantregion)
FcgRIIIa ImmunoglobulingammaFcreceptorRIIIa
FcRn NeonatalFcreceptor
FDA FoodandDrugAdministration
FFF Field flowfractionation
FID Freeinductiondecay
FIX
BloodclottingfactorIX
FL Fluorescence
FT-ICR Fouriertransformioncyclotronresonance
FTIRorFT-IR Fouriertransforminfraredspectroscopy
fuc Fucose
FUV-CD
FVIII
Farultravioletcirculardichroism
BloodclottingfactorVIII gal Galactose
GlcNAc N-acetylglucosamine
GLP
GMP
H/DX-MSorHDX-MS
HSA
HCH
HCl
HCLF
HDC
HDX
Goodlaboratorypractices
Goodmanufacturingpractices
Hydrogen/deuteriumexchangemassspectrometry
Humanserumalbumin
Humangrowthhormone
Hydrochloricacid
Highconcentrationliquidformulation
Hydrodynamicchromatography
Hydrogen/deuteriumexchange
HF5 Hollow fiber flow field fl owfractionation
HGH
hI
HIC
HILIC
HMQC
Humangrowthhormone
Hydrophobicityindex
Hydrophobicinteractionchromatography
Hydrophilicinteractionchromatography
Heteronuclearmultiplequantumcoherencespectroscopy
HMW Highmolecularweight
HOS
HPLC
HRR-DSC
HSQC
Higher-orderstructure
Highperformanceliquidchromatography
Highrampratedifferentialscanningcalorimetry
Heteronuclearsinglequantumcoherencespectroscopy
HT Hightension
HX
Hydrogenexchange
ICH Internationalconferenceonharmonizationoftechnicalrequirementsforregistrationofpharmaceuticalsforhumanuse
icIEF
Imagingcapillaryisoelectricfocusing
IDP Intrinsicallydisorderedprotein
IDR Intrinsicallydisorderedregion
IEC
Ion-exchangechromatography
IEF Capillaryisoelectricfocusing
IF Intrinsic fluorescence
IFNIFNb orIFNb1a
IgG1
Interferon-b-1a
Immunoglobulingamma1orimmunoglobulinG1
ILP Integerlinearprogramming
IM Ionmobility
ITC Isothermaltitrationcalorimetry
IUP Intrinsicallyunstructuredprotein
IUR Intrinsicallyunstructuredregion
IV Intravenousinjection
kD DiffusionInteractionParameter
LC/MS
Liquidchromatography/massspectrometry
LMW Lowmolecularweight
LNPs lipidnanoparticles
LO Lightobscuration
LOQ Limitofquantitation
LS Lightscattering
mAb Monoclonalantibody
MALDI Matrix-assistedlaserdesorption/ionization
MALLSorMALS Multianglelaserlightscattering
man Mannose
MD Moleculardynamics
MEM Maximumentropymethod
MEMS Micro-Electro-MechanicalSystems
MFI Micro- flowimaging
MHz Megahertz
MREor[M.R.E]
Meanresidueellipticity
mRNA Messengerribonucleicacid
MS Massspectrometry
MS/MS Massspectrometry/massspectrometryortandemmassspectrometry
MW Molecularweight
NEM N-ethylmalemide
NIBS Noninvasivebackscatteringtechnique
nIEF Nativeisolelectricfocusing
NIST NationalInstituteofScienceandTechnology
NMR Nuclearmagneticresonanceornuclearmagneticresonancespectroscopy
NNLS Nonnegativeleastsquares
NOE NuclearOverhauserEffect
NOESY
NuclearOverhauserEffectspectroscopy
NTA Nanoparticletrackinganalysis
OCD Orientedcirculardichroism
OD Opticaldensity
OQ Operationqualification
OS-GAGs Oversulfatedglycosaminoglyclans
PBS Phosphatebufferedsaline
PCA Principalcomponentanalysis
PDA Photodiode-array
PDB Proteindatabank
PDI Polydispersityindex
PEG Polyethyleneglycol
PEM Photoelasticmodulator
PFG Pulsed fieldgradient
PFGE Pulsed fieldgradientecho
Phe Phenylalanine
pI Isoelectricpoint
PK Pharmacokinetic
PL Pathlength
PLIMSTEX
Protein ligandinteractionsbymassspectrometry,titration,andH/Dexchange
PMT Photomultipliertube
PPC ProcedurePerformanceCriterion
PPI
Protein-proteininteractions
PPQ
ProcedurePerformanceQualification
PQ Performancequalification
Pr
PTMorPTMs
Probability
Posttranslationalmodification(s)
QA QualityAnalysis
QbD Qualitybydesign
QToF Quadrupoletimeof flight
QTPP Qualitytargetproductpro file
RDCs
Residualdipolarcouplings
RF Radio-frequency
rhGM-CSF
Recombinanthumangranulocytecolonystimulatingfactor
rhuEPO Recombinanthumanerythropoietin
RI Refractiveindex
rmAb Recombinantmonoclonalantibody
RMM Resonantmassmeasurement
RP-HPLC,RPLCorrpLC Reversed-phasehighperformanceliquidchromatographyorreversed-phaseliquid chromatography
RPS Resistivepulsesensing
RS Referencestandard
RT Roomtemperatureorretentiontime
Sor s Standarddeviation
S/N Signaltonoise
SANS Smallangleneutronscattering
SAP SpatialAggregationPropensity
SAXS SmallangleX-rayscattering
SCorSubQ Subcutaneousinjection
SDS-PAGE
SE-AUC
Sodiumdodecylsulfatepolyacrylamidegelelectrophoresis
Sedimentationequilibriumanalyticalultracentrifugation
SEC Size-exclusionchromatography
SE-HPLCorHP-SEC Size-exclusionhighperformanceliquidchromatographyorhigh-performancesizeexclusionchromatography
SEM Scanningelectronmicroscopy
SFC Supercritical fluidchromatography
SIC Self-interactionchromatography
SIMCA Softindependentmodelingofclassanalogy
SIMSTEX Self-associationinteractionsbymassspectrometry,self-titration,andH/DX
SLS Staticlightscattering
SMP Submicronparticles
SMR Suspendedmicrochannelresonator
SPE Solid-phaseextraction
SRCD Synchrotronradiationcirculardichroism
STEM Scanningtransmissionelectronmicroscopy
SUPREX Stabilityofunpuri fiedproteinsfromratesofH/Dexchange
SV-AUC Sedimentationvelocityanalyticalultracentrifugation
SVD Singularvaluedecomposition
SVP Subvisibleparticles
T1 Longitudinalrelaxationtimeconstant
T2 Transverserelaxationtimeconstant
TCEP-HCl
Tris(2-carboxyethyl)phosphinehydrochloride
TDA
TEM
TIC
TM-DSCorMT-DSC
TMU
TPP
Taylordispersionanalysis
Transmissionelectronmicroscopy
Totalioncurrent
Temperature-modulateddifferentialscanningcalorimetry
Targetmeasurementuncertainty
Targetproductpro file
Try Tyrosine
TSP
Trimethylsilylpropionate
Tyr Tryptophan
UPLCorUHPLC
USP
Ultrahighperformanceliquidchromatographyorultra-performanceliquid chromatography
UnitedStatesPharmacopeia
UV Ultravioletlight
UV VIS Ultraviolet visiblespectroscopy
VH
Immunoglobulingammaheavychainvariabledomain
VIS Visiblelight
VL
VLPorVLPs
Immunoglobulingammalightchainvariabledomain
Virus-likeparticle(s)
WAXS WideangleX-rayscattering
WCX
WSD
XIC
Weak-cationexchangechromatography
Weightedspectraldifference
Extractedionchromatogram
Z Netcharge
Thecomplexityofproteinstructure andthechallengesitposesin developingbiopharmaceuticals StevenA.Berkowitza,DamianJ.Houdeb
aConsultant,Sudbury,MA,UnitedStates; bBiomolecularDiscovery,RelayTherapeutics, Cambridge,MA,UnitedStates
1.1Thebasicsofproteinhigherorderstructure(HOS) Proteinsareanimportantclassoflargebiologicalmoleculesthatareclassifiedmoregenerallyasmacromoleculesorpolymers.However,giventheirbiologicalorigin,theseunique moleculesareoftenreferredtoasbiomacromoleculesorbiopolymers.Theyaretrulycomplex,particularlywhencomparedtosynthetic(man-made)polymersandevenothertypes ofbiopolymers,e.g.,DNA.Oneofthemainreasonsforthiscomplexityarisesfromtheirbasic buildingblocks,whichinsyntheticpolymerchemistryarereferredtoasmonomerunits.In thecaseofmostsyntheticpolymers,thechemicalcompositionconsiststypicallyofonlyone typeofmonomer(althoughsomesyntheticpolymerscalledcopolymersorblock-copolymers arecomposedoftwoorpossiblymoredifferentmonomerunits).Proteinsmadeinnaturevia aprocesscalledtranslationutilizingthegeneticcodearecomposedofnotone,two,oreven threedifferentmonomerunits,butratherarecomposedofasmanyas20different “naturally” occurringmonomerunitscalled aminoacids.These20aminoacids(orproteinogenic aminoacids,whichdoesnotincludetheotherknow,butrareproteinogenicaminoacidsselenocystineorpyrrolysine)arereferredtoasthestandardaminoacids.Althoughnotallproteinscontainall20aminoacids,mostdo.Thepresenceofsuchalargediversityinchemical composition,invirtuallyeveryprotein,isakeyelementfortheirstructuralcomplexity,which inturngivesrisetotheirdiversefunctionality.Indeed,thischemicalcomplexity,coupled withthelargenumberofaminoacidunitsor residues (N)presentinproteins(thatcannumber inthethousands),andtheuniquenessoftheaminoacidslinearsequentialarrangement (whichinproteinchemistryiscalledthe primary (1 )structure,see Fig.1.1A),enablesa
FIG.1.1 (A)Thelinearsequentialorderingofaminoacids(representedbytherectangularblackdashedboxes)in aproteinisreferredtoasitsprimarystructure.Theextremeleftaminoacidcorrespondstotheamino-terminus,while theextremerightaminoacidcorrespondstothecarboxyl-terminusendoftheproteinchain.Thegrayshadedarea correspondstothepeptidebondsthatlinkalltheaminoacidunitsinaprotein,yieldingthepolypeptidebackbone(or chain)indicatedbythered(grayinprintversion)dottedrectangle.(B)Anillustrationoftheplanarstructureoftwo adjacentamideplanes(eachresultingfromthedoublebondcharacter,duetoresonance,ofthepeptidebondshown asblackdashes),correspondingtothelightblue(lightgrayinprintversion)shadedareasin(A),wherethebottom amideplaneisformedfromthepeptidebondbetweenthecarboxylgroupofaminoacid1(containingR1)andthe aminogroupofaminoacid2(containingR2)andthetopamideplaneisformedfromthepeptidebondformed betweenthecarboxylgroupofaminoacid2andtheaminogroupofaminoacid3(containingR3).Duetostericissues, angularrotationaroundCaN(expressedby F,phi)andCCa (expressedby J,psi)bondsarelimited.(C)Arepresentationofacommonsecondarystructure,the a-helix.Thesmallrectangleoutlinedinblackdashescorrespondsto asmallsectionofthehelicalarrangementsoftheamideplanes,shownin(B).
staggeringnumberofdifferentpossibleproteins,20N,tobemade.Giventheenormousarray ofdifferentproteinsthatcanbemade,thecellhasexploitedthisdiversityinproteinstructure tocreateproteinstoperformnearlyeveryfunctionalandstructuralroleneededforits existence.
Inproteins,theaminoacidunitsarelinkedtogetherthroughauniquechemicalbond calledthe peptidebond,whichisalsoreferredtoastheamidelink,see Fig.1.1A.Thecollection ofthesepeptidebondsinagivenproteinformacommonelementfoundinallproteinscalled the polypeptidebackbone or chain,see Fig.1.1A.Auniquefeatureofthepeptidebondisthe planarstructurethatitformsbetweenthecarbonyloxygen,carbonandthe a-carbons(Ca oralphacarbon)ofoneaminoacidandtheamidenitrogen,hydrogenand a-carbonsofan adjacentaminoacid.Theresultingplanarfeatureoftheselinkedatomsarisesasaresultof thepartialdoublebondcharacterthatexistsbetweenthecarbonylcarbon(C)andtheamide nitrogen(N)atomsduetothepresencesofresonancestructures,see Fig.1.1B.Thisplanar structureanditsattributesplayanimportantroleinaprotein’sstructure,asitspresences confinesthepolypeptidebackbonetoonlycertaincon figurations,viastericeffects,whichrestrictstheangularrangeofbondrotationaroundtheCa N (expressedby F,phi)andC-Ca (expressedby J,psi)bonds.Theserestrictionshavebeensummarizedina2-dimensional graphicalplotcalledtheRamachandranplot,developedbyRamachandranandothersin 1963 [1].Suchaplotgraphicallyshowshowcertainstructuralfeaturesofproteinscanonly existwithinalimitedrangeofanglescharacterizedby J and F,e.g., a-helix,see Fig.1.1C. Theserestrictionsplayanimportantroleinthedevelopmentofprotein’sspatialstructure or higherorderstructure (HOS).
1.1.1ThelevelsofproteinHOS Indevelopingproteinbiopharmaceuticalsandinstudyingproteinsingeneral,themost importantconceptis “structure”.Intheprevioussection,webrieflydiscussedthemostbasic componentofaprotein’sstructure,itslinearsequenceofaminoacids,orprimarystructure. However,thefocusofthisbookisconcernedwithaprotein’ s three-dimensional (3D)orspatial structure ,alsoreferredtoasits conformation orHOS.Ultimately,whenconsideringthestructuresofproteins,itistheHOSinconcertwithitsprimarystructure(whichalsoincludesall theprimarychemicalbondmodificationsthatoccurtoitsaminoacidunits,see Section1.1.4) thatenablesaproteintoproperlyfunctionor,aswewillalsodiscussinlattersections, malfunction.
IntermsofproteinHOS,therearethreedifferentlevelsthathavebeendefined.Thesethree levelsinclude: secondary (2 ), tertiary (3 ),and quaternary (4 )structure,see Fig.1.2.The first twostructurallevelsareconcernedwithasinglepolypeptidechain,whilethelatterisassociatedwithproteinstructuresthatinvolvetheinteractionoftwoormorepolypeptidechains. Aprotein’s2 structurereferstothelocalfoldingpatternsofaprotein’spolypeptidechain,in whichthe a-helix(see Fig.1.2A),the b-sheet,turns,andrandomcoilsarethemostprominent resultingstructuralelementsthatareformed.Theselocalfoldedelementscanfurtherparticipateinhigherlevelsoffoldingthatinvolveanarrayofsecondarystructuralelementsthat giverisetothe final3Dstructureofaproteinreferredtoas3 structureofaprotein;see Fig.1.2B.Thesummationof2 ,3 and(ifpresent)4 structure,alongwithitsentire1 structure,iswhatgivesaproteinitsuniquestructure,chemicalandphysicalpropertiesand thereforeitsuniquefunction.Indeed,itisthisrelationshipbetweenstructureandfunction thatisthegenesisoftheprotein “structure-function” concept,whichstatesthataprotein’ s structuredeterminesitsfunction.
FIG.1.2 Illustrationofthethreelevelsofaprotein’sHOS.(A)Representativesecondarystructuralelement,as illustratedbyaribbonrepresentativestructureofan a-helix.(B)Acartoonrepresentationofthefoldingofallthe secondarystructuralelementsinapolypeptidechain,whichgivesrisetothepolypeptide’stertiarystructure.(C)A cartoonrepresentationofthequaternarystructureofaprotein,whichariseswhenthe finalproteinstructureinvolves theassociationofmorethanonepolypeptidechaintoformthe finalfoldedproteinstructure(alsosee Fig.1.3).
Althoughthefoldingandinteractionsofthesecondarystructuralelementscangiveriseto anenormousarrayofdifferentproteintertiarystructures,eachwithuniquepropertiesand functions,it’snotuncommonto findthatthe3 structureofaproteinoftenconsistsofone ormorecommonlyfoldedpatternscalled motifs, super-secondarystructures,or complexfolds [2 4].Thesecommonlyfoldedstructurescontainseveralfoldedsecondaryelements involvingonlyaportionoftheentirepolypeptidechainofaprotein,whichcanblursome ofthedistinctionbetweenaprotein’s2 and3 structure.Hence,onemightlookatmotifs, super-secondarystructures,orcomplexfoldsas “local3 structure”,whilereferringtothe 3 structureoftheentireproteinmoleculeasits “global3 structure”
Anotherstructuralelementthatfurthersubclassifiesthestructurallevelofaprotein betweenwhatwecallaprotein’s2 and3 structureistheconceptof domain [5,6].Domains aretypicallyamuchlargercollectionoffoldedstructuralelementsthanmotifs,supersecondarystructures,orcomplexfolds.Intermsoftheglobalstructureofaprotein,domainscorrespondtooneormoreindependentcompactregionofaprotein’spolypeptidechain,as
FIG.1.3 DifferentrepresentationsoftheHOSofamonomericIgG1antibody.Thetwoheavychainsarecolorcodedinblue(lightgrayinprintversion)andgray,whilethetwolightchainsarebothcolor-codedinred(dark grayinprintversion).(A)AribbonmodelofanIgG1antibody(PDB:1HZH).Theblackcirclecorrespondstothe variabledomainononeoftheIgG1lightchain(VL).(B)AsimplifiedcartoonofthemonomericIgG1antibody indicatingthevarioussectionsofindividualdomainspresent.Theblacklineslinkingthevariousinterchaindomains correspondtoareaswherecovalentlinkagesexist(disulfidebonds)betweendifferentpolypeptidechainsintheIgG1 molecule.TheblackcirclecorrespondstothesameVL domainintheIgG1moleculeasshownin(A).(C)AspacefillingstructuralmodelofthemonomericIgG1antibody.Theblackcircledregionagaincorrespondstothesame VL domainintheIgG1antibodyasshownin(A).(D)AlineardepictionofamonomericIgG1structureshowingall thevariouscovalentlinkages(disulfidebonds)presentintheIgG1antibody.Thosedisulfidebondspresentwithin thesamepolypeptidechainarereferredtoas intrachain disulfidebonds,whilethosedisulfidebondsthatlinktwo differentpolypeptidechainsarereferredtoas interchain disulfidebonds.
indicatedbytheblackcirclesshownin Fig.1.3A C.Proteinscontainingtwoormoredomainsarefrequentlyreferredtoasmultidomainproteins.Intheseproteins,thedomains arechemicallylinkedbyshortsectionsofthepolypeptidechainthataretypicallyhighly flexible,calleda “linker”,butneverthelessexistasstableandindependentfoldedunits.Incertain cases,commondomainstructurescanalsobefoundinproteinsmuchlikethatobservedwith motifs,super-secondarystructures,orcomplexfolds.
Whatisinterestingaboutthesefoldedelementsisthatthereisacertainamountofchange inthe1 structurethatcanbetoleratedwhilestillarrivingat,effectively,thesamefolded structure.Thisobservationexplainsthecommonpresenceofsimilarsecondary,supersecondary,andevendomainstructuresseenindifferentproteinswithdifferentsequences. Hence,theformationofthesebasicfoldingelementscandisplaysomelevelofdiscrepancy intermsoftherequiredorallowableaminoacidsequencevariationsandstillgiveriseto
thesamefunctioningprotein.Thisfeatureplaysanimportantroleinbiologicalevolution,in generatingHOSbuildingblocks,andincontrollingandregulatinggroupsofproteinsthat performverysimilarfunctionsindifferentbiochemicalpathways [7 9].Nevertheless,itis importanttomentionthatinproteins,thereexistmanysequenceregionswhereevenaslight change,i.e.,oneaminoacidchangeoraminorchemicalmodification(e.g.,oxidation,deamidation),cansignificantlyalteraprotein’sstructureandthereforeitsfunction [10,11].
Formanyproteins,however,theuniquefoldedstateofitspolypeptidechainisnotthelast stepinattaininga finaloverall3Dstructure.Manyproteinsarecomposedofmorethanone polypeptidechains,whichmaybeidenticalornonidentical,givingtheseproteinsanadded levelofstructuralcomplexity,4 structure;see Fig.1.2C.
Whenreferringtoaprotein’s4 structure,alackofclarityorconfusioncanunfortunately arise.Anexampleisillustratedin Fig.1.3.Inthis figure,amonomericintactIgG1antibodyis shown.However,thisproteincouldbereferredtoasaproteindimer(madeoftwoidentical proteinunits)oraproteintetramermadeoffourseparatepolypeptidechains,whichinthis casearechemicallycrosslinkedviacovalentbondscalleddisulfidebonds(whichisthemost commonprimarybondusedinnaturetocross-linkpartsofpolypeptides).Suchachoiceof descriptivewordsunfortunatelycanleadtosomeconfusion.Asaresult,somecareshouldbe takenwhendescribingthebasicstructureofaprotein.Inthecaseofthe4 structureofIgG1 molecule,asshownin Fig.1.3,theuseofatetramerinthecontextofits4 structurewouldbe correct.However,inthecontextofacompletefunctioningunit(initslowestcompleteform) themoleculeisamonomer.
1.1.2StabilizingtheHOSofproteins Inallthreelevelsofaprotein’sHOS(i.e.,2 ,3 ,and4 ),variouschangesintheconformationofthepolypeptidechain(s)occurasaproteinfoldstoreachits finalnativestructure. Thesechangesaretypicallyaccompaniedbyanincreaseinoverallstructuralorder,which impartsasignificantreductionintheprotein’sentropythatbyitselfishighlyunfavorable, intermsoftheoverallfree-energychange.However,asaproteinfolds,variousweaknoncovalent(secondary)bondsformviaionic,dipoles(hydrogenbonds),nonpolar(hydrophobic effect),andvanderWaalsinteractions.Theseweakbondsinvolvetheinteractionsof aminoacidsidechains,aswellaselementsofthepolypeptidebackbone,particularlythe amidehydrogen.Whileindividuallytheseinteractionsareweak,duringthefoldingprocess theirlargenumberandthecooperativewaytheyformprovidethenecessaryenthalpicand entropicdrivingforces(releaseofstructuredwaterviathehydrophobiceffect)tooverridethe largeunfavorabledecreaseinentropythatoccursasaproteinfoldsintoitsnative(moreordered)conformation.Thestabilizationofthefoldedprotein,however,isonlymarginal. Comparingthelevelofstabilizationagainsttheaveragethermalenergycontentofaprotein molecule(whichisequalto kT,where k ¼ Boltzmannconstantand T ¼ temperature)and thedistributionofthisenergy,intermsoftheamountofthermalenergypermolecule,avariety oftheseweaksecondarybondscanbebrokenasafunctionoftime.Suchspatialandtemporal rupturingoftheseweaksecondarybondsenablesaproteintodisplaydynamicstructuralpropertiesinitsconformation(sometimesreferredtoas proteinbreathing).Thisdynamicproperty canplayanimportantroleinaprotein’sfunction [12 15] andstability [16,17].Thisdynamic property,however,canalsoconstituteaweaknessforproteinbiopharmaceuticals,giventhe
widerangeofstressfulenvironmentsanaveragebiopharmaceuticalmustendureduringits biosynthesis,purification,formulation,packaging/storage,patienthandling,anditsadministration.Hence,insearchingforagoodtherapeuticbiopharmaceutical,scientistslookformoleculeswithhighstability,suchthatthedynamicpropertiesoftheproteindonotresultinloss ofactivityoradversestructuralchanges.Proteinsthathavesuchattributesaresaidtohave gooddevelopabilityproperties.
Inadditiontoweaksecondarybonds,stabilizationoftheHOSofaproteincanbeachieved throughprimarybondsformedbetweenfoldedelementswithinaprotein.Asalready mentioned,themostcommonsuchbondisthedisulfidebond,see Fig.1.3D.Althoughthe numberofdisulfidebondsfoundinagivenproteintypicallyamountstoonlyafewsuch bondsperproteinmolecule(andmaynotevenexistwithinsomeproteins),theyoftenplay importantrolesinaprotein’soverallstructure-functionandstability [18].Disul fidebonds canoccurbothwithinasinglepolypeptidechain(wheretheyarereferredtoas intrachain disulfidebonds;see Fig.1.3C)andbetweentwodifferentpolypeptidechainsinthesameprotein(wheretheyarereferredtoas interchaindisulfidebonds;see Fig.1.3C).Disulfidebonds alsooccurbetweentwodifferentproteinmoleculeswheretheyfunctiontostabilizelarge complexmultiproteinsupramolecularstructures [19].Unfortunately,however,theformation ofdisulfidebondscangoastrayleadingtoalteredHOSstructuresoraggregatesviadisulfide scramblingorexchangebetweenotherdisulfidebondsorfreecysteineresiduesinthesame proteinordifferentproteins.Thesemodesofproteindegradation [20 27] areanotherreason whythebiopharmaceuticalscientistneedtoconstantlyscrutinizethestructureofthebiopharmaceuticalduringitsdevelopment.
1.1.3DynamicspropertiesofaProtein’sHOS TheHOSofvirtuallyallproteinsisprimarilyheldtogetherbyalargearrayofrelatively weakbonds.Inthecontextofaprotein’sthermalenergycontent,thesebondscanbreak enablingvariouslevelsof fluctuationswithinaprotein’sHOSthatcanspananenormous timerange,from10 15 stotensofsecondsandevenlonger [12,28].Again,the fluctuations inaprotein’sconformationessentiallyoccurbecauseoftheopeningorbreakingofvarious weaksecondarybonds.Theextentofthese fluctuationsintermsofamplitudeandlocation isverydependentonmanyfactors,e.g.,environmentalconditions,thestrengthofeachsecondarybond,thedistributionofthesebondswithintheprotein,aswellasthedistributionof thermalenergywithintheprotein.Variationsinthese(andother)factorswilldeterminethe locationofwhichsecondarybondswillbreakinaprotein’sHOSandtherefore,thenatureof theconformationalchange(s)andthepopulationofproteinmoleculesinaspeci ficconformationasafunctionoftime.Whilethesechangesareforthemostpartcontainedtoaregion wherethesecondarybond(s)break,changesmightalsoextendtootherareasoftheprotein, viaallostericeffects.Duetotherandomnatureofthethermalenergy fluctuationswithina protein,arangeofdifferentconformationsandpopulationsofdifferentconformationalstates willexistatanyonetime.Forthemostpart,theextentofchangeinaprotein’sHOSaretypicallynotthatlargeandareoftenreversibleallowingthealteredproteinstructuretoreturnto itsmorestableconformations.
Consequently,insolutionproteinsexistasan ensemble ofdifferentconformations,rather thanasasingle fixeduniqueconformation.Thisensembleislimitedandcontrolledbythe
1.Thecomplexityofproteinstructureandthechallengesitposesindevelopingbiopharmaceuticals
interplayoftheoverallstructureoftheproteinanditsphysicochemicalenvironment.However,underappropriateconditions,involvingsomeformofstressorsubtlechangesinaprotein’schemicalstructure,changesinconformationmaycauseaproteintodisplaydifferent physicochemicalproperties.Inthecaseofaproteinbiopharmaceutical,changesinitsphysicochemicalpropertiescouldalterthedrug’sabilitytobindwithitstherapeutictargetor enableittobindtodifferentmaterialsitencounters,e.g.,variouscontainerclosuresurfaces [29 33].Otherpossibleadverseeventsincludetheformationofaggregatesthatarenonfunctionaland/orevenmoreconcerning,immunogenic [34 36].Itshouldbenotedthatthe formationofaggregatesandtheirassociatedlinktolossofproteinfunctionand/orimmunogenicitycorrespondstooneofthemostcommonformsofproteindegradationthatisclosely monitoredinthebiopharmaceuticalindustry.
1.1.4Finerstructuralalterationofproteins Onceaproteinissynthesized,orasitisbeingsynthesized,additionalprimarystructural changescanoccurinvivo.Inmostcases,thesechangesareduetoadditionalenzymaticprocessingreactionsinvolvingamultitudeofpotentialchemicalmodificationstovariousamino acids,aswellaschangesinvolvingcleavageorcross-linkingreactions.Thesereactionsmayor maynotplayanimportantroleinthenormalfunction/activityofaprotein,butrathermay representalterationsthatplayouttothedetermentofthecelloreventheorganismduetoan immunogenicresponse.Generally,mostmodificationsareconfinedtotheprotein’ssurface. However,modificationscanalsooccurtotheprotein’sinteriorduetothedynamicproperties ofitsstructure(whichexposestheseburiedinternalareas)orduringitssynthesiswhenthese normallyburiedinternalareashadnothadachancetoproperlyfold.Suchalterationscan leadtochangesinthelocalorglobalHOSoftheprotein.Ingeneral,thesemodifications arereferredtoas posttranslationalmodifications (PTMs).Principally,PTMsoccurinvivoand thenumberofdifferentPTMsthataproteincanexperienceisquitelarge [37].Ineukaryotes, oneofthemorecommon(andbiopharmaceuticallyrelevant)PTMsisglycosylation.This modificationinvolvestheenzymaticadditionofcarbohydrate(alsocalledglycanorsugar) unitstoaproteinatspeci ficasparagine(wheretheyarecalledN-linkedglycan)orserine orthreonine(wheretheyarecalledO-linkedglycan)aminoacid [38].WhilemostPTMsoccur invivo(insidethecell),PTMscanalsooccurinvitro(outsidethecell).TheselatterPTMs, however,typicallyrepresentformsofproteindegradationthatoccurduetodirectphysical orchemicalinteractions(e.g.,oxidation,deamidation,glycation,etc.)andarealsoofgreat concerninthebiopharmaceuticalindustryastheyareoftenlinkedtoinstabilityleadingto loseofdrugactivityandadverseeffects [3,39 44].
1.2Thesearchforhowproteinsattaintheircorrect HOS:theproteinfoldingproblem Inthe1950sand1960s,biophysicalresearchledscientiststotherealizationthataprotein’ s HOSiseffectivelydictatedbyitsprimarysequence.ChristianAnfinsenwasthekeyscientist whoformalizedthisidea,andin1972wasawardedtheNobelPrizeinchemistryforhiscontributions [45].Inthescientificliterature,thisideahasbeenfrequentlyreferredtoasthe
I.Proteinsandbiophysicalcharacterizationinthebiopharmaceuticalindustry
“An finsendogma” orthe “thermodynamichypothesis”.Thefoldingpathaproteintakesto achieveitscorrectfunctionalHOSisintrinsicallydictatedbyits1 structure(whichmayalso includePTMs).Howthefoldingprocessadvancessoef ficiently,incombinationwiththeway aproteinissynthesizedinvivo,inthespeci ficphysicochemicalenvironmentwithinthecell, hasfascinatedscientistsformanyyears [46].Thisfascinationstemsfromtherealizationthat proteinsachievetheircorrectHOSwithinamatterofmillisecondstoseconds!
Inthe1960s,CyrusLevinthalprosedthefollowinginterestingandsimpleproblemconcerningproteinfolding.Foraproteinconsistingof100aminoacidsinaninitiallyunfolded state,howlongwouldittakethisproteinto find,throughacompletelyrandomprocess, itsappropriatenativeHOSgivenitsphysicochemicalenvironment [28]?Thisproblemis nicelyrestatedinthewordsofAmitKesselandNirBen-Talintheirbook “Introductionto Proteins:Structure,FunctionandMotion” [47] asfollows:
Assumingthattheproteinfoldingprocessinvolvesthefreesamplingofallpossibleconformationsofthe protein(i.e.,ofeachresidueindependently),andthateachresiduehasatleastthreestates,thenthefoldingofa 100-residueproteinisexceptedtosample3100 ¼ 5 1047 conformations.Nowifweassumethatittakesa protein1picosecondtosampleasingleconformation,thenthetimeittakestosampleallpossibleconformationsinorderto findtherightoneshouldbe3100 10 12 s ¼ 5 1035 s ¼ 1.6 1028 years.Thisperiodof timeisabout1018 timeslongerthantheageoftheuniverse!!
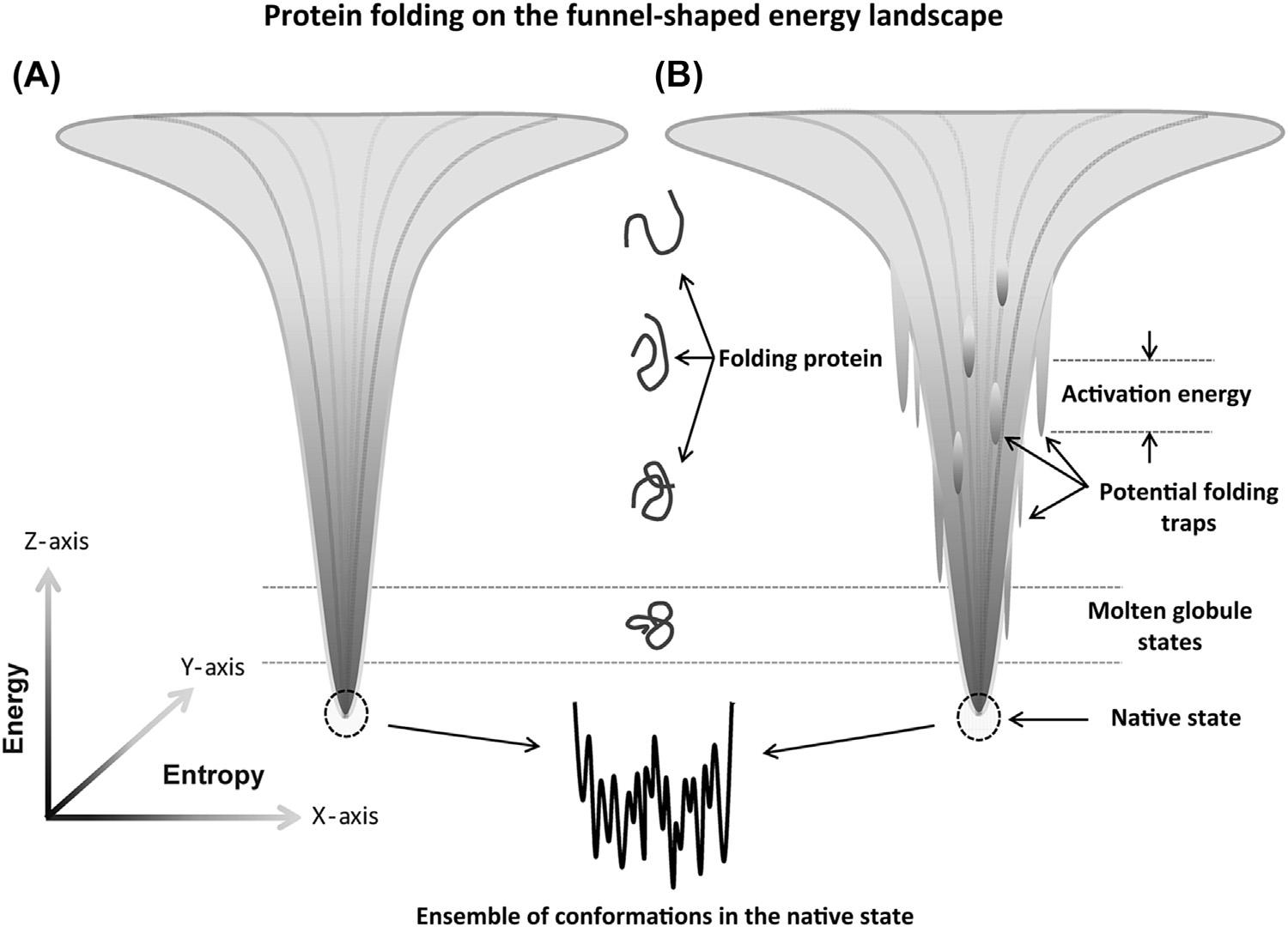
ThissimpleproblemproposedbyLevinthaliscalled “Levinthal’sParadox” andwasasignificantdrivingforceforthegeneratingwhatiscalled “theproteinfoldingproblem”.Clearly, thenatureofproteinfoldingisnowhereassimpleasstartingwiththecompletelysynthesized andunstructured(denaturedorrandomcoil)formofaprotein,whichisthenallowedtoundergoacompletelyrandomsamplingprocessofconformationalspace.Proteinfoldingmust proceedviaaprocessthatisenormouslymoreefficient,buthow!!?Answerstothisproblem appeartoliewithintheideaofa “funnel-shapedfoldingenergylandscape ” [48 52],see Fig.1.4,whichmightpossiblytakeadvantageofthewayproteinsaremadeinvivoalong withaconceptof “divideandconquer”.Inthisprocessaproteinproceedstofoldthrough ahierarchyofsubassemblyunitscalleda “foldon” [53,54].Theseunitscanfoldsomewhat independentofeachotherinparalleltoformrelativelylocalhigherorderstructuresthat caneventuallycollapseintothe finalnativeHOSoftheprotein.
Ingeneral,thefunnelingprocessofproteinfoldingislikelynotassimpleasthatportrayed in Fig.1.4A.Rather,itisexpectedtobemorecomplexandtreacherous,asindicatedin Fig.1.4B.Inthelatterscenario,afoldingproteincouldencounterconformationalstates thatarenotasoptimallyfoldedasitsnativestateandcontainhighactivationenergybarriers thatinhibititssearchto findthemoststableconformation.Hence,theproteininthesestates would finditselftrapped,duetothehighenergyofactivationneededtotransitionthemisfoldedstatebackintoamoreunfoldedstatesoitcan finditsmorestableandnativeform. Althoughthesemisfoldedproteinformsmaybeencounteredatverylowlevelsundernormal conditions,thesituationcouldescalateunderstressedconditions,suchasforcingacellto producealargequantityofoneproteininaveryshortperiod.Forsuchasituation,ahigher frequencyofmisfoldedormetastablefoldedproteinstatescouldbeencounteredleavingthe biopharmaceuticalscientistwithamoredifficultpurificationprocessthatresultsinalower proteindrugyield. 1.2ThesearchforhowproteinsattaintheircorrectHOS:theproteinfoldingproblem
FIG.1.4 Agraphicalviewofthethree-dimensionalfunnel-shapedenergylandscapeforproteinfolding.Thetopof eachfunnelcorrespondstothecompletelyunfoldedprotein.Thebottomofeachfunnelplotcorrespondstothefully foldedproteinmoleculeinitsnativestate,whichundercloserscrutinyactuallyconsistsofalargearrayofslight differentenergeticallyfoldedstates(conformations)thatdifferinmostcasesbyasmallamountoffreeenergythus enablingthenativeproteintoexistinsolutionasanensembleofdifferentconformations.(A)Afoldingprocessfreeof situationswhereitcanbetrappedinincompleteorpartialfoldedstate.(B)Afoldingprocessthatenablespartially foldedproteinstobepotentiallytrappedduetothepresenceofsmallershapedfoldingfunnelswithrelativelylarge energyofactivationthatmustbeovercomeinordertoescapeand findits finalnativestate.
1.2.1Invivoproductionofproteins:revisitingtheproteinfoldingproblem
Anotheruniqueattributeofproteinsisthecomplexmannerwithwhichtheyaremade invivo.Proteinsynthesisinvolvesacomplexarrayofcellularmachinery,themaincomponentofwhichistheribosome.Invivo,proteinsaresynthesizedfromtheN-terminusto theC-terminusinasequentialmanneratarateof50 300aminoacids/min [55,56].Thespecificorderingandchemicalcouplingoftheaminoacidsforagivenproteinisachievedbya processcalled translation,whichcontrolstheproteinsynthesisprocessdictatedbythegenetic codinginformationstoredinaspecificmessengerRNA(m-RNA).Asthenascentprotein chainissynthesizedandexposedtothecellmatrix,itcanbegintofold.However,itshould benotedthatthe first50 60aminoacidsinthegrowingpolypeptideareinitiallylimitedto someextentintheirabilitytofreelyfold,duetothephysicalrestrictions(sterichindrance)of theenvironmentwithintheribosome [57].Thisideaofconcurrent,invivo,proteinsynthesis andfoldingarereferredtoas cotranslationalproteinfolding [58] andlikelyplaysanimportant roleinthefoldingofnewlysynthesizedpolypeptide.
1.2ThesearchforhowproteinsattaintheircorrectHOS:theproteinfoldingproblem
Theimportanceofcotranslationalproteinfoldingmostlikelyarisesbecauseonlythe growingpolypeptidechainthathasadvancedbeyondtheribosometunnelwillbeableto fullyparticipateinthefoldingprocess.Thisallowsonlyaportionofthegrowingprotein chaintofoldwithouttheinterferencefromotherpartsoftheproteinthathaseithernot beensynthesizedorislocatedintheribosometunnel.Asaresult,thisshouldimprovethe efficiencyofthesequentialfoldingofthelocalhigherorderstructuralelementscharacterized asfoldonunitstoproceedinamoreorderlymanner.SuchfoldonunitsmostlikelycorrespondtolocalHOSelementsthatarepresentinthe finalnativeprotein.Nevertheless,as theselocalhigherorderstructuralelementsareformed,theymustsearchoutandundergo higherlevelsoffoldingastheproteinchaincontinuestogrow.Asaresult,thesevarioushierarchyoffoldedstructuralelementsareprobablynotarrangedorpackedoptimally(asthey areintheprotein’ s finalnativestate)untiltheentireproteinisfullysynthesizedandrelease fromtheribosome.Oncethishappens,whatremainingloosearrangementoffolded(or partiallyfolded)structuralelementsthatstillexistmustcollapseintothe finalnativestructure oftheprotein.This finalconsolationoffoldedorpartiallyfoldedstructuralelementsmost likelyproceedsthroughtheinteractionsofkeyaminoacidsidechainstomakethe finalfunctionalprotein(notwithstandinganyadditionalchangesinHOSresultingfromPTMsandthe formationofquaternarystructures).
Consequently,cotranslationalproteinfoldingconstrainsandtosomeextentguidesthe overallproteinfoldingprocess.Bylimitingthenumberoffoldingpathways(speci fically badfoldingpathways,whichwouldsignificantlyincreasetheamountoftimerequiredto findthecorrectnativeconformation)availabletoaprotein,relativetothesituationwhere foldingonlybeginsoncetheproteinisfullysynthetizedandreleasefromtheribosome,could maketheroleofcotranslationalproteinfoldingtrulyanimportantattributeinthesuccessful foldingofaprotein.
1.2.2Invivoproductionofproteins:avoidingandeliminatingfoldingerrors viatheuseofchaperones
Invivo,therearemechanismsinvolvingotherproteins,called chaperones,thathelpproteinsthatarefoldingavoidthesituationofbeingmisfolded.Thistaskisachievedviathe chaperone’sabilitytoassistafoldingproteintoavoidfoldingtrapsbyparticipatinginthe proteinfoldingprocessthroughprotein proteininteractions [59 63].Inadditiontochaperones,therealsoexistsinvivocellularmachinerywhosefunctionistoidentifythepresencesof misfoldedproteinsandeliminatethemviaproteolytichardwareexistingwithinthecell [64]. However,thesesystemsarenotperfect,andfailuretoremoveorpreventtheseerroneously foldedproteinsfromaccumulatingwithinthecellcanalterthecell,causingadverseeffects thatcouldeventuallyleadtoitsdeath.Inthecaseofproducingaproteinbiopharmaceutical, onceamisfoldedproteinisreleasedintothecellculturemedia,itthenbecomestheproblem fortheprocessscientisttodevelopappropriatepuri ficationstrategiestoremovethemisfoldedproteinfromthe finalproteindrugproduct.Iftheseerroneouslyfoldedproteinsare notremoved,theycouldleadtoadverseeffectswhenthe finaldrugproductisadministered toapatient.Hencebiophysicalanalysisofthebiopharmaceutical’sHOSagainbecomesan importantactivityindevelopingproteinbiopharmaceuticalswithminimallevelsofthese misfoldedformsinthe finaldrugproduct.
1.3Surprisesintheworldofproteinfolding:intrinsicallydisorderedor unstructuredproteins(anapparentchallengetotheprotein Structure Functionparadigm)
Withinthepasttwodecades,ithasbeenrealizedthatmanyproteins,especiallyineukaryotesormulticellularorganism,existwithinthecellwithnodefinedHOS [28].Rather,these proteinsappeartobedisorderedorunstructured,approachingwhatmightbecalleda randomcoilstructure,anomaloustowhatisfrequentlyseenwithsyntheticpolymersor denaturedproteins.However,whentheseproteinsinteractwiththeirtargetmolecule(s) theycommonlyappeartotakeonaleveloforganizedHOS.Hence,thisstructuraldisorder istransientinmanycasesandadisorder-to-ordertransitionoccursduringtheirfunctioning (i.e.,interactingwiththeirbindingtarget).Suchbehaviorcouldplayanimportantrolein allowingtheseproteinstobindtoanarrayofdifferentpartnermoleculesbytakingadvantage oftheplasticityoftheirpolypeptidechain’ s flexibility [28].Thisprocessisliabletobemodulatedbyotherfactorswithinthecell,whichcontrolandregulatethebindingpartnersthey interactwith.Indeed,thelevelofdisorderedproteinsishigherineukaryotesormulticellular organism,incomparisontoprokaryotes,wherehighlevelsofsignalingandregulationis required.Thisuniqueclassofproteinshasbeenreferredtoasintrinsicallydisorderedproteins(IDPs)orintrinsicallyunstructuredproteins(IUPs) [65,66].Theexistenceofthese IDPswouldappeartopresentachallengetotheparadigmofstructure functiondiscussed earlierinthischapter.
WiththerealizationoftheexistenceofIDPs,manyofthelargerandomcoil-likeregionsof proteinsconsistingof20 30ormoreaminoacidsinlengtharenowbeingreferredtoas intrinsicallydisorderedorunstructuredregions(IDRsorIURs) [66].Thesestructuralelementsarecommonlyseenaslinkersbetweenorderedproteinregionssuchasdomainswhere theyarethoughttoalsoplayimportantrolesinprovidingprotein flexibility,allowingproper foldingortofacilitatedomain domaininteractionsordomainbindingtofunctioningbindingtargets.Atpresent,IDPshavenotmadetheirwayintothebiopharmaceuticalindustry, althoughitisprobablyonlyamatteroftimeuntilsuchaproteindrugwillappear.
1.4Proteinsandthebiopharmaceuticalindustry:problemsand challenges Althoughproteinscanbechemicallysynthesizedexternaltothecell,theirhighcost(which isafunctionofproteinsize),aswellastheiroverallcomplexityleadstopooreconomicsfor buildingaviablecommercialdrugbusinessviathisapproach.Overtheyears,however,scientistshave figuredouthowtogetcellstoproducesignificantlylargeamountsofaspecific protein,bymanipulatingtheirDNAviarecombinantDNAtechnology.Thedevelopmentof thiscapabilitywasthekeyinenablingthebiopharmaceuticaldrugindustryto flourish.Overallthisprocessofmakingproteinbiopharmaceuticaldiffergreatlyfromtheclassicalprocess usedtomakesimpleorganicdrugmoleculescalledpharmaceuticals,see Fig.1.5.Cellularand molecularbiologistcannowproduceproteinbiopharmaceuticalsatconcentrationsexpressed intheculturemediavolumeasgreatas10g/L [67].Nevertheless,thechallengesofdoingthis
FIG.1.5 Asimplecomparisonillustratingdifferencesintheprocessofmakingapharmaceuticalversusmakinga biopharmaceutical:(A)Coarseoutlineofthesequentialchemicalreactionsformakingapharmaceutical,usingaspirin asanexampleand(B)acoarseoutlineofthebasicstepsformakingabiopharmaceutical,whichconsistsof first synthesizingapieceofDNAcontainingthecorrectnucleotidesequencecodeformakingthedesiredbiopharmaceutical’spolypeptidechain(s),theinsertionofthisDNAintoaninitialsmallcollectionofcells(themicroscale factoriesformakingthebiopharmaceutical)usingrecombinantDNAtechnology,thelarge-scalegrowthofthesecells duringwhichthecell’sinternalproteinsynthesizingnano-machine(theribosome,acomplexcellularorganelle composedofmanyproteinsandseveralpiecesofRNA)aredirectedtosynthesizethetargetbiopharmaceutical, illustratedhereaseitherinterferonbeta-1a(IFNb)oramonoclonalantibody(mAb).Notethatthespace-filling molecularmodelsofaspirin,IFNb andmAbhaveallbeendisplayedroughlyonthesamearbitraryscaletohelp providethereaderwithanapproximateperspectiveonhowtheywouldrelativelycomparetoeachotheronthebasis ofsize.ThedashedcirclehighlightingpartofthestructureofIFNb correspondstothecarbohydrate-containing portionofthisbiopharmaceuticalthatplaysadominantroleingivingrisetoitsmicroheterogeneityshownin chapter2inFigure2.3whencoupledwithotherposttranslationalmodifications(PTMs). Reproducedwithpermission fromBerkowitzSA [116]
successfullyaresigni ficant.Forcingacelltoproduceunusuallylargeamountsofasingle proteinpresentsuniqueproblemstothecell.Particularlyintermsofmakingsurethatall theproteinmoleculesareproperlyfoldedandhaveconsistentphysical,chemical,andbiologicalproperties.Hence,toachievethisgoalrequirestheconstantanddiligentmonitoringand characterizationoftheproteinbiopharmaceutical’sHOS.
Theprocessof finding,developing,andobtainingregulatoryapprovalofaprotein biopharmaceuticalthatismadeusingrecombinantDNAtechnologyproceedsthrougha sequenceofkeyactivitiesorbasicphasesofactivitythatisoutlinedin Fig.1.6.Successof
FIG.1.6 Acoarseoverviewofthebasicareasandsequenceofmajoractivitiesinvolvedincommercializinga proteinbiopharmaceutical.Therelativelengthofeachblockarrowisroughlyassociatedwiththelengthoftime typicalspentateachstage,fromresearchthroughcommercialization.Overall,thecostinthisprocesscaneasilybein excessofoneormorebilliondollarsandcanrequiremorethanandecadetodevelop [68].Thesenumberscanvary significantlyfromcompanytocompanyandfromdrugtodrug.Asaresulttheyareonlyapproximate. 1.Thecomplexityofproteinstructureandthechallengesitposesindevelopingbiopharmaceuticals
thisprocessishighlydependentonbiophysicalcharacterizationworkassociatedwithmonitoringtheconsistencyofthephysicochemicalpropertiesoftheproteindrug,confirmingthe absenceofchangesinthedrug’sHOS(whichmightgiverisetosmallunwantedsubpopulationsofalteredmolecules),andinassessingthepotentialimpactthatPTMsmighthaveona drug’sstructure.
Duringthe firstpartofthischapterwehavedealtwiththeverybasicpropertiesofprotein structure.Intheremainingsections,wewilldiscusshowthesepropertiesareresponsiblefor manyofthepotentialproblemsthatareofgreatconcerntoalargerangeofbiopharmaceuticalscientists.Inaddition,wewillbrieflylookatsomeofthemorenoveltypesofprotein biopharmaceuticalsthathaveandarebeingdevelopedthatfurtherchallengethetaskof biophysicallycharacterizingthesecomplexdrugs.
1.4.1ImpactofPTMsontheHOSofproteinbiopharmaceuticals Thecomplexchemicalcompositionofproteins,consistingof20chemicallydifferentnaturallyoccurringbuildingblocks(i.e.,aminoacids)effectivelyempowerthecellwiththe neededcomponents(chemistryset)tomakethenecessaryarrayofproteinsitneedstoproperlyfunction.However,theseaminoacidsalsoofferarangeofchemicallydifferenttargets thatcanundergochemicalchanges,viadirectchemicalreactionsorthroughtheparticipation ofvariousenzymaticreactions.Thechemicalchangesthataproteinbiopharmaceuticalcan incurofferopportunitiestoaltertheHOSofthesemolecules,impactingtheconsistencyof manufacturingorworse,causeadverseeventswhenadministratedtoapatient.As mentionedin section1.1.4,thesechemicalchanges,whethertheyoccurinvivoorinvitro, arecollectivelyreferredtoasPTMs.ManyPTMsplayrolesinthebiologicalfunctionofaproteininvivo,whileothersarearesultofnormaldegradationoraging.Hence,theideathata givenproteinexistsasasingledefineduniquechemicalentityismisleading.Infact,nearlyall proteinbiopharmaceuticalsexistasacollectionofhighlysimilarvariantforms.Therangeof thesevariantsandtheiramountsinthe finalbiopharmaceuticaldrugproductisdetermined bythenatureofthecell-lineused,thecellcultureconditions(e.g.,rawmaterialsandholdtimeswithinthebioreactor),theresolutionpropertiesofthepurificationprocess,aswellas ourabilitytodetectandcharacterizethem.Thecollectionofhighlysimilarproteins, variant forms or proteoforms [69] ofeffectivelythesameproteinthatcharacterizeabiopharmaceutical