BiochemistryofLipids, Lipoproteinsand Membranes
SeventhEdition
Editedby
NealeD.Ridgway
DepartmentofPediatrics,DepartmentofBiochemistry&Molecular Biology,DalhousieUniversity,Halifax,NS,Canada
RogerS.McLeod
DepartmentofBiochemistry&MolecularBiology, DalhousieUniversity,Halifax,NS,Canada
ElsevierScience
Radarweg29,POBox211,1000AEAmsterdam,Netherlands TheBoulevard,LangfordLane,Kidlington,OxfordOX51GB,UnitedKingdom 50HampshireStreet,5thFloor,Cambridge,MA02139,UnitedStates
Copyright 2021ElsevierB.V.Allrightsreserved.
Nopartofthispublicationmaybereproducedortransmittedinanyformorbyanymeans, electronicormechanical,includingphotocopying,recording,oranyinformationstorage andretrievalsystem,withoutpermissioninwritingfromthepublisher.Detailsonhowto seekpermission,furtherinformationaboutthePublisher’spermissionspoliciesandour arrangementswithorganizationssuchastheCopyrightClearanceCenterandtheCopyright LicensingAgency,canbefoundatourwebsite: www.elsevier.com/permissions .
Thisbookandtheindividualcontributionscontainedinitareprotectedundercopyrightby thePublisher(otherthanasmaybenotedherein).
Notices
Knowledgeandbestpracticeinthis fieldareconstantlychanging.Asnewresearchand experiencebroadenourunderstanding,changesinresearchmethods,professionalpractices,ormedicaltreatmentmaybecomenecessary.
Practitionersandresearchersmustalwaysrelyontheirownexperienceandknowledgein evaluatingandusinganyinformation,methods,compounds,orexperimentsdescribed herein.Inusingsuchinformationormethodstheyshouldbemindfuloftheirownsafety andthesafetyofothers,includingpartiesforwhomtheyhaveaprofessionalresponsibility.
Tothefullestextentofthelaw,neitherthePublishernortheauthors,contributors,or editors,assumeanyliabilityforanyinjuryand/ordamagetopersonsorpropertyasamatter ofproductsliability,negligenceorotherwise,orfromanyuseoroperationofanymethods, products,instructions,orideascontainedinthematerialherein.
LibraryofCongressCataloging-in-PublicationData
AcatalogrecordforthisbookisavailablefromtheLibraryofCongress
BritishLibraryCataloguing-in-PublicationData
AcataloguerecordforthisbookisavailablefromtheBritishLibrary
ISBN:978-0-12-824048-9
ForinformationonallElsevierSciencepublicationsvisitour websiteat https://www.elsevier.com/books-and-journals
Publisher: StacyMasucci
AcquisitionsEditor: LinsleyB.Peter
EditorialProjectManager: PatGonzalez
ProductionProjectManager: SwapnaSrinivasan
CoverDesigner: MarkRogers
TypesetbyTNQTechnologies
Contributors
KhosrowAdeli,MolecularMedicine,ResearchInstitute,theHospitalforSick Children,andDepartmentofLaboratoryMedicineandPathobiology,Universityof Toronto,Toronto,ON,Canada
ValentinBlanchard,DepartmentofMedicine,CentreforHeartLungInnovation, UniversityofBritishColumbia,Vancouver,BC,Canada
MichaelB.Boffa,DepartmentofBiochemistry,SchulichSchoolofMedicine& Dentistry,UniversityofWesternOntario,London,ON,Canada
MikhailBogdanov,DepartmentofBiochemistryandMolecularBiology,TheUniversityofTexasHealthScienceCenter,McGovernMedicalSchool,Houston,TX, UnitedStates
LauraM.Bond,DepartmentofBiochemistry,UniversityofWisconsin-Madison, Madison,WI,UnitedStates
AndrewJ.Brown,SchoolofBiotechnologyandBiomolecularSciences,The UniversityofNewSouthWales(UNSWSydney),Sydney,NSW,Australia
HudsonW.Coates,SchoolofBiotechnologyandBiomolecularSciences,The UniversityofNewSouthWales(UNSWSydney),Sydney,NSW,Australia
PaulA.Dawson,DivisionofPediatricGastroenterology,HepatologyandNutrition, DepartmentofPediatrics,SchoolofMedicine,EmoryUniversity,Atlanta,GA, UnitedStates
FangDing,KeyLaboratoryofRNABiology,InstituteofBiophysics,ChineseAcademyofSciences,Beijing,China;UniversityofChineseAcademyofSciences, Beijing,China
WilliamDowhan,DepartmentofBiochemistryandMolecularBiology,TheUniversity ofTexasHealthScienceCenter,McGovernMedicalSchool,Houston,TX,United States
GuangweiDu,DepartmentofIntegrativeBiologyandPharmacology,TheUniversity ofTexasHealthScienceCenteratHouston,Houston,TX,UnitedStates
SarahFarr,MolecularMedicine,ResearchInstitute,theHospitalforSickChildren, andDepartmentofLaboratoryMedicineandPathobiology,UniversityofToronto, Toronto,ON,Canada
EricA.Fisher,DepartmentofBiochemistry&MolecularBiology,Dalhousie University,Halifax,NS,Canada
GordonA.Francis,DepartmentofMedicine,CentreforHeartLungInnovation, UniversityofBritishColumbia,Vancouver,BC,Canada
AnthonyH.Futerman,DepartmentofBiomolecularSciences,WeizmannInstituteof Science,Rehovot,Israel
JessicaM.Gullett,DepartmentofInfectiousDiseases,St.JudeChildren’sResearch Hospital,Memphis,TN,UnitedStates
JoachimHerz,DepartmentofMolecularGeneticsandCenterforTranslationalNeurodegenerationResearch,UTSouthwesternMedicalCenter,Dallas,TX,United States
VictoriaHiggins,MolecularMedicine,ResearchInstitute,theHospitalforSick Children,andDepartmentofLaboratoryMedicineandPathobiology,Universityof Toronto,Toronto,ON,Canada
MurrayW.Huff,RobartsResearchInstituteandDepartmentsofMedicineand Biochemistry,TheUniversityofWesternOntario,London,ON,Canada
ElinaIkonen,MinervaFoundationInstituteforMedicalResearch,Helsinki,Finland; DepartmentofAnatomyandStemCellsandMetabolismResearchProgram,FacultyofMedicine,UniversityofHelsinki,Helsinki,Finland
MarlysL.Koschinsky,DepartmentofPhysiologyandPharmacology,SchulichSchool ofMedicine&Dentistry,UniversityofWesternOntario,London,ON,Canada; RobartsResearchInstitute,SchulichSchoolofMedicine&Dentistry,Universityof WesternOntario,London,ON,Canada
KristinaKuhbandner,DepartmentofMolecularGeneticsandCenterforTranslational NeurodegenerationResearch,UTSouthwesternMedicalCenter,Dallas,TX,United States
RichardLehner,GrouponMolecularandCellBiologyofLipids,Universityof Alberta,Edmonton,AB,Canada;DepartmentsofPediatricsandCellBiology, UniversityofAlberta,Edmonton,AB,Canada
SarahA.Lewis,DepartmentofBiochemistry,UniversityofWisconsin-Madison, Madison,WI,UnitedStates
GaryF.Lewis,DivisionofEndocrinology,DepartmentofMedicine,Universityof Toronto,Toronto,ON,Canada
JunLiu,DepartmentofBiochemistryandMolecularBiology,MayoClinicCollegeof MedicineandScience,Rochester,MN,UnitedStates
ZhaojinLiu,DepartmentofBiochemistry,UniversityofWisconsin-Madison,Madison,WI,UnitedStates
AndreaF.Lopez-Clavijo,LipidomicsFacility,BabrahamInstitute,Babraham ResearchCampus,Cambridge,UnitedKingdom
RogerS.McLeod,DepartmentofBiochemistry&MolecularBiology,Dalhousie University,Halifax,NS,Canada
MakotoMiyazaki,DepartmentofMedicine,DivisionofRenalDiseasesandHypertension,UniversityofColorado,AnschutzMedicalCampus,Aurora,CO,United States
JamesM.Ntambi,DepartmentofBiochemistry,UniversityofWisconsin-Madison, Madison,WI,UnitedStates;DepartmentofNutritionalSciences,Universityof Wisconsin-Madison,Madison,WI,UnitedStates
VesaM.Olkkonen,MinervaFoundationInstituteforMedicalResearch,Helsinki, Finland
LucasM.O’Neill,DepartmentofBiochemistry,UniversityofWisconsin-Madison, Madison,WI,UnitedStates
TheresaPohlkamp,DepartmentofMolecularGeneticsandCenterforTranslational NeurodegenerationResearch,UTSouthwesternMedicalCenter,Dallas,TX,United States
ArielD.Quiroga,InstituteofExperimentalPhysiology(IFISE-CONICET),Facultyof BiochemicalandPharmaceuticalSciences,Rosario,SantaFe,Argentina
KateyJ.Rayner,UniversityofOttawaHeartInstitute,DepartmentofBiochemistry, Microbiology&Immunology,FacultyofMedicine,UniversityofOttawa,Ottawa, ON,Canada
MarilynD.Resh,CellBiologyProgram,MemorialSloanKetteringCancerCenter, NewYork,NY,UnitedStates
NealeD.Ridgway,DepartmentofPediatrics,DepartmentofBiochemistry& MolecularBiology,DalhousieUniversity,Halifax,NS,Canada
CharlesO.Rock,DepartmentofInfectiousDiseases,St.JudeChildren’sResearch Hospital,Memphis,TN,UnitedStates
KatherineM.Schmid,DepartmentofBiology,ButlerUniversity,Indianapolis,IN, UnitedStates
LauraJ.Sharpe,SchoolofBiotechnologyandBiomolecularSciences,TheUniversity ofNewSouthWales(UNSWSydney),Sydney,NSW,Australia
BebianaC.Sousa,LipidomicsFacility,BabrahamInstitute,BabrahamResearch Campus,Cambridge,UnitedKingdom
JenniferTaher,MolecularMedicine,ResearchInstitute,theHospitalforSick Children,andDepartmentofLaboratoryMedicineandPathobiology,Universityof Toronto,Toronto,ON,Canada
JenniferK.Truong,DivisionofPediatricGastroenterology,HepatologyandNutrition, DepartmentofPediatrics,SchoolofMedicine,EmoryUniversity,Atlanta,GA, UnitedStates
MichaelJ.O.Wakelam,LipidomicsFacility,BabrahamInstitute,BabrahamResearch Campus,Cambridge,UnitedKingdom
ChangtingXiao,DepartmentofAnatomy,PhysiologyandPharmacology,Collegeof Medicine,UniversityofSaskatchewan,Saskatoon,SK,Canada
HongyuanYang,SchoolofBiotechnologyandBiomolecularSciences,TheUniversity ofNewSouthWales,Sydney,NSW,Australia
DakaiZhang,DepartmentofIntegrativeBiologyandPharmacology,TheUniversityof TexasHealthScienceCenteratHouston,Houston,TX,UnitedStates
Preface
The36yearssincethepublicationofthefirsteditionofthistextbookhaswitnessedremarkableadvancesinourunderstandingofthecellandmolecular biologyoflipids,andtheever-broadeningrolethattheseessentialmolecules playinhumanphysiologyandpathology.Nowinits7thedition,thetextbook summarisesandcontextualisesrecentadvancesintopicsrangingfrommembranebiophysicsandanalyticallipidanalysistothecellbiologyoflipidmetabolismandlipidmacromolecularassembliesinthecellandcirculation.Thetext alsoexploresnewevidencefortheroleofdifferentlipidspeciesinhumanpathologies,suchascardiovasculardisease,diabetes,lipidstoragedisorders, neuropathiesandimmunedisorders.Eachchapterprovidesaconciseandhighly accessibleintroductiontothediverseareasoflipidresearch,withinsightsthat canonlybeprovidedbyauthorswhoareleadersintheirrespectivefields.The contentisstructuredtoprovidethereaderwiththeessentialhistoricalperspectiveandbackgroundknowledgetofacilitatetheunderstandingofcurrentand complexproblemsinlipidbiochemistryandphysiology.Theauthorshave purposelyprovidedchaptersthatarenotcomprehensive,criticalreviewsbut insteadprovideessentialinformationandcontextforthereadertoventuredeeper intoeachresearchtopicattheirownpace.Thisisfacilitatedbyafocusontopics withimportantimplicationstofundamentalandappliedresearch.Aseditors,we havedeliberatelyaskedforandreceivedalimitedreferencelistthathighlights definitivereviewsandimpactfulrecentresearchstudies.Thisformatwillbenefit researchersneedinganintroductiontospecificfields,andthereferencestodelve further,aswellasundergraduateandgraduatestudentsseekingaresourcefor course-andresearch-relatedstudies.Thevisualappealandclarityofthisedition hasbeenenhancedbyfullcolourfiguresandstandardisedtables.
Wewouldliketoacknowledgethetimeandeffortthatnewandreturning authorsdevotedtoeachoftheirchapters.Thiswasespeciallydifficultsince authorsacceptedinvitationstoparticipatejustastheyhadtodealwithdisruption andshutdownoftheirlaboratoriesresultingfromtheSARS-CoV-2pandemic. ThiseditionisdedicatedtothememoryofDr.MichaelJ.O.Wakelam.
xxviii Preface
Thecontributorsandeditorsassumefullresponsibilityforthecontentherein, andwouldappreciateanycommentsonthecontentandlayoutfortherefinement offutureeditions.
NealeD.RidgwayandRogerS.McLeod
Halifax,NovaScotiaCanada
April2021
4 BiochemistryofLipids,LipoproteinsandMembranes
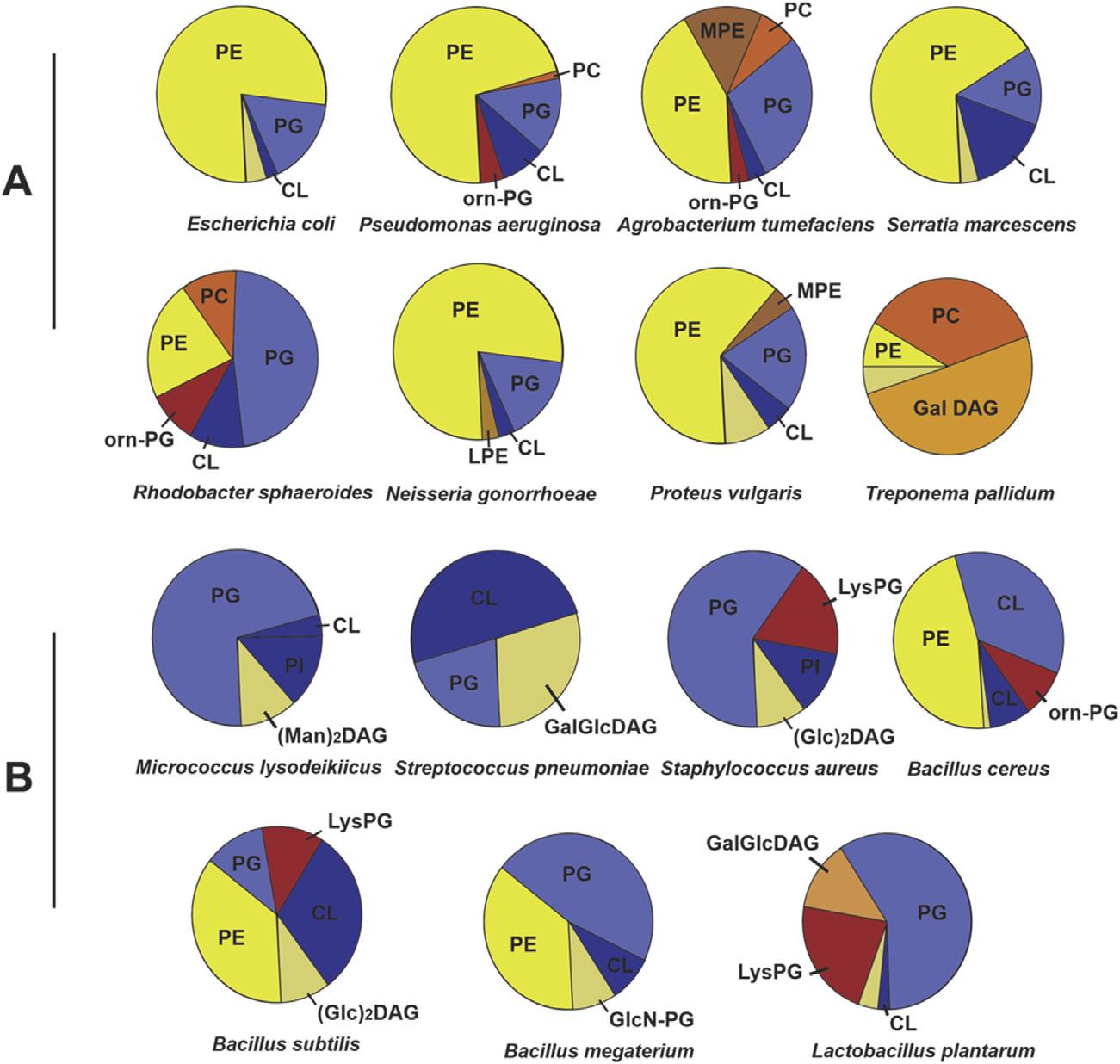
FIGURE1.2 Phospholipidprofilesofgram-negativebacteria(A)andgram-positivebacteria (B)areshownaspie-charts.Compositionisgiveninmol%. (Man)2 DAG,dimannosylDAG; CL, cardiolipin; GalDAG,monogalactosylDAG; GalGlcDAG,galactosylglycosylDAG; GlcDAG, monoglucosyldiacylglycerol; GlcGlcDAGor(Glc)2,diglucosylDAG; GlcN-PG,glucosaminyl PG; LPE,lysoPE; LysPG,lysylPG; MPE,monomethylPE; Orn-PG,phosphorus-freeornithine PG; PA,phosphatidicacid; PC,phosphatidylcholine; PE,phosphatidylethanolamine; PG, phosphatidylglycerol; PI,phosphatidylinositol; PS,phosphatidylserine. Constructedfromdata fromSohlenkampandGeigerFEMS.MicrobiolRev2016;40:133 159withpermission.
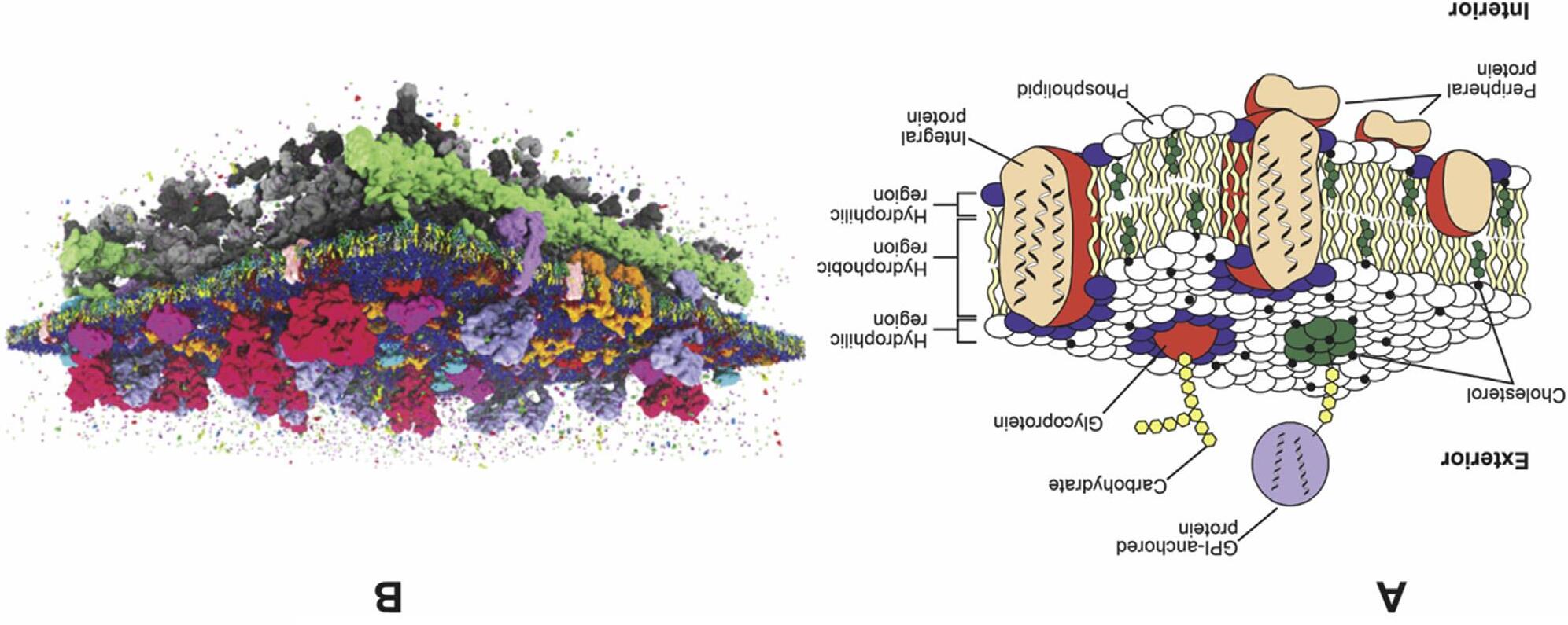
Thelipidbilayerprovidesthesolventforintegralmembraneproteins (MPs)whosetransmembranedomains(TMDs)spanthebilayerandare integratedintoahighlyhydrophobicinteriorboundedbythehydrophilicand chargedlipidheadgroups.Thelatteractsasascaffoldforassociationof peripheralMPs(Figure1.1).Thelipidandproteincomponentsofthemembranearenotheldtogetherbycovalentinteractionsandthereforearein dynamicequilibriumundergoingtransientinteractionswithinthesupramolecularmembranestructure.Aswillbediscussedfurther,theinterfacialregion atthemembranesurfaceorganiseswaterandcounterionsinamannerdifferent fromthecellaqueousphase,whichimpartsdistinctpropertiestotheaqueous layerinclosecontactwithmembranes.
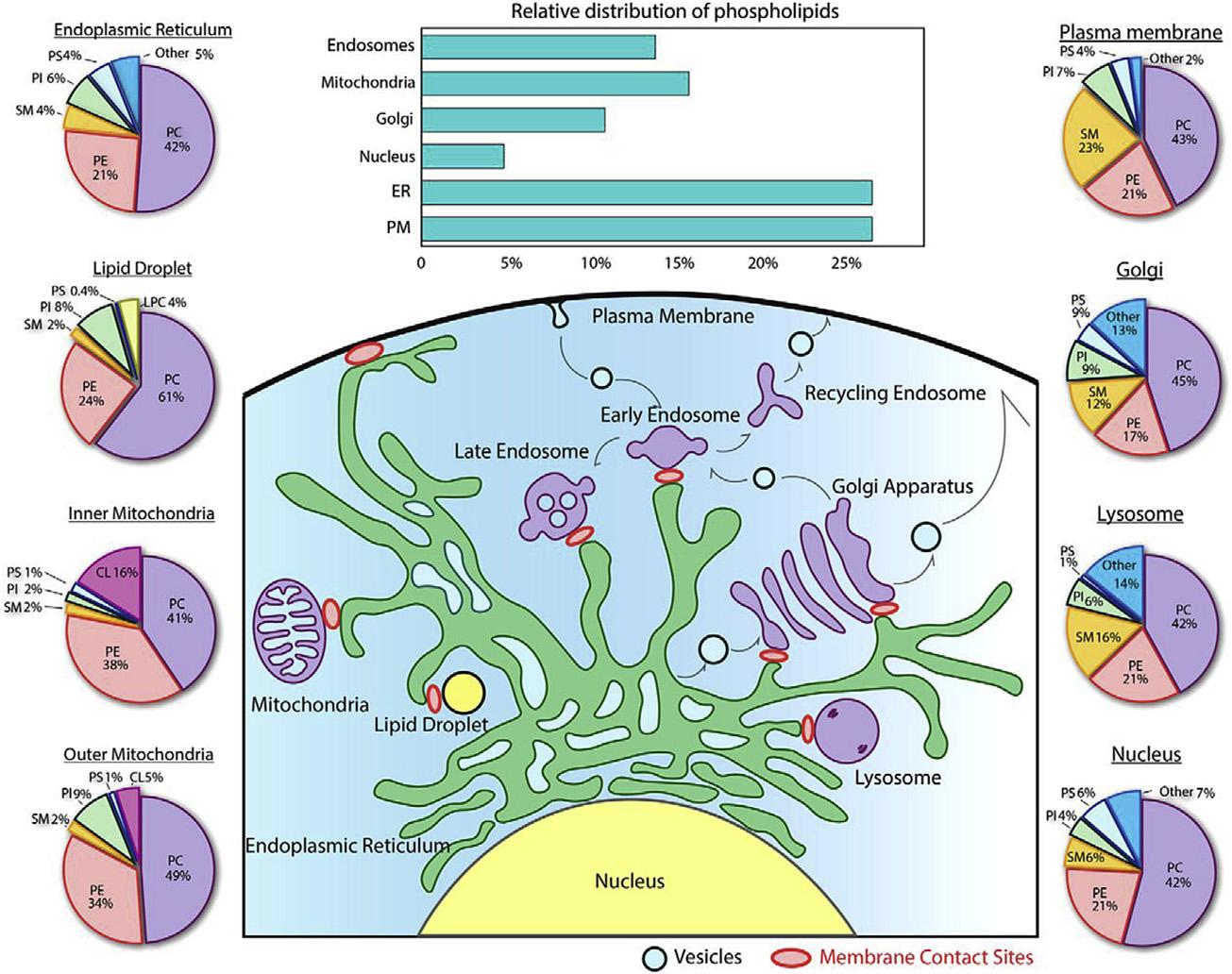
FIGURE1.3 Distributionoflipidsineukaryoticorganellemembranes.Shownisthedistribution ofindividualphospholipidsandsphingomyelin(SM)incellularorganelles,aswellasthedistributionoftotalphospholipidsinthesecompartments. AdaptedwithpermissionfromYang,etal.J BiolChem2018;293:6230 6240.
Determiningthefunctionofindividuallipidsandthecollectiveproperties ofthemembranelipidbilayerhasbeenchallenging.Manylipidfunctionshave beendefinedbytheirserendipitouseffectonproteinsandotherbiological componentsobservedinvitrowithlittleverificationinvivo.Genetic approaches,whichhavebeensuccessfulindefiningproteinfunctions,are complicatedbythefactthatlipidsareproducedbycomplexbiosynthetic pathwaysratherthanbydirectgeneticdetermination.Inordertoalterlipid compositioninvivo,mutationsmustbemadetoenzymesinthesebiosynthetic pathways,whichmayaffectmultiplelipidprecursorsandproducts.Since lipids,unlikeproteins,arenotlocalisedtoaspecificsiteincells,genetic manipulationoflipidsynthesishasglobaleffects.However,combininggenetic studieswithbiochemicalcharacterisationofinvivophenotypesandstudiesin definedreconstitutedsystemsinvitrohasuncoveredspecificandgeneralroles formembranelipidsincellfunction.
Thischapterwilloutlinethediversestructure,chemistryandsupramolecularpropertiesoflipids.Geneticapproachestostudylipidfunctioninvivo willbesummarised.Finally,howthephysicalandchemicalpropertiesof lipidsrelatetotheirmultiplefunctionsinlivingsystemswillbereviewedto
provideamolecularbasisforthediversityoflipidstructuresinnatural membranes.
2.Diversityinlipidstructure
Lipidswereoriginallydefinedasthosebiologicalmoleculesreadilysolublein organicsolventssuchaschloroform,etherortoluene.However,manypeptides andsomehydrophobicproteinsarealsosolubleinorganicsolvents,andlipids withlargehydrophilicdomainsarenotreadilysolubleinthesesolvents.Here wewillconsiderthoselipidsthatcontributesignificantlytomembrane structureorhavearoleindeterminingproteinstructureorfunction.TheLIPID MAPSconsortium(http://www.lipidmaps.org)intheUnitedStates,Lipid Bank(http://www.lipidbank.jp)inJapanandtheLipidomicNet(http://www. lipidomicnet.org)inEuropehavecooperatedtodeviseclassificationsystems andmethodologyforthebenefitofresearchers.
2.1Glycerolipids
Thediacylglycerolbackboneineubacteriaandeukaryotesis sn-3-glycerol (L-glycerol)esterifiedatthe1-and2-positionwithlong-chainfattyacids (Figure1.4A).
Ineubacteria,fattyacidchainlengthsvaryfrom12to18carbonsandcan befullysaturatedorcontaindoublebonds(See Chapter3).Somegrampositivebacteriacontainodd-numbered,branchedchainfattyacidsrather thanunsaturatedfattyacids.Eukaryoticlipidscontainfattyacidchainsupto 26carbonsinlengthwithmultipleornodoublebonds.Theheadgroupsofthe phospholipids(Figure1.4A)extendthediversityoflipids.In Archaea,sn-1glycerol(D-glycerol)formsthebackbone,andthehydrophobicdomainis composedofphytanyl(saturatedisoprenyl)groupsinetherlinkageatthe2and3-position(anarchaeol)(Figure1.4B).Inaddition,two sn-1-glycerol groupsareconnectedinetherlinkagebytwobiphytanylgroups(dibiphytanyldiglycerophosphatetetraether)(Figure1.4C),orbiphytanyldiglycerol diether(Figure1.4D),toformacovalentlylinkedbilayer.Many Archaea phospholipidscontainheadgroupsofglycerol,serine,ethanolamine, myo-inositol,aswellasglyceromethylphosphateandacardiolipin(CL) analogue. Archaea alsohaveneutralglycanlipidderivativesinwhichmonoanddisaccharides(glucoseorgalactose)aredirectlylinkedtothe sn-1position ofarchaeol(Figure1.4E). Archaea membranesarehighlyresistanttotheharsh acidicandhyperthermicenvironmentoftheseextremophilicorganisms. Furtherstabilityofthelipidbilayerof Archaea comesfrommanyofthe hydrocarbonchainsspanningthemembranewithcovalentlylinkedheadgroupsateachend.Someeubacteria(mainlyhyperthermophiles)havedialkyl (long-chainalcoholsinetherlinkage)phospholipidsandsimilaretherlinkages arefoundintheplasmalogensofeukaryotes(see Chapter7).
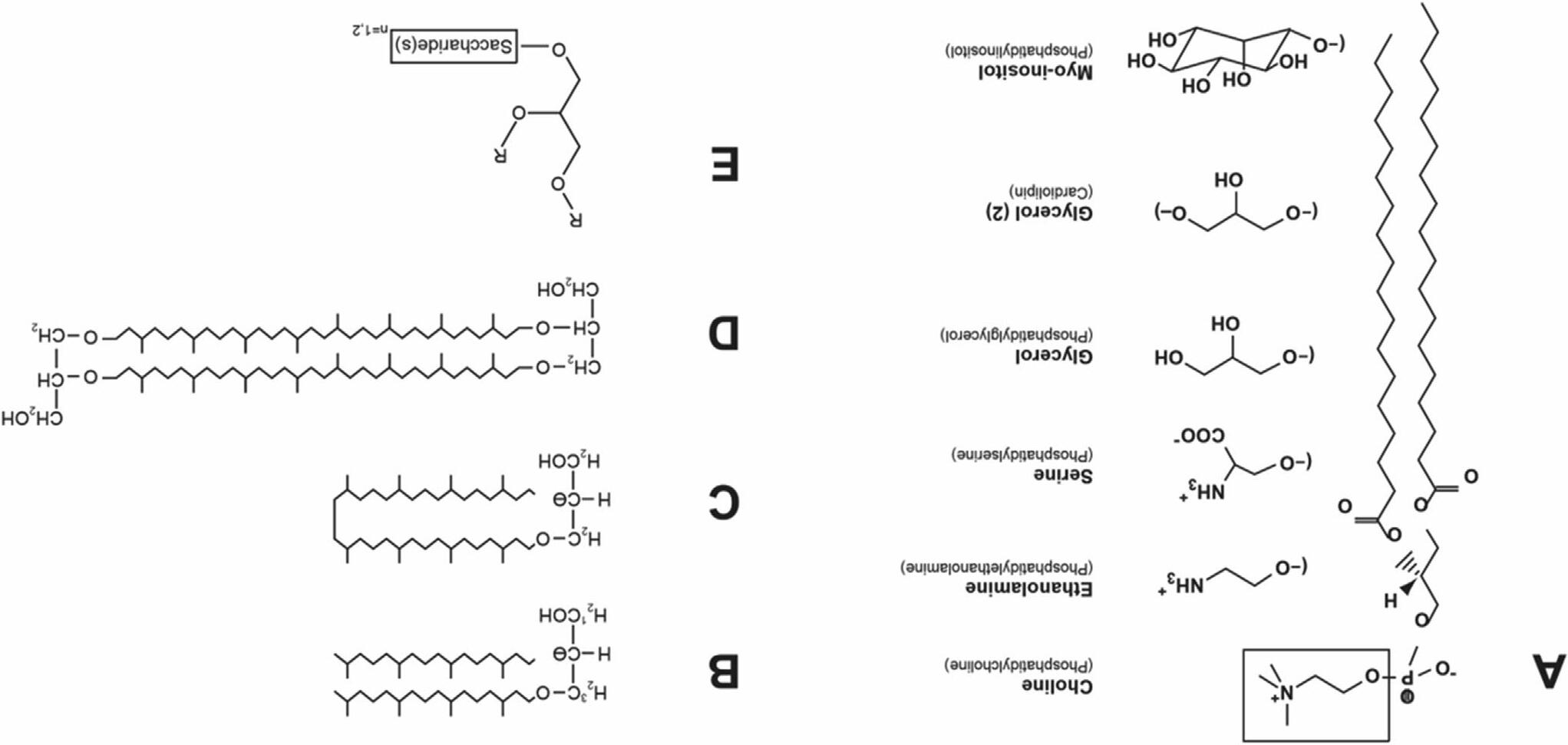
FIGURE1.4 Structureofglycerophosphate-basedlipids.(A)Thelipidstructureshownis1,2distearoyl-sn-glycerol-3-phosphocholineorphosphatidylcholine (PC).Substitutionofcholine(boxed)withotherheadgroupsproducestheindicatedphospholipids.(B)StructureofArchaealdialkylglycerolwith phytanylchainsin etherlinkagetothe2-and3-positionsof sn-1-glycerol(archaeol).Shownisdiphytanylglycerol(C20 C20diether)withthestereochemistryofglycerolindicated. (C)Cyclicbiphytanyl(C40)diether.(D)Biphytanyldiglyceroldiether.(E)Aglyceroglycanwitheitheramono-ordisaccharide(glucoseorgalactose)atthe 1-positionof sn-1-glycerol.TheRgroupsareether-linkedphytanylchains.Similarglyceroglycansarefoundineubacteriaandplantswitha sn-3-glycerolbackbone andester-linkedfattyacidchainsatthe1-and2-position.
8 BiochemistryofLipids,LipoproteinsandMembranes
Themajorityoftheinformationonthechemicalandphysicalpropertiesof lipidscomesfromstudiesonthemajorphospholipidclassesofeubacteriaand eukaryotes.Thebiosyntheticpathwaysandthegeneticsoflipidmetabolism havealsobeenextensivelystudiedineubacteria(see Chapter3)andeukaryotes(see Chapter7).Howthephysicalpropertiesofthemorecommonly studiedlipidschangewithenvironmentwillbediscussedlater.
2.2Saccharolipids
TheOMofgram-negativebacteriacontainslipopolysaccharide(LPS),or endotoxin,whichismadeupofabackbonederivedfromglucosaminephosphateratherthanglycerophosphate(Figure1.5A).
Thecorelipid(LipidA)of E.coli containstwoglucosaminegroupsin b 1 6linkagethataredecoratedatpositions2,3,20 and30 withR-3hydroxymyristicacid(C14)andatpositions1and40 withphosphates (Figure1.5B).Furthermodificationatposition60 withaKDOdisaccharide (two3-deoxy-D-manno-octulosonicacidsina1 3linkage)resultsinKDO2LipidAthatisfurthermodifiedbyaninnercore,anoutercore,andthe O-antigen.StudiesofLipidAareofclinicalimportancebecauseitisthe primaryantigenresponsiblefortoxicshocksyndrome.
ThecoreLipidAformstheoutermonolayeroftheOMbilayerofgramnegativebacteriawiththeinnermonolayerbeingmadeupofphospholipids (about90%PE)(Figure1.5A).LPSismodifiedpost-assemblyinresponseto environmentalfactors,suchasgrowthmedia,temperature,ionicpropertiesand antimicrobialagents,anddisplaysadditionaldiversityamongentericandnonentericgram-negativebacteria.
ThehighlyasymmetrictransmembranearrangementofLPS(outerleaflet) andPE(periplasmicleaflet)intheOMisexpectedtobeinanon-equilibrium thermodynamicstatethatismaintainedinnormallygrowingcells.However, stressfulconditions(acidicpH,chelatingagentsanddetergents)displaceLPS fromouterleafletandpromotetheaberranttranslocationofphospholipidsinto theouterleafletofOMreplacingLPSmoleculesthathavebeenshed.
2.3Sphingolipids
Alleukaryoticcellscontainsphingolipidsderivedbythecondensationof palmitoyl-CoAandserinetoformalong-chainbase,followedbyconversion tothecoreceramidemolecule(see Chapter9)(Figure1.6A).Inhigher eukaryotes,thelong-chainbasederivedfrompalmitatecanhaveadditional doublebondsandhydroxylgroupsaswellasconsiderablediversityinthe amide-linkedfattyacid,whichcanbeupto26carbonsinlength.
Yeastcellscontainmainlyderivativesofphytoceramide(4-hydroxyceramide)andC26fattyacidchainsinamidelinkage.Themajorclassesof sphingolipidsaregroupedaccordingtotheheadgroupesterifiedattheprimary
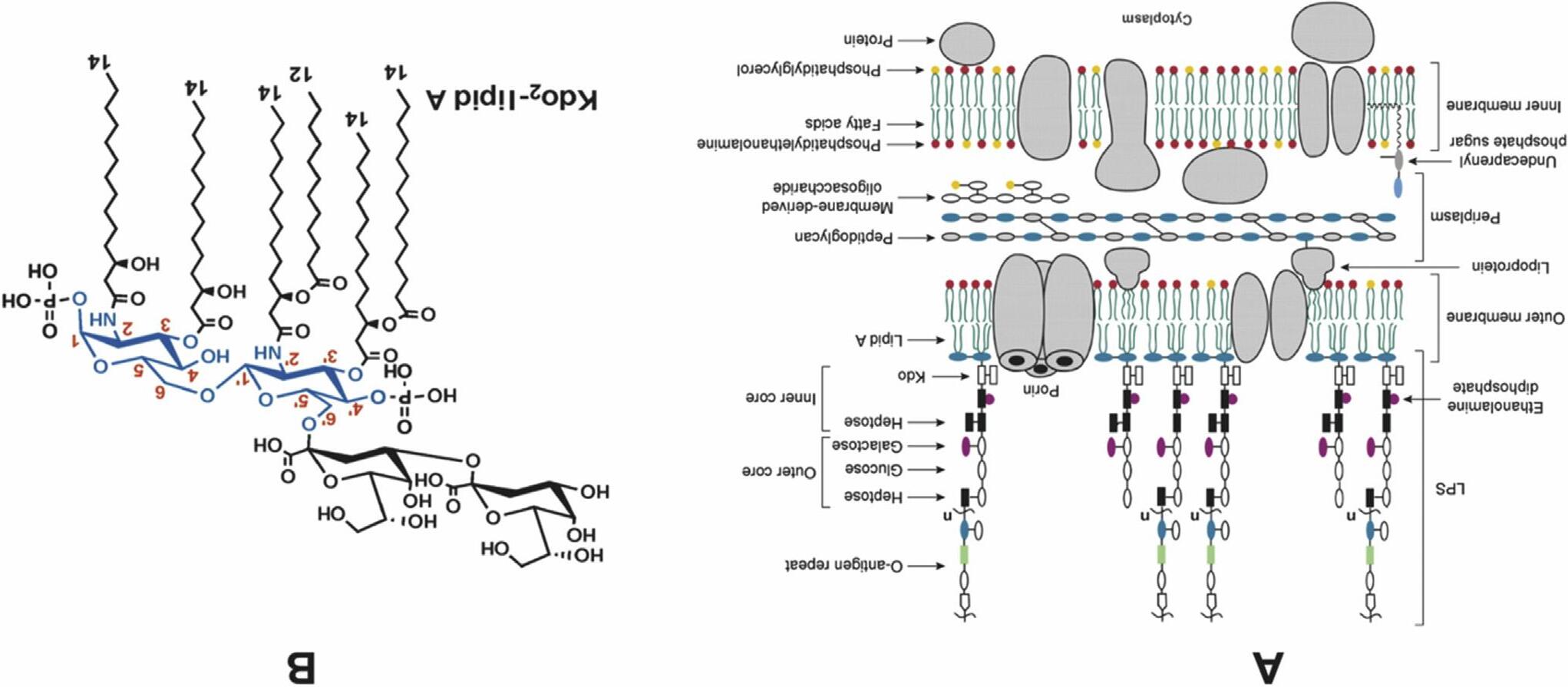
FIGURE1.5 The E.coli double-membraneenvelope.(A)Gram-negativebacteriaareenvelopedbytwolipidbilayers,separatedbytheperiplasmicspace containingthepeptidoglycancellwall.Theasymmetricphospholipidoutermembrane(OM)ofgram-negativebacteriaservesasthefirstlineofdefenceagainst cytotoxicsubstancesincludingantibiotics.TheOMiscomposedofaninnermonolayerofphospholipidandanoutermonolayeroftheLipidAportionof lipopolysaccharide(LPS)thatislocatedexclusivelyintheouterleafletoftheOM.Theperiplasmicspacecontainsmembrane-derivedoligosaccharide(MDO)that isacomponentoftheosmolarityregulatorysystem.Theaminoacid sugarcross-linkedpeptidoglycangivesstructuralrigiditytothecellenvelope.(B)Structureof KDO2-LipidA.LipidAisadisaccharideofglucosaminephosphatethatismultiplyacylatedwithbothamideandesterlinkagestofattyacidsofthechainlengths indicated. AdaptedfromRaetzetal.,Discoveryofnewbiosyntheticpathways:thelipidAstory,J.Lipid.Res.2009,50:S103 S108
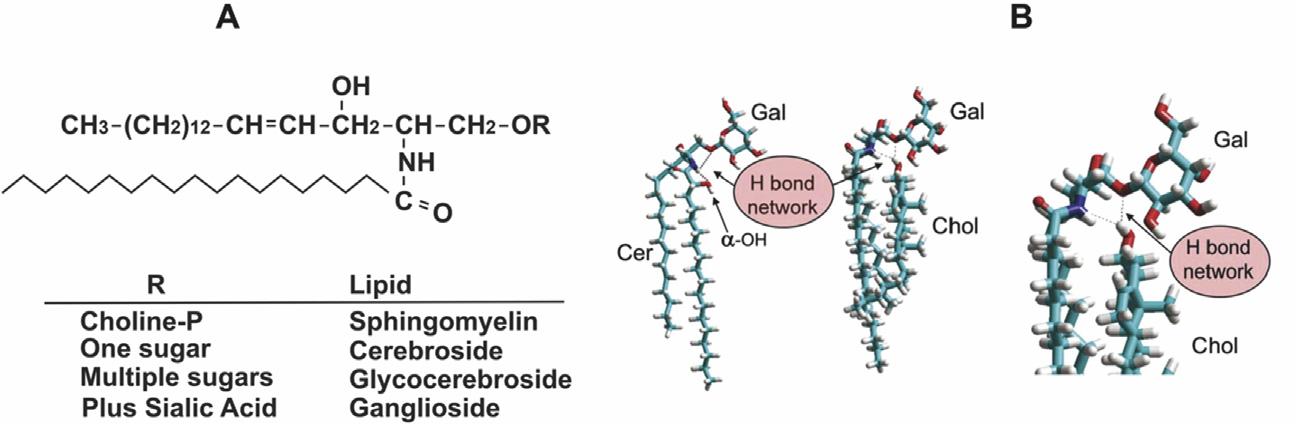
FIGURE1.6 Sphingolipidstructure.(A)Sphingosineisan18-carbonaminoalcoholwithan unsaturatedhydrocarbonchain.Ceramideiscomposedofsphingosineandan N-acylatedfatty acid.Headgroups(R)areaddedtoceramidetoformtheindicatedsphingolipids.(B)Molecular modellingsimulationsofgalactosylceramide(GalCer)complexedwithcholesterol,wherethe galactoseheadgroupismaintainedthroughanetworkofH-bondsinvolvingthehydroxylof cholesterol(donorgroup),thenitrogenatomofsphingosineandtheoxygenatomoftheglycosidic bond(bothacceptorgroups). AdaptedwithpermissionfromYahi,etal.PLoSOne2010;5:e9079.
b-hydroxylofceramide(Figure1.6A).Sphingomyelinhascholinephosphate atthispositionwhiletheglycosphingolipidshaveoligosaccharidesofvarious lengths.Theacidicglycosphingolipids,foundprimarilyinmammaliancells, containeithersulphatedsugars(sulfatide)orsialicacid(gangliosides)inthe terminalsugarposition.Yeastsphingolipidscontaininositolphosphateand mannoseinositolphosphatelinkedatthishydroxyl.Theamidebondandthe hydroxylgroupofsphingolipidsallowspecificH-bondinginteractionswith otherlipidsincludingcholesterol(Figure1.6B).
3.Propertiesoflipidsinsolution
Thematrixthatdefinesabiologicalmembraneisalipidbilayercomposedofa hydrophobiccoreexcludedfromwaterandanionicsurfacethatinteractswith wateranddefinesthehydrophobic hydrophilicinterface.Muchofour understandingofthephysicalpropertiesoflipidsinsolutionandthedriving forcefortheformationoflipidbilayerscomesfromtheconceptofthe ‘hydrophobicbond’asdescribedbyWalterKauzmanninthecontextofthe forcesdrivingproteinfolding,whichwaslaterextendedtothe‘hydrophobic effect’byCharlesTanfordtoexplainself-associationoflipidswithinbiological systems[1].The‘fluidmosaic’modelformembranestructurefurtherpopularisedtheseconcepts[2].Althoughthismodelstimulatedresearchinthearea ofMPs,itrelegatedlipidstoamonolithicroleasafluidmatrixwithinwhich MPsresideandfunction.Aswillbediscussedbelow,ourcurrentunderstanding oftheroleoflipidsincellfunctionisasspecificanddynamicasthatofproteins.
3.1Whydomembranesform?
Polarlipidsareamphipathicinnature,containinghydrophobicdomains, whichdonotinteractwithwater,andhydrophilicdomainsthatreadilyinteract
withwater.Thebasicpremiseofthehydrophobiceffectisthatthehydrocarbondomainsdistortthestablehydrogen-bondedstructureofwaterby inducingorderedcage-likestructuresaroundtheapolardomains[1].Selfassociationofthehydrophobicdomainsminimisesthetotalsurfaceareain contactwithwaterresultinginentropy-drivenrelaxationofwaterstructureand anenergyminimumforthefinalself-associatedmolecularorganisation.The polardomainsoflipidsinteracteitherthroughhydrogenbondingorionic interactionwithwaterorotherlipidheadgroupsandthereforeareenergetically stableinanaqueousenvironment.Thestructuralorganisationthatapolarlipid assumesinwaterisdeterminedbyitsconcentrationandthelawofopposing forces,i.e.hydrophobicforcesdrivingself-associationofhydrophobic domainsversusstericandionicrepulsiveforcesofthecloselyassociatedpolar domainsopposingself-association.Atlowconcentrations,amphipathic moleculesexistasmonomersinsolution.Astheconcentrationofmolecules increase,theirstabilityinsolutionasamonomerdecreasesuntiltheunfavourablerepulsiveforcesofcloselypackedpolardomainsareoutweighedby thefavourableself-associationofthehydrophobicdomains.Atthispoint, termedthecriticalmicelleconcentration(CMC),afurtherincreaseinconcentrationresultsintheformationofincreasingamountsofself-associated monomersinequilibriumwithaconstantamountoffreemonomer.Dueto theincreasedhydrophobiceffect,alargerhydrophobicdomainresultsina lowerCMC.However,increasingtheeffectivesizeofthepolardomain,either duetosterichindranceofneutraldomainsorchargerepulsionforionicdomains,increasestheCMC.TheCMCofamphipathicmoleculeswithanet chargeisloweredbyincreasingtheionicstrengthofthesolutionphasedueto dampeningofthechargerepulsioneffect.
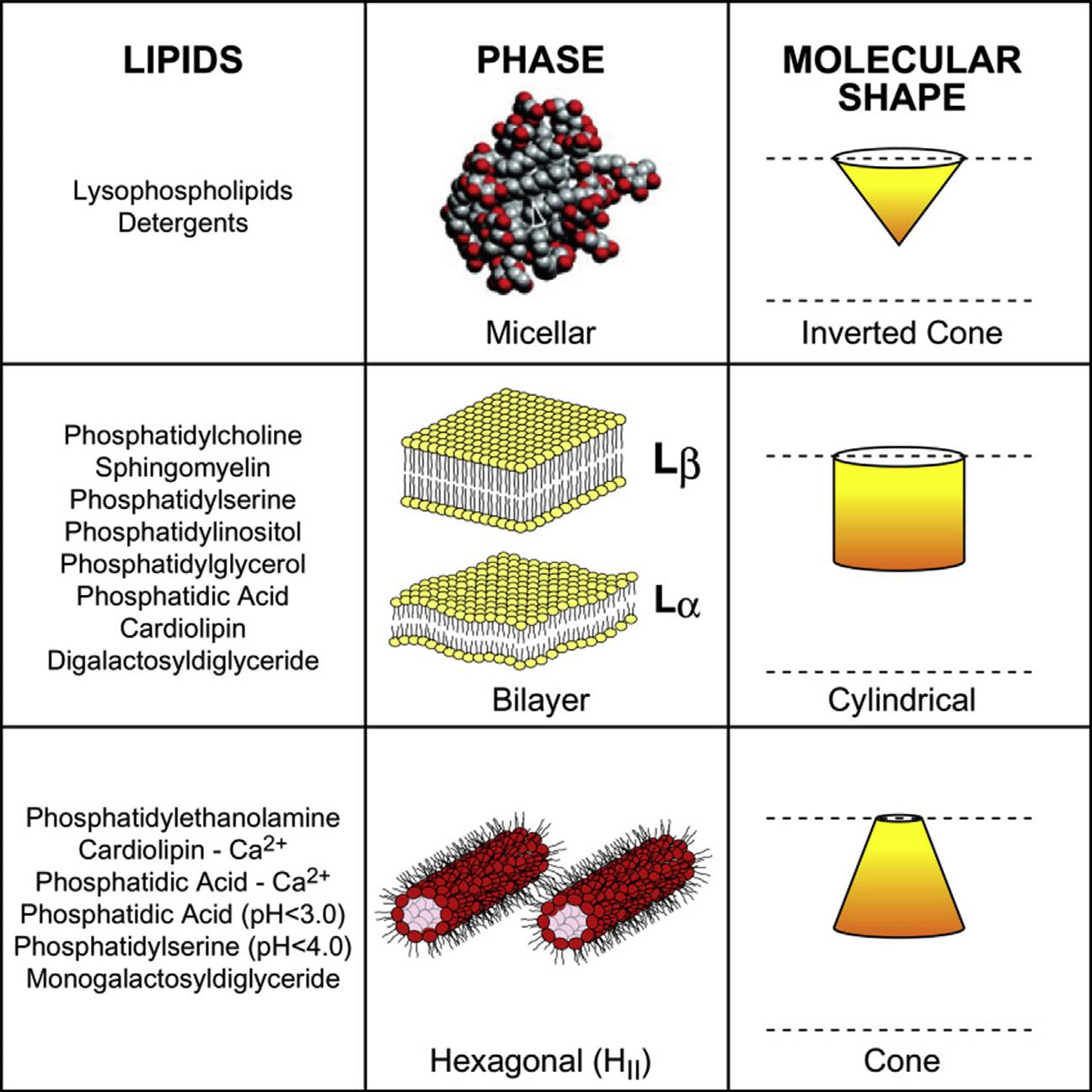
Thesephysicalpropertiesandtheshapeofamphipathicmoleculesdefine threesupramolecularstructuralorganisationsofpolarlipidsanddetergentsin solution(Figure1.7).
Detergents,detergent-likelysophospholipids(containingonlyonealkylor acylchain),andphospholipidswithshortalkyloracylchains(8orfewer carbons)haveaninvertedconeshape(largeheadgrouprelativetoasmall hydrophobicdomain)andself-associateabovetheCMCwithasmallradiusof curvaturetoformmicellarstructureswithahydrophobiccoreexcludingwater. Themicellesurface,ratherthanbeingasmoothsphericalorellipticalstructure withthehydrophobicdomainscompletelysequesteredinsideashellofpolar residuesthatinteractwithwater,isaveryroughsurfacewithmanyofthe hydrophobicdomainsexposedtowater.Theoverallstructurereflectsthe optimalpackingofamphipathicmoleculesatanenergyminimumasgoverned bythelawofopposingforces.TheCMCformostdetergentsrangesfrom micromolartomillimolar.LysophospholipidsalsoformmicelleswithCMCs inthemicromolarrange.However,phospholipidswithchainlengthsof16 carbonsself-associateataconcentrationaround10 10 M.Thecylindrical shapeofmostphospholipidswithlongacylchainsatphysiological