Diagnostic
https://ebookmass.com/product/diagnostic-imaging-oncology-2nd-editionakram-m-shaaban/
ebookmass.com
ADVISORYBOARDS
DavidRodríguez-Lázaro
Loong-TakLim
MichaelEskin
IsabelFerreira
CrispuloGallegos
Se-KwonKim
KeizoArihara
SERIESEDITORS
GEORGEF.STEWART(1948–1982)
EMILM.MRAK(1948–1987)
C.O.CHICHESTER(1959–1988)
BERNARDS.SCHWEIGERT(1984–1988)
JOHNE.KINSELLA(1989–1993)
STEVEL.TAYLOR(1995–2011)
JEYAKUMARHENRY(2011–2016)
FIDELTOLDRÁ(2016– )
AcademicPressisanimprintofElsevier
50HampshireStreet,5thFloor,Cambridge,MA02139,UnitedStates 525BStreet,Suite1650,SanDiego,CA92101,UnitedStates TheBoulevard,LangfordLane,Kidlington,OxfordOX51GB,UnitedKingdom 125LondonWall,London,EC2Y5AS,UnitedKingdom
Firstedition2022
Copyright©2022ElsevierInc.Allrightsreserved.
Nopartofthispublicationmaybereproducedortransmittedinanyformorbyanymeans,electronic ormechanical,includingphotocopying,recording,oranyinformationstorageandretrievalsystem, withoutpermissioninwritingfromthepublisher.Detailsonhowtoseekpermission,further informationaboutthePublisher’spermissionspoliciesandourarrangementswithorganizationssuch astheCopyrightClearanceCenterandtheCopyrightLicensingAgency,canbefoundatourwebsite: www.elsevier.com/permissions.
Thisbookandtheindividualcontributionscontainedinitareprotectedundercopyrightbythe Publisher (otherthanasmaybenotedherein).
Notices
Knowledgeandbestpracticeinthisfieldareconstantlychanging.Asnewresearchandexperience broadenourunderstanding,changesinresearchmethods,professionalpractices,ormedical treatmentmaybecomenecessary.
Practitionersandresearchersmustalwaysrelyontheirownexperienceandknowledgeinevaluating andusinganyinformation,methods,compounds,orexperimentsdescribedherein.Inusingsuch informationormethodstheyshouldbemindfuloftheirownsafetyandthesafetyofothers,including partiesforwhomtheyhaveaprofessionalresponsibility.
Tothefullestextentofthelaw,neitherthePublishernortheauthors,contributors,oreditors,assume anyliabilityforanyinjuryand/ordamagetopersonsorpropertyasamatterofproductsliability, negligenceorotherwise,orfromanyuseoroperationofanymethods,products,instructions,orideas containedinthematerialherein.
ISBN:978-0-323-99082-0
ISSN:1043-4526
ForinformationonallAcademicPresspublications visitourwebsiteat https://www.elsevier.com/books-and-journals
Publisher: Zoe Kruze
AcquisitionsEditor: SamMahfoudh
DevelopmentalEditor: JhonMichaelPeñano
ProductionProjectManager: VijayarajPurushothaman
CoverDesigner: MatthewLimbert
TypesetbySTRAIVE,India
Contributors
AlexiosAlexopoulos
LaboratoryofAgronomy,DepartmentofAgriculture,UniversityofthePeloponnese, Kalamata,Messinia,Greece
MikelAn ˜ ibarro-Ortega
CentrodeInvestigac ¸aodeMontanha(CIMO),InstitutoPolitecnicodeBraganc ¸a,Braganc ¸a, Portugal
LillianBarros
CentrodeInvestigac ¸ a ˜ odeMontanha(CIMO),InstitutoPolitecnicodeBraganc ¸a,Braganc ¸a, Portugal
EdmundoBrito-delaFuente
Product&ProcessEngineeringCentre,FreseniusKabiDeutschlandGmbH,BadHomburg, Germany
AdrianoGomesdaCruz
DepartmentofFood,FederalInstituteofEducation,ScienceandTechnologyofRiode Janeiro(IFRJ),RiodeJaneiro,RJ,Brazil
IsabelDian ˜ ez
ChemicalProcessandProductTechnologyResearchCentre(Pro2TecS),Departamentode Ingenierı´aQuı´mica,UniversidaddeHuelva,Huelva,Spain
N.A.MichaelEskin
DepartmentofFood&HumanNutritionalSciences,UniversityofManitoba,Winnipeg, MB,Canada
IsabelC.F.R.Ferreira
CentrodeInvestigac ¸ a ˜ odeMontanha(CIMO),InstitutoPolitecnicodeBraganc ¸a,Braganc ¸a, Portugal
JoseM.Franco
ChemicalProcessandProductTechnologyResearchCentre(Pro2TecS),Departamentode Ingenierı´aQuı´mica,UniversidaddeHuelva,Huelva,Spain
Crı´spuloGallegos
Product&ProcessEngineeringCentre,FreseniusKabiDeutschlandGmbH,BadHomburg, Germany
Loong-TakLim
DepartmentofFoodScience,UniversityofGuelph,Guelph,ON,Canada
BrunaMarchesanMaran
DepartmentofChemicalEngineeringandFoodEngineering,FederalUniversityofSanta Catarina,TechnologyCenter,Floriano ´ polis,SantaCatarina,Brazil
EmanueliMarchesanMaran
DepartmentofChemicalEngineeringandFoodEngineering,FederalUniversityofSanta Catarina,TechnologyCenter,Floriano ´ polis,SantaCatarina,Brazil
InmaculadaMartı ´ nez
ChemicalProcessandProductTechnologyResearchCentre(Pro2TecS),Departamentode Ingenierı´aQuı´mica,UniversidaddeHuelva,Huelva,Spain
LeticiaMora
InstitutodeAgroquı´micayTecnologı´adeAlimentos(CSIC),Paterna,Spain
RuchiraNandasiri
DepartmentofFood&HumanNutritionalSciences,UniversityofManitoba;Richardson CentreforFunctionalFoods&Nutraceuticals,Winnipeg,MB,Canada
SpyridonA.Petropoulos
DepartmentofAgriculture,CropProductionandRuralEnvironment,Universityof Thessaly,Volos,Greece
MilenaDutraPierezan
DepartmentofFoodScienceandTechnology,AgriculturalSciencesCenter,Federal UniversityofSantaCatarina,Floriano ´ polis,SantaCatarina,Brazil
TatianaColomboPimentel
FederalInstituteofParana ´ ,Paranavaı´,Parana,Brazil
JosePinela
CentrodeInvestigac ¸aodeMontanha(CIMO),InstitutoPolitecnicodeBraganc ¸a,Braganc ¸a, Portugal
DavidRodrı´guez-La ´ zaro
MicrobiologyDivision,FacultyofSciences;ResearchCentreforEmergingPathogensand GlobalHealth,UniversityofBurgos,Burgos,Spain
RachelSiqueiradeQueirozSimo˜es
InstituteofTechnologyinImmunobiologicals,Bio-Manguinhos,OswaldoCruz Foundation,Fiocruz,Manguinhos,RiodeJaneiro,Brazil;MicrobiologyDivision,Facultyof Sciences;ResearchCentreforEmergingPathogensandGlobalHealth,UniversityofBurgos, Burgos,Spain
FidelToldra ´
InstitutodeAgroquı´micayTecnologı´adeAlimentos(CSIC),Paterna,Spain
SilvaniVerruck
DepartmentofFoodScienceandTechnology,AgriculturalSciencesCenter,Federal UniversityofSantaCatarina,Floriano ´ polis,SantaCatarina,Brazil
AmrZaitoon
DepartmentofFoodScience,UniversityofGuelph,Guelph,ON,Canada
HongfeiZhang
DepartmentofFoodScienceandTechnology,NationalUniversityofSingapore,Singapore, Singapore
WeibiaoZhou
DepartmentofFoodScienceandTechnology,NationalUniversityofSingapore,Singapore, Singapore
Preface
Theseries AdvancesinFoodandNutritionResearch hasreachedVolume100, andthisrepresentsasignificantmilestoneaftermorethan70yearsofthe series’history.Thisserieswasinitiallynamed AdvancesinFoodResearch, andthefirstvolumewaspublishedin1948.Ithad459pagesdistributed across10chaptersandwaseditedbyEmilM.Mrak(editoruntil1987) andGeorgeF.Stewart(editortill1982).Thelinktothisvolumeis https://www.sciencedirect.com/book s eries/advances-in-food-research/ vol/1/suppl/C .Aneweditor,C.O.Chichesterwasappointedin1959, whoremaineduntil1988.Thenameoftheserieswaschangedto Advances inFoodandNutritionResearch in1989withVolume33andJohnE.Kinsella whowastheeditoruntil1993.Atthispoint,theserieswasnotpublished onayearlybasis;however,in2001,itstartedwithoneortwovolumesbeing publishedeveryyear.In1995,SteveL.Taylorwastheeditor,whoremained until2011,followedbyJeyakumarHenryfrom2011to2016.Then,Fidel Toldra ´ tookoverastheeditorin2016withVolume76.Duetoitssuccess, theseriesmoveduptothreevolumesayearin2009,thenfourvolumesayear in2018,andcurrentlyapprovedtoreachfivevolumesayearin2023andsuccessiveyears.SuccesscontinuesasScienceDirectusageexperienced anoticeableincreaseof68%intheperiod2016–2020.Thenumberofcitationsisalsoincreasing,withaCiteScoreof4.7in2019thatincreasedupto7.0 in2020,forthisseriesinQ1,ranking32outof310publicationsinFood Sciencewith89thpercentilebasedonCiteScoredatainScopus.
Thisvolume(Volume100)containseightchapterswrittenbyaninternational boardofauthors—someofthemwerepreviousguesteditorsof serialthematicvolumesincludingthisseries’editor—andreportsthelatest developmentsinrelevantandinterestingtopicssuchastheuseofpeptidomicstoolsandtheirapplicationsinbioactivepeptides,thecontrolled releaseofbioactivesinfood,theroleofcanololasanantioxidantandanticanceragent,theapplicationsofSolanaceaeinfoodsandnutraceuticals, thelatestdevelopmentsin3Dprintingoffoods,thesafeuseofrawmilk intheprocessingofdairyfoods,thenewthreatsofentericviruses,and, finally,theuseoflow-energyX-rayforinnovativenonthermalfood processing.
Thefirstchapterdealswiththeuseofpeptidomicsandthemethodologiesinvolvedinthestudyofbioactivepeptides,includingtheiridentification
andquantitation,andinsilicoapproaches.Ananalysisofposttranslational modificationsinpeptidesthatcanmodifytheirphysicalandchemicalpropertiesisalsopresented,andthespecificapplicationsofpeptidomicsin bioactivepeptidesarefinallyreported.Thesecondchapterprovidesanoverviewofthebasiccontrolledandtriggeredreleaseconceptsrelevanttofood andactivepackagingapplications.Itpresentsapproachestoencapsulatebioactivecompounds,theirmodeofrelease,themasstransportprocesses,and theinteractivecarriersthatareresponsivetocertainspecificstimuli.The thirdchapterfocusesonthenatureofcanololanditsapplicationsinfood andmedicine.Canololisauniquephenolformedduringprocessingthat hasantioxidantandanticancerproperties.Specificextractabilityrequirementsfromoilseedusingdifferentprocessingconditionsandcanololderivatives,includingdimersandtrimers,arediscussed.Thefourthchapter discussesthenutritionalvalueofthemostimportantSolanaceaespecies commonlyusedfortheirediblefruit,aswellasthoseusedinthedevelopmentoffunctionalfoodsandnutraceuticalsduetotheirbioactiveconstituents.Thetoxicandpoisonouseffectsaswellasthesustainablemanagement practicesimplementedtoincreasetheaddedvalueofsuchcropsarealsodiscussed.Thefifthchapterreviewsthecurrentadvancesinextrusion-based 3Dfoodprintingsystems,presentinginnovative3Dfoodprintingsystems basedongellingand/ormixing.Thefutureofextrusion-based3Dfood printinganditsapplicationsintheproductionofcustomizedfoodsforspecific needssuchascontrollednutritionalorrheologicalpropertiesarealsodiscussed.Thesixthchapterreviewsthecontaminantsthatmaybepresentin rawmilk,especiallyconventionalandemergingpathogens,antimicrobialresistantpathogensandcommensalstrainsfoundinmilkanddairyproducts, andbiologicaltoxicsubstancesformedbymicroorganismslikebiogenic aminesandmycotoxins.Thechapteralsoreportsotherchemicalcontaminantssuchasantibioticresidues,heavymetals,pesticides,polycyclicaromatic hydrocarbons,melamine,dioxins,polychlorinatedbiphenyls,plasticizers,and additives.Theseventhchapterdealswiththethreadsofentericvirusesdueto theirhighcapacityforinfectionandpreservationinfoodenvironmentsand theirdifficultyforcorrectandsensitivedetection.Thechapterdiscussesthe currenteffortstowardprioritizingdifferentaspectsoftheirdetection,epidemiology,andcontrolaswellastheneedtocalibratethecurrentdisinfection proceduresinthefoodindustry.Theeightandlastchapterfocusesontheuse oflow-energyX-rayirradiationoffoodsfornonthermalmicrobialinactivation.Thistechnologyprovidesahighermicrobialinactivationefficacythan otherirradiationtechniques,and,therefore,low-energyX-rayirradiation
constitutesanattractivealternativefoodpreservationtechnique.Furthermore, thecurrentapplicationsoflow-energyX-ray,consumeracceptance,and itslimitationsarediscussedinthischapter.
Insummary,thisvolumepresentsthecombinedeffortsof27professionalswithdifferentbackgroundswhoaredevelopingtheirresearchina varietyofcountrieslikeCanada,Brazil,Singapore,Portugal,Greece, Germany,andSpain.TheeditorthanksJhonMichaelPen ˜ ano(developmentaleditor),SamMahfoudh(acquisitioneditor),theproductionstaff, andallthecontributorsforsharingtheirexperiencesandformakingthis bookpossible.
FIDEL TOLDRA ´ Editor
Peptidomicsasausefultool inthefollow-upoffoodbioactive peptides
FidelToldrá∗ andLeticiaMora
InstitutodeAgroquı´micayTecnologı´adeAlimentos(CSIC),Paterna,Spain
Mechanismsofenzymatichydrolysisoffoodproteins
5. Releaseofbioactivepeptidesinfoodsbyendogenouspeptidases
Abstract
Thereisanintenseresearchactivityonbioactivepeptidesderivedfromfoodproteinsin viewoftheirhealthbenefitsforconsumers.However,theiridentificationisquitechallengingasaconsequenceoftheirsmallsizeandlowabundanceincomplexmatrices suchasfoodsorhydrolyzates.Recentadvancesinpeptidomicsandbioinformaticsare gettingimprovedsensitivityandaccuracyandthereforesuchtoolsarecontributingto thedevelopmentofsophisticatedmethodologiesfortheidentificationandquantificationofpeptides.Thesedevelopmentsareveryusefulforthefollow-upofpeptides releasedthroughproteolysiseitherinthefooditselfthroughtheactionofendogenous peptidasesduringprocessingstageslikefermentation,dryingorripening,orfromfood proteinshydrolyzedbycommercialpeptidasesormicroorganismswithproteolytic activity.Thischapterispresentingthelatestadvancesinpeptidomicsanditsusefor theidentificationandquantificationofpeptides,andasausefultoolforcontrolling theproteolysisphenomenainfoodsandproteinhydrolyzates.
1.Introduction
Ithasbeenwidelyreportedthatfoodproteinsconstituteagoodsource ofbioactivepeptidesthatremaininactivewhileformingpartoftheparent protein,althoughtheymayturnactiveifreleasedthroughenzymatichydrolysis.Foodbioactivepeptidesmayexerthealthpromotingbeneficialeffects suchasantioxidant,antihypertensive,antiinflammatory,hypoglycemic, hypocholesterolemic,antimicrobialandantitumoractivities(Toldra ´ , Reig,Aristoy,&Mora,2018).Suchbioactivityinformationisavailable inopenaccessdatabasesthatreportdataaboutchemicalandstructuralcharacteristicsofbioactivepeptidesandproteinoforigin,itsIC50,andreferences (Minkiewicz,Iwaniak,&Darewicz,2019).Thebeneficialeffectsofbioactivepeptidesnaturallygeneratedinfoods,preventinginfectionanddiseases isofgreatinterestinanagingpopulation.Thismakessuchfoodsrichinbioactivepeptidesanattractiveoptionindailydiet.Furthermore,bioactive peptidesproducedthroughenzymatichydrolysisinlargeamountsmake themattractiveasfoodsupplementsandasfunctionalcomponentstoregulatehealth.
Peptidomicstechniquesaremainlyfocusedonthestudyofpeptides, includingtheiridentificationandquantitation.Thetargetpeptidesmayhave beengeneratedindifferentwaysspeciallywhentheycomefromfoodmatrices(see Fig.1).So,bioactivepeptidesmaybereleasedduringkeystagesin
Fig.1 Schemeofpeptidesgenerationfromfoodproteinsandmaindisciplinesforthe studyofproteinsandpeptides.
foodprocessinglikefermentation,drying,andripeningofmeatanddairy products,wine,saucesamongother(Corr^ ea etal.,2014; Mohanty, Mohapatra, Misra,&Sahu,2016; Moraetal.,2015).Bioactivepeptides may bealsoproducedthroughcontrolledenzymatichydrolysis,withcommercialpeptidasesormicroorganisms,ofproteinsobtainedfromdifferent typesoffoodwasteandby-productsresultingfromslaughterhouses,fisheries,whey,fruitspeels,etc.(Mora,Reig,&Toldra ´ , 2014; Ryder,Bekhit, McConnell, &Carne,2016),andalsohydrolyzatesfromegg,soybean and peanutproteins,amongother(DeOliveiraetal.,2015; Ji,Sun, Zhao, Xiong,&Sun,2014).Commercialpeptidasesareadequatetobe used infoodandcanbeobtainedfromdifferentoriginssuchasmicroorganisms,vegetablesourcesoranimalorgans.Finally,gastrointestinaldigestion mustbetakenintoaccountbecausepepsininthegastricstepandtrypsin, chymotrypsinandpancreaticenzymesintheintestinalsteparealsoable toreleasebioactivepeptidesfromtheingestedproteinsoreveninactivate somepeptidesduetofurtherhydrolysis(Capriottietal.,2015; Pepe et al.,2016).Furthermore,anintenseproteolyticactivityduetotheaction of brushborderintestinalepitheliumproteasesandbloodstreamenzymes thatcompleteproteindigestionactingasexopeptidasesandreleasing dipeptidesandfreeaminoacids.
Therecentimprovementsinsensitivityandaccuracyachievedin peptidomicsinstrumentationandadvancesinbioinformaticstoolsare allowingthedevelopmentofmoresophisticatedmethodologiesforthe identificationofpeptides.Theknowledgeofstructureandfunctionof bioactivepeptidesnaturallygeneratedinfoodsandinhydrolyzatestobe usedinfoodsupplementsandnutraceuticals,isessentialfortheoptimization ofprocessingandqualitycontrol.
Thischapterispresentingthelatestadvancesinpeptidomicsasauseful toolforabetterunderstandingandcontroloftheproteolysisphenomena andthereforeasanadequatetoolforfollowingthegenerationofbioactive peptidesanditsquantitationinfoodsandhydrolyzates.
2.Peptidomicsinthecharacterizationofpeptides
Peptidomicstechniqueshavebeensuccessfullyusedforabetter understandingofgastrointestinaldigestionprocess,thecontributionof endogenousandmicrobialenzymesindifferentfoodprocessessuchas fermentation(Li&Wang,2021; Yu,Yu,&Jin,2021),curing(Gallego,
Peptidomics Biomarkers
Authentification, OGMs, allergens, pathogensor toxins
Bioactivities: antioxidant, antihypertensive, hypoglycemic, hypercholesterolemic, etc.
Structure-activity prediction
Fig.2 Majorareasofpeptidomicsapplicationsinfoods.
Mora,&Toldra ´ ,2016),aging(Kominami,Hayashi,Tokihiro,&Ushio, 2021; Renzone,Novi,Scaloni,&Arena,2021),theidentificationofbioactive peptides,andthesearchofpeptidemarkersinfoodsystems(see Fig.2).Arecentinterestinpeptidomicsisgrowingintheareaoffoodwaste and by-productsvalorizationbecausesuchpeptidomicstoolscontributeto abetterunderstandingoftheproteolysisphenomenaandthereforea bettercontrolandsteeringoftheprocesses(Martini,Solieri,Cattivelli, Pizzamiglio, &Tagliazucchi,2021; Martini,Solieri,&Tagliazucchi,2021).
Peptidomics, inconjunctionwithotherdisciplineslikeproteomics, genomics,transcriptomics,andmetabolomics,contributestoabetter understandingofthefoodsystems.However,therearesomedifficultiesthat makethestudyoftheproteomeverycomplicatedsuchasthelowabundanceofpeptides,theirheterogeneityandvarietyinsize,charges, characteristics,andthecomplexityofbiologicalmatriceswhentheyare distributed.Forthisreason,ahighamountofextraction,separation,isolation,identification,andquantitationtechniqueshavebeendevelopedand usedinthelastyears.
Thedescriptionofmainstepsusedtosimplify,reducecomplexityand eliminatepotentialinterferencesinfoodsamples,beforetheidentification
Fig.3 Descriptionofmainstepsusedtosimplify,reducecomplexity,andeliminate potential interferencesinfoodsamplesbeforetheidentificationandquantitationof bioactivepeptides.
andquantitationofbioactivepeptidesisshownin Fig.3.Thepretreatment step isusedtoremovepotentialinterferences,aswellastofacilitatethe extractionandconcentrationofpeptides.Currenttendenciesaretheuse ofgreenandcost-effectivetechnologiessuchasultrasounds,microwaves, pulsedelectricfields,orhighhydrostaticpressure,althoughchemical pretreatmentshavebeenalsodescribedtoresultveryeffectiveinthelater extractionofpeptidesorproteins(Franca-Oliveira,Fornari,&Herna ´ ndezLedesma,2021).Theextractionprocedureofpeptidesisdoneusingwater or organicsolventsdependingontheexpectedcharacteristicsofthegenerated peptides.Polarpeptidesareeasilyextractedusingaqueousandslightly acidicsolventswhereashydrophobicsequencesareextractedwithorganic phases.Theextractionofpeptidesmayalsocarrypartofproteinfraction, especiallywhenaqueoussolventsareused,andthereforealaterstepof deproteinizationbyprecipitationandcentrifugationisoftenneeded.Thenext stepistheuseofoneoracombinationofdifferentseparationtechniquesin ordertofractionatethepeptideextractatthesametimethatconcentrate thebiologicallyactivefraction.ThemostusedmethodologiesareTricine SDS-PAGEelectrophoresisthatallowstheseparationofoligopeptidesand longpeptidesupto2500Da(Sch€agger,2006)althoughwhentheinterest isintheseparationofsmallerpeptides,chromatographytechniquesarethe
methodofchoice.Size-exclusionchromatographyisbasedontheseparation ofpeptidefractionsaccordingtotheirmolecularweight,andtheresolutionof separationdependsonparticlesize,poresize,flowrate,columnlengthand diameter,andsamplevolume.Generally,thehighestpossibleresolutionis obtainedusingmoderateflowrates,long,narrowcolumns,andsmallparticle sizegels,usingsameeluentinsampleandmobilephase.Molecularweight standardsareusedincolumncalibrationinordertoknowthemolecular weightsofunknownpeptidesintheextract.Alsohighresolutionliquidchromatography(HPLC)isafrequentlyusedmethodologyintheseparationof peptidesaccordingtotheiraminoacidcompositionandcharacteristics(charge, hydrophobicinteraction,andmolecularsize).Inthissense,reversed-phase followedbyhydrophilicinteractionchromatography,andion-exchangechromatographyarethemostusedmethodsforpeptidemixturesseparations. Reversed-phaseC18stationaryphasesarethemostpopular,andtheelution occursbyincreasingthehydrophobicityofthemobilephase.However,this separationnotalwaysresultsadequatewhenhighlypolarandsmallpeptidesare inthemixture,andhydrophilicinteractionchromatography(HILIC)have resultedinverygoodresultstoseparatesmallpolarcompoundsonpolar stationaryphases.Thismethodcanbecomplementarytoreversed-phaseas theorderofelutionofthepeptidesareoppositetothatfoundinreversedphase.Theuseofion-exchangechromatographyintheanalysisofpeptides islimitedtothoseapplicationsthatpermitsalteliminationinlaterstepssince moderateconcentrationsofsaltscouldinhibittheionizationusingelectrospray ionization(ESI)priormassspectrometryanalysis.Afterthechromatographic separationusingHPLC,elutingfractionsarecollectedforsubsequentpurificationstepssuchasultrafiltrationwithdifferentmolecularcut-offmembranes ortobedirectlyanalyzedbymassspectrometry.
Massspectrometrymethodologieshavebeenwidelydevelopedduring thelastdecadeand,currently,theyhavebecomethebestchoiceforthe identificationandquantificationofbioactivepeptides(Panchaud,Affolter, & Kussmann,2012).Inthiscontext,theadvancesinmassspectrometry applications andtheprogressiveimprovementintheresolutionoftheinstrumentshaveincreasedthedevelopmentofpeptidomicsapplicationsinthe foodfield,especiallyregardingtheidentificationandquantitationofbioactivepeptides.Alsothelatestdevelopmentsinbioinformaticsarenecessaryto obtainthemaximumbenefitfromthegenerateddata,includingtheuseof insilico tools,structure–activityrelationshipmodels,chemometrics,protein andpeptidedatabasesandsearchengines,andstatisticstomanagethe enormousamountofdatagenerated.
2.1Identificationofpeptides
Traditionally,proteomicshasbeenusedtoidentifyandquantifyproteins fromdifferenttypesoffoodmatrices,obtainingthecharacterizationof theproteinprofileofthesample.Themostfrequentlyusedstrategyhas beentheseparationofproteinsusinggelelectrophoresisandthesubsequent digestionoftheisolatedproteinswithproteaseswithcleavagesitesincluded inthedatabasessuchastrypsin,Arg-CorAsp-N.Theobtainedhydrolyzates areanalyzedbymassspectrometryusingspecializedtoolstoobtainalistof peakstobeprocessedusingproteindatabases.MainapproachesfortheidentificationofproteinsarebasedonPeptideMassFingerprint(PMF),where theexperimentaldataarealistofpeptidemassvaluesfromthedigestionofa proteinbyaspecificenzymeaspreviouslyindicated,andonMS/MSions search,wheretheidentificationisbasedonrawMS/MSdatafromone ormorepeptides.Despitetheseapproachesareveryusefulfortheidentificationofthepresenceofcertainproteins,theexperimentaldesignneeds tobeadaptedwhentheobjectiveistoanalyzebioactivepeptidesobtained fromeithernaturalprocessessuchasfermentation,curing,orgastrointestinaldigestion,orcommercialhydrolysissuchasthetreatmentoffood by-products.
Bioactivepeptidescanexertdifferenttypeofbioactivitiesdependingon differentfactorssuchastheirphysico-chemicalcharacteristics,amino acidssequence,structuralconformation,etc.Infact,bioactivepeptideshave beendescribedtobeabletomodifyormodulatedifferentphysiologicalsystemssuchasthecardiovascular,digestive,endocrine,immune,muscular, nervous,renal,reproductive,respiratory,andskeletalsystems,duetotheir potentialtoexertantihypertensive,antidiabetic,anticholesterolemic,antioxidant,immunomodulatory,amongotheractivities. Fig.4 showsthemain
Fig.4 Diagramshowingmainpercentagesofthemostreportedbioactivepeptides available inBIOPEPdatabase;accessedonFeb,2022(Minkiewiczetal.,2019).
bioactivitiesstudiedaccordingtoBIOPEPdatabase(Minkiewiczetal., 2019),showingACE-inhibitoryactivityasthemostreportedbiological activityfollowedbyantioxidantandantimicrobialactivity.
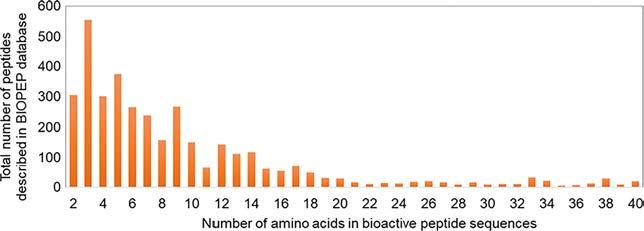
Thesepeptidesareverydifferentandcanshowdifferentcharacteristics suchaschargestate,hydrophobicityorlength,includinglarge,medium, andsmallpeptides.However,smallbioactivepeptides(from2to10amino acidslength)representbyfarthelargestcategoryasindicatedin Fig.5 (obtainedfromBIOPEPbioactivepeptidesdatabase),probablyduetotheir smallsizemakesthemmoreadequatetocrosstheintestinalbarrierand achieveintactthebloodstreamandtargetorganstodeveloptheirbiological activity.Thus,despitethenumerousdifficultiesthatinvolvetheanalysisof smallpeptides,morethanhalfofthepeptidesdescribedintheliterature andincludedintheBIOPEPdatabase(Minkiewiczetal.,2019)aresmaller than6aminoacidslength(see Fig.5).
Thestrategyfortheidentificationofbioactivepeptideswillbedifferent consideringthedifferentphysicalandchemicalpropertiesoftargetpeptides. Inthissense,separationmethodsusedinmassspectrometryinstrumentsand informaticsanalysiswilldependonthecharacteristicsandsizeofpeptides, beingintheinterfacebetweenproteomicsandmetabolomicsapproaches. Inthisregards,dataanalysisresultsverychallengingasthereisanestimation offalsepositives100timeshigherforpeptidomicsthanforproteomics becausepeptidomicsreliesonasinglepeptideidentificationandrequires veryhighqualityspectraandfrequent denovo identification.
2.1.1Identificationofdi-,tri-,andtetrapeptides
Verysmallpeptidesareofparticularinteresttobeusedasbioactivepeptides, buttheanalyticalstrategyfortheiridentificationmustbecarefullydesigned
Fig.5 NumberofaminoacidsofthebioactivepeptidesavailableinBIOPEPdatabase; accessedon17Feb,2022(Minkiewiczetal.,2019).
consideringthattheanalysisofthesesmallpeptidesarebetweentheanalysis ofpeptidesandsmallmolecules.
Firststepsofisolationandpurificationneedtobeplannedinorderto avoidlossesinpartofthetargetpeptides.Infact,whereassomeofthese peptidescontainhydrophobicaminoacidsandareproperlyretainedin C18columns,othersaremainlyhydrophilicandneedothertypesofstationaryphasestoberetained.Thus,liquidchromatographicseparationof peptidesbeforetheirinjectionintothemassspectrometryanalysis shouldhavetakenintoconsiderationthephysico-chemicalcharacteristics oftheexpectedpeptides,thepotentialmixtureofdifferentsmallpeptides inthesampleextract,andthechoiceofthemostappropriateseparation techniques,forinstance,acombinationofdifferentseparationtechniques suchasreversedphaseandhydrophilicinteractionchromatography. Concentrationstepsarenecessarytoincreasetheabundanceofthesepeptidesthatfrequentlyfoundatverylowconcentration,butthissteprequires specialattentionbecauseitmayoftenleadtothelossofsomeofthe sequencesinthewashing/cleaningstep.
Onceretentionandisolationisachieved,theanalysisofpeptidesby massspectrometryisanotherverycriticalstep.Theselectionofsuitablemass analyzerdependsontheapplicationsofinterest.Whenanalyzingsamples withcomplexmatrices,aquadrupolewouldbeusefultoreducetheinterferencesfromthematrices.Forfurtherquantitativeanalysisoftracecomponentsincomplexsamples,QqQorQ-Orbitrapwouldbeagoodchoicedue totheirhighsensitivityandquantitativeability.Orbitrapisalsousedto obtaintheaccuratemassofthetargetedmolecules,whichrevealsthe elementalcompositionsofthemolecules(Changetal.,2021).Intheidentification ofdi-andtripeptides,itisnecessarytousehighmassaccuracyand highresolutioninstrumentssuchasQuadrupoleTime-of-Flight(Q-ToF)or TripleQuadrupole(QQQ)analyzers,althoughQTrapcanrealizeMSn analysis,whichisbeneficialtoqualitativeanalysis.Inthisregards,themain difficultyistheoptimizationofthecollisionenergynecessaryforthefragmentationofthedi-andtripeptides.Inlongerpeptides,itispossibletoget severalionfragmentsanddifferentiatetheprecursor,whereasinsmall peptidestheamountsandqualityoffragmentsarelower.
Infact,intheidentificationofdipeptidesbymassspectrometryitisnecessarytoconsiderretentiontime,precursormass,andionfragmentsinorder toestablishanadequatecorrelationbetweenthisinformationandthe sequenceofthepeptide.Inthissense,moststudiesfocusedontheidentificationofsmalldiandtripeptidesalsocoverthequantitationofthe molecules.
Dipeptidesequencescanberepeatedmanytimesinproteinsequencesso theyarenotunique.Forthisreason,dipeptidesidentificationbymatching theobtainedspectrumwithproteindatabasessuchasUniProtandNCBInr isnotfeasible.Furthermore,currentsearchalgorithmsareoftenlimitedto peptidescontaining4ormoreaminoacids,assmallerpeptideswouldresult inunacceptablescoresbecausethesearchenginesarenotpreparedfor thisuse(Tang,Li,Lin,&Li,2014).Thus, de novo identificationofthe sequencesbyhighly-experiencedpersonnelisfrequentlyneeded,which canbedifficultandtime-consuming,mostlyforuntargetedanalysis (Moraetal.,2017).However,thishasbeenthemethodofchoiceofmany researchersalthoughalimitedamountofspectracanbeanalyzed.
ThemainproteomicapproachusedfortheidentificationandquantitationofsmallpeptidesistheMultipleReactionMonitoring(MRM)using QQQinstruments.Quadrupoleisoftenplacedinfrontofothermass analyzerstopre-selecttargetedions.Interferencesfrommatrixionsare removed,significantlyreducingchemicalnoises.Ingeneral,QqQmass spectrometerhasthehighestsensitivity.TheQqQisthemostcommonly usedmassanalyzerforquantitativedetectionduetoitshigheraccuracy. MRMalsopresentscertaindifficulties,asitneedstobepreviouslyoptimized,andrequirestrainedpersonnelandexpensiveinstruments.
Ontheotherhand,massspectrometryapproachesbasedonmatrixassistedlaserdesorption/ionization(MALDI)massspectrometryhavethe advantageofconvenience,speed,andaccuracy,butalsopresentgood resolution,robustness,andsensitivity.Relativeandabsolutequantification canbeperformedusingMALDIapproachesbasedonmassspectrometryin tandem(MS/MS)(Wang&Giese,2017).However,thismethodologyis not verypopularfortheidentificationofsmallpeptidesmainlyduetothe potentialinterferencesoflow-molecularpeptideswithMALDImatrices usedforionizationalthoughthehighsensitivityofMALDI-MSandits toleranceagainstinterferencebysaltsandothercomponentsallowedfor thedetectionofnativepeptidesfromcomplexbiologicalmixtureswith reducedsamplepreparationefforts.Recently,aMALDITime-of-Flight (ToF)approachwasdevelopedtoachievethefastdetectionofdipeptides indry-curedhampeptideextractswiththeadvantageofusingverylow amountsofsample,whilereducingtimeandcost(Heres,Saldan ˜ a, Toldra ´ , &Mora,2021; Heres,Yokoyama,etal.,2021).Thecharacteristic matrix ionsappearingbetween200and300 m/z wereeliminatedbysubtractionusingbioinformaticstools,andanexampleofMALDI-ToFMS spectraisshownin Fig.6.Later,theidentificationofthedipeptideswas confirmed byQQQ(Fig.7).
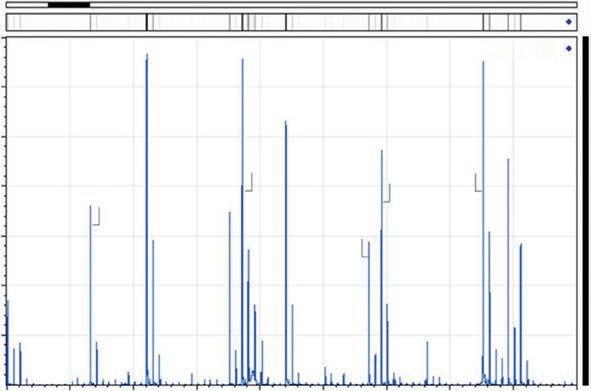
Fig.6 MALDI-ToFMSspectraofpeptidemixture(Mix).Peptidesequencesaregivenas aminoacidone-lettercode. Source:ReproducedfromAlejandroHeres,CeliaSaldaña,Fidel Toldrá,LeticiaMora,(2021).IdentificationofdipeptidesbyMALDI-ToFmassspectrometryin long-processingSpanishdry-curedham,FoodChemistry:MolecularSciences,3,100048, withpermissionfromElsevier.
2.1.2Pentapeptidesorlongerpeptidesidentification
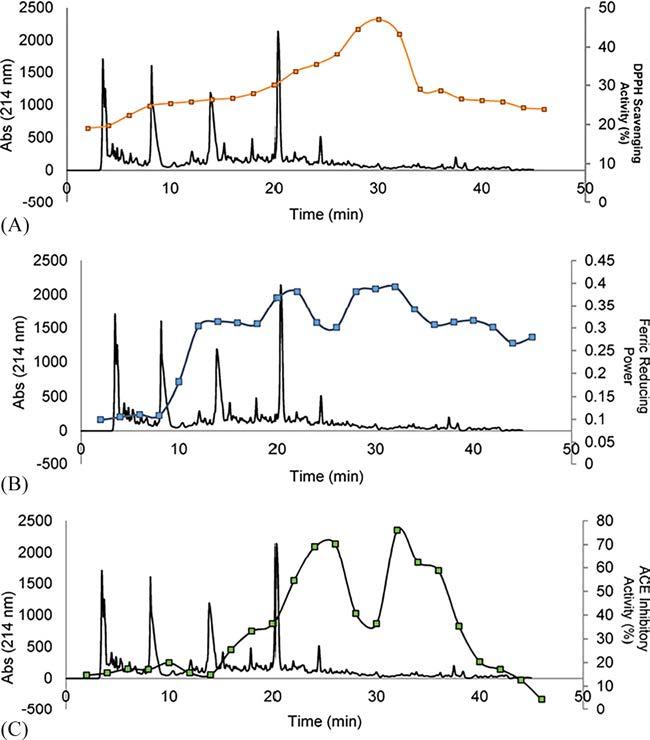
Bioactivepeptidesbetween5and30aminoacidslengthcanbegenerated eithernaturallyduetotheactionofendogenous/digestiveenzymesor duetotheactionofcommercialenzymes.Theextractionandpurification strategywilldependonthecharacteristicsofthepeptidesofinterestandit wouldbeoptimizedaccordingtotheirphysico-chemicalprofilealthough theuseofC18reversedphasecolumnsisthemostpopularseparation methodologyusedinthistypeofpeptides(seeanexamplein Fig.8).
However,whereaspeptidesobtainedfromthehydrolysiswithcommercialenzymescanbemoreabundantduetohydrolysisconditionsandhave beenoptimizedtoobtainahighrecoveryinbioactivepeptides,thosepeptidesnaturallygeneratedbyendogenousorgastrointestinalenzymeswill beatverylowconcentrationswithincomplexbiologicalmatrices,and thereforetheanalyticalmethodsusedfortheiridentificationhavetobevery sensitiveandrelyonaspecificsamplepreparationstrategy.Regardingthe identificationbymassspectrometryintandem,traditionalproteomicsprocedurescouldbeusedsincethesizeofthepeptidesissimilartothosesizes obtainedintraditionalproteomicsprotocolsusingtrypsinbutwithacertain
Fig.7 ESI-QQQspectraofthedipeptidesAH,AL,DD,EVandVFidentifiedin18months dry-curedhamextracts. Source:ReproducedfromAlejandroHeres,CeliaSaldaña,Fidel Toldrá,LeticiaMora,(2021).IdentificationofdipeptidesbyMALDI-ToFmassspectrometry inlong-processingSpanishdry-curedham,FoodChemistry:MolecularSciences,3,100048, withpermissionfromElsevier.
adjustmentofthebasictechniquesinordertoidentifyalargernumberof endogenouspeptides.Bestoptionfortheidentificationofthesepeptides istheuseoftandemmassspectrometryinstrumentswithhighresolution. IonizationsourcesusedareMALDIorESIthatareusuallycoupledtotandemmassspectrometerssuchasquadrupole-iontrap,commonlyusedto
Fig.8 Reversed-phasechromatographicseparationafter invitro gastrointestinaldigestionofanAlcalasehydrolysateoforangeby-productsfortheproductionofbioactive peptidesaftergastrointestinaldigestion.Thefigureshowsthefractionsfrom44to 54obtainedafterSECseparation.Thefractionswereautomaticallycollectedand assayedfortheirDPPHscavengingactivity(A),ferricreducingcapacity(B),ACEIinhibitoryactivity(C) α-amylaseinhibitoryactivity. ReproducedfromMazloomi,S.N.; Mahoonak,A.S.;Mora,L.;Ghorbani,M.;Houshmand,G.;Toldrá,F.PepsinHydrolysisof OrangeBy-ProductsfortheProductionofBioactivePeptideswithGastrointestinal ResistantProperties.Foods2021,10,679.
conductMSnfragmentationoftheparentpeptides,quadrupole/time-offlight(Q/ToF),ortime-of-flight/time-of-flight(ToF/ToF)(Panchaud etal.,2012).Inthisregards, Fig.9 showsthedistributionofpeptidesidentifiedindry-curedhambeforeandaftergastrointestinaldigestionaccording totheirproteinoforigin.Inthisexample,chromatographicandmass
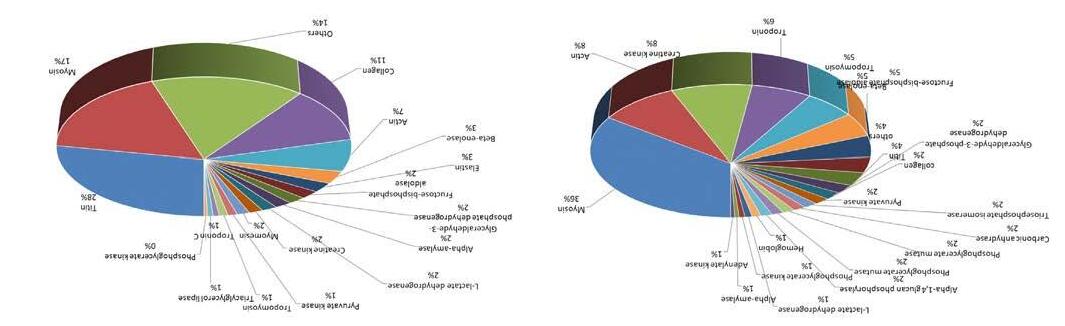
Fig.9 DistributionofthepeptidesidentifiedbynLC-MS/MSaccordingtotheiroriginproteinsin(A)undigestedand(B)digesteddry-cured hamsamplessimulating invitro gastrointestinaldigestion.Theidentificationofpeptideswasperformedbynanoliquid chromatography-tandemmassspectrometry(nLC-MS/MS)usingaNano-LCUltra1DPlussystem(EksigentofABSciex,CA,USA)coupled tothequadrupole/time-of-flight(Q/ToF)TripleTOF® 5600+system(ABSciexInstruments,MA,USA)withananoelectrosprayionization source(nESI). ReproducedfromGallego,M.,Mauri,L.,Aristoy,MC.,Toldrá,F.,Mora,L.(2020).Antioxidantpeptidesprofileindry-curedham asaffectedbygastrointestinaldigestion,JournalofFunctionalFoods,69,103956,withpermissionfromElsevier.
spectrometryanalysisrevealedchangesinthepeptideprofilesandevidenced thedegradationofproteins,mainlytitin,bythedigestiveenzymes(Gallego, Mauri, Aristoy,Toldra ´ ,&Mora,2020).
The dataanalysisapproachhastobeadaptedaccordingtotheoriginof thepeptides.CurrentsearchenginessuchasMascothasbeendesignedto workwithspectradatacorrespondingtopeptidesofsizeslongerthan4 aminoacids.However,massspectrometryinstrumentparameters,peaklist parameters,anddatabasesearchparametershavetobecarefullyselectedin ordertogetthemaximumamountofidentificationswithhighscore.The cleavagesitesofbioactivepeptidesarenotincludedinanyofthedescriptions includedinthesearchenginessofrequentlythechooseofno-enzymeis theuniqueoption.Thisfactincreasesthesearchtimesanddecreasesthe specificityintheidentificationbutgoodresultscouldbeobtainedifall parametersarewelladjusted.Itisrecommendedapreviousoptimization oftheparametersusingacontrolledproteindigestionsuchasBSA.Inorder tovalidateautomaticallyobtainedresultsusingasearchenginelikeMascot (https://www.matrixscience.com/search_form_select.html), severaltools havebeendevelopedsuchastherepetitionofthesearchusingidentical searchparameters,againstadatabaseinwhichthesequenceshavebeen reversedorrandomized.Then,itisnotexpectedtogetanytruematches fromthe“decoy”databasesothenumberofmatchesthatarefoundisan excellentestimationofthenumberoffalsepositivesthatarepresentin theresultsfromtherealor“target”database(Chen,Zhang,Xing,& Zhao,2009; Elias,Haas,Faherty,&Gygi,2005).
2.2Quantificationofpeptides
Thereisalackofinformationabouttheconcentrationofthebioactivepeptidesgeneratedduringfoodprocessing,theamountofpeptidesbioavailable inthebloodstreamaftergastrointestinaldigestion,andeventhenecessary concentrationstoestablishanadequatedosetobeusedasingredients.Infact, thequantificationofbioactivepeptidesfacesuptoseveraldifficultiesdueto theirsmallmolecularsize,thatisbetweenmetabolomicsandproteomicsdisciplines,andthecomplexityoffoodmatrices(Fig.10).Thus,theuseof advanced massspectrometrytechniquesiscriticaltoidentifythesequence ofthesesmallpeptidesandtodevelopfast,preciseandsensitivemethodologiesfortheirquantitation.Inpeptidomics,thestudyofpeptidesoften includestheirrelativequantificationwithlabeledorlabel-freemethods. Therelativequantificationofpeptidesisfocusedontheassessmentof peptideamountsintwoormoresamples,andtheresultsaregivenbythe comparisonoftheseamountsbetweensamples.Thesemethodologies
Fig.10 Proteomicsvspeptidomics.Maindifferencesareinthegenerationandidentificationofpeptides.Differentapproachesareneededwhenobjectivesaretheidentificationofproteinbiomarkersandtheidentificationofprotein-derivedbioactive peptides. ReproducedfromMora,L.,Gallego,M.,Reig,M.,Toldrá,F.(2017).Challenges inthequantitationofnaturallygeneratedbioactivepeptidesinprocessedmeats,Trends inFoodScience&Technology,69,PartB,306–314,withpermissionfromElsevier.
provideasimple,reliable,versatile,andcost-effectivequantificationthatcan bebasedonpeakintensitymeasurementsorspectralcounting(Bantscheff, Schirle,Sweetman,Rick,&Kuster,2007; Zhu,Smith,&Huang,2010). Theseapproachesresultveryinterestinginthecomparisonofpeptides profilesbetweendifferenthydrolyzatesaswellastoelucidatethemainproteinsresponsibleforthegenerationofbioactivepeptides.Inthisregards, Fig.11 showstheresultsofaPCAanalysisofthepeptidesprofileobtained fromdry-curedhamby-productscookedanddigested,andthislabel-free analysisallowedtheidentificationandrelativequantitationofthosepeptides responsiblefortheantioxidantactivitydetectedinsamplesafter1hof cooking,whichwerecollagenpeptides(Gallego,Mora,Hayes,Reig,& Toldra ´ ,2017).
Ontheotherhand,otherapproachessuchasmultiplereactionmonitoring(MRM)providethemostaccuratequantitativevaluesforbioactivepeptides,buttheyrequirecomplexexperimentalprotocols.MRMisahighly sensitiveandselectivemethodofdirectedMSthatusesthreequadrupoles fortheanalysisofshortsequencesandlowconcentrationpeptidesincomplexmixtures(Huetal.,2011; Nakashimaetal.,2011; Priyantoetal.,2015; Rawendraetal.,2014).Mainimportantchallengeofthisprocedureisthe
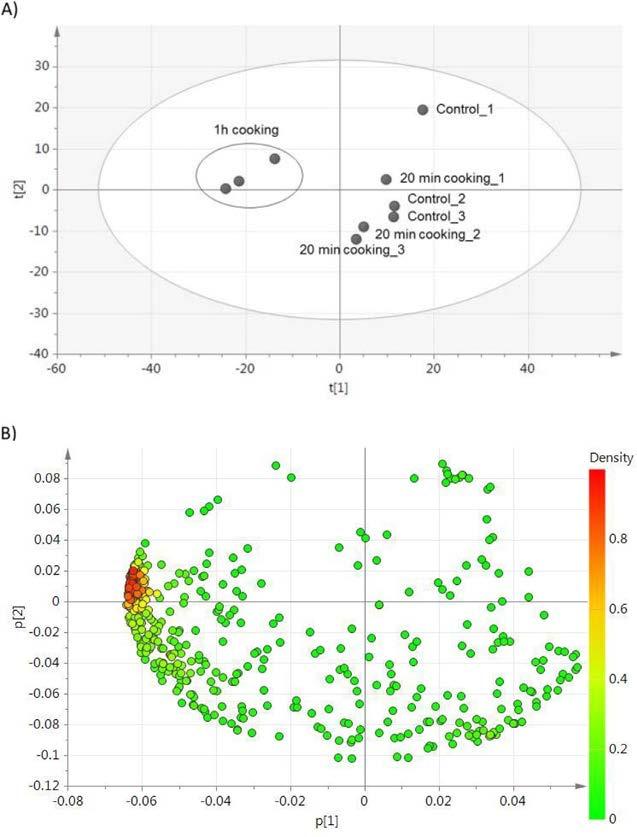
Fig.11 (A)PrincipalComponentAnalysis(PCA)scoreplottoassessthevarianceamong allthepeptidesofdigestedsamplesH2O,100 °C20min,and100 °C1hinthreereplicates(n ¼ 3).Discriminantcomponent1(t[1])anddiscriminantcomponent2(t[2]) explaineda52.9%and20%ofvariabilityinthedataset,respectively.(B)PCAloading plotshowingtheproteinsoforiginofthosepeptidesmoreresponsibleformaindifferencesbetweentheuncookedandcookedsamplesafterdigestion. Reproducedfrom Gallego,Mora,Hayes,Reig,Toldrá,Effectofcookingand invitro digestionontheantioxidantactivityofdry-curedhamby-products,FoodResearchInternational,97,2017, 296–306,withpermissionfromElsevier.
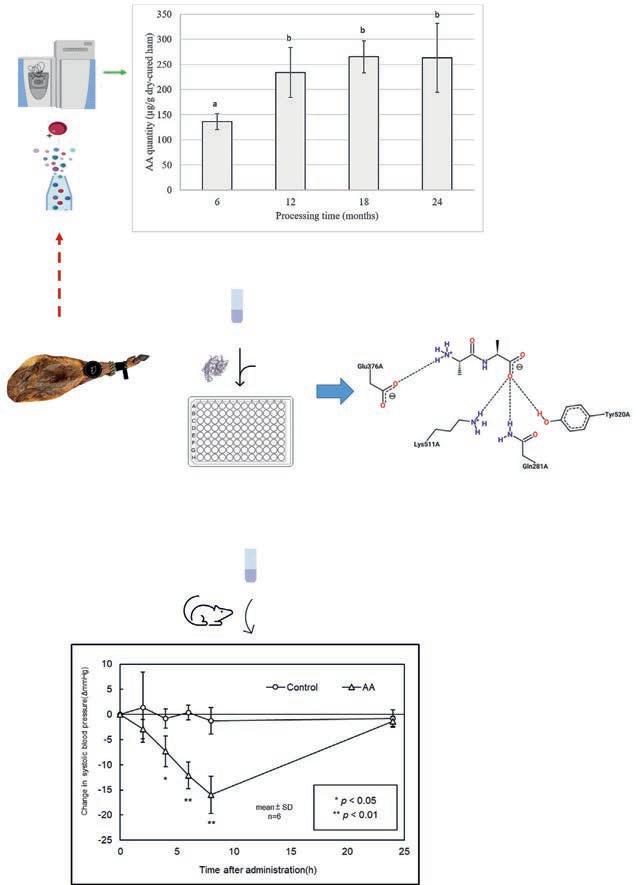
Fig.12 Graphicalabstractincludingthewholeprocedurefortheidentification,quantitationandconfirmationofAla-Aladipeptideasantihypertensivepeptide. Reproduced fromHeres,Yokoyama,Gallego,Toldrá,Arihara,Mora,(2021)Antihypertensivepotential ofsweetAla-Aladipeptideanditsquantitationindry-curedhamatdifferentprocessing conditions.JournalofFunctionalFoods,87,104818,withpermissionfromElsevier.
optimizationoftheMRMparametersoflowabundantpeptidesorsmall sizepeptidestogetafinalaccurateandsensitivequantificationwithoutinterferencesfromotherpeptidesorsignalsuppression(Capriotti,Cavaliere, Piovesana,Samperi,&Lagana,2016). Fig.12 includesmainresultsobtained
afterthequantificationoftheantihypertensivedipeptideAAatdifferent timesofprocessingofdry-curedhamusingatriplequadrupoleinstrument.
3. Insilico approaches
Anexampleofaclassicalempiricalapproachtoidentifyandconfirm thepresenceofbioactivepeptidesisshownin Fig.13.Thisincludesthe hydrolysis steptoobtainbioactivepeptidesfollowedbytheirextraction andlaterchromatographicseparation.Frequently,severalchromatographic techniquesareusedforabetterisolationofthepeptidesofinterest.Infact, theprevioussteptomassspectrometryidentificationusuallyconsistsof liquidchromatographyseparationofthealreadyisolatedpeptides.After identificationbymassspectrometry,thosepeptidesequencesapparently responsibleforthebiologicalactivitiesaresynthesizedandthebioactivity isconfirmed invitro and invivo.
of origin
Food matrix
HydrolysisExtraction
1st fractionation
Isolation of bioactive fractions (SEC, CE, LC, GF-IEF,...)
2nd fractionation
Purification of peptides of interest (HPLC)
In vitro test In vitro test
Identification by MS/MS
In vitro test
Synthesis of peptides
Bioactivity test in vitro and in vivo Confirmation
Fig.13 Schemeofthetraditionalempiricalprocedurefortheidentificationandconfirmation ofbioactivepeptidesfromfoodmatrices.SEC,size-exclusionchromatography; CE,capillaryelectrophoresis;LC,liquidchromatography;IEF,isoelectricfocusing;HPLC, high-performanceliquidchromatography;MS/MS,massspectrometryintandem. ReproducedfromMora,Gallego,Toldrá,F.(2018)ACEI-InhibitoryPeptidesNaturally GeneratedinMeatandMeatProductsandTheirHealthRelevance.Nutrients,10,1259.
Protein
Peptides