COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses Muhammad Adnan Shereen
https://ebookmass.com/product/covid-19-infectionorigin-transmission-and-characteristics-of-humancoronaviruses-muhammad-adnan-shereen/

Download more ebook from https://ebookmass.com
More products digital (pdf, epub, mobi) instant download maybe you interests ...

Pathogenic Coronaviruses of Humans and Animals: SARS, MERS, COVID-19, and Animal Coronaviruses with Zoonotic Potential Lisa A. Beltz
https://ebookmass.com/product/pathogenic-coronaviruses-of-humansand-animals-sars-mers-covid-19-and-animal-coronaviruses-withzoonotic-potential-lisa-a-beltz/

Coronavirus Disease: From Origin to Outbreak Adnan I. Qureshi
https://ebookmass.com/product/coronavirus-disease-from-origin-tooutbreak-adnan-i-qureshi/

The Environmental Impact of COVID-19 Deepak Rawtani
https://ebookmass.com/product/the-environmental-impact-ofcovid-19-deepak-rawtani/

Molecular immune pathogenesis and diagnosis of COVID-19 Xiaowei Li
https://ebookmass.com/product/molecular-immune-pathogenesis-anddiagnosis-of-covid-19-xiaowei-li/

COVID-19: The Essentials of Prevention and Treatment Jie-Ming Qu
https://ebookmass.com/product/covid-19-the-essentials-ofprevention-and-treatment-jie-ming-qu/

Organising Care in a Time of Covid-19 Palgrave
https://ebookmass.com/product/organising-care-in-a-time-ofcovid-19-palgrave/

Covid-19: Biomedical Perspectives Charles S. Pavia
https://ebookmass.com/product/covid-19-biomedical-perspectivescharles-s-pavia/

COVID-19 and the Future of Higher Education In India
Saraswathi Unni
https://ebookmass.com/product/covid-19-and-the-future-of-highereducation-in-india-saraswathi-unni/

Mental Health Effects of COVID-19 Ahmed A. Moustafa (Editor)
https://ebookmass.com/product/mental-health-effects-ofcovid-19-ahmed-a-moustafa-editor/

JournalofAdvancedResearch24(2020)91–98
Contentslistsavailableat ScienceDirect
journalhomepag e:www.elsevier.c om/locate/jare

COVID-19infection:Origin,transmission,andcharacteristicsofhuman coronaviruses
MuhammadAdnanShereen a,b,1,SulimanKhan a,1,⇑,AbeerKazmi c,NadiaBashir a,RabeeaSiddique a
a TheDepartmentofCerebrovascularDiseases,TheSecondAffiliatedHospitalofZhengzhouUniversity,Zhengzhou,PRChina
b StateKeyLaboratoryofVirology,CollegeofLifeSciences,WuhanUniversity,Wuhan,PRChina
c CollegeofLifeSciences,WuhanUniversity,Wuhan,PRChina
graphicalabstract

articleinfo
Articlehistory:
Received15March2020
Accepted15March2020
Availableonline16March2020
Keywords: Coronaviruses
COVID-19
Origin Outbreak Spread
PeerreviewunderresponsibilityofCairoUniversity.
⇑ Correspondingauthor.
abstract
Thecoronavirusdisease19(COVID-19)isahighlytransmittableandpathogenicviralinfectioncausedby severeacuterespiratorysyndromecoronavirus2(SARS-CoV-2),whichemergedinWuhan,Chinaand spreadaroundtheworld.GenomicanalysisrevealedthatSARS-CoV-2isphylogeneticallyrelatedtosevereacuterespiratorysyndrome-like(SARS-like)batviruses,thereforebatscouldbethepossibleprimary reservoir.Theintermediatesourceoforiginandtransfertohumansisnotknown,however,therapid humantohumantransferhasbeenconfirmedwidely.Thereisnoclinicallyapprovedantiviraldrugor vaccineavailabletobeusedagainstCOVID-19.However,fewbroad-spectrumantiviraldrugshavebeen evaluatedagainstCOVID-19inclinicaltrials,resultedinclinicalrecovery.Inthecurrentreview,wesummarizeandcomparativelyanalyzetheemergenceandpathogenicityofCOVID-19infectionandprevious humancoronavirusessevereacuterespiratorysyndromecoronavirus(SARS-CoV)andmiddleeastrespiratorysyndromecoronavirus(MERS-CoV).Wealsodiscusstheapproachesfordevelopingeffectivevaccinesandtherapeuticcombinationstocopewiththisviraloutbreak.
2020THEAUTHORS.PublishedbyElsevierBVonbehalfofCairoUniversity.Thisisanopenaccessarticle undertheCCBY-NC-NDlicense(http://creativecommons.org/licenses/by-nc-nd/4.0/).
E-mailaddress: suliman.khan18@mails.ucas.ac.cn (S.Khan).
1 Contributedequally(M.A.SandS.K).
Introduction
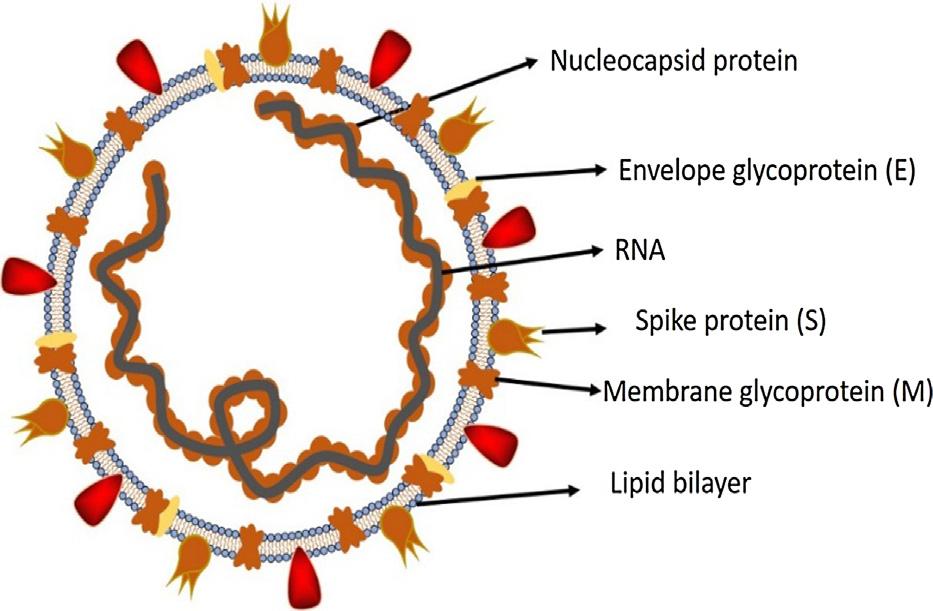
CoronavirusesbelongtotheCoronaviridaefamilyintheNidoviralesorder.Coronarepresentscrown-likespikesontheoutersurfaceofthevirus;thus,itwasnamedasacoronavirus.
https://doi.org/10.1016/j.jare.2020.03.005 2090-1232/ 2020THEAUTHORS.PublishedbyElsevierBVonbehalfofCairoUniversity. ThisisanopenaccessarticleundertheCCBY-NC-NDlicense(http://creativecommons.org/licenses/by-nc-nd/4.0/).
Coronavirusesareminuteinsize(65–125nmindiameter)and containasingle-strandedRNAasanucleicmaterial,sizeranging from26to32kbsinlength(Fig.1).Thesubgroupsofcoronaviruses familyarealpha(a),beta(b),gamma(c)anddelta(d)coronavirus. Thesevereacuterespiratorysyndromecoronavirus(SARS-CoV), H5N1influenzaA,H1N12009andMiddleEastrespiratorysyndromecoronavirus(MERS-CoV)causeacutelunginjury(ALI)and acuterespiratorydistresssyndrome(ARDS)whichleadstopulmonaryfailureandresultinfatality.Theseviruseswerethought toinfectonlyanimalsuntiltheworldwitnessedasevereacuterespiratorysyndrome(SARS)outbreakcausedbySARS-CoV,2002in Guangdong,China [1].Onlyadecadelater,anotherpathogenic coronavirus,knownasMiddleEastrespiratorysyndromecoronavirus(MERS-CoV)causedanendemicinMiddleEasterncountries [2]
Recentlyattheendof2019,Wuhananemergingbusinesshub ofChinaexperiencedanoutbreakofanovelcoronavirusthatkilled morethaneighteenhundredandinfectedoverseventythousand individualswithinthefirstfiftydaysoftheepidemic.Thisvirus wasreportedtobeamemberofthe b groupofcoronaviruses. ThenovelviruswasnamedasWuhancoronavirusor2019novel coronavirus(2019-nCov)bytheChineseresearchers.TheInternationalCommitteeonTaxonomyofViruses(ICTV)namedthevirus asSARS-CoV-2andthediseaseasCOVID-19 [3–5].Inthehistory, SRAS-CoV(2003)infected8098individualswithmortalityrateof 9%,across26contriesintheworld,ontheotherhand,novelcorona virus(2019)infected120,000induvidualswithmortalityrateof 2.9%,across109countries,tilldateofthiswriting.Itshowsthat thetransmissionrateofSARS-CoV-2ishigherthanSRAS-CoVand thereasoncouldbegeneticrecombinationeventatSproteininthe RBDregionofSARS-CoV-2mayhaveenhanceditstransmission ability.Inthisreviewarticle,wediscusstheoriginationofhuman coronavirusesbriefly.WefurtherdiscusstheassociatedinfectiousnessandbiologicalfeaturesofSARSandMERSwithaspecialfocus onCOVID-19.
Comparativeanalysisofemergenceandspreadingof coronaviruses
In2003,theChinesepopulationwasinfectedwithaviruscausingSevereAcuteRespiratorySyndrome(SARS)inGuangdongprovince.TheviruswasconfirmedasamemberoftheBetacoronavirussubgroupandwasnamedSARS-CoV [6,7].Theinfected patientsexhibitedpneumoniasymptomswithadiffusedalveolar injurywhichleadtoacuterespiratorydistresssyndrome(ARDS).
SARSinitiallyemergedinGuangdong,Chinaandthenspread rapidlyaroundtheglobewithmorethan8000infectedpersons and776deceases.Adecadelaterin2012,acoupleofSaudiArabian
nationalswerediagnosedtobeinfectedwithanothercoronavirus. Thedetectedviruswasconfirmedasamemberofcoronaviruses andnamedastheMiddleEastRespiratorySyndromeCoronavirus (MERS-CoV).TheWorldhealthorganizationreportedthatMERScoronavirusinfectedmorethan2428individualsand838deaths [8].MERS-CoVisamemberbeta-coronavirussubgroupandphylogeneticallydiversefromotherhuman-CoV.TheinfectionofMERSCoVinitiatesfromamildupperrespiratoryinjurywhileprogressionleadstosevererespiratorydisease.SimilartoSARScoronavirus,patientsinfectedwithMERS-coronavirussufferpneumonia,followedbyARDSandrenalfailure [9].
Recently,bytheendof2019,WHOwasinformedbytheChinesegovernmentaboutseveralcasesofpneumoniawithunfamiliaretiology.TheoutbreakwasinitiatedfromtheHunanseafood marketinWuhancityofChinaandrapidlyinfectedmorethan 50peoples.TheliveanimalsarefrequentlysoldattheHunanseafoodmarketsuchasbats,frogs,snakes,birds,marmotsandrabbits [10].On12January2020,theNationalHealthCommissionofChina releasedfurtherdetailsabouttheepidemic,suggestedviralpneumonia [10].Fromthesequence-basedanalysisofisolatesfromthe patients,theviruswasidentifiedasanovelcoronavirus.Moreover, thegeneticsequencewasalsoprovidedforthediagnosisofviral infection.Initially,itwassuggestedthatthepatientsinfectedwith WuhancoronavirusinducedpneumoniainChinamayhavevisited theseafoodmarketwhereliveanimalsweresoldormayhaveused infectedanimalsorbirdsasasourceoffood.However,further investigationsrevealedthatsomeindividualscontractedtheinfectionevenwithnorecordofvisitingtheseafoodmarket.These observationsindicatedahumantothehumanspreadingcapability ofthisvirus,whichwassubsequentlyreportedinmorethan100 countriesintheworld.Thehumantothehumanspreadingof thevirusoccursduetoclosecontactwithaninfectedperson, exposedtocoughing,sneezing,respiratorydropletsoraerosols. Theseaerosolscanpenetratethehumanbody(lungs)viainhalationthroughthenoseormouth(Fig.2) [11–14].
Primaryreservoirsandhostsofcoronaviruses
Thesourceoforiginationandtransmissionareimportanttobe determinedinordertodeveloppreventivestrategiestocontainthe infection.InthecaseofSARS-CoV,theresearchersinitiallyfocused onraccoondogsandpalmcivetsasakeyreservoirofinfection. However,onlythesamplesisolatedfromthecivetsatthefood marketshowedpositiveresultsforviralRNAdetection,suggesting thatthecivetpalmmightbesecondaryhosts [15].In2001the sampleswereisolatedfromthehealthypersonsofHongkongand themolecularassessmentshowed2.5%frequencyrateofantibodiesagainstSARS-coronavirus.Theseindicationssuggestedthat SARS-coronavirusmaybecirculatinginhumansbeforecausingthe outbreakin2003 [16].Lateron,Rhinolophusbatswerealsofound tohaveanti-SARS-CoVantibodiessuggestingthebatsasasourceof viralreplication [17].TheMiddleEastrespiratorysyndrome (MERS)coronavirusfirstemergedin2012inSaudiArabia [9] MERS-coronavirusalsopertainstobeta-coronavirusandhaving camelsasazoonoticsourceorprimaryhost [18].Inarecentstudy, MERS-coronaviruswasalsodetectedinPipistrellusandPerimyotis bats [19],profferingthatbatsarethekeyhostandtransmitting mediumofthevirus [20,21].Initially,agroupofresearcherssuggestedsnakesbethepossiblehost,however,aftergenomicsimilarityfindingsofnovelcoronaviruswithSARS-likebatviruses supportedthestatementthatnotsnakesbutonlybatscouldbe thekeyreservoirs(Table1) [22,23].FurtheranalysisofhomologousrecombinationrevealedthatreceptorbindingspikeglycoproteinofnovelcoronavirusisdevelopedfromaSARS-CoV(CoVZXC21 orCoVZC45)andayetunknownBeta-CoV [24].Nonetheless,to

Fig.2. Thekeyreservoirsandmodeoftransmissionofcoronaviruses(suspectedreservoirsofSARS-CoV-2areredencircled);only a and b coronaviruseshavetheabilityto infecthumans,theconsumptionofinfectedanimalasasourceoffoodisthemajorcauseofanimaltohumantransmissionofthevirusandduetoclosecontactwithan infectedperson,thevirusisfurthertransmittedtohealthypersons.Dottedblackarrowshowsthepossibilityofviraltransferfrombatwhereasthe solidblackarrow representtheconfirmedtransfer.(Forinterpretationofthereferencestocolourinthisfigurelegend,thereaderisreferredtothewebversionofthisarticle.)
Table1
ComparativeanalysisofbiologicalfeaturesofSARS-CoVandSARS-CoV-2.
FeaturesSARS-CoVSARS-CoV-2Reference
EmergencedateNovember2002December2019 [37,79–81] AreaofemergenceGuangdong,ChinaWuhan,China DateoffullycontrolledJuly2003Notcontrolledyet KeyhostsBat,palmcivetsandRaccondogsBat [22,82,83] Numberofcountriesinfected26109 [84]
EntryreceptorinhumansACE2receptorACE2receptor [22,55,85] Signandsymptomsfever,malaise,myalgia,headache,diarrhoea,shivering, coughandshortnessofbreath
Cough,feverandshortnessofbreath [12,23,85]
DiseasecausedSARS,ARDSSARS,COVID-19 [85,86]
Totalinfectedpatients8098123882 [84]
Totalrecoveredpatients732267051 Totaldiedpatients776(9.6%mortalityrate)4473(3.61%mortalityrate)
eradicatethevirus,moreworkisrequiredtobedoneintheaspects oftheidentificationoftheintermediatezoonoticsourcethat causedthetransmissionofthevirustohumans.
Keyfeaturesandentrymechanismofhumancoronaviruses
AllcoronavirusescontainspecificgenesinORF1downstream regionsthatencodeproteinsforviralreplication,nucleocapsid andspikesformation [25].Theglycoproteinspikesontheouter surfaceofcoronavirusesareresponsiblefortheattachmentand entryofthevirustohostcells(Fig.1).Thereceptor-binding domain(RBD)islooselyattachedamongvirus,therefore,thevirus mayinfectmultiplehosts [26,27].Othercoronavirusesmostlyrecognizeaminopeptidasesorcarbohydratesasakeyreceptorfor entrytohumancellswhileSARS-CoVandMERS-CoVrecognize exopeptidases [2].Theentrymechanismofacoronavirusdepends uponcellularproteaseswhichinclude,humanairwaytrypsin-like protease(HAT),cathepsinsandtransmembraneproteaseserine2 (TMPRSS2)thatsplitthespikeproteinandestablishfurtherpenetrationchanges [28,29].MERS-coronavirusemploysdipeptidyl peptidase4(DPP4),whileHCoV-NL63andSARS-coronavirus requireangiotensin-convertingenzyme2(ACE2)asakeyreceptor [2,26]
SARS-CoV-2possessesthetypicalcoronavirusstructurewith spikeproteinandalsoexpressedotherpolyproteins,nucleopro-
teins,andmembraneproteins,suchasRNApolymerase,3chymotrypsin-likeprotease,papain-likeprotease,helicase,glycoprotein,andaccessoryproteins [30,31].Thespikeproteinof SARS-CoV-2containsa3-DstructureintheRBDregiontomaintain thevanderWaalsforces [32].The394glutamineresidueinthe RBDregionofSARS-CoV-2isrecognizedbythecriticallysine31 residueonthehumanACE2receptor [33].Theentiremechanism ofpathogenicityofSARS-CoV-2,fromattachmenttoreplicationis wellmentionedin Fig.3.
GenomicvariationsinSARS-CoV-2
ThegenomeoftheSARS-CoV-2hasbeenreportedover80% identicaltotheprevioushumancoronavirus(SARS-likebatCoV) [34].TheStructuralproteinsareencodedbythefourstructural genes,includingspike(S),envelope(E),membrane(M)andnucleocapsid(N)genes.The orf1ab isthelargestgeneinSARS-CoV-2 whichencodesthepp1abproteinand15nsps.The orf1a gene encodesforpp1aproteinwhichalsocontains10nsps [34–36] Accordingtotheevolutionarytree,SARS-CoV-2liesclosetothe groupofSARS-coronaviruses [37,38] (Fig.5).Recentstudieshave indicatednotablevariationsinSARS-CoVandSARS-CoV-2such astheabsenceof8aproteinandfluctuationinthenumberof aminoacidsin8band3cproteininSARS-CoV-2 [34] (Fig.4).Itis alsoreportedthatSpikeglycoproteinoftheWuhancoronavirus
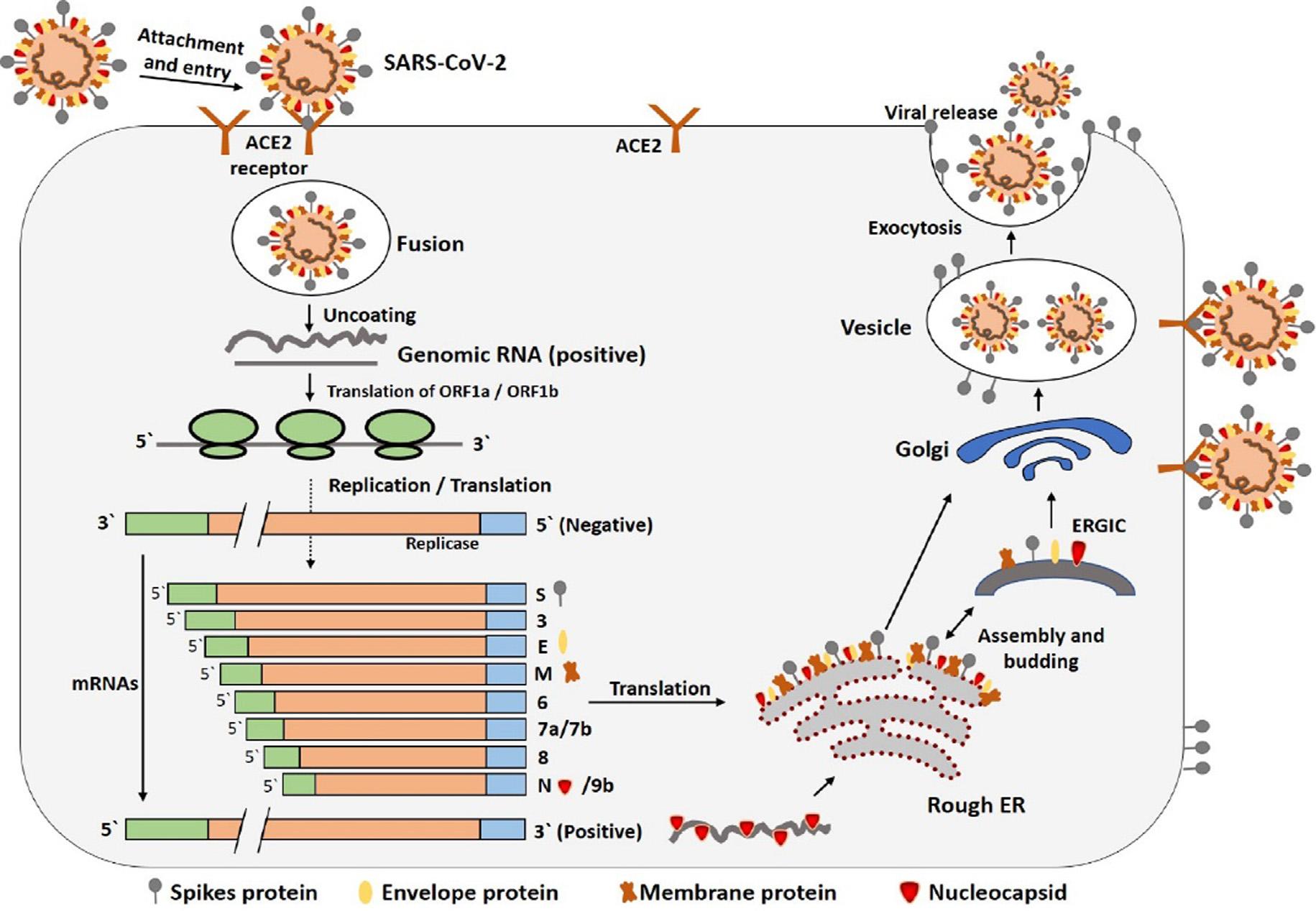
Fig.3. ThelifecycleofSARS-CoV-2inhostcells;beginsitslifecyclewhenSproteinbindstothecellularreceptorACE2.Afterreceptorbinding,theconformationchangein theSproteinfacilitatesviralenvelopefusionwiththecellmembranethroughtheendosomalpathway.ThenSARS-CoV-2releasesRNAintothehostcell.GenomeRNAis translatedintoviralreplicasepolyproteinspp1aand1ab,whicharethencleavedintosmallproductsbyviralproteinases.Thepolymeraseproduces aseriesofsubgenomic mRNAsbydiscontinuoustranscriptionandfinallytranslatedintorelevantviralproteins.ViralproteinsandgenomeRNAaresubsequentlyassembled intovirionsintheER andGolgiandthentransportedviavesiclesandreleasedoutofthecell.ACE2,angiotensin-convertingenzyme2;ER,endoplasmicreticulum;ERGIC,ER–Golgiintermediate compartment.
ismodifiedviahomologousrecombination.Thespikeglycoprotein ofSARS-CoV-2isthemixtureofbatSARS-CoVandanotknown Beta-CoV [38].Inafluorescentstudy,itwasconfirmedthatthe SARS-CoV-2alsousesthesameACE2(angiotensin-converting enzyme2)cellreceptorandmechanismfortheentrytohostcell whichispreviouslyusedbytheSARS-CoV [39,40].Thesingle N501TmutationinSARS-CoV-2’sSpikeproteinmayhavesignificantlyenhanceditsbindingaffinityforACE2 [33].
Themajorobstacleinresearchprogress
Animalmodelsplayavitalroletouncoverthemechanismsof viralpathogenicityfromtheentrancetothetransmissionand designingtherapeuticstrategies.Previously,toexaminethereplicationofSARS-CoV,variousanimalmodelswereusedwhich showedthesymptomsofsevereinfection [43].Incontrastto SARS-CoV,noMERS-CoVpathogenesiswasobservedinsmallanimals.MicearenotvulnerabletoinfectionbyMERS-coronavirus duetothenon-compatibilityoftheDPP4receptor [44].Asthe entiregenomeofthe2019-novelcoronavirusismorethan80% similartotheprevioushumanSARS-likebatCoV,previouslyused animalmodelsforSARS-CoVcanbeutilizedtostudytheinfectious pathogenicityofSARS-CoV-2.ThehumanACE2cellreceptorisrecognizedbybothSARSandNovelcoronaviruses.Conclusively, TALENorCRISPR-mediatedgeneticallymodifiedhamstersorother smallanimalscanbeutilizedforthestudyofthepathogenicityof novelcoronaviruses.SARS-CoVhasbeenreportedtoreplicateand causeseverediseaseinRats(F344),wherethesequenceanalysis revealedamutationatspikeglycoprotein [45].Thus,itcouldbe
anothersuitableoptiontodevelopspikeglycoproteintargeting therapeuticsagainstnovelcoronaviruses.Recently,micemodels andclinicalisolateswereusedtodevelopanytherapeuticstrategy againstSARS-CoV-2inducedCOVID-19 [46,47].Inasimilarstudy, artificialintelligencepredictionwasusedtoinvestigatetheinhibitoryroleofthedrugagainstSARS-CoV-2 [48].SARS-CoV-2infected patientswerealsousedtoconductrandomizedclinicaltrials [46,49,50].Itisnowimportantthatthescientistsworldwidecollaboratethedesignasuitablemodelandinvestigatetheinvivo mechanismsassociatedwithpathogenesisofSARS-CoV-2.
PotentialtherapeuticstrategiesagainstCOVID-19
Initially,interferons-a nebulization,broad-spectrumantibiotics,andanti-viraldrugswereusedtoreducetheviralload [49,51,52],however,onlyremdesivirhasshownpromisingimpact againstthevirus [53].Remdesivironlyandincombinationwith chloroquineorinterferonbetasignificantlyblockedtheSARSCoV-2replicationandpatientsweredeclaredasclinicallyrecovered [46,50,52].Variousotheranti-viralsarecurrentlybeingevaluatedagainstinfection.Nafamostat,Nitazoxanide,Ribavirin, Penciclovir,Favipiravir,Ritonavir,AAK1,Baricitinib,andArbidol exhibitedmoderateresultswhentestedagainstinfectionin patientsand in-vitro clinicalisolates [46,48,50,52].Severalother combinations,suchascombiningtheantiviralorantibioticswith traditionalChinesemedicineswerealsoevaluatedagainstSARSCoV-2inducedinfectioninhumansandmice [46].Recentlyin Shanghai,doctorsisolatedthebloodplasmafromclinicallyrecoveredpatientsofCOVID-19andinjecteditintheinfectedpatients
M.A.Shereenetal./JournalofAdvancedResearch24(2020)91–98

Fig.4. Betacoronavirusesgenomeorganization;TheBetacoronavirusforhuman(SARS-CoV-2,SARS-CoVandMERS-CoV)genomecomprisesofthe50 -untranslatedregion(50UTR),openreadingframe(orf)1a/b(greenbox)encodingnon-structuralproteins(nsp)forreplication,structuralproteinsincludingspike(blue box),envelop(maroonbox), membrane(pinkbox),andnucleocapsid(cyanbox)proteins,accessoryproteins(lightgrayboxes)suchasorf3,6,7a,7b,8and9bintheSARS-CoV-2genome,andthe30untranslatedregion(30 -UTR).ThedotedunderlinedinredaretheproteinwhichshowskeyvariationbetweenSARS-CoV-2andSARS-CoV.Thelengthofnspsandorfsarenot drawninscale.(Forinterpretationofthereferencestocolourinthisfigurelegend,thereaderisreferredtothewebversionofthisarticle.)

Fig.5. Phylogenetictreeofcoronaviruses(contentinredisthelatestadditionofnewlyemergedSARS-CoV-2andWSFMPWuhan-Hu-1isusedasareferenceinthetree);The phylogenetictreeshowingtherelationshipofWuhan-Hu-1(denotedasred)toselectedcoronavirusisbasedonnucleotidesequencesofthecompletegenome.Theviruses aregroupedintofourgenera(prototypeshown):Alphacoronavirus(skyblue),Betacoronavirus(pink),Gammacoronavirus(green)andDeltacoronavirus(lightblue). Subgroupclustersarelabeledas1aand1bfortheAlphacoronavirusand2a,2b,2c,and2dfortheBetacoronavirus.ThistreeisbasedonthepublishedtreesofCoronavirinae [3,41] andreconstructedwithsequencesofthecompleteRNA-dependentRNApolymerase-codingregionoftherepresentativenovelcoronaviruses(maximumlikelihood methodusingMEGA7.2software).severeacuterespiratorysyndromecoronavirus(SARS-CoV);SARS-relatedcoronavirus(SARSr-CoV);theMiddleEastrespiratory syndromecoronavirus(MERS-CoV);porcineentericdiarrheavirus(PEDV);Wuhanseafoodmarketpneumonia(Wuhan-Hu-1).BatCoVRaTG13Showedhighsequence identitytoSARS-CoV-2 [42].(Forinterpretationofthereferencestocolourinthisfigurelegend,thereaderisreferredtothewebversionofthisarticle.)
whoshowedpositiveresultswithrapidrecovery [54].Inarecent study,itwasidentifiedthatmonoclonalantibody(CR3022)bindswiththespikeRBDofSARS-CoV-2.Thisislikelyduetotheantibody’sepitopenotoverlappingwiththedivergentACE2
receptor-bindingmotif.CR3022hasthepotentialtobedeveloped asatherapeuticcandidate,aloneorincombinationwithother neutralizingantibodiesforthepreventionandtreatmentofCOVID-19infection [55].
VaccinesforSARS-CoV-2
ThereisnoavailablevaccineagainstCOVID-19,whileprevious vaccinesorstrategiesusedtodevelopavaccineagainstSARS-CoV canbeeffective.RecombinantproteinfromtheUrbani(AY278741) strainofSARS-CoVwasadministeredtomiceandhamsters, resultedintheproductionofneutralizingantibodiesandprotectionagainstSARS-CoV [56,57].TheDNAfragment,inactivated wholevirusorlive-vectoredstrainofSARS-CoV(AY278741),significantlyreducedtheviralinfectioninvariousanimalmodels [58–63].DifferentotherstrainsofSARS-CoVwerealsousedtoproduceinactivatedorlive-vectoredvaccineswhichefficiently reducedtheviralloadinanimalmodels.Thesestrainsinclude, Tor2(AY274119) [64,65],Utah(AY714217) [66],FRA(AY310120) [59],HKU-39849(AY278491) [57,67],BJ01(AY278488) [68,69], NS1(AY508724) [70],ZJ01(AY297028) [70],GD01(AY278489) [69] andGZ50(AY304495) [71].However,therearefewvaccines inthepipelineagainstSARS-CoV-2.ThemRNAbasedvaccinepreparedbytheUSNationalInstituteofAllergyandInfectiousDiseasesagainstSARS-CoV-2isunderphase1trial [72].INO-4800DNAbasedvaccinewillbesoonavailableforhumantesting [73] ChineseCentreforDiseaseControlandPrevention(CDC)working onthedevelopmentofaninactivatedvirusvaccine [74,75].Soon mRNAbasedvaccine’ssample(preparedbyStermirnaTherapeutics)willbeavailable [76].GeoVax-BravoVaxisworkingtodevelop aModifiedVaccinaAnkara(MVA)basedvaccine [77].WhileClover Biopharmaceuticalsisdevelopingarecombinant2019-nCoVSproteinsubunit-trimerbasedvaccine [78].
Althoughresearchteamsallovertheworldareworkingto investigatethekeyfeatures,pathogenesisandtreatmentoptions, itisdeemednecessarytofocusoncompetitivetherapeuticoptions andcross-resistanceofothervaccines.Forinstance,thereisapossibilitythatvaccinesforotherdiseasessuchasrubellaormeasles cancreatecross-resistanceforSARS-CoV-2.Thisstatementof cross-resistanceisbasedontheobservationsthatchildreninchina werefoundlessvulnerabletoinfectionascomparedtotheelder population,whilechildrenarebeinglargelyvaccinatedformeasles inChina.
Conclusionandperspective
ThenovelcoronavirusoriginatedfromtheHunanseafoodmarketatWuhan,Chinawherebats,snakes,raccoondogs,palmcivets, andotheranimalsaresold,andrapidlyspreadupto109countries. ThezoonoticsourceofSARS-CoV-2isnotconfirmed,however, sequence-basedanalysissuggestedbatsasthekeyreservoir.DNA recombinationwasfoundtobeinvolvedatspikeglycoprotein whichassortedSARS-CoV(CoVZXC21orCoVZC45)withtheRBD ofanotherBetaCoV,thuscouldbethereasonforcross-species transmissionandrapidinfection.Accordingtophylogenetictrees, SARS-CoVisclosertoSARS-likebatCoVs.Untilnow,nopromising clinicaltreatmentsorpreventionstrategieshavebeendeveloped againsthumancoronaviruses.However,theresearchersareworkingtodevelopefficienttherapeuticstrategiestocopewiththe novelcoronaviruses.Variousbroad-spectrumantiviralspreviously usedagainstinfluenza,SARSandMERScoronaviruseshavebeen evaluatedeitheraloneorincombinationstotreatCOVID-19 patients,micemodels,andclinicalisolates.Remdesivir,Lopinavir, Ritonavir,andOseltamivirsignificantlyblockedtheCOVID-19 infectionininfectedpatients.ItcanbecocludedthatthehomologusrecombinationeventattheSproteinofRBDregionenhanced thetransmissionabilityofthevirus.Whilethedecisionofbring backthenationalsfrominfectedareabyvariouscountriesand poorscreeningofpassengers,becometheleadingcauseofspreadingvirusinotherscountries.
Mostimportantly,humancoronavirusestargetingvaccinesand antiviraldrugsshouldbedesignedthatcouldbeusedagainstthe currentaswellasfutureepidemics.Therearemanycompanies workingforthedevelopmentofeffectiveSARS-CoV-2vaccines, suchasModernaTherapeutics,InovioPharmaceuticals,Novavax, VirBiotechnology,StermirnaTherapeutics,Johnson&Johnson, VIDO-InterVac,GeoVax-BravoVax,CloverBiopharmaceuticals,CureVac,andCodagenix.Butthereisaneedforrapidhumanand animal-basedtrailsasthesevaccinesstillrequire3–10months forcommercialization.Theremustbeacompletebanonutilizing wildanimalsandbirdsasasourceoffood.Besidethedevelopment ofmostefficientdrug,astrategytorapidlydiagnoseSARS-CoV-2in suspectedpatientisalsorequired.Thesignsandsymptomsof SARS-CoV-2inducedCOVID-19areabitsimilartoinfluenzaand seasonalallergies(pollenallergies).Personsufferingfrominfluenzaorseasonalallergymayalsoexhibittempraturewhichcan bedetectedbythermo-scanners,hencethepersonwillbecome suspected.Therefore,anaccurateandrapiddiagnostickitormeter fordetectionofSARS-CoV-2insuspectedpatientsisrequired,as thePCRbasedtestingisexpensiveandtimeconsuming.Different teamsofChinesedoctorsshouldimmediatelysenttoEurpean andothercountries,especiallyspainandItalytocontroltheover spreadofCOVID-19,becauseChinesedoctorshaveefficientlycontrolledtheoutbreakinchinaandlimitedthemortalityratetoless than3%only.ThetherapeuticstrategiesusedbyChinese,should alsobefollowedbyothercountries.
Acknowledgments
TheauthorsacknowledgethePostdoctoralgrantfromTheSecond AffiliatedHospitalofZhengzhouUniversity(forS.K).
DeclarationofCompetingInterest
Theauthorsofthismanuscriptdeclarenoconflictofinterest.
References
[1] ZhongN,ZhengB,LiY,PoonL,XieZ,ChanK,etal.Epidemiologyandcauseof severeacuterespiratorysyndrome(SARS)inGuangdong,People’sRepublicof China,inFebruary,2003.TheLancet2003;362(9393):1353–8
[2] WangN,ShiX,JiangL,ZhangS,WangD,TongP,etal.StructureofMERS-CoV spikereceptor-bindingdomaincomplexedwithhumanreceptorDPP4.CellRes 2013;23(8):986
[3] CuiJ,LiF,ShiZ-L.Originandevolutionofpathogeniccoronaviruses.NatRev Microbiol2019;17(3):181–92
[4] LaiC-C,ShihT-P,KoW-C,TangH-J,HsuehP-R.Severeacuterespiratory syndromecoronavirus2(SARS-CoV-2)andcoronavirusdisease-2019(COVID19):theepidemicandthechallenges.IntJAntimicrobAgents2020;105924
[5]OrganizationWH.Laboratorytestingforcoronavirusdisease2019(COVID-19) insuspectedhumancases:interimguidance,2March2020.WorldHealth Organization,2020.
[6] PeirisJ,GuanY,YuenK.Severeacuterespiratorysyndrome.NatMed2004;10 (12):S88–97
[7] PyrcK,BerkhoutB,VanDerHoekL.Identificationofnewhuman coronaviruses.ExpertReviewofAnti-infectiveTherapy2007;5(2):245–53
[8] RahmanA,SarkarA.Riskfactorsforfatalmiddleeastrespiratorysyndrome coronavirusinfectionsinSaudiArabia:analysisoftheWHOLineList,2013–2018.AmJPublicHealth2019;109(9):1288–93
[9] MemishZA,ZumlaAI,Al-HakeemRF,Al-RabeeahAA,StephensGM.Family clusterofMiddleEastrespiratorysyndromecoronavirusinfections.NEnglJ Med2013;368(26):2487–94
[10] WangC,HorbyPW,HaydenFG,GaoGF.Anovelcoronavirusoutbreakofglobal healthconcern.TheLancet2020
[11] PhanLT,NguyenTV,LuongQC,NguyenTV,NguyenHT,LeHQ,etal. Importationandhuman-to-humantransmissionofanovelcoronavirusin Vietnam.NEnglJMed2020
[12]RiouJ,AlthausCL.Patternofearlyhuman-to-humantransmissionofWuhan 2019novelcoronavirus(2019-nCoV),December2019toJanuary2020. Eurosurveillance.2020;25(4).
[13]ParryJ.Chinacoronavirus:casessurgeasofficialadmitshumantohuman transmission.BritishMedicalJournalPublishingGroup;2020.
[14] LiQ,GuanX,WuP,WangX,ZhouL,TongY,etal.Earlytransmissiondynamics inWuhan,China,ofnovelcoronavirus–infectedpneumonia.NEnglJMed 2020
[15] KanB,WangM,JingH,XuH,JiangX,YanM,etal.Molecularevolutionanalysis andgeographicinvestigationofsevereacuterespiratorysyndrome coronavirus-likevirusinpalmcivetsatananimalmarketandonfarms.J Virol2005;79(18):11892–900.
[16] ZhengBJ,GuanY,WongKH,ZhouJ,WongKL,YoungBWY,etal.SARS-related viruspredatingSARSoutbreak,HongKong.EmergInfectDis2004;10(2):176
[17] ShiZ,HuZ.AreviewofstudiesonanimalreservoirsoftheSARScoronavirus. VirusRes2008;133(1):74–87
[18] PadenC,YusofM,AlHammadiZ,QueenK,TaoY,EltahirY,etal.Zoonotic originandtransmissionofMiddleEastrespiratorysyndromecoronavirusin theUAE.ZoonosesPublicHealth2018;65(3):322–33
[19] AnnanA,BaldwinHJ,CormanVM,KloseSM,OwusuM,NkrumahEE,etal. Humanbetacoronavirus2cEMC/2012–relatedvirusesinbats,Ghanaand Europe.EmergInfectDis2013;19(3):456
[20] HuynhJ,LiS,YountB,SmithA,SturgesL,OlsenJC,etal.Evidencesupportinga zoonoticoriginofhumancoronavirusstrainNL63.JVirol2012;86 (23):12816–25
[21] LauSK,LiKS,TsangAK,LamCS,AhmedS,ChenH,etal.Genetic characterizationofBetacoronaviruslineageCvirusesinbatsrevealsmarked sequencedivergenceinthespikeproteinofpipistrellusbatcoronavirusHKU5 inJapanesepipistrelle:implicationsfortheoriginofthenovelMiddleEast respiratorysyndromecoronavirus.JVirol2013;87(15):8638–50
[22] LuR,ZhaoX,LiJ,NiuP,YangB,WuH,etal.Genomiccharacterisationand epidemiologyof2019novelcoronavirus:implicationsforvirusoriginsand receptorbinding.TheLancet2020
[23] ChanJF-W,YuanS,KokK-H,ToKK-W,ChuH,YangJ,etal.Afamilialclusterof pneumoniaassociatedwiththe2019novelcoronavirusindicatingperson-topersontransmission:astudyofafamilycluster.Lancet2020.
[24] ChanJF-W,KokK-H,ZhuZ,ChuH,ToKK-W,YuanS,etal.Genomic characterizationofthe2019novelhuman-pathogeniccoronavirusisolated fromapatientwithatypicalpneumoniaaftervisiting.Wuhan.Emerging Microbes&Infections2020;9(1):221–36
[25] vanBoheemenS,deGraafM,LauberC,BestebroerTM,RajVS,ZakiAM,etal. Genomiccharacterizationofanewlydiscoveredcoronavirusassociatedwith acuterespiratorydistresssyndromeinhumans.MBio2012;3(6): e00473–e512
[26] RajVS,MouH,SmitsSL,DekkersDH,MüllerMA,DijkmanR,etal.Dipeptidyl peptidase4isafunctionalreceptorfortheemerginghumancoronavirus-EMC. Nature2013;495(7440):251–4
[27] PerlmanS,NetlandJ.Coronavirusespost-SARS:updateonreplicationand pathogenesis.NatRevMicrobiol2009;7(6):439–50
[28] GlowackaI,BertramS,MüllerMA,AllenP,SoilleuxE,PfefferleS,etal.Evidence thatTMPRSS2activatesthesevereacuterespiratorysyndromecoronavirus spikeproteinformembranefusionandreducesviralcontrolbythehumoral immuneresponse.JVirol2011;85(9):4122–34
[29] BertramS,GlowackaI,MüllerMA,LavenderH,GnirssK,NehlmeierI,etal. Cleavageandactivationofthesevereacuterespiratorysyndromecoronavirus spikeproteinbyhumanairwaytrypsin-likeprotease.JVirol2011;85 (24):13363–72
[30] WuF,ZhaoS,YuB,ChenY-M,WangW,SongZ-G,etal.Anewcoronavirus associatedwithhumanrespiratorydiseaseinChina.Nature2020;1–5
[31] ZhouP,YangX,WangX,HuB,ZhangL,ZhangW,etal.Apneumoniaoutbreak associatedwithanewcoronavirusofprobablebatorigin.Nature[Internet]. 2020;Feb:3
[32] XuX,ChenP,WangJ,FengJ,ZhouH,LiX,etal.Evolutionofthenovel coronavirusfromtheongoingWuhanoutbreakandmodelingofitsspike proteinforriskofhumantransmission.ScienceChinaLifeSciences2020;63 (3):457–60
[33] WanY,ShangJ,GrahamR,BaricRS,LiF.Receptorrecognitionbynovel coronavirusfromWuhan:ananalysisbasedondecade-longstructuralstudies ofSARS.JVirol2020
[34] WuA,PengY,HuangB,DingX,WangX,NiuP,etal.Genomecompositionand divergenceofthenovelcoronavirus(2019-nCoV)originatinginChina.Cell HostMicrobe2020
[35] LuR,ZhaoX,LiJ,NiuP,YangB,WuH,etal.Genomiccharacterisationand epidemiologyof2019novelcoronavirus:implicationsforvirusoriginsand receptorbinding.Lancet2020;395(10224):565–74
[36] ChenY,LiuQ,GuoD.Emergingcoronaviruses:genomestructure,replication, andpathogenesis.JMedVirol2020
[37]HuiDS,IAzharE,MadaniTA,NtoumiF,KockR,DarO,etal.Thecontinuing 2019-nCoVepidemicthreatofnovelcoronavirusestoglobalhealth—Thelatest 2019novelcoronavirusoutbreakinWuhan,China.InternationalJournalof InfectiousDiseases.2020;91:264–6.
[38]LiB,SiH-R,ZhuY,YangX-L,AndersonDE,ShiZ-L,etal.DiscoveryofBat CoronavirusesthroughSurveillanceandProbeCapture-BasedNextGenerationSequencing.mSphere.2020;5(1).
[39] GralinskiLE,MenacheryVD.Returnofthecoronavirus:2019-nCoV.Viruses 2020;12(2):135
[40] XuX,ChenP,WangJ,FengJ,ZhouH,LiX,etal.Evolutionofthenovel coronavirusfromtheongoingWuhanoutbreakandmodelingofitsspike proteinforriskofhumantransmission.SciChinaLifeSci2020;1–4
[41] CormanVM,ItheteNL,RichardsLR,SchoemanMC,PreiserW,DrostenC,etal. RootingthephylogenetictreeofmiddleEastrespiratorysyndrome
coronavirusbycharacterizationofaconspecificvirusfromanAfricanbat.J Virol2014;88(19):11297–303
[42]ZhouP,YangX-L,WangX-G,HuB,ZhangL,ZhangW,etal.Discoveryofa novelcoronavirusassociatedwiththerecentpneumoniaoutbreakinhumans anditspotentialbatorigin.bioRxiv;2020.
[43] GretebeckLM,SubbaraoK.AnimalmodelsforSARSandMERScoronaviruses. CurOpinVirol2015;13:123–9.
[44] CockrellAS,PeckKM,YountBL,AgnihothramSS,ScobeyT,CurnesNR,etal. Mousedipeptidylpeptidase4isnotafunctionalreceptorforMiddleEast respiratorysyndromecoronavirusinfection.JVirol2014;88(9):5195–9
[45] NagataN,IwataN,HasegawaH,FukushiS,YokoyamaM,HarashimaA,etal. Participationofbothhostandvirusfactorsininductionofsevereacute respiratorysyndrome(SARS)inF344ratsinfectedwithSARScoronavirus.J Virol2007;81(4):1848–57
[46] SheahanTP,SimsAC,LeistSR,SchäferA,WonJ,BrownAJ,etal.Comparative therapeuticefficacyofremdesivirandcombinationlopinavir,ritonavir,and interferonbetaagainstMERS-CoV.NatCommun2020;11(1):1–14
[47] WangZ,ChenX,LuY,ChenF,ZhangW.Clinicalcharacteristicsandtherapeutic procedureforfourcaseswith2019novelcoronaviruspneumoniareceiving combinedChineseandWesternmedicinetreatment.BioScienceTrends2020
[48] RichardsonP,GriffinI,TuckerC,SmithD,OechsleO,PhelanA,etal.Baricitinib aspotentialtreatmentfor2019-nCoVacuterespiratorydisease.TheLancet 2020
[49] NgCS,KasumbaDM,FujitaT,LuoH.Spatio-temporalcharacterizationofthe antiviralactivityoftheXRN1-DCP1/2aggregationagainstcytoplasmicRNA virusestopreventcelldeath.CellDeathDiffer2020:1–20
[50] HolshueML,DeBoltC,LindquistS,LofyKH,WiesmanJ,BruceH,etal.Firstcase of2019novelcoronavirusintheUnitedStates.NEnglJMed2020
[51] WangBX,FishEN.editors.Globalvirusoutbreaks:Interferonsas1st responders.Seminarsinimmunology.Elsevier;2019
[52] WangM,CaoR,ZhangL,YangX,LiuJ,XuM,etal.Remdesivirandchloroquine effectivelyinhibittherecentlyemergednovelcoronavirus(2019-nCoV) invitro.CellRes2020;1–3
[53] AgostiniML,AndresEL,SimsAC,GrahamRL,SheahanTP,LuX,etal. Coronavirussusceptibilitytotheantiviralremdesivir(GS-5734)ismediated bytheviralpolymeraseandtheproofreadingexoribonuclease.MBio2018;9 (2):e00221–e318
[54] DerebailVK,FalkRJ.ANCA-associatedvasculitis—refiningtherapywithplasma exchangeandglucocorticoids.MassMedicalSoc2020
[55]TianX,LiC,HuangA,XiaS,LuS,ShiZ,etal.Potentbindingof2019novel coronavirusspikeproteinbyaSARScoronavirus-specifichumanmonoclonal antibody.bioRxiv;2020.
[56] BishtH,RobertsA,VogelL,SubbaraoK,MossB.Neutralizingantibodyand protectiveimmunitytoSARScoronavirusinfectionofmiceinducedbya solublerecombinantpolypeptidecontaininganN-terminalsegmentofthe spikeglycoprotein.Virology2005;334(2):160–5
[57] KamYW,KienF,RobertsA,CheungYC,LamirandeEW,VogelL,etal. AntibodiesagainsttrimericSglycoproteinprotecthamstersagainstSARS-CoV challengedespitetheircapacitytomediateFccRII-dependententryintoBcells invitro.Vaccine2007;25(4):729–40
[58] YangZ-y,KongW-p,HuangY,RobertsA,MurphyBR,SubbaraoK,etal.ADNA vaccineinducesSARScoronavirusneutralizationandprotectiveimmunityin mice.Nature2004;428(6982):561–4
[59] StadlerK,RobertsA,BeckerS,VogelL,EickmannM,KolesnikovaL,etal.SARS vaccineprotectiveinmice.EmergInfectDis2005;11(8):1312.
[60] KapadiaSU,RoseJK,LamirandeE,VogelL,SubbaraoK,RobertsA.Long-term protectionfromSARScoronavirusinfectionconferredbyasingle immunizationwithanattenuatedVSV-basedvaccine.Virology2005;340 (2):174–82
[61] BishtH,RobertsA,VogelL,BukreyevA,CollinsPL,MurphyBR,etal.Severe acuterespiratorysyndromecoronavirusspikeproteinexpressedby attenuatedvacciniavirusprotectivelyimmunizesmice.ProcNatlAcadSci 2004;101(17):6641–6
[62] BuchholzUJ,BukreyevA,YangL,LamirandeEW,MurphyBR,SubbaraoK,etal. Contributionsofthestructuralproteinsofsevereacuterespiratory syndromecoronavirustoprotectiveimmunity.ProcNatlAcadSci2004;101 (26):9804–9
[63] BukreyevA,LamirandeEW,BuchholzUJ,VogelLN,ElkinsWR,StClaireM, etal.MucosalimmunisationofAfricangreenmonkeys(Cercopithecus aethiops)withanattenuatedparainfluenzavirusexpressingtheSARS coronavirusspikeproteinforthepreventionofSARS.Lancet2004;363 (9427):2122–7
[64] SeeRH,ZakhartchoukAN,PetricM,LawrenceDJ,MokCP,HoganRJ,etal. Comparativeevaluationoftwosevereacuterespiratorysyndrome(SARS) vaccinecandidatesinmicechallengedwithSARScoronavirus.JGenVirol 2006;87(3):641–50
[65] WeingartlH,CzubM,CzubS,NeufeldJ,MarszalP,GrenJ,etal.Immunization withmodifiedvacciniavirusAnkara-basedrecombinantvaccineagainst severeacuterespiratorysyndromeisassociatedwithenhancedhepatitisin ferrets.JVirol2004;78(22):12672–6.
[66] SpruthM,KistnerO,Savidis-DachoH,HitterE,CroweB,GerencerM,etal.A double-inactivatedwholeviruscandidateSARScoronavirusvaccinestimulates neutralisingandprotectiveantibodyresponses.Vaccine.2006;24(5):652–61
[67] TakasukaN,FujiiH,TakahashiY,KasaiM,MorikawaS,ItamuraS,etal.A subcutaneouslyinjectedUV-inactivatedSARScoronavirusvaccineelicits systemichumoralimmunityinmice.IntImmunol2004;16(10):1423–30
[68] TangL,ZhuQ,QinE,YuM,DingZ,ShiH,etal.InactivatedSARS-CoVvaccine preparedfromwholevirusinducesahighlevelofneutralizingantibodiesin BALB/cmice.DNACellBiol2004;23(6):391–4
[69] QinE,ShiH,TangL,WangC,ChangG,DingZ,etal.Immunogenicityand protectiveefficacyinmonkeysofpurifiedinactivatedVero-cellSARSvaccine. Vaccine2006;24(7):1028–34
[70] ZhouJ,WangW,ZhongQ,HouW,YangZ,XiaoS-Y,etal.Immunogenicity, safety,andprotectiveefficacyofaninactivatedSARS-associatedcoronavirus vaccineinrhesusmonkeys.Vaccine2005;23(24):3202–9
[71] QuD,ZhengB,YaoX,GuanY,YuanZ-H,ZhongN-S,etal.Intranasal immunizationwithinactivatedSARS-CoV(SARS-associatedcoronavirus) inducedlocalandserumantibodiesinmice.Vaccine2005;23(7):924–31
[72]McKayBLP.Drugmakersrushtodevelopvaccinesagainstchinavirusthewall streetjournal.[cited202028January];Availablefrom:<https://www. wsj.com/articles/drugmakers-rush-to-develop-vaccines-against-china-virus11579813026>
[73]InovioIP.Inovioselectedbycepitodevelopvaccineagainstnewcoronavirus inovio.[cited202029January];Availablefrom:<http://ir.inovio.com/newsand-media/news/press-release-details/2020/Inovio-Selectedby-CEPI-toDevelop-Vaccine-Against-NewCoronavirus/default.aspx>.
[74]J.-H.Z.W.LeeLZ.Chinesescientistsracetodevelopvaccineascoronavirus deathtolljumps:Southchinamorningpost.[cited202029January];Available from:<https://www.scmp.com/news/china/society/article/3047676/ numbercoronavirus-cases-china-doubles-spread-rate-accelerates>.
[75]CheungE.Chinacoronavirus:Hongkongresearchershavealreadydeveloped vaccinebutneedtimetotestit,expertreveals:Southchinamorningpost. [cited202029January];Availablefrom: https://www.scmp.com/news/ hongkong/health-environment/article/3047956/china-coronavirus-hongkong-researchers-have
[76]Xinhua.Chinafast-tracksnovelcoronavirusvaccinedevelopmentxinhua. [cited20229January];Availablefrom: http://www.xinhuanet.com/english/ 2020-01/28/c_138739378.htm.
[77]Geo-Vax.Geovaxandbravovax(wuhan,china)tocollaborateondevelopment ofcoronavirusvaccine.[cited20203March];Availablefrom: https:// www.geovax.com/news/geovax-and-bravovax-wuhan-china-to-collaborateondevelopment-of-coronavirus-vaccine
[78]CloverB.Cloverinitiatesdevelopmentofrecombinantsubunit-trimervaccine forwuhancoronavirus(2019-ncov).[cited20206March];Availablefrom: http://www.cloverbiopharma.com/index.php?m=content&c=index&a=show& catid=11&id=40
[79]HuangY.TheSARSepidemicanditsaftermathinChina:apoliticalperspective. LearningfromSARS:Preparingforthenextdiseaseoutbreak;2004.p.116–36.
[80] HolmesKV.SARScoronavirus:anewchallengeforpreventionandtherapy.J ClinInvestig2003;111(11):1605–9
[81] HuangC,WangY,LiX,RenL,ZhaoJ,HuY,etal.Clinicalfeaturesofpatients infectedwith2019novelcoronavirusinWuhanChina.TheLancet2020 [82] PerlmanS.Anotherdecade,anothercoronavirus.MassMedicalSoc2020 [83] BollesM,DonaldsonE,BaricR.SARS-CoVandemergentcoronaviruses:viral determinantsofinterspeciestransmission.CurrentOpinVirol2011;1 (6):624–34.
[84]VaraV.Coronavirusoutbreak:Thecountriesaffected.11MARCH2020; Availablefrom: https://www.pharmaceutical-technology.com/ features/coronavirus-outbreak-the-countries-affected/
[85] ShiY,YiY,LiP,KuangT,LiL,DongM,etal.Diagnosisofsevereacute respiratorysyndrome(SARS)bydetectionofSARScoronavirusnucleocapsid antibodiesinanantigen-capturingenzyme-linkedimmunosorbentassay.J ClinMicrobiol2003;41(12):5781–2
[86]DongN,YangX,YeL,ChenK,ChanEW-C,YangM,etal.Genomicandprotein structuremodellinganalysisdepictstheoriginandinfectivityof2019-nCoV,a newcoronaviruswhichcausedapneumoniaoutbreakinWuhan,China. bioRxiv.2020.
MuhammadAdnanShereen isaPhDresearcheratWuhanUniversity,workingon Zikavirusandcoronavirusintheaspectsofpathogenesis,drugscreeningand molecularmechanisms.Heisanauthorin8articlespublishedinjournalswith impactfactormorethan5includingtherecentlyacceptedpaperinNaturemicrobiology.
SulimanKhan hascompletedhisPhDdegreefromChineseAcademyofSciences andcurrentlyworkingatsecondaffiliatedhospitalofZhengzhouuniversityas postdoctoralscientist.Hehaspublishedmorethan25articlesand5onSARS-CoV-2 inwellreputedjournalsincludingClinicalmicrobiologyandinfection(CMI)and Journalofclinicalmicrobiology(ASM-JCM)asfirstandcorrespondingauthor.
AbeerKazmi isaPhDstudentatWuhanUniversity.
NadiaBashir isaPhDstudentatWuhanUniversityworkingoncoronaviruses.She isanauthorinmorethan5paperspublishedoracceptedinrenownedjournals.
RabeeaSiddique isaPhDstudentatZhengzhouuniversity.Shehaspublishedmore than10papersinwellreputedjournalsasfirstorcoauthor.
Another random document with no related content on Scribd:


