SawfliesoutofGondwana:phylogeneticsand biogeographyofArgidae(Hymenoptera)
LEONARDOA.MALAGÓN-ALDANA
1,2 ,ARNR.JENSEN 2,3 , DAVIDR.SMITH 4 ,AKIHIKOSHINOHARA 5 and LARSVILHELMSEN 2
1 MuseoentomológicoUNAB,GrupoSistemáticadeInsectosAgronomíaSIA,FacultaddeCienciasAgrarias,UniversidadNacional deColombia,Bogotá,Colombia, 2 NaturalHistoryMuseumofDenmark,Science,UniversityofCopenhagen,Copenhagen,Denmark, 3 DepartmentofBiologyandBiotechnologies‘CharlesDarwin’,SapienzaUniversityofRome,Rome,Italy, 4 SystematicEntomology Laboratory,AgriculturalResearchService,U.S.DepartmentofAgriculture,c/oNationalMuseumofNaturalHistory,Smithsonian Institution,Washington,DistrictofColumbia,U.S.A.and 5 DepartmentofZoology,NationalMuseumofNatureandScience, Tsukuba,Japan
Abstract. Wepresentthefirstphylogenetichypothesisforthecosmopolitanfamily Argidaebasedonbothmolecularandmorphologicaldata.Furthermore,wepresent abiogeographicscenariobasedonadatedphylogenyandinterprettheevolutionary historyofthefamily.InformationfromsequencesofeightgenesisanalysedtoreconstructthephylogenyoftheArgidaebasedonmaximumlikelihoodandBayesianinference.TotalevidenceBayesiananalysesarealsoconducted,combiningthemolecular datasetwithdatafromadultandlarvalmorphology.Forthehistoricalbiogeographic reconstruction,divergencetimesareestimatedusingnodedatingwithuncorrelated relaxed-clockanalysis,andancestralbiogeographicaldistributionsareestimatedapplyingaDispersalExtinctionCladogenesismodelandaBayesianbinarymodel.Argidae s.s.isretrievedasmonophyleticinallanalysesandthecladeZenargidae + (Argidae + Pergidae)isbettersupportedwhencombiningmolecularandmorphologicalcharacters,andwhenexcludingthesaturatedthirdcodonpositioninthemolecularanalysis. WithinArgidae,twolargecladescorrespondingtothesubfamiliesArginaeandSterictiphorinaesensuBensonareretrievedasmonophyletic.Theancestraldistributionof ArginaeandSterictiphorinaeisestimatedtobetheAustralianandNeotropicalregions. DivergenceofArgidae-PergidaeandArginae-Sterictiphorinaeisestimatedtooccurin themiddle-upperJurassicbeforeorduringtheeast-westGondwanabreakup.DiversificationofArgidaemaybeassociatedwiththeradiationofangiospermsintheEarly Cretaceous,especiallyfortheNeotropicalSterictiphorinaeaftertheseparationofSouth AmericaandAfrica.Arginaewereprobablyintroducedtothenorthernhemisphere bydispersaltoAfricaand/orIndiaandsubsequentcontinentalcollisionwithEurasiaintheCenozoic.TheoccurrenceofSterictiphorinaeinthenorthernhemisphereis moredifficulttoexplain.MaternalcareandbroodguardingbehaviourevolvedindependentlyinArgidaeandPergidae,withasingleorigininasubclade(Dielocerini)of Sterictiphorinae.
Correspondence:LeonardoA.Malagon-Aldana,MuseoentomológicoUNAB,GrupoSistemáticadeInsectosAgronomíaSIA,Facultadde CienciasAgrarias,UniversidadNacionaldeColombia,Cra.30#45-03,Bogotá,D.C.,Colombia.E-mail:landresmalagon@gmail.com
Introduction
ArgidaeisthesecondlargestfamilyintheTenthredinoideaor truesawflies.TheTenthredinoideainturnisthelargestclade amongtheprimarilyherbivorous‘Symphyta’,whichareparaphyleticinrelationtothepredominantlycarnivorousApocrita (Vilhelmsen&Turrisi,2011).Argidaearedistributedworldwide,beingabsentonlyfromAntarctica,Madagascar,GreenlandandNewZealand.Unlikemostotherfamiliesoftrue sawflies,theyhaveastrongrepresentationinthesouthernhemisphere,especiallyintheNeotropics.ThecloselyrelatedPergidaeisalmostexclusivelyGondwananindistribution,occurring mostlyinAustraliaandSouthAmerica,withonlythegenus Acordulecera extendingintoNorthAmerica.Argidae + Pergidaehavebeenplacedassistergroupsinvirtuallyallcomprehensivephylogenetictreatmentsof‘Symphyta’(Vilhelmsen,1997, 2001;Schulmeister etal.,2002;Schulmeister,2003a;Schulmeister,2003b;Ronquist etal.,2012a;Malm&Nyman,2015). ThesplitbetweenPergidaeandArgidaehasbeenestimatedto haveoccurredatapprox.133Ma(116–158Ma),aroundthetime oftheGondwanabreakup(Schmidt&Walter,2014;Nyman etal.,2019).Untilrecently,nocomprehensivephylogenetic hypothesiswasavailableforArgidaeandnobiogeographical scenarioinacladisticframeworkhasbeenproposed.
Malagón-Aldana etal. (2021a,2021b)analysedcomprehensivedatasetsofArgidaebasedonadult(all56extantgenera represented)andlarvalmorphology(21generaincluded).Their mainfindingwasthatthemonotypicAustraliangenus Zenarge (Fig.1K)thatwaspreviouslyplacedinArgidaedidnotgroup withtheremainderofthefamily(Argidaes.s.),butinsteadwas placedaseithersistertoPergidae,ormostfrequentlysisterto (Argidaes.s. + Pergidae);regardlessofitsposition, Zenarge wasalwaysinamonophylumwithArgidaes.s.andPergidae (theZAPclade).Consequently,Malagón-Aldana etal. (2021a) placed Zenarge inaseparatefamily,theZenargidae.Thiscontrastswiththeresultsofthefewpreviousmolecularanalysesof‘Symphyta’thatincluded Zenarge intheirtaxonsample(Malm&Nyman,2015),wherethegenuswasretrievedas sistertoArgidaes.s.;comparativelyfewexemplarsofArgidae wereincludedintheseanalyses.Malagón-Aldana etal. (2021a, 2021b)retrievedtwomajorcladeswithinArgidaes.s.,correspondingtothesubfamiliesArginae(Fig.1A–J)andSterictiphorinae(Fig.2)asdefinedbyBenson(1963)(Table1).Other classificationschemesforArgidaerecognizingmoresubfamiliesandtribeshavebeenproposed(e.g.,Smith,1992;Taeger etal.,2010).Malagón-Aldana etal. (2021a)wereunabletocorroboratethese,althoughsomepreviouslysuggestedtribes(e.g., Trichorhachini,Atomacerini,DieloceriniandSterictiphorini) wereretrievedasmonophyletic.RevisingthetribalclassificationofArgidaes.s.willrequireadensertaxonsamplingthan wasobtainedinMalagón-Aldana etal. (2021a).
ThefossilrecordofArgidaeincludesonlysevenrepresentatives(Nel,2004),allofthemrelativelyyoungandfrom thenorthernhemisphere:thecompressionfossil Sterictiphora konowi (Rohwer)fromtheFlorissantformationofColorado (37.2–33.9Ma,upperEocene–lowerOligocene);theDominicanamberfossils Didymiadominicana Smith, D.davisi Smith,
D.protera Smith, D.poinari Smithand D.ebena Smith (20.4–13.6Ma,lowerMiocene),andthecompressionfossil Mioargeazari Nel,fromvolcano-sedimentarymaarof Mont-CharayinFrance(7.2–5.3Ma,uppermostMiocene) (Rohwer etal.,1908;Smith&Poinar,1992;Nel,2004;Paleobiologydatabase,fossilworks.org).
SomeNeotropicalgeneraofArgidaes.s.(Themos, Dielocerus, Pachylota, Digelasinus and Sericoceros)presentacharacteristic subsocialbehaviour,wherethefemalesguardtheeggsatleast untiltheyhatch,defendingthemfrompredators.Thelarvaestay together,feedinggregariouslyandmakeamasscocoonwhen theypupate.However,thisgeneralbehaviourincludesvariation withinseveraltraitsamongthesegenera(defencemechanisms ofthefemale,hostplantspecies,egglocationanddistributionon leaves,totalperiodofguarding,masscocoonstructures,etc.) (Benson,1938;deSouzaDias,1975;deSouzaDias,1976; Smith,1992;Ciesla,2002;Smith&Janzen,2003;Boraschi etal.,2005).Asimilarmaternalguardingbehaviourisalso presentinsomePergidae(Perga, Pseudoperga, Philomastix, Cladomacra and Syzygonia)(Benson,1938;Shinohara,1986; Schiff,2004;Schmidt etal.,2006).LarvaeofArgidaes.s.Feed onadiversevarietyofangiosperms.Althoughthelarvaeof mostargidsremainunknown,theyalsopresentvariousdegrees ofgregariousness,chemicaldefence,feedingbehaviourand morphologicaladaptationstotheirhostplants(Smith,1992; Schmidt etal.,2000;Boevé etal.,2018).Contrarytothe knownPergidaeandArgidaes.s.,thelarvaeof Zenarge feed ongymnosperms,includingthenativeAustralian Callitris spp. (Cupressaceae)(Moore,1963a,1963b).
Hereweaimtoreconstructaphylogenetichypothesisforthe Argidae,basedonmolecularcharactersincludingmitochondrial,ribosomalandnucleargenefragments,forthehighest numberofargidterminalssequencedsofar.Wealsoanalyse thecombinedmorphological(adultsandlarvae)andmoleculardatasetsinatotalevidenceapproachtofurtherexplorethe phylogenyofArgidae,includingnodedatingcalibrationofthe phylogenyusingavailablefossilinformationforthefamilyand outgroup(otherTenthredinoideafamilies).Finally,weattempt toelucidatethebiogeographicalandecologicalpatterns(e.g., broodguardingbehaviour)forArgidae.
Methods
Specimensexamined
Adultandlarvalspecimensof45speciesand18different generaofArgidaeandfiveoutgrouptaxaofTenthredinoidea (Pergidae,Tenthredinidae)weresuccessfullyextractedfor DNA(TableS1).Themajorityofthespecimenswerepreservedin96%ethanol,whilesomeofthemweredry-pinned specimens.Exemplarswereobtainedfromthefollowingcollections:InstitutodeInvestigacionesBiológicasAlexander vonHumboldt,Colombia(IAvH);ColeccióndeZoología, InstitutodeCienciasNaturalesICN,UniversidadNacionalde Colombia;Bogotá,D.C.(ICN-MHN);MuseoJaverianode HistoriaNatural,Colombia(MPUJ);StatensNaturhistoriske
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
Fig.1. (A–C)Head,frontalviewofmales,subfamilyArginae.(A) Cibdelajanthina.(B) Pampsilotadahomeyanus.(C) Spinargefulvicornis (D–K)Lateralhabitusoffemales.(D–J)SubfamilyArginae.(D) Antargidiumdentivalve.(E) Scobinalepida.(F) Pampsilotaafer .(G) Triargenigra (H) Athermantusimperialis.(I) Tanyphatnideamicrocephala.(J) Spinargefulvicornis.(K)FamilyZenargidae, Zenargeturneri
Museum,UniversityofCopenhagen,Denmark(NHMD); NationalMuseumofNaturalHistory,SmithsonianInstitution, WashingtonD.C.,U.S.A.(NMNH);NationalMuseumof NatureandScience,Tsukuba,Japan(NMNS);Senckenberg DeutschesEntomologischesInstitut,Müncheberg,Germany (SDEI);MuseoentomológicoUNABFacultaddeCiencias AgrariasBogota,UniversidadNacionaldeColombia(UNAB).
DNAextraction,amplificationandsequencing
Onehindleg(orpieceoflarvaltissue)wasremovedfromeach specimen(TableS1),breakingitatthecoxaandtryingtoinclude partsofthebasalextrinsicmusclesoftheappendage.Thedissectionforcepswerecleanedwithbleach(10%)andethanol (70%)priortoworkingwithanewspecimen.Thesampleswere
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
Fig.2. Head,frontalviewofmales,subfamilySterictiphorinae.(A) Trichorhachusnitidus.(B) Aprocerosleucopoda.(C) Atomaceracoerulescens HabitusoffemalesofArgidae,subfamilySterictiphorinae.(D) Trichorhachusnitidus.(E) Atomaceracoerulescens.(F) Sterictiphorafurcata.(G) Ptenos leucopodus.(H) Duckeanaprodiga.(I) Pachylotaaudouinii.(J) Schizocerellapilicornis.(K) Didymiamartini
transferredtoseparatevialsandthenstoredin96%ethanol. DNAextractionandamplificationwerecarriedoutintheGeogeneticslaboftheGlobeInstitute,UniversityofCopenhagen, Denmark.TotalDNAwasextractedusingtheDNeasy® Blood andTissuekitsusingspincolumnsmanufacturedbyQiagen(QIAGENAarhus,Denmark)withaslightmodification
entailingsoakingthesamplesovernightforaround12hat56∘ C withgentleagitation.Toremovesurfacecontaminationfromold dry-pinnedspecimens,thelegswereremovedandinitiallyputin atissuelysisbuffer(ATLbuffer + proteinaseK)for1hat56∘ C withgentleagitation.Thelysisbufferwasthendiscarded,and freshlysisbufferwasadded.Thesampleswherethenincubated
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
Table1. ListoftaxaexaminedspecimensofArgidaeandZenargidaetaxa.
Examinedspecies(ormorphospecies)
FamilyGenusSpeciesMolecularMorphology
Argidae(Arginae) †Mioarge Nel,2005101
Antargidium Morice,1919632
Arge Schrank,1802395 a 2310
Asiarge Gussakovskij,1935301
Athermantus Kirby,18822 a 11
Cibdela Konow,18991411
Kokujewia Konow,1902311
Pseudarge Gussakovskij,1935601
Pseudosinarge Saini,Thind&Kaur,1998101
Scobina Lepeletier&Serville,182856 a 52
Sinarge Forsius,1934101
Sjoestedtia Konow,1907201
Spinarge Wei,199813 a 51
Styphelarge Benson,1938101
Tanyphatnidea Rohwer,1912201
Triarge Forsius,19319 a 12
Zhuhongfuna Wei&Nie,1999101
Argidae(Sterictiphorinae)
Acrogymnia Malaise,19411102
Acrogymnidia Malaise,1955401
Adurgoa Malaise,1937401
Aproceros Malaise,193110 a 11
Aprosthema Konow,189958 a 41
Atomacera Say,183635 a 32
Brachyphatnus Konow,1906301
Brevisceniana Wei,2005101
Dacrogymnia Smith&Malagón-Aldana,20205 a 11
Didymia Lepeletier&Serville,182826 a 13
Dielocerus Curtis,18446 a 11
Digelasinus Malaise,1937201
Duckeana Malaise,1941401
Durgoa Malaise,1937402
Eriglenum Konow,1900401
Mallerina Malaise,1941101
Manaos Rohwer,191212 a 11
Neoptilia Ashmead,18981001
Neurogymnia Malaise,1937702
Ortasiceros Wei,1997601
Pachylota Westwood,1841201
Pampsilota Konow,189912 a 14
Pseudaprosthema Gussakovskij,1935303
Ptenos Norton,187232 a 35
Ptilia Lepeletier,18237 a 22
Schizocerella Forsius,1927521
Sericoceros Konow,19052112
Sphacophilus Provancher,188852 a 13
Sterectiphora Billberg,182045 a 42
Subsymmia Malaise,1955201
Tanymeles Konow,190623 a 12
Themos Norton,18671312
Topotrita Kirby,1882301
Trailia Cameron,1878201
Trichorhachus Kirby,1882402
Triptenus Malaise,1937201
Trochophora Konow,1905211
Yasumatsua Togashi,1970301
Zynzus Smith,19921901
Sterictiphorinaemales,unknowngenus-10
Genussp.-01
Zenargidae Zenarge Rohwer,1918111 a Generawithatleastonemolecularsequenceproducedinthisstudy.
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
Table2. Primersandannealingtemperatureusedforgenefragmentamplification.
GenePrimerNamePrimersequence(5′ –3′ )ReferencesAnnealingT∘ C 16SAHym(F)TRACTGTRCAAAGGTAGCSchulmeister(2003b)47–50 BHym(R)TTAATTCAACATCGAGGTCSchulmeister(2003b)47–50 28SD2F(F)CGGGTTGCTTGAGAGTGCAGCSchmidt&Walter(2014)47 D2Ra(R)CTCCTTGGTCCGTGTTTCSchmidt&Walter(2014)47 CAD743nF-ino(F)GGIGTIACIACIGCITGYTTYGARCCJohanson&Malm(2010)55–60 CADhym2F(Fi)GGNAGYTCNATGAARAGYGTNGGMalm&Nyman(2015)55–60 1028R-ino(R)TTRTTIGGIARYTGICCICCCATJohanson&Malm(2010)55–60 COICOIfly-C1-J-1514(F)CAAATCATAAAGATATTGGFarrell(2001);Malm&Nyman(2015)56 sym-C1-J-1718(Fi)GGAGGATTTGGAAAYTGAYTAGTWCCNyman etal. (2006)52 A2590(R)GCTCCTATTGATARWACATARTGRAAATGNormark etal. (1999)52–56 GLNGLN1F(F)GTNTAYTTYTGGCARGGRegier etal. (2008)51 GLNhym2F(Fi)GCNGGVAAYATGGGHTGGYTNACRegier etal. (2008)56 GLN4R(R)CYCCANCCRTGRAARCAYTTMalm&Nyman(2015)51–56 PGDPGDhym3F(F)TGGTRCACAAYGGMATHGARTAYGGMalm&Nyman(2015)60 PGDhymintRb(R)ATRATRCANCCDCCYCKCCACATMalm&Nyman(2015)60 Forward(F = external,Fi = internal)andReverse(R)sequences.
overnightorover2daysforaround12–48hat56∘ Cwith gentleagitation.DNAextractswerestoredinafreezerat 20∘ C.DNAconcentrationofeachsamplewasmeasuredwith aQubit™4FluorometerandaQubitdsDNAHighSensitivity AssayKit.
Thefollowingsixgeneregionswereamplified:Mitochondrialribosomal16S,nuclearribosomal28S(Schulmeister,2003b;Schmidt&Walter,2014),mitochondrial cytochromeoxidaseI(COI)andthenuclearprotein-coding genescarbamoyl-phosphatesynthetase(CAD),gelsolin(GLN) andphosphogluconatedehydrogenase(PGD)(Malm& Nyman,2015).PCRwasperformedusing5× HOTFIREPol® BlendMasterMix(10mMMgCl2 )manufacturedbySolisBioDyne(Tartu,Estonia),withafinalvolumeof25 μL,composed of:17 μLoftheHOTFIREPol® BlendMasterMix,4 μLof molecularbiologygradeH2 O,1 μLofDimethylsulfoxide DMSO,0.5 μLofforwardandreverseprimereachand1 μLof thesampleDNAextract.PCRprofilesandprimersequencesfor COI,CAD,PGDandGLNfollowedMalm&Nyman(2015) exceptthatthenumberofcycleswasincreasedto50andthe annealingtemperatureforeachgenewasmodified(Table2). Inthecaseoftwoforwardprimers(externalandinternal)we usedprimarilytheexternal(Table2),whiletheinternalprimer wasusedonlywhentheexternaldidnotworkforthespecific sample.For16Sand28SweusedtheprimersfromSchmidt& Walter(2014)butmodifyingthePCRprofileasfollows:denaturationat94∘ Cfor1min,followedby40(16S)or30(28S)cycles ofdenaturationat94∘ Cfor40s,annealingfor40s,andextensionat72∘ Cfor1min;thelastcyclewasfollowedbyafinal extensionstepat72∘ Cfor1min.ThePCRproductswerevisualizedunderaUVchamberafterseparatingthefragmentswithgel electrophoresisusing2%agarosewithGelRed® (MerckMillipore,Darmstadt,Germany)anda100bpladder.PCRproducts thatshowedsharpvisualizedbandsweresentforpurification andSangersequencingatMacrogenEurope(Amsterdam, Netherlands).
Additionalsequences
GenBankdatabaseshttps://www.ncbi.nlm.nih.gov/genbank/ (Benson etal.,2017)andBarcodeofLifeDataSystem http://www.boldsystems.org/index.php/(Ratnasingham& Hebert,2007)wereemployedtoobtain216additional sequences,including83Argidae(32species,17genera) and133ofoutgrouptaxa(33species,23genera)(TableS1) forthesixdifferentmarkerswesequencedourselves,aswell assequencesofthegenesTPI(triose-phosphateisomerase)and IDH(isocitratedehydrogenase),whichweresuggestedtobe phylogeneticallyrelevantbyMalm&Nyman(2015)(TableS1). The83downloadedArgidaesequencesweredistributedacross genesasfollows:COI(29),GLN(8),TPI(8),CAD(9),PGD (9),IDH(6),16S(7)and28S(7).Thesesequenceswere producedbySchulmeister etal. (2002),Schulmeister(2003b), Linnen&Farrell(2008),Heraty etal. (2011),Leppänen etal. (2012),Boevé etal. (2013),Klopfstein etal. (2013),Isaka &Sato(2014),Liston etal. (2014),Schmidt&Walter(2014), Malm&Nyman(2015),Schmidt etal. (2017),Shinohara etal. (2016),Endara etal. (2018),Smith etal. (2019)and Prous etal. (2019).ThetotalnumberofArgidaespeciesor morphospecies(i.e.,producedinthisworkandobtainedfrom databases)includedinthemolecularanalyseswere70in25 genera,foratotalof72terminals(TableS1,Fig.3).For thoseoutgroupterminalswithseveralspeciesrepresentedin thesequencesample(chimeras),thecombinedterminaltaxon wasgivenasthegenusname,e.g., Athalia spp.*consistsof sequencesfrom Athaliacircularis (COI,GLN,IDH,andTPI) and A.scutellariae (28S,CADandPGD).
Alignmentandassemblingofsequences
ThesequencesproducedwerefirstsubmittedtotheBasic LocalAlignmentSearchTool(BLAST®)ontheGenBank
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
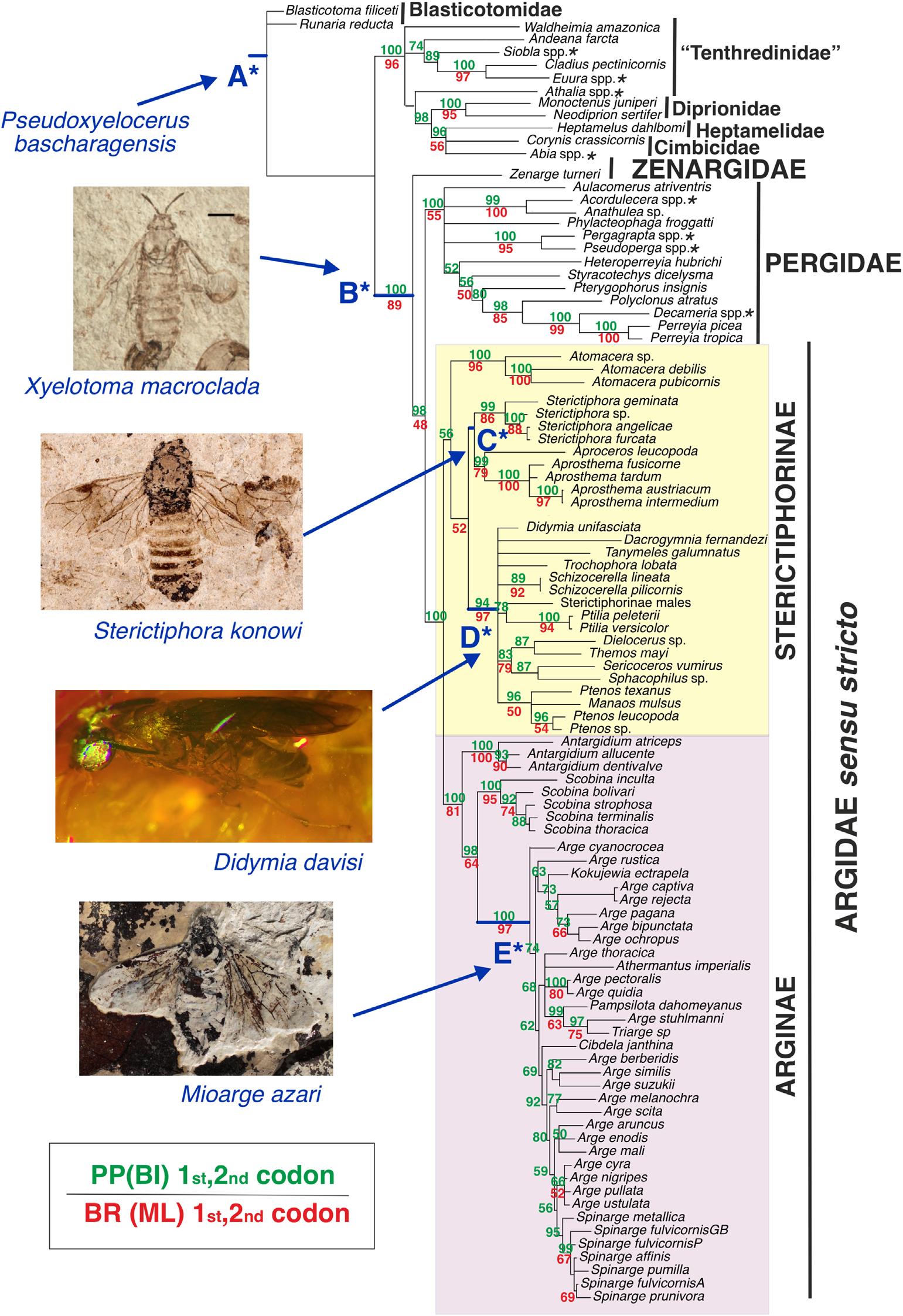
Fig.3. BayesiantreereconstructionofArgidaes.s.Basedonthecombinedmolecularinformationoffirstandsecondcodonpositionsofproteincodon genesusingMaximumlikelihood(ML)andBayesianinference(BI)analyses.Capitallettersfollowedbyanasteriskshowthefivecalibrationpoints usedinthecombinedmorphologicalandmoleculardatasetbasedonthefossilspeciestotheleft.Asteriskafterthenameofaterminal(e.g., Athalia spp.)indicatesthatthistaxonisachimera,i.e.,themolecularinformationfromvariousmarkerswasobtainedfromdifferentspeciesofthesamegenus. PP = posteriorprobability,BR = Bootstrapresamplingsupport.
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
websitetocross-checktheidentificationsanddetectpotentialcontaminations(familyorgenuslevel).Chromatograms ofsequenceswerevisualizedusingfinchtv1.5.0where low-qualityendsweretrimmed.Forwardandreversetrimmed sequencechromatogramsofthesamesamplewerealigned, editedandassembledusingcodoncodealignersoftware 9.0.1(CodonCodeCorporation).Low-qualitysequences(forward,reverse,orboth)wereexcludedandamplifiedagain. TheassembledsequencesofeachgeneperspeciesweresubmittedtoGenBank ® (TableS1).Sequencesfromthesamplesandtheadditionaldownloadedsequences(TableS1) wereputtogetherandfirstalignedforeachgeneinmega XformacOS(Stecher etal.,2020)usingtheintegrated multiplesequencealignmentalgorithmclustalw(Larkin etal.,2007).Fortheprotein-codinggenes(COI,PGD,GLN, CAD),theaminoacidalignmentswerealsocheckedforstop codons.Intronsweredetectedbyeyeasambiguouslyaligned regionswithseveralgapsorinconsistenttranslationsandtheir start-endwasassessedbyatypicalnucleotidepattern‘AG/GT (start)–AG/CAG(stop)’thatsignifiessplicingsites(Klopfstein&Ronquist,2013;Malm&Nyman,2015).Theidentified intronswereindividualizedandseparatedfromtheexons.Preliminaryalignmentsoftheprotein-codinggeneswererealigned usingmafftversion7onlineservice(Katoh etal.,2019)and aniterativerefinementG-INS-imethodwithglobalhomology.The16Sand28SrDNAsequenceswererealignedusing T-Coffee(Notredame etal.,2000),whichcombinesalocal andglobalpairwisealignment,improvingthefinalalignment forthosesequenceswithsecondarystructureinformation.All finalgene-alignmentswereconcatenatedusingsequencematrix1.8.0(Vaidya etal.,2011).
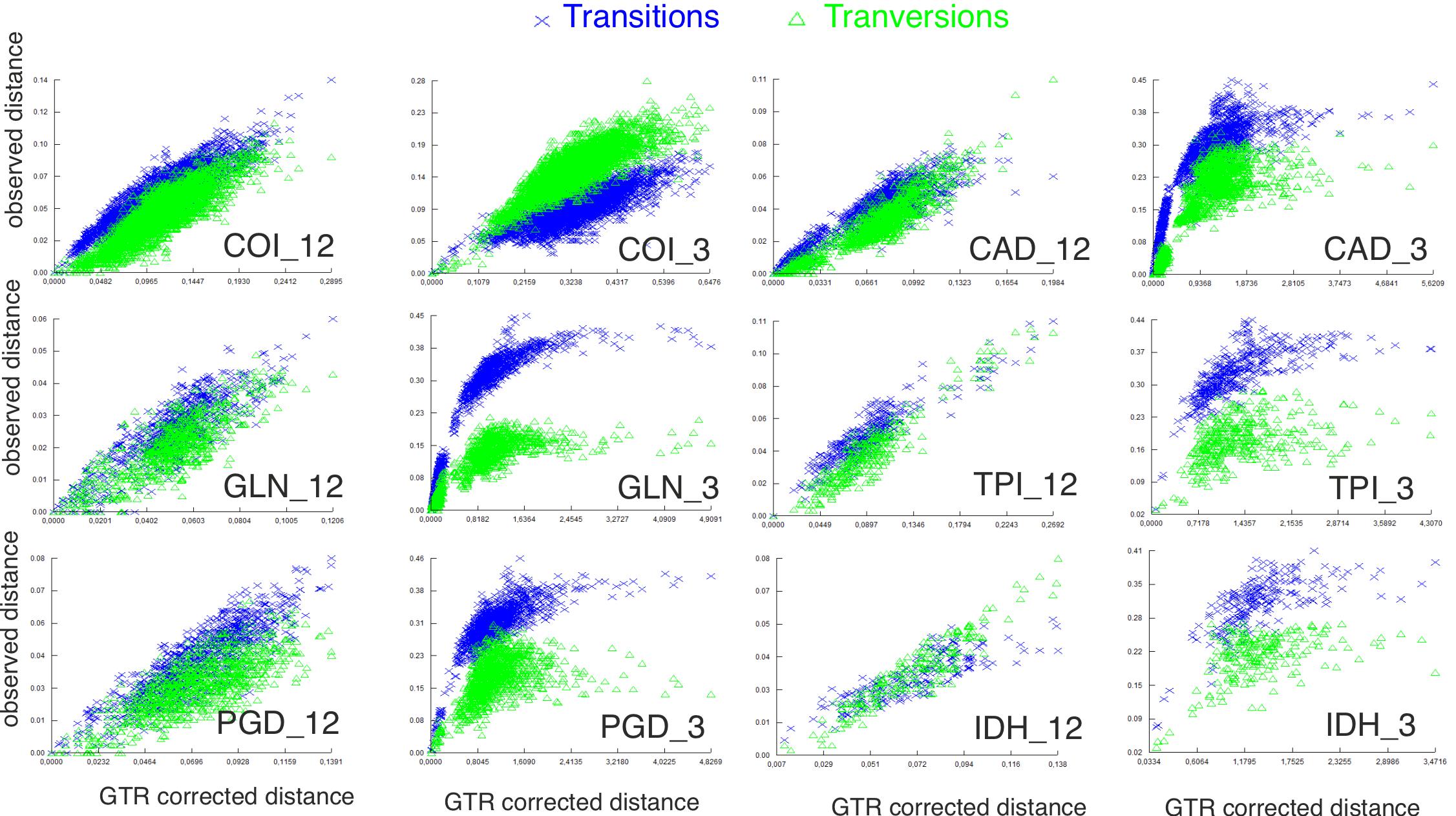
Saturationsubstitutionrate
Thelevelofsubstitutionsaturationfortheprotein-coding geneswasassessedforthefirstandsecondcodonposition together,andthethirdcodonpositionseparatelyusingdambe7 (Xia&Xie,2001).First,wevisuallyinspectedtheplotofthe p-distanceintermsoftransitionsandtransversions(observations/lengthofsequence)againstthecorrectedgeneticdistance pairwisecomparisonusingGTRsubstitutionmodel(=GTRdistance)(Salemi,2009).Inaddition,foreachgeneweestimated therandomsaturationsubstitutionindex(Iss)andthecorrected substitutionindex(Iss.c)(Xia etal.,2003).
Molecularphylogeneticinference
TheconcatenatedalignmentswereanalysedunderMaximum Likelihood(ML)andBayesianInference(BI)toreconstructthe phylogenetichypothesesandexaminetopologicalcongruence betweenthemethods.Gapsweretreatedasmissingdata.The treeswererootedon Blasticotomafiliceti (Blasticotomidae) inallanalyses,afamilythathasbeenrecoveredassister groupofallotherTenthredinoideainmostpreviousstudies (Vilhelmsen,2001;Schulmeister,2003a;Schulmeister,2003b; Malm&Nyman,2015).
Dataoftheconcatenatedalignmentswereprimarily partitionedpergene,i.e.,16S,28S,CAD,GLN,TPI,COI, PGDandIDH.Inaddition,theprotein-codinggenes(CAD, GLN,TPI,IDH,COIandPGD)werepartitionedbycodon position:Firstandsecondcodonpositiontogetherandthethird codonpositionseparately.Tocomparetheeffectofthethird codonpositioninthephylogeneticreconstructions,weranall analysesincludingandexcludingthiscodonpositionforthe protein-codinggenes.Similarly,wecomparedtheinfluenceof theadditionorexclusionoftheTPIandIDH(TableS1)inthe reconstructedphylogeny.
MLtreeswereinferredusingtheWebinterfaceof W-IQ-TREE(Nguyen etal.,2015;Trifinopoulos etal.,2016). Thebestsubstitutionandbestpartitionschememodelfor eachpartitionwasestimatedinthesameprogrammeusingthe W-IQ-TREEModelfinderalgorithm(Chernomor etal.,2016; Kalyaanamoorthy etal.,2017)andselectedaccordingtothe Bayesianinformationcriterionscores(Schwarz,1978),with an‘edgeunlinked’partitionmodelallowingadifferentset ofbranchlengthsforeachpartition.Bootstrapresampling support(BR)wascalculatedwith100standardnonparametric replicates.MLtreeswerevisualizedintheInteractivetreeof life(ITOL)v4portal(Letunic&Bork,2019)andeditedwith corelphoto-paintx7(CorelCorporation).
FortheBI,werantheanalysesintheCIPRESScience GatewayV.3.3(Miller etal.,2012)portal.Thebestsubstitutions modelsandbestpartitionschemeforBIwereestimatedin PartitionFinder2(Lanfear etal.,2017)usingthe‘greedy’ searchalgorithm,‘unlinkedbranchlengths’option(Lanfear etal.,2012)andwereselectedbasedonthecorrectedAikaike InformationCriterion.ABItreeforthecombinationofallgenes wasreconstructedusingMrBayes3.2.2(Ronquist etal.,2012b), whiletheparametersofsubstitutionmodelweresetaccording tothePartitionFinder2estimation.TworunswithfourMarkov chainsMonteCarlo(MCMC)chains(onecoldandthree heated)wererunfor20milliongenerations;diagnosticswere savedevery1000thgenerationand25%ofthesampleswere discarded(burn-infraction)priortothesummarystatistics.The summarizedparameterresultsofeachrunwerevisualizedand examinedintracer1.7.1plottingtheloglikelihoodvalues againstthegenerationtime.ConvergenceoftheMarkovchains MonteCarlochainswithinandamongrunswasconsidered sufficientbywhentherewasacorrectmixingbehaviourin tracerplots,thepotentialscalereductionfactorwascloseto1, theaveragestandarddeviationofsplitfrequencieswasbelow 0.01andtheEffectiveSampleSizesforallparameterswas higherthan200.Themajorityruleconsensustreewasvisualized usingfigtreev1.4.4(http://tree.bio.ed.ac.uk/software/figtree/) andeditedincorelphoto-paintx7(CorelCorporation).
Totalevidencephylogeneticanalysis:Molecules andmorphology
AnadditionaltotalevidenceapproachusingBayesiantree inferencewasperformedcombiningthedatamatrixof223adult anatomicalcharacters(117taxa)(Malagón-Aldana etal.,2021a) ©2021TheRoyalEntomologicalSociety,
and85larvalcharacters(48taxa)(Malagón-Aldana etal.,2021b)withthemoleculardataset.Fortheadultmorphologicalcharactermatrix,sevencharacters(41,45,87,89, 161,162and175)weretreatedasorderedinaccordancewith Malagón-Aldana etal. (2021a),andthreeadditionalcharacters (5,27,39)basedontheresultpatternsofthelarvalmorphology dataset(Malagón-Aldana etal.,2021b).Themolecularand morphologicaldatasetswereassembledusingmesquite3.6 (Maddison&Maddison,2019).Forsomegenerarepresentedby differentspeciesinthemolecularandmorphologicaldatasets wecomposedaterminaltaxon(taxondenotedasgenusname + spp.*)iftherewasevidenceformonophylyofthegenusin previousstudies(Malagón-Aldana etal.,2021a).Treeswere rootedon B.filiceti (Blasticotomidae).Theparametersettings forthemolecularpartitionwereidenticalwiththeabove;for themorphologicaldatasetastandarddiscretemodelwasused withcodingsetto‘variable’andratesto‘gamma’.TheBI analysisforthecombineddatasetwasrunusingmrbayes3.2.2 (Ronquist etal.,2012b)intheCIPRESScienceGatewayV. 3.3(Miller etal.,2012)portal.TworunswithfourMarkov chainsMonteCarlo(onecoldandthreeheated)wererunfor 20milliongenerations,andremainingparametersandtestof convergencewereidenticaltotheabove-describedmolecularBI analysis.Themajorityruleconsensustreewasvisualizedusing figtreev1.4.4andeditedincorelphoto-paintx7(Corel Corporation).
Molecularclockcalibrationanddivergencedating
TodatethedivergencetimeswithinArgidaeandPergidae, weusedanodedating(treecalibrationbyemployingpriorson nodeagesbasedontheoldestknownfossilsforcorrespondingclades)approach(Ronquist etal.,2012a;Zhang,2017). Fivecalibrationnodeswereassigned(Fig.3)basedonfossil informationonwellsupportednodes(PP = 90–100)andconstrainedasmonophyletic.WeusedanoffsetexponentialprobabilitydistributionassuggestedbyRonquist etal. (2012a), withameanageof235Ma(maximumoldestfossilageof Hymenoptera, Triassoxyela Rasnitsyn)andaminimumagefor eachselectednodeaccordingtothefossil(Fig.3):(A)Forthe root(=treeage) Pseudoxyelocerusbascharagensis Nel etal. (183Ma,oldestfossilofTenthredinoidea);(B)forthenode ZAP: Zenarge + (Argidae + Pergidae), Xyelotomamacroclada Gao etal. (164.7Ma,oldestfossilclosesttoArgidae + Pergidae)(Ronquist etal.,2012a);(C)forthenode Sterictiphora,the fossil S.konowi Rohwer(37.2Ma)(Zhelochovtzev&Rasnitsyn,1972);(D)forthenodeof‘NeotropicalSterictiphorinae’the fossil Didymiadavisi Smith&Poinar(20.4Ma)and(E)forthe Arginae(except Antargidium and Scobina)thefossil Mioarge azari Nel(7.2Ma)(Malagón-Aldana etal.,2021a). Forthenodedatingofthecombineddataset,weusedmrbayes 3.2.2(Ronquist etal.,2012b).Theanalyseswererunin CIPRESScienceGatewayV.3.3.,withanIGR(independent gammarateateachbranchwithmean1.0)model(Ronquist etal.,2012a).TheremainingparametersfollowedRonquist etal. (2012a)forHymenoptera:Lognormal( 7.08069,1)and
prsetigrvarpr = exp(37.12).Weusedtwodifferentpriorsto comparetheireffectontheageestimation,anuniformtree prior(clock:uniform)andabirth-deathprior(clock:birth-death). Forthelatter,thefollowingsettingswereused:Speciation prior = exponential(10),extinctionpriorbeta(1,1)and extantsampleproportionfixedto0.1(oursampleofArgidae taxawasabout10%ofalldescribedspecies).Substitution modelswereassignedtoeachgenepartitionaccordingto themodelsestimatedpreviouslywithPartitionFinder2.The analysishadtworuns,fourchainsfor20milliongenerations andamajorityruleconsensustreewasestimated,withthesame parametersandtestofconvergenceandeffectivesamplesize usedfortheabove-describedBayesianphylogeneticanalysis (seeSection2.6).
Biogeographicreconstruction
Thegeographicrecordsofgeneraandspecieswereobtained fromtheElectronicWorldCatalogueofSymphyta(Taeger etal.,2010,2018),labelsoftheexaminedspecimens,literature (Smith,1992;Saini,2009;Koch etal.,2015)andtheWaspWeb (vanNoort,2020).Worldbiogeographicregionswereassigned accordingtothezoogeographicrealmsofHolt etal. (2013) withsomemodificationsfromMorrone(2015)(i.e.,theMediterraneanandAfricanorthofSaharawereconsideredaspartsof thePalearcticregion;NewGuineaconsideredpartoftheAustralianregion).Theancestralbiogeographicalstateswerereconstructedusingrasp10.15(Yu etal.,2015)applyingS-BGB(StatisticalBioGeoBEARS)andBBM(BayesianBinaryMCMC) methods,estimatedovertheultrametriccalibratedconsensus treeofthenodedatinganalysistocomparedirectlywiththe divergencetimeestimations.RegardingS-BGB(Matzke,2013a, 2013b,2014),weusedtheDispersalExtinctionCladogenesis (DEC)approachmodelthatimplementsaLagrange(LikelihoodAnalysisofGeographicRangeEvolution)sourcecode (Ree&Smith,2008)andestimateslikelihoodsofancestral statesatcladogenesiseventsconstrainingdispersalmultipliers betweenregionsaccordingtocertaingeologicalevents(Brown &Lomolino,1998;SanMartin&Ronquist,2004;Hall&Sevastjanova,2012;Müller etal.,2019)from0to1inamatrix (FileS1).Weusedtwodispersalrates,0.1and1(e.g.,dispersalbetweenAfrotropicalandtheNeotropicalregionswas setas1beforetheirbreakup,around110Maand0.1after it).Anadditionalsampleof1000treesderivedfromthenode datingBayesiananalysiswasusedfortheS-BGBanalysis. TheBayesianBinaryAnalysis(BBM)(Ronquist&Huelsenbeck,2003;Yu etal.,2015)wasrunwith10chainsand500000 generations,statesamplingevery100generations;andapplyingaF81modelwithasiteratevariationofgamma,which isthemostcomplexmodelimplementedinrasp10.15.The maximumnumberofancestralstateswassettothreeinboth analyses.BothS-BGBandBBManalysesacceptedpolytomies intheinputconsensustree.ForthecompositeArgidaeterminal taxa = *genus + spp.(twospeciesinthesamegenusfromdifferentdatasets)thedistributionstatewasassignedasacombinationofboth,e.g., Aprosthematardum (Palearctic) + Aprosthema ©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
brunniventre (Nearctic) = Aprosthema spp.*(Palearcticand Nearctic).
Results
Sequenceassemblageandsaturationofsubstitutionrate
Thefinalmolecularassemblageofalignedsequencesincluded 99terminaltaxa(72sequencesand70speciesofArgidaeand 27sequencesofoutgrouptaxa)(TableS1,Fig.3)and5350 charactersortotalsites,including2296parsimonyinformative sites,490singletonsand2564constantsites.ThemostcompletelysampledgeneswereCOI(82%),28S(69%)and16S (61%),whileTPI(27%)andIDH(19%)weretheleastsampled(Table3),thelattertwoconsistingexclusivelyofsequences fromtheGenBankdatabase.Therewere46%missingdata. Sixtysequencescontainedmorethan50%gapswhenincludingIDHandTPI.Thesuccessfulamplificationofmitochondrial andribosomalDNA(COI,16Sand28S)washigherthanfor thenucleargenes.rDNA16Salignmentwashighlyincongruentamongthedifferentalignmentmethodsduetoabundanceof ambiguoussectionsinitsstructure.Thelengthsofthealigned generegionswere1015bp(COI),711bp(IDH),513bp(PGD), 834bp(GLN),783bp(CAD),453bp(TPI),397bp(16S)and 644bp(28S),respectively.Fortheprotein-codinggenesCOI, IDHandPGD,nointronregionsweredetected,whilefor CAD,GLNandTPI,oneormoreintronswerepresentand excluded.Wewereabletocorrectoneerroneousidentification fromtheGenBankdatabase,thespecies Sericoceros nr. tannuus (KC975751.1,ZSMHYMAE086,Schmidt&Walter,2014; Schmidt etal.,2017;Boevé etal.,2018)wasrecoveredinside thegenus Atomacera inalltrees;aftercheckingthephotographs ofthevoucherspecimeninthehttp://www.boldsystems.org/,we concludethatthisspecimenisan Atomacera sp.Weprovidethe firstknownsequencesfortheSterictiphorinaegenera Manaos (28S,COI,andPGD), Dielocerus (16S,CAD,COI,GLN,PGD andTPI), Sphacophilus (16S,CAD,COI,GLN,PGDandTPI), Tanymeles (16S)andfortheArginaegenera Athermantus (16S, COI), Pampsilota (16S,28S,CADandCOI), Triarge (28S,16S, andCOI);aswellasforseveralspecies(TableS1).
Amongtheprotein-codinggenestherewasacleardifference inthevariationofsitesamongthefirstandsecondcodon positionsrelativetothethird(Table3),thethirdcodonhaving ahighproportionofuniqueandparsimonyinformativesites (>98%, >90%)andfewinvariantsites(= highvariation)(<8%). TherDNAgeneswereintermediatelyconservedintermsof uniqueandinvariantsites,whilefirstandsecondcodonpositions ofcodinggeneswerethemostconservedpartitions(Table3). Thesubstitutionsaturationindex(Iss)(Table3)showslittle saturation(Iss < Iss.c)inthefirstandsecondcodonposition ofallgenes,exceptforCOI,whichissaturatedaccording tothisindexinallthreecodonpositions.Asimilarpattern wasobservedinthegraphicsofthesubstitution’ssaturation (Fig.S1).Heretheasymptoticbehaviouroftheobservedthird codonofp-distanceagainstthecorrectedgeneticdistance indicatesthatthesubstitutionrateissaturated,whileforthefirst
andsecondposition,itshowsalineargrowthandlowsaturation inallgenes,includingCOI.
Genetrees
WhenanalysingindividualMLgenetreesandallcodonpositions(Fig.S2),themonophylyofArgidaes.s.andPergidaewas recoveredonlyforCAD,GLN,COI,PGDandIDH.Therelationship(Zenarge + Argidaes.s.)wasretrievedwhenanalysing GLN;(Zenarge + Pergidae)fortheindividualanalysesofCOI, PGDandCAD. Zenarge + (Pergidaes.s. + Argidaes.s.)was recoveredforIDH.The16Sgenetreedidnotrecoverthemonophylyofanyofthesefamilies,whileforthegene28Sonlythe familyPergidaewasretrievedasmonophyleticandforTPIthe familyArgidaes.s.wasretrievedasmonophyletic(Fig.S2). Thesefamiliesweregenerallynotrecoveredwhenusingonly thefirstandsecondcodonforeachindividualgenetree.Anadditionalexampleofcontrastingresultsbetweenthephylogenetic reconstructionofindividualgeneswasobservedforthegenus Atomacera,whichwasrecoveredeitherasbeingthesistergroup ofArginaeforCAD,GLNandIDH,orassistergroupofSterictiphorinaeforPGDand28S(seebelow).
Combinedmoleculardataset
ThebranchsupportwaslowerinallcasesintheMLtrees (BR)relativetotheBItrees(posteriorprobability),evenwith similartopology(Fig.S3).TheBItreesreconstructedwith onlythefirstandsecondcodonpositionwerelessresolved andsupportedtospecieslevel(e.g., Arge)(Fig.3),whilethe inclusionofthethirdcodonpositionintheanalysesimproved theresolutionandsupport(>50%)atthislevel.Ontheother hand,excludingthesaturatedthirdcodonpositionimprovedthe supportofdeepnodes(e.g.,atthebaseoftheZAPclade,see below)(Fig.S3).
TheMLandBIshoweddifferenttopologiesforArgidaes.s., Pergidaeand Zenarge relationships,althoughinallcasesthey formedpartofthesamecladewithhighsupport(BR orposterior probability PP = 89–100)(Fig.S3).(Zenarge + Argidaes.s.) wasretrievedwhenusingMLandBIwithallcodonpositions (Fig.S3:A,C).Ontheotherhand, Zenarge + (Pergidae + Argidaes.s.)wasobtainedinbothanalyseswhenusingonlythe firstandsecondcodonpositions(Fig.S3:B,D).
RegardingtheinternalrelationshipsofArgidaes.s.,inthe combinedmoleculardatasetusingeitherallcodonpositions oronlythefirstandsecondcodonpositionwerecoveredtwo maincladesthatcorrespondtothesubfamiliesArginaeandSterictiphorinaes.Benson(1963)(Fig.3);thecladeswithineach subfamilyarealsohighlycongruentwiththemorphological hypotheses(Malagón-Aldana etal.,2021a,2021b).ForSterictiphorinae, Atomacera wasretrievedassistergroupofthe remaininggenera(Fig.3).Othercladesrecoveredwithinthe SterictiphorinaewerethesisterrelationshipbetweentheHolarcticclade Sterictiphora + (Aproceros + Aprosthema)anda Neotropicalcladecomprisinganumberofgenerabutwhose
Table3. Descriptiveparametersofalignedsequenceswithcorrespondingsubstitutionmodelassigned(IQ-treeandpartitionfinder).
Gene
16s6139770.544.141.8GTR
28s6964462.339.346.4SYM
COI128367748.926.063.8TIM
COI38333897.689.14.4TIM + F + G4TIM + G1.650.690.000
IDH122047429.315.677.0TIM3
IDH32023798.793.73.0TNe
PGD124534240.119.073.4SYM + I + G4GTR + I + G0.390.690.000
PGD34517198.892.47.6K2P
TPI122930243.019.564.2K2P + I + G4SYM +
TPI32915199.395.43.3TPM3
CAD125452250.618.471.5TIM3
CAD35426199.293.95.7TPM2
GLN124855645.714.271.9GTR
GLN34827898.289.94.7HKY
internalrelationshipswerenotresolved(Fig.3).Concerning Arginae,theAustraliangenus Antargidium wasrecoveredasthe sistergroupoftheremaininggenera.TheNeotropicalgenus Scobina isthesistergroupoftheremainingArginae,minus Antargidium.
Totalevidenceanalysisandphylogeneticrelationships
Thecombinedmorphologicalandmoleculardatasetcomprised164terminaltaxa(52generaofArgidaeoutof55)and 5658characters(FileS2).Thegenera Pseudosinarge, Zhuhongfuna and Mioarge (fossil)wereexcludedfromthemorphologicalmatrixandtheanalysesduetothelownumberofscored charactersandtheirnegativeeffectintheresolutionofArginae. WhencombiningmolecularandmorphologicaldatausingBI andallcodonpositions,weretrieved Zenarge + (Pergidae + Argidaes.s.)(Figs4,5).PergidaeandArgidaewererecovered asmonophyleticsistergroups,bothwithhighbranchsupport (PP = 100).WithregardtoPergidae(Fig.5),thenewworld subfamilyAcordulecerinaeisretrievedasthesistergroup oftheremaininggenera,whichinturnaredividedintwo mainclades(Fig.5).OnecladeincludesPhylacteophaginae + (Perginae + Parasyzygoninae)(PP = 96),whilethesecondclade hasLoboceratinae(Aulacomerus)(PP = 89)assistergroup oftheremainingsubfamilies:(Styracotechynae + Heteroperreyia) + ((Pterygophorinae + (Philomastiginae + (Euryinae + (Syzygoninae + Perreyinae)).Remarkably,theSouthAmerican Perreyinaewasrecoveredasparaphyletic, Heteroperreyia being outsidethesubfamily.
InsideArgidae,twomaincladeswereretrievedforallthedifferentreconstructedphylogenies.TheycorrespondtothesubfamiliesArginae(PP = 100)andSterictiphorinae(PP = 66) (Figs4,5),asintheabovemolecularanalysesandpreviousmorphologicalphylogenetichypotheses(Malagón-Aldana etal.,2021a,2021b).ForthesubfamilyArginae(Fig.4),the Australiangenus Antargidium (Fig.4,cladeI)wasretrieved
asthesistergroupoftherest,followedbytheNeotropical Scobina (Fig.4,cladeII).CladeIII(Fig.4)comprises exclusivelyAfricantaxa(PP = 100),i.e.,thegenus Triarge, Pampsilota andAfrican Arge spp.IncladeIII, Pampsilotaafra wasrecoveredassistergroupoftheremaining taxa. Pampsilota isparaphyletic, P.afra beingapartfromthe otherspecies, P.dahomeyanus,whichisinacladewith Arge congrua and A.fenestralis Triarge isrecoveredasmonophyleticandsistergroupof Argemontana and A.stuhlmanni CladeIIIincludestwosubclades,oneconsistingof Athermanthus + Sinarge (Oriental-Sino-japanese)andasecondcomprising Cibdela + (Tanyphatnidea + Sjoestedtia)(Oriental-African) (Fig.4).CladeIIIwasrecoveredasthesistergroupofthelast majorcladeofthesubfamilythatcomprisesthemajorityofthe ubiquitousgenus Arge (Fig.4,cladeV),withanumberofother generaembeddedintheclade(Asiarge, Kokujewia, Brevisceniana, Pseudarge and Spinarge).
InSterictiphorinae(Fig.5),theAustralian Trichorhachus + Styphelarge (cladeVI)wereretrievedassister groupoftheremaininggenera,followedby Atomacera (clade VII)fromtheNearcticandNeotropicalregions.Athird cladeincludestheHolarcticgenera(cladeVIII),amongthem Sterictiphora wasthesistergroupoftheremaininggenera Aprosthema + (Pseudaprosthema, Ortasiceros and Yasumatsua + Aproceros).Finally,theremainderoftheSterictiphorinae wereplacedinalarge,predominantlyNeotropicalclade (cladeIX)withmaleshavingtheflagellumfurcate, Brachyphatnus (southernSouthAmerica)beingthesistergroupof theremaininggenera.Thedeeperrelationshipsofthelatter arepoorlyresolved,butsomesubcladeshavehighsupport (PP ≥ 70)(Fig.5).Thesesubcladesare:cladeX, Ptenos (paraphyletic), Manaos and Zynzus;cladeXI, Acrogymnidia + (Acrogymnia + Trailia);cladeXII,the‘Dielocerinis.l.’ clade(Smith,1992)including Sericoceros, Subsymmia, Durgoa, Neoptilia, Themos, Mallerina, Topotrita, Pachylota, Dielocerus and Digelasinus andcladeXIII, Schizocerella, Didymia, Ptilia, Trochophora, Triptenus and Neurogymnia + Eriglenum,where
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
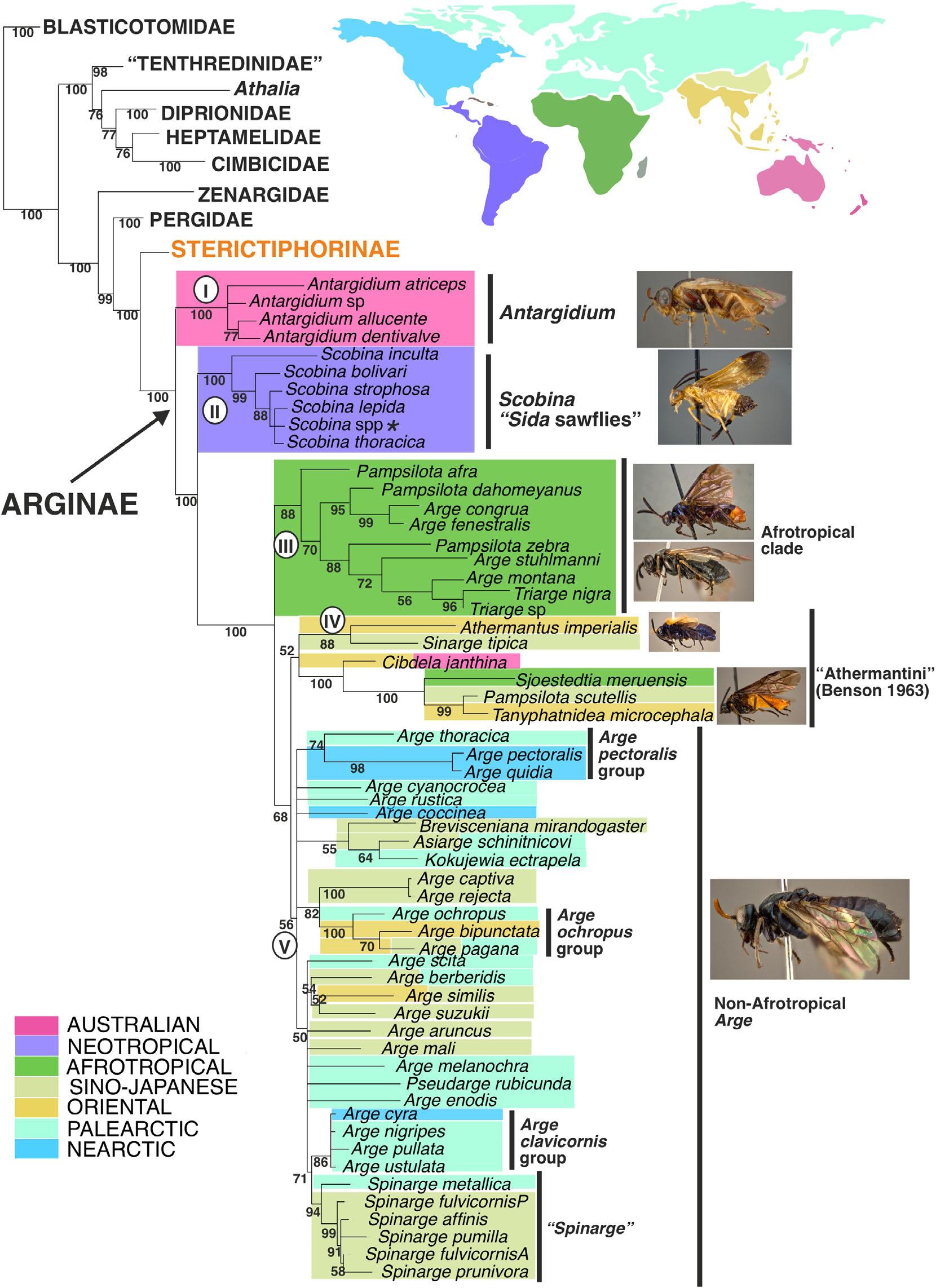
Fig.4. TotalevidenceBayesiantreeofsubfamilyArginae(Argidae)highlightingthecurrentbiogeographicaldistribution.Maincladesarelabelled withnumbersincircles.Asteriskafterthenameofaterminal(e.g., Athalia spp.)indicatesthatthistaxonisachimera,i.e.,themorphologicaland molecularinformationwasobtainedfromdifferentspeciesofthesamegenus.
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
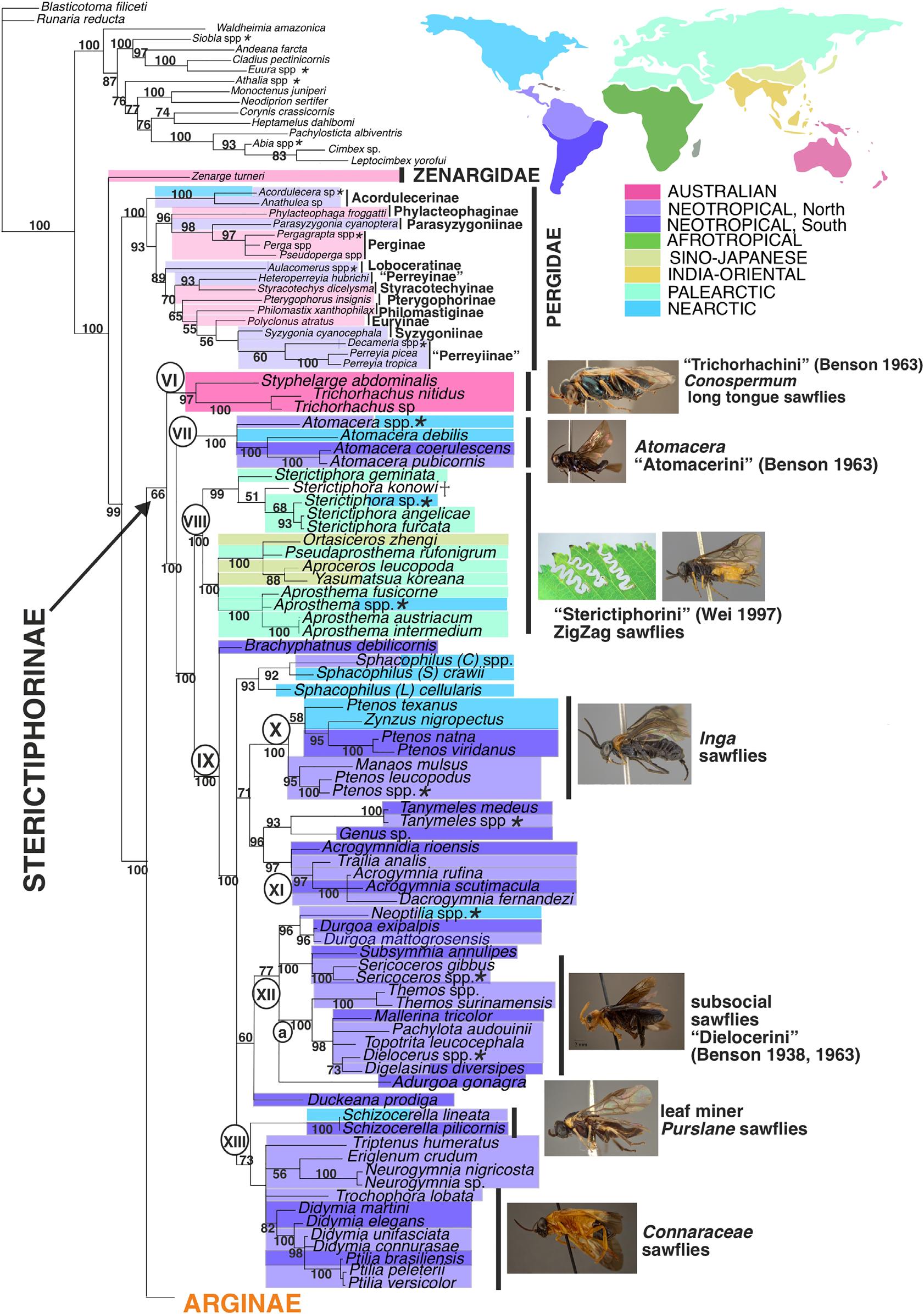
Fig.5. TotalevidenceBayesiantreeofsubfamilySterictiphorinae(Argidae)highlightingthecurrentgeographicaldistribution.Maincladesarelabelled withnumbersincircles.Asteriskafterthenameofaterminal(e.g., Athalia spp.)indicatesthatthistaxonisachimera,i.e.,themorphologicaland molecularinformationwasobtainedfromdifferentspeciesofthesamegenus.ColoursofoccurrenceinPergidaeandZenargidae(Neotropical,Australian andNearcticregion)arelighterthaninlegendtoemphasizethedistributioninArgidaes.s.
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
thegenus Didymia wasnotrecoveredmonophyletic.Wewere unabletoproperlyresolvetheexactpositionof Sphacophilus
Nodedatingandancestraldistribution(Figs6,7)
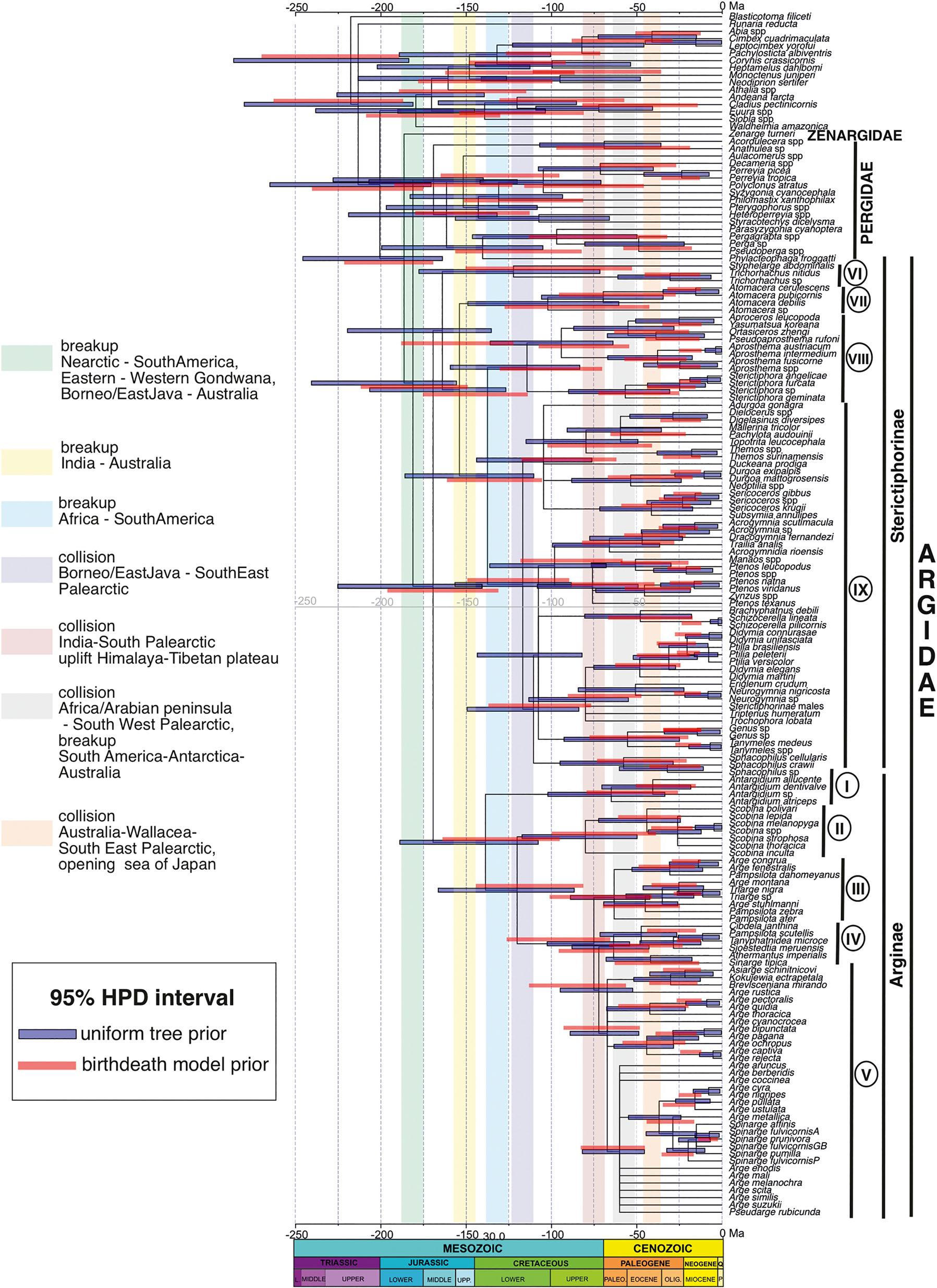
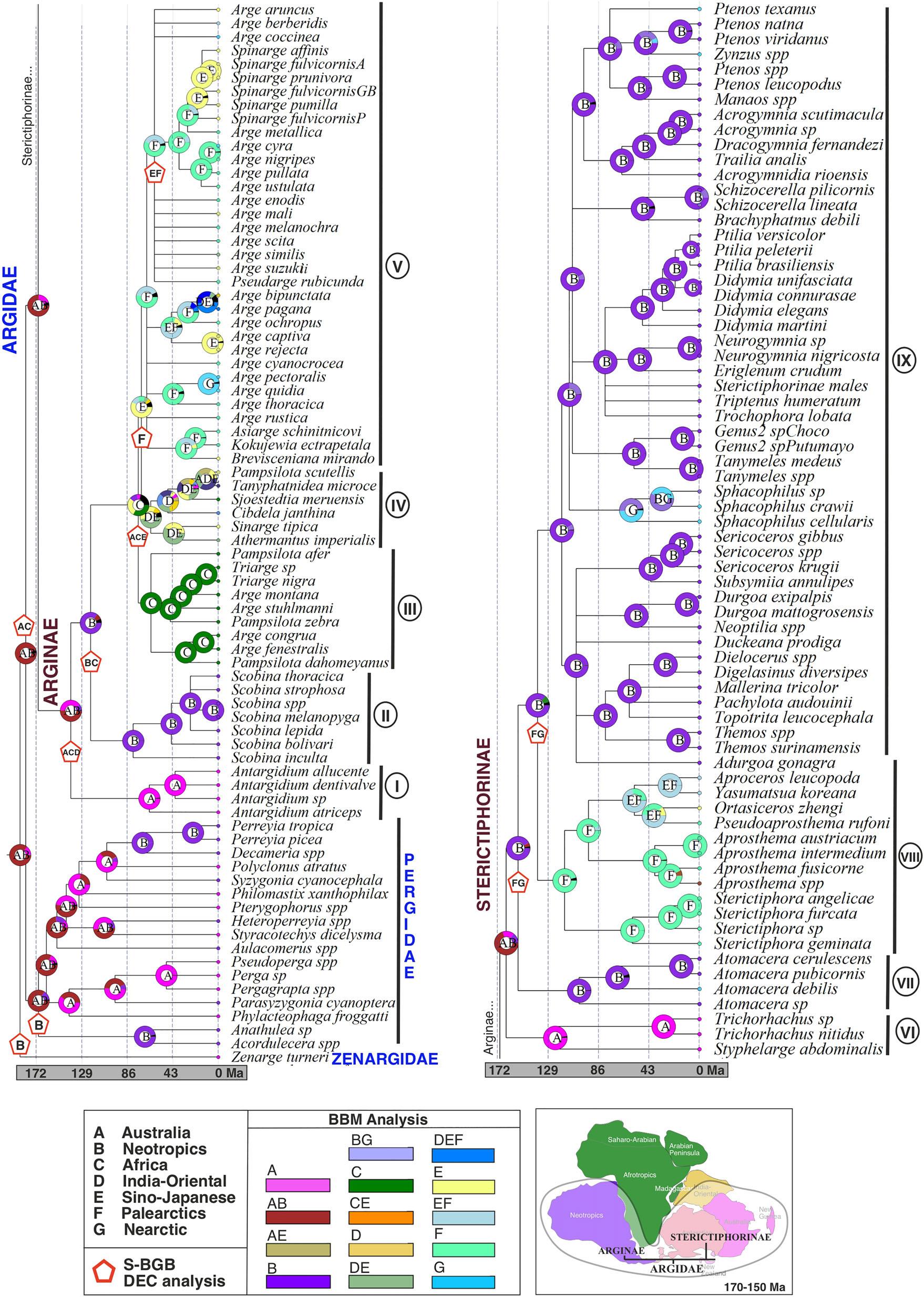
Theestimatedmeanageofthecladeswasslightlyolderwhen usingtheuniformtreepriorthanwiththebirth-deathprior.In thesamewaythe95%highestposteriordensity(HPDinterval) waswiderfortheuniformtreeprior(Fig.6;Table4);herewe describetheresultsusingtheshorterHPDinterval.Theageof theZAPcladewasestimatedtobe177.1Ma(164.7–215.8), e.g.,earlyJurassic;thisagreeswiththeestimateofRonquist etal. (2012a).Thebiogeographicalreconstructions(both S-BGB/DECandBBM)indicateanancestraldistributionof theZAPancestorintheNeotropicsorAustralia + Neotropics (Fig.7).ThedivergencebetweenPergidaeandArgidaes.s. occurredapprox.168.5Ma(144.9–205.4).Thisintervalof timeoverlapswithotherestimates(Schmidt&Walter,2014; Nyman etal.,2019)andiscontemporarywiththebeginningof thebreakupbetweenwestandeastGondwana.Thefirstsplit withinArgidaes.s.isdatedfromthemiddletolateJurassic around155.1Ma(127.4–191.1Ma).TheageoftheSterictiphorinaecrowngroupisestimatedtobearound148.2Ma (118.6–183.2Ma)inthebeginningoftheCretaceous,significantlyolderthancrowngroupArginaeatapprox.123.3Ma (92.1–159.5).TheancestralareaofArgidaes.s.wasinferred tobeAustralian/Neotropical(BBM)orAustralian/African (S-BGB/DEC);foreachsubfamilytheancestraldistributionwasestimatedalsoasAustralian/Neotropical(BBMand S-BGB/DEC)(Fig.7).
Discussion
Althoughformanytaxatheoverallgenesamplingwaslow (TableS1),especiallyforthenuclearprotein-codinggenes,the phylogeneticrelationshipsretrievedwiththeavailablemoleculardata(Fig.3)arehighlycongruentwithprevioushypotheses,e.g.,monophylyofArgidae,Pergidae,Sterictiphorinaeand Arginae.However,formorethanhalfofthegeneraofArgidae, molecularinformationisstillabsent,andthereforetherelationshipsofthesetaxarelyexclusivelyonmorphologicalevidence.
Saturationofsubstitutionrate,additionaldataandtheir phylogeneticeffect
Ourresultsshowhowthetypeofdatadifferentiallyaffect theplacementoftaxa,withspecialeffectonisolatedtaxa, e.g., Zenargeturneri (Zenargidae)(Fig.S3).Theplacement of Zenarge washighlyinfluencedbythethirdcodonposition, andwhenexcludingit,thephylogeneticpositionof Zenarge wasmoresimilarinthemolecularandmorphologicaltrees (Malagón-Aldana etal.,2021a,2021b), Zenarge beingthe sistergroupofPergidae + Argidaes.s.However,formore distalnodesinthetopology,e.g.,within Arge,theresolution
decreasedsubstantiallywhenexcludingthethirdcodonposition (Fig.3).Thisindicatesthatwhilethethirdcodonposition mighthaveachievedsaturationforthedeepernodes,thismay notbethecaseforthemoredistalnodes;asimilareffect occursinimpliedweightsanalyses,wheredifferent k -values mightresolvesomepartsofthephylogenybetterthanothers do.Usingthehighlysaturatednucleotidesinphylogenetic analysesisstillcontroversial(Källersjö etal.,1999;Strimmer &vonHaeseler,2009;Breinholt&Kawahara,2013).Malm &Nyman(2015)retrieved Zenarge + Argidaes.s.evenif excludingthethirdcodonpositionformostoftheprotein-coding genestheyused,exceptforCADandGLN.
Additionally,whenwecombinedthemolecularandmorphologicaldatasets,includingallcodonpositions, Zenarge was recoveredassistergroupofArgidae + Pergidae.Thisshowsthat morphologicalcharacters,inspiteoftheirsignificantlylower numberrelativetomolecularsites,canstillinfluencetheresults ofthecombinedanalyseswhenusingfewgenes.Thesampling ofadditionalfreshmaterialof Zenarge (wereliedonGenBank sequences),togetherwithaphylogenomicapproachwouldhelp totestitscurrentplacement.Fornow,basedonthemorphological(adultandlarvalstages)andmolecularevidenceanalysedhere,weupholdplacing Zenarge inaseparatefamily,the Zenargidae(Malagón-Aldana etal.,2021a).
Zenargidae
ThecombinedanalysissupportsthetopologyZenargidae + (Argidae + Pergidae)orZAPclade(Figs4,5). Zenarge turneri,thesolerepresentativeofZenargidae,onlyoccursin NewSouthWalesinAustralia.ItistheonlytaxonoftheZAP cladethatfeedsongymnosperms,theendemicCypresspine Callitris aswellastheintroduced Cupressus (Cupressaceae) (Moore,1963a,1963b;Smith,1992).Thepositionof Zenarge as sistertotherestoftheZAPcladeandourestimationoftheage ofthesplitbetweenZenargidaeandArgidae + Pergidaearound 180MaintheearlyJurassic,priortotheGondwanabreakup (Fig.6),isapparentlyinconflictwiththeinferenceofNyman etal. (2019)thatthecommonancestoroftheZAPclade(indeed, ofallTenthredinoidea)wasfeedingonangiospermsandthe feedinghabitsof Zenarge aresultofasecondaryshifttogymnosperms.TheoriginoftheZenargidaeisaccordingtoourestimatesofsimilarageastheearliestputativeangiospermflower fossil(Nanjinganthusdendrostyla;Fu etal.,2018of174Ma), olderthantheageestimate(120Ma,i.e.,EarlyCretaceous)of thesplitbetween Zenarge andtheremainingArgidaeaccordingtoNyman etal. (2019).Differentlinesofevidencesupportthepre-Cretaceousoriginofangiospermplants(Hochuli& Feist-Burkhardt,2013;Fu etal.,2018);however,fossilevidence indicatesthatthemajorradiationofangiospermsdidnottake offuntiltheEarlyCretaceous(Silvestro etal.,2015;Herendeen etal.,2017;Nyman etal.,2019).TheevolutionoftheTenthredinoideaseemstodisplayasimilarpattern(Nyman etal.,2019) andtheymaystillhaveco-evolvedwithangiospermsevenif theircommonancestorintheearlyJurassicwasfeedingongymnosperms.However,theestimatedageof Callitris,thecurrent
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
Fig.6. NodedatingchronogramofArgidaeusingthecombineddataset(morphologyandmolecules).Horizontalbarsshowthe95%highestposterior densityinterval(HPD)oftheestimatednodeage,inblueforthecombineddatasetandredforthemoleculardata.Verticalbarsshowrelevantgeological eventsinthattransitionperiodoftime.Maincladesarelabelledwithnumbersincircles.
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
Fig.7. BiogeographicancestralreconstructionofArgidae,subfamilyArginae(left)andSterictiphorinae(right)showingBayesianBinaryAnalysis (BBM)estimationsincolourcharts,andS-BGB/DECDispersalExtinctionCladogenesis(DEC)inwhitepentagons.Maincladesarelabelledwith numbersincircles.
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
Table4. Divergencetimesofmainclades(Fig.8)basedonnodedatingBayesiananalysis,usingauniformtreeprior(UN)andabirth-deathprior (BD).
NodeAge(Ma)UN
95%Highest posteriordensity (HPD)(Ma)UN Age (Ma)BD
(Ma)BD
Zenarge + (Argidae + Pergidae)187.3164.7–246.9177.1164.7–215.8 Argidae + Pergidae182.1156.4–241.8168.5144.9–205.4 Sterictiphorinae + Arginae170.2141.3–226.3155.1127.4–191.1
I + remaining139.4108.3–189.8123.392.1–159.5
II + remaining120.787.1–167.1109.878.7–140.4
III + remaining75.554.5–103.188.263.1–122.6
IV + remaining72.652.7–95.277.854.1–109.8
V + remaining67.649.1–89.566.946.2–90.1
VI + remaining164.8135.9–220.6148.2118.6–183.2
VII + remaining154.7127.4–207.5135.7110.5–170.7
VIII + remaining138.2110.9–186.8124.5102.3–156.9 IX117.488.8–157.3112.586.7–145.2
hostplantof Zenarge,indicatesthatgymnospermfeedingin thelattermightbesecondaryratherthanancestral. Callitris is agenusofdrought-resistantplantsthatevolvedaround30Ma, probablyinresponsetoclimatechangeinthemiddleCenozoic asAustraliadriftednorthwards(Pittermann etal.,2012;Larter etal.,2017).The Zenarge lineagecannothaveswitchedto Callitris earlierthanthis,whetheritwasfromanangiosperm oragymnospermhost.
Pergidae
ThereconstructionofancestralbiogeographicareaofPergidae(Fig.7,BBM,S-BGB/DEC)iscongruentwiththehypothesisofGondwananorigin(Neotropicaloracombinationof NeotropicalandAustralia)withameanageof150Maforthe crowngroup,similartotheestimation(140Ma)ofSchmidt& Walter(2014).Ourresultssuggest,inaccordancewithSchmidt &Walter(2014),apatternofrepeateddispersalsbetween SouthAmericaandAustraliathroughAntarcticabeforethefinal separationofthethreecontinentsaround35Ma(Figs6,7). Schmidt&Walter(2014)suggestedthatseveralpergidlineages (Perginae,Phylacteophaginae,andPterygophorinae)radiatedon Myrtaceae(Eucalyptus andtheirrelatives)astheAustralianclimatebecamemorearidduringtheCenozoicera,whileother lineagesexperiencedreduceddiversityorwentextinct.
Thecombinedanalysis(Fig.5)placedthenewworldsubfamilyAcordulecerinae(Acordulecera + Anathulea),assistergroupoftheremaininggenera;thisincontrasttoSchmidt &Walter(2014)thatplacedLoboceratinae(Aulacomerus) assistertoallotherPergidae.Althoughbothsubfamilies haveasimilardistributioninSouthAmerica,Acordulecerinae haveahigherdiversity,especiallywithinthegenus Acordulecera thatalsoextendsintoNorthAmerica.Previousstudies (Smith,1990;Schmidt&Walter,2014;Malm&Nyman,2015; Malagón-Aldana etal.,2021a)suggestedthatthesubfamily AcordulecerinaebelongsinacladewithPhylacteophaginaeand PerginaefromAustralia.RegardingLoboceratinae,allstudiesto
datehaveonlyincludedonegenus, Aulacomerus,andthemonophylyofthesubfamilyitselfremainsuncertain.
ThetwomaincladesofPergidaeretrieved(Fig.5)aremostly congruentwithpreviousstudies(Schmidt&Walter,2014; Boevé etal.,2018)intermsofsubfamilycomposition,although theirrelationshipspartlydiffer;thismightbecausedbydifferencesintaxonandsequencesampling.Remarkably,we retrievedtheAustralian Heteroperreyia assistergroupofthe Australian Styracotechys (Styracotechyinae);thismakesthe NeotropicalsubfamilyPerreyinaeparaphyleticsince Heteroperreyia isplacedapartfrom Perreyia + Decameria,similartowhat wasfoundinlarvalmorphologicalanalysis(Malagón-Aldana etal.,2021b);theydiffersubstantiallyintheovipositorstructure (welldevelopedin Heteroperreyia,reducedin,e.g., Perreyia and Decameria)(Smith,1990;Malagón-Aldana etal.,2021a) andlarvallifehistory(saprophagous/mycophagousin Perreyia/Decameria,phytophagousin Heteroperreyia)(Schmidt& Walter,2014;Smith etal.,2019).
TheNeotropicalParasyzygoniinaewasretrievedassister groupofPerginae(Australia),and Syzygonia (Syzygoninae) assistergroupofPerreyinae(Neotropics).Parasyzygoniinaeis amonotypicsubfamilywithonlytwospecies(Parasyzygonia cyanoptera and P.pallidor) fromVenezuelaandBrazil,which hasbeenpreviouslysuggestedtobeclosetoPerginaeby Smith(1990)andMalagón-Aldana etal. (2021a).Withregard totheNeotropicalSyzygoninae,Schmidt&Walter(2014) includedonlythegenus Lagideus,andtheyretrieveditassister groupofPhilomastiginae(Philomastix).Themonophylyofthe subfamilySyzygoninae(e.g., Syzygonia and Lagideus)hasnot beenproperlytested.
Argidaerelationships
Ourphylogeneticresults(Figs4,5),basedonthecombined analysis,arehighlysimilartothemorphologicalphylogenetic hypotheses(Malagón-Aldana etal.,2021a,2021b)withtwo largeclades,i.e.,thesubfamiliesArginaeandSterictiphorinae.
Ingeneralterms,Arginaecorrespondtothetaxawheremales haveanundividedantennalflagellum(Fig.1A–C)andSterictiphorinaetothosewheremaleshaveafurcateantenna (Fig.2A,B).Thegenus Atomacera istheonlygenuswherethe antennaofmalesissimple(Fig.2C)butrecoveredinsidethe Sterictiphorinae,seebelow.
WithinArginae,thepositionof Antargidium (cladeI)and Scobina (cladeII)assuccessivesistergroupstotherestof thesubfamilyisstableandwellsupportedbybothmolecular andmorphologicaldata(Figs3,4);thesetwogeneraretain someplesiomorphies,includingthepresenceofacomplete cupulainthemalegenitaliaandtwoseparatedsuprapedal lobesintheirlarvalstage(Malagón-Aldana etal.,2021a, 2021b).CladesIandII(Fig.4)arecongruentwithclades 1and2oftheadultmorphologyanalysis(Malagón-Aldana etal.,2021a)andpartlycongruentwithcladeAofthelarval analysis(Malagón-Aldana etal.,2021b).Weretrieveacladeof Africantaxa(Pampsilota, Triarge and Arge)(cladeIII)thatisin conflictwiththecurrentgenusclassificationandquestionsthe diagnosticvalueofsomeofthemorphologicalcharactersthat thisclassificationisbasedon,e.g.,theoccurrenceofpreapical tibialspurs(absentin Pampsilota,presentin Arge)andthe presenceofthe2r-mveinintheforewing(absentin Triarge, presentin Arge).Accordingtotheadultmorphologicalanalysis ofMalagón-Aldana etal. (2021a,fig.20H),thepresenceoflong volsellarteethonthedigitusofthemalescouldbeconsidereda putativesynapomorphyforcladeIII;moreAfrotropicalArginae needtobeexaminedtoconfirmthis.
ThecladesIIIandIV(Fig.4)arepartlycongruentwith clades6aand6bofMalagón-Aldana etal. (2021a)andthe tribeAthermantiniofBenson(1963).Thegeneraofclade IVlackthepreapicaltibialspurinmidandhindtibiaeand arerestrictedtotheOriental/Sino-Japaneseregion,exceptfor Sjoestedtia (Afrotropics).CladeIVincludesasubcladewith twosmallgenera, Athermantus (twospecies)and Sinarge (one species),forwhichwewereonlyabletosequencemitochondrialmarkers(COIand16S)for Athermantus.InthesecondsubcladewithincladeIV, Cibdela isretrievedasthesistergroupoftheremaininggenera. Pampsilotascutellis is retrievedinside Sjoestedtia + Tanyphatnidea;thethreetaxa haveparapenislobesinthemalegenitalia,thethirdvalvulaeoffemalesstronglyangulateddorsolaterallyandanepistomalsulcuspresentbutweak(Malagón-Aldana etal.,2021a). Additionally, P.scutellisalsohastheanalcellpresentin thehindwing,asin Tanyphatnidea.Thegenera Tanyphatnidea and Pampsilota (recoveredparaphyletichere)havebeen defineddifferentlybytaxonomists(Togashi,1975;Wei,1997a; Saini,2009);however,in Pampsilota theparapenislobesof malesandepistomalsulcusareabsent,anddorsolateralcornersofthethirdvalvulaeoffemalesarenotangulated.In addition, Tanyphatnidea isrestrictedtosouthernChina,Taiwan andtheOrientalregion,while Pampsilota speciesareendemic totheAfrotropics(Liston etal.,2017).Wedidnotexamine theChinesespecies Pampsilotacenchra,althoughthedescriptionbyWei(1997a)suggestedhighsimilaritywith P.scutellis RegardinghostplantsofcladeIV, Cibdelajanthina larvaefeeds on Rubusalcifolius (Rosacae)(Florens etal.,2017),whereas
Tanyphatnideasinensis feedson Polygonum sp.(Polygonaceae) (Togashi,1980).
Theremaining,non-Afrotropical Arge spp.(cladeV,Fig.4) areplacedinamonophylumthatincludesseveralothersmall generafromthePalearcticregion.Ofthese, Kokujewia and Spinarge havebeensampledforbothmolecular(onlyCOIfor Kokujewia) andmorphologicaldata; Asiarge, Brevisceniana and Pseudarge onlyformorphology. Kokujewia, Asiarge and Brevisceniana formacladeprimarilydefinedbytheabsenceof preapicaltibialspursandtheshortapicalmaxillarypalpomere. Duetothelargenumber(approx.400)ofdescribedspeciesof Arge,itisstandardpracticetodefinespeciesgroupstofacilitate itsclassification(e.g.,Smith,1989);weretrievedsomeofthese asmonophyletic(Fig.4):The pectoralis group, ochropus group and clavicornis group.Malagón-Aldana etal. (2021a)retrieved Argeochropus assistertoalargecladecomprisingthemajority oftheArginaeincluded,whichcouldbeduetotheretention ofplesiomorphicconditionsintheovipositorwithasimple, flatthirdvalvula,fromwhichmorecomplexshapesofthethird valvulamighthaveevolved.However,herethe Argeochropus group(Fig.4)(Smith,1989;Yan etal.,2009)isinapolytomy withotherspeciesof Arge thathaveahighlymodifiedovipositor sheath.HostplantsassociationswithincladeVseemtofollow theoligophagouspatternsofothertemperateherbivores(Haack &Mattson,1993)withpreferenceforwoody,shrubbyand herbaceousfoodplantsbelongingpredominantlytothefamilies Rosaceae,Betulaceae,Fagaceae,EricaceaeandSalicaceae (Boevé etal.,2018);Someexamplesofargidswithhighhost specificityare Kokujewiaectrapetala on Rumex spp.(Polygonaceae), Argeberberidis on Berberidis spp.(Berberidaceae) andthe Argeochropus groupon Rosa (Rosaceae).
Alargeamountofevidencesupportstheinclusionof Spinarge in Arge (Figs3,4,cladeV)(Boevé etal.,2018;Malagón-Aldana etal.,2021a,2021b).However,acleardelimitationof Arge s.s. isstillnecessary.SinceAfrotropicalspeciesof Arge aremore closelyrelatedtoothergenera(Triarge and‘Pampsilota’)than totherestof Arge species,itfurthercomplicatesthetaxonomic delimitationofthegenus.Onealternativecouldbetodefine Arge s.s.asthenon-Afrotropicalspecies(cladeV,Fig.4).Alarger sampleofAfrotropicalspeciesof Arge thanours(fouroutof 125species)(Taeger etal.,2010)isneededtocorroboratethis.
Thebranchsupport(PP = 66)fortheSterictiphorinaeislower thanforArginae(PP = 100).Thismightbecausedbythe lackofmoleculardatafor Trichorhachus and Styphelarge,and theretentionofseveralplesiomorphiccharacterstatesinadult andlarvalmorphologyin Trichorhachus (e.g.,preapicaltibialspurspresentinadultsandsimplesetaeinepipharynxand laciniaoflarva)(Malagón-Aldana etal.,2021a,2021b).Nevertheless,thesetwogeneraformcladeVI(Fig.5)whichis congruentwithclade7ofMalagón-Aldana etal. (2021a)and thetribeTrichorhachiniofBenson(1963).CladeVIisthesistergroupoftheremainingSterictiphorinae.TheNeotropical genus Atomacera (cladeVII,Fig.5)isrecoveredwithinSterictiphorinaeassuggestedbyBenson(1963)(tribeAtomacerini) andMalagón-Aldana etal. (2021a,2021b),althoughinsome individualgenetreesitwasplacedclosertoArginae.Itwas retrievedassistergroupofArginae + SterictiphorinaeinMalm
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
&Nyman(2015).Incontrasttotheputativereversaltoasimple flagellumantennainthemales,anumberofcharactersjustify theplacementof Atomacera inSterictiphorinae,e.g.,thepresenceofsetaemultiplebranchedinthelaciniaandepipharynxof larva(Malagón-Aldana etal.,2021b).Theconflictingevidence fromindividualgenetrees(Fig.S2)aswellasadultandlarval anatomyindicatesthattheexactpositionof Atomacera needs furtherclarification.
CladeVIII(Fig.5)iswellsupported(PP = 100)andretrieved withmolecularandtotalevidence(Figs3,5),aswellas previousmorphologicalanalyses(clade8ofMalagón-Aldana etal.,2021a,cladeDofMalagón-Aldana etal.,2021b). CladeVIIIispartlycongruentwiththetribeSterictiphotiniof Wei(1997b)butincludesthegenus Ortasiceros aswell.For thesmallgenera Pseudaprosthema (threespecies), Ortasiceros (sixspecies)and Yasumatsua (threespecies)onlytypematerial exists,sotheamountofinformationthatcanbeextractedfrom themislimited.Acharacteristiclarvalfeedingbehaviourhas beendescribedforlarvaeofsomespeciesofcladeVIII;they makesinusoidalchannelsorzig-zagfeedingtracksontheir hostplants,whichare Prunus spp.(Rosaceae)[Sterictiphora serotina, S.prunivora]and Ulmus spp.(Ulmaceae)[Aproceros leucopoda](Blank etal.,2014;Eiseman,2015).Incontrast, Aprosthematardum doesnotdisplaythisbehaviourwhen feedingonherbaceousLeguminosae(Liston etal.,2018).
ForcladeIX(Fig.5)(PP = 100)thatincludesalltheNeotropicalSterictiphorinaeexcept Atomacera,internalresolutionand supportisweak,evenifmorphologicalsynapomorphiescan beinferredforsomesubclades(Malagón-Aldana etal.,2021a, 2021b).ThisNeotropicalcladecomprisesmorethanhalfof allgeneraofArgidae(29outof55)whilemolecularinformation(mostlyCOI)isonlyavailablefor12genera.Thisis notenoughtoresolvethepolytomyofwellsupportedinternalsubclades(Fig.5,cladesX,XI,XIIandXIII).Within subcladesofcladeIX,somehostplantassociationspatterns areevident.ForcladeX,larvaeareassociatedmainlywith theFabaceae Inga + Zygia clade(Smith,1970;Smith,1992; Smith&Janzen,2003;Smith etal.,2013;Endara etal.,2018; Malagon-Aldana etal.,2019).Endara etal. (2018)suggesteda coevolutionaryarmsracebetween Ptenos and Inga,where Inga defencechemistryplayedaroleincospeciationwith Ptenos IncladeXIII,thelarvaefeedonmembersofConnaraceae (Kimsey&Smith,1985;Smith,1992;Smith&Janzen,2003; Smith etal.,2013).
BiogeographyofArgidae
TheancestralbiogeographicreconstructionofextantArgidae s.s.iscongruentwithaGondwanandistributionwiththesame probabilityfortheAustralianandtheNeotropicalregion(Fig.7, S-BGB/DECandBBM)duringtheearlybreakupbetween eastandwestGondwanainthemiddle-upperJurassic(around 155Ma,crowngroupage,Fig.6).Whilethiscorrespondstothe patterndisplayedbythePergidae(Schmidt&Walter,2014),it ignoresthedistributionalinformationfrom Xyelotomamacroclada,afossiltaxonpossiblycloselyrelatedtotheZAPclade
andusedtocalibratethisnodeinthedivergencedatinganalyses (seeMethods).ThisfossilisfromthelateJurassic(164.7Ma) andoccursintheDaohugouFormationfromChina,aswell asKazakhstan(seehttp://fossilworks.org/?a=taxonInfo&taxon_ no=139523),i.e.,thenorthernhemisphere(Gao etal. 2009).We havenotbeenabletostudythisfossilandintegrateitinour morphologicaldataset;theestimateofitsphylogeneticpositionisfromRonquist etal. (2012a).However,evenif Xyelotomamacroclada wasincludedinourbiogeographicanalyses, theotherwisestrongGondwanansignalinthedeepernodesof theZAPcladewouldprobablynotbeoverturned.
TheradiationofArgidaes.s.intheEarlyCretaceouswas probablyrelatedtoradiationoftheangiospermsatthesame time(Sun etal.,1998;Soltis etal.,2008),asithasbeen suggestedforothersawflies(Nyman etal.,2019).Inside eachsubfamily,thesameancestraldistributioninGondwanaisinferred(Fig.7),eitherAustralian/Neotropics(BBM) orAustralian/Africa/India-Oriental(S-BGB/DEC).Inboth ArginaeandSterictiphorinae,thesistergroupsoftheextant genera,cladesIandVII, Antargidium (Arginae)and Trichorhachus + Styphelarge (Sterictiphorinae),respectively, areendemictoAustralia,whilethenextcladestobranch off,i.e.,II(Arginae: Scobina)andVII(Sterictiphorinae: Atomacera)(Figs4,5),respectively,areNeotropical. Trichorhachus + Styphelarge arerestrictedtoWesternAustralia, while Antargidium ismainlypresentinEasternAustralia;itis possiblethatthehabitatoftheirancestorscontractedsubstantiallyduringtheincreasedaridityintheinteriorofAustralia, whentheAustralianplatedriftedtowardstheEquatorinthe Miocene(Wilford&Brown,1994).Someofthefirstextant generatobranchoffhaveascurrenthostplantstaxathatevolved muchmorerecentlythantheancestorsofthesawflies. Antargidium feedontheAustralianSapindaceae Arytera and Alectryon (20–30Ma;Muellner-Riehl etal.,2016). Trichorhachus feed onflowersoftheendemicAustralianProteaceae Conospermum (70Ma;Lamont&He,2012)fromfire-adaptedvegetation inwesternAustralia;theadultshavealongproboscisand probablyuseittoreachthenectarof Conospermum small flowersatthebaseofthecalyxtube,similartosomecolletid bees(Houston,1989).UnlikesomeAustralianPergidaethat haveapparentlyexperiencedmodestdiversityincreaseasaconsequenceoffeedingonMyrtaceae(Schmidt&Walter,2014), theendemicAustralianargidgenerahavenotswitchedtothis family,anotherpossiblecauseoftheircurrentlowdiversity.
TheearlydiversificationintheNeotropicsofbothArginae andSterictiphorinaepresentasimilarpattern,occurringinthe lowerCretaceous,i.e.,ancestorsof Scobina (Arginaeclade II,78.6–140.35Ma,Fig.6)and Atomacera (Sterictiphorinae cladeVII,86.7–145.2Ma).ThisiscongruentwiththeisolationofSouthAmericafromAfricaaround110Ma(SanMartin &Ronquist,2004)inavicarianceevent.Bothgeneraare associatedwithMalvaceae(Smith,1992),whichpresumably alsoaroseinGondwanainthelatelowerCretaceousor earlyupperCretaceous(Areces-Berazain&Ackerman,2017; Hernandez-Gutierrez&Magallon,2019). Atomacera apparently dispersedfromtheNeotropicaltotheNearcticregion,although theestimatedageof A.debilis,theNearcticspeciesinour
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
20 L.A.Malagon-Aldana etal.
sample,ismucholderthantheformationofthePanamian Isthmus(13–15Ma)(Montes etal.,2015).However,thecurrenthostgeneraoftheseargidtaxaaremuchyoungerthan theancestorsofcladeIIandVII: Scobina larvaefeedonthe pantropical Sidarhombifolia (30–40Ma)and Atomacera on Hibiscus (30–50Ma)(Areces-Berazain&Ackerman,2017).
ThemajorradiationofSterictiphorinaeoccurredinthe NeotropicalcladeIX(Figs5–7)afterdivergingfromtheancestorsofthePalearcticcladeVIIIinthelowerCretaceous.This probablyhappenedaftertheisolationofSouthAmericafrom Africaaround110Ma.SouthAmericacanbeconsideredanarea ofendemismforSterictiphorinae,whosegenerahaveadiverse rangeofhostplantfamilies(Smith,1992),andtheangiosperm radiationmighthaveinfluencedthediversificationofSterictiphorinae.
TheearlyevolutionaryhistoryofArgidaeandPergidaeare similar(Fig.7),beingrestrictedtoGondwana(Schmidt& Walter,2014).Themaindifferencebetweenthetwofamiliesis thediversificationofArgidaeintherestoftheworld,thenorthernhemisphereinparticular.HowArgidaecolonizedEurasia issomewhatdifficulttoexplain,andthecorrelationofsplittingeventsinthefamilywithtectoniceventsishamperedby thewideHPDintervalsfortheestimateddivergencetimesfor theformer(Fig.6).Thedispersaltothenorthernhemisphere mighthavehappenedthroughdispersalanddiversificationin Africaand/orIndiaintheCretaceous,whichseparatelydrifted northwardstocollidewithEurasiaduringtheearlyEocene (India)andOligocene(Africa-ArabianPeninsula)(Brown& Lomolino,1998).ThebreakupbetweenWest(SouthAmerica, Arabia,andAfrica)andEastGondwana(Australia,India,and Antarctica)beganaround160Ma,severalMaaftertheestimateddivergenceofArgidae + Pergidae.Subsequently,ancestorsofextantlineagesofArgidaes.s.mighthavedispersedto Indiaand/orAfrica.However,accordingtotheancestralbiogeographicreconstructionsofArgidae(Figs7,8),theAfrotropical and/orIndian/Orientaldistributioncanonlybeinferredforthe ancestorsofArginaebutnotforSterictiphorinae,e.g.,cladesI, II,IIIandIV(Fig.7).AlthoughtheAfricancontinentcollided withthePalearcticregion(throughtheArabianPeninsula)later thanIndia(Brown&Lomolino,1998),botheventsmighthave influencedthedispersalofcladeVtoEurasia,sinceeitherofthe twocontinentsmighthavehousedArginaeancestors(Fig.8). AccordingtotheHPDintervalofthedivergencetimes (Fig.6),theancestorsofcladeI(92.1–159.5Ma)andclade II(78.7–140.3Ma)couldprecedetheseparationofAfrica fromSouthAmericaandtheseparationofIndiafromAfrica (130-110Ma),aswellastheirdispersiontothesecontinents. However,theancestorsofcladesIIIandIV(distributedcurrentlyinAfricaandIndia/Oriental,respectively)mighthave evolvedafterIndiaandAfricaseparatedfromtherestofGondwana,sincetheirestimatedageisyoungerthanthoseevents (75–63Ma).AssumingthatallextantAfrotropical Arge (125 species)belongtothisclade,theradiationoftheexclusively AfrotropicalcladeIIIcouldberelatedtothediversification oftheAfricanaustro-temperateandaridflorasinthePaleogeneandEocene,althoughtheirancestorsmighthaveevolved togetherwiththeradiationoflowlandforestangiospermflora
intheCretaceous(Linder,2014).IncladeIII, Pampsilota and Afrotropical Arge spp.haveawidedistributioninsub-Saharan Africa(Koch etal.,2015;Liston etal.,2017),whilethegenus Triarge isrestrictedtoSouthAfricaanditsaustro-temperate flora.Hostplantsfor Triarge areunknown,whilelarvaeof Afrotropicalspeciesgroupsof Arge feedonplantsofGeraniaceaefoundindifferentnaturalbiomesofshrub,bushand grasslands(Koch etal.,2015).ContrarytotheAustralkingdomhypothesissuggestedmainlybyphytogeographers(Linder,2014;Morrone,2015),wedidnotfindtaxawithaffinities acrosssouthernAfrica(e.g., Triarge),southernSouthAmerica andAustralia.
TheextantArginaeoccurringintheIndia-Orientalregion (cladeIV)arerestrictedtonorthernIndia,adjacentandparalleltotheHimalayas(Fig.S4)tothenorth.PossibleexplanationsforthecurrentabsenceofcladeIVincentraland southernIndiaare:(i)Climaticvariation:theirancestorsin southernIndiamovednorthwardsorwentextinctduetoa substantialclimaticchangetomuchdrierconditionsincentralandsouthernIndia,shapedbyitsnorthwarddriftfrom GondwanaandtheformationoftheHimalayas(Chatterjee etal.,2017)intheearlyEocene;(ii)Cretaceousinterchange: theirancestorscouldhavedispersedfromnorthernAfrica (andtheArabianPeninsula)throughalandbridgethatconnectonlytonorthernIndiafromthelateCretaceousbefore IndiacollidedwithEurasia(Oman-Kohistan-Ladakh[OKL]arc, Fig.8C)(Reinert,1970;Chatterjee&Bajpai,2016;Chatterjee etal.,2017);(iii)Mio-Plioceneinterchange:theirancestors couldhavedispersedmuchmorerecentlyfromAfricaduring aMioceneinterchangeaftertothecollisionoftheArabianplate withEurasiaaround30–15Maandbefore/duringthearidificationofSahara,asithasbeensuggestedforotherorganisms withsimilardistribution(Bibi,2011;Portik&Papenfuss,2015). GiventheabsenceofanyevidencefortheoccurrenceofArginae inIndiasouthoftheIndo-GangeticPlain,itisperhapsmost likelythattheyarrivedinthesubcontinentfromthewestsometimeintheTertiary.Nonetheless,accordingtoourdivergence timeestimation,ancestorsofcladeIV(mostlyOrientaltaxa) wouldbeolder(54.1–109.8Ma)thantheMioceneinterchange scenario.RecordsofArgidaeintheArabianPeninsularegion areunknown.Hypotheses2and3abovecouldalsopartly explainthesisterrelationshipbetweentheAfrotropical Sjoestedtia andtheOriental(Pampsilotascutellis + Tanyphatnidea)in cladeIV.
RegardingthedistributionpatternofcladeIVsouthandparalleltotheHimalayanchain(Fig.S4),itispossiblethatthe upliftofthismountainrangefunctionedasanaturalbarrier fordispersaloftheancestorsofthisclade.OrientalrepresentativesofcladeIVareusuallycollectedover3000mofelevation(Saini,2009),indicatingthattheyhaveadaptedtocooler conditions.ThisisincontrasttotheoccurrenceofArgidaein theNeotropics(e.g.,Colombia,Ecuador),wheretheyareonly recordedbelow2500m(Malagon-Aldana etal.,2019).The genus Cibdela (cladeIV,Figs6–8G)istheonlyIndian-Oriental genusthatreachestheAustralianregioninNewGuinea,which couldbeamorerecentdispersalevent(e.g.,duringthecollision ofAustralia-Wallacea-SEAsiaintheearlyMioceneorduring
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
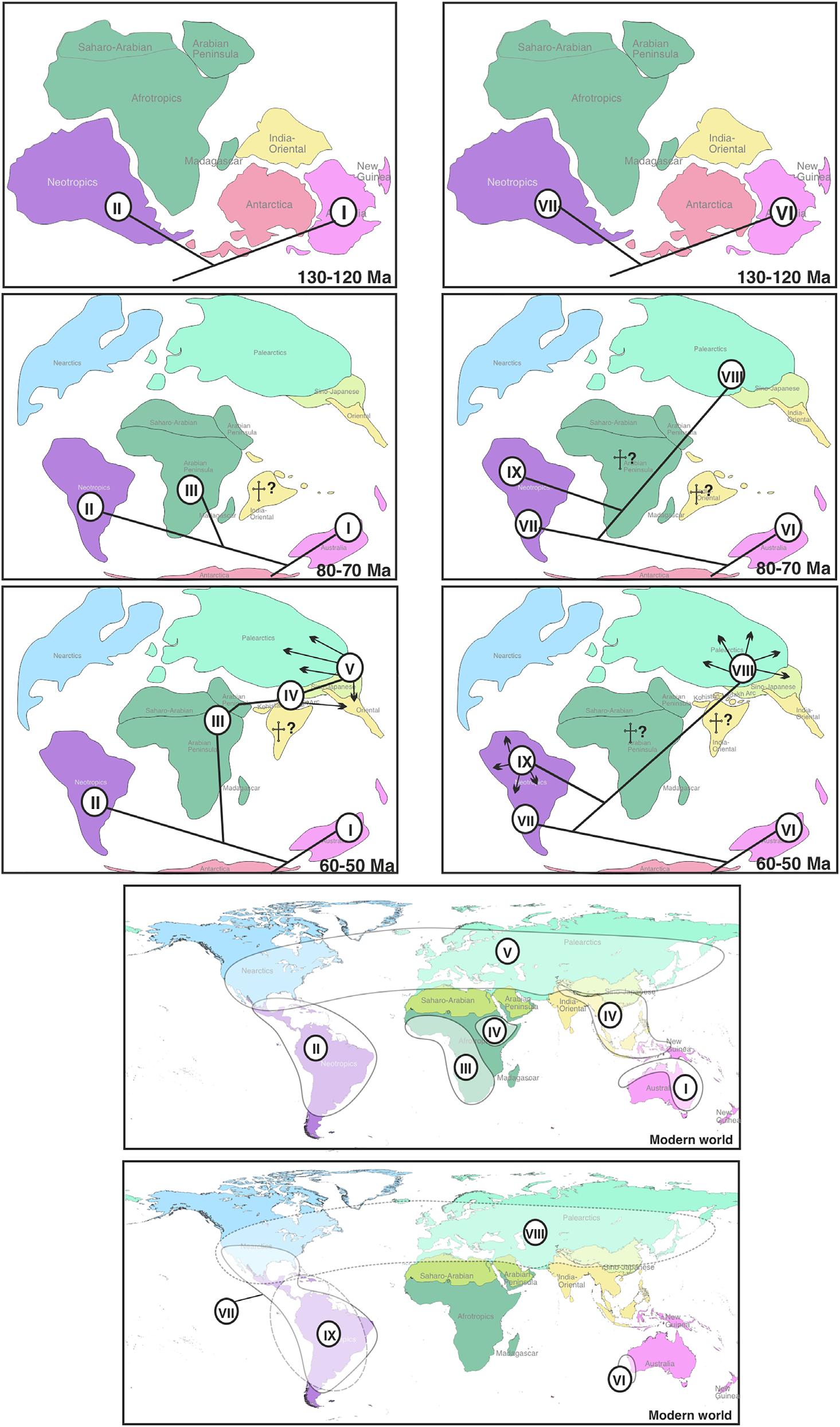
Fig.8. HypotheticsequenceofancestralnodedistributionforsubfamiliesArginae(A–C)andSterictiphorinae(D–F)ofArgidaeindifferentgeologic times.(G)CurrentdistributionofsubfamilyArginae.(H)CurrentdistributionofsubfamilySterictiphorinae.Maincladesarelabelledwithnumbersin circles.
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
Fig.9. MasscocoonsconstructedbyindividualsofSterictiphorinaespecies.(A) Sericocerosdimidiatus Konow,1908(NMNH).(B) Themosolfersii (Klug,1834)(NMNH).(C) Dielocerusdiasi Smith,1975(NMNH).(D) Digelasinusdiversipes (Kirby,1882)(NMNH).(E) Pachylotaaudouinii Westwood,1841(NHMUK).NMNH = NationalMuseumofNaturalHistory,SmithsonianInstitution,WashingtonD.C.,USA,NHMUK=Natural HistoryMuseum,London,UK.
theglaciationsofPleistocenecausedbyfallingsealevelsand expositionofglaciallandbridges)(Brown&Lomolino,1998; Hall&Sevastjanova,2012),sincetheirancestraldistributionisestimatedtobeIndian-Oriental(S-BGB/DEC,BBM, Figs4,7).
TheradiationofthesubfamilyArginaeismostprevalentinthe non-Afrotropical Arge (cladeV)(+250species)inthePalearcticandSino-Japaneseregionsandoccurredbetween38and 61Ma,aroundthetimeestimationofthecollisionbetween IndiaandAsia,45–50Ma,thatformedtheHimalaya-Tibetan plateau(Fig.6)(Chatterjee etal.,2017)andproducedsevere climaticshiftsintheOrientalregion.CladeVhasacurrentdistributioninnorthernIndiasimilartocladeIV,althoughwith asubstantiallowernumberofspecies(20taxa,13endemic) (Saini,2009)thanintheSino-Japaneseregionwherethere aremorethan200species(Wei etal.,2006;Hara&Shinohara,2018;Taeger etal.,2018).Thisincludesseveral Spinarge spp.thatdiversifiedmainlyinJapanaftertheopeningofthe Japanesesea(Fig.6)around15.3–33.7Ma.Morerecently, Arge spp.ofcladeVreachedtheNearctic,atleastintwodifferentdispersalevents,representedby Argecyra (15.7Ma) and A.quidia + A.pectoralis (16.2Ma)(Figs4,7),probablythroughoneoftheBeringlandbridgesthatoccurredat
differenttimesintheCenozoic(Hopkins,1959;SanMartin etal.,2001).
InSterictiphorinae,noAfricanorIndiantaxahavebeen foundthatcanlinktheprimarilyNeotropicalcladesVIIand IXandthemostlyPalearcticcladeVIII.Onlyonespeciesof SterictiphorinaehasbeenrecordedfromtheOrientalregion, Aprocerossikkimensis Saini&Thind,1992(Saini,2009);this speciesoccursinnorthernmostIndiaandwasnotincludedhere. NoreliablerecordsofSterictiphorinaeexistfromtheAfrotropicalregion,exceptforoneoftheotherwiseNeotropical Sphacophilus (recordedas Sterictiphoraafra byPasteels,1963); thisrecordisprobablybasedonamislabelledspecimen(Liston etal.,2017).ThevirtualabsenceofSterictiphorinaefrom theAfrotropicalandOrientalregionsmayhavebeencaused byextinctionorfailuretocolonizetheseregions.Eitherway, itisnotpossibletoestablishaconnectionbetweenNeotropicalandEurasianmembersofthesubfamily.TheancestraldistributionofcladeVIIIisestimatedtobePalearctic(Figs7, 8).ThecladeisdistributedthroughouttheHolarcticandcomprisesmorethan100species,primarilyinthegenera Sterictiphora (41spp.)and Aprosthema (55spp.). Ortasiceros (6 spp.)and Yasumatsua (3spp.)areendemictotheSino-Japanese region,asare8outof10speciesof Aproceros;thespecies
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
Aprocerosleucopoda hasbeenintroducedtoEurope(Blank etal.,2014). Sterictiphora and Aprosthema reachedtheNearctic intheMiocene(Figs5,8),presumablythroughaBeringian connection.
Subsocialbehaviourandmaternalcare
Accordingtoourresults,thesubsocialbehaviouroffemales withinArgidaeandPergidaeevolvedindependently;this behaviourcannotbeascribedtothegroundplanofeither family.InPergidae,subsocialityoccursingeneraindifferent subfamilies(Perginae,Philomastiginae,PerreyinaeandSyzygoninae)representingatleasttwodifferentlineages(Schmidt etal.,2006).InArgidae,itoccursonlyincladeXII(Fig.5), correspondingtothetribeDieloceriniofBenson(1938)or subfamilyDielocerinaeofSmith(1992);thebehaviourisnot developedtothesamedegreeinallgenera.Accordingtoour phylogeny,cladeXIIincludesseveralsubcladeswithunresolvedrelationships.Maternalcarehasnotbeenrecordedfor subcladeXIIa, Neoptilia + Durgoa,althoughthelarvaeofsome speciesdisplaysignsofsubsocialbehaviour,e.g., Durgoamattogrossensis and Neoptiliatora aregregariousfeeders(Smith etal.,2013)andalsobuildsmallirregularmasscocoons(D.R. Smith,pers.observation).InsubcladeXIIb, Sericoceros shows maternalcare(Ciesla,2002),whileforitssistergroup Subsymmia thelifehistoryisunknown.InsubcladeXIIc, Themos clearlyshowsmaternalcareandsubsocialbehaviour,together withthegenera Pachylota, Dielocerus and Digelasinus,while for Topotrita and Mallerina,thebiologyisunknown(deSouza Dias,1975;deSouzaDias,1976;Smith,1992;Ciesla,2002; Smith&Janzen,2003;Boraschi etal.,2005).
In Sericoceros thematernalcarebehaviourismoreweakly developedthaninothergeneraofsubcladeXIIcthatshow thistrait(Ciesla,2002;Smith&Janzen,2003).Theless developedsubsocialbehaviourof Sericoceros (constructionof smallandirregularmasscocoons,lowlevelsofaggressiveness whenguardingtheeggsbythefemaleandshorterdurationof maternalcare)mightsuggestthatitdisplaystheinitialstepsof evolvingthisbehaviourcomplex.However,withincladeXIIc thesamecharactersdonotseemtofollowatransformation series(numberofdaysofguardinganddegreeofdefence aggressivenessbythefemale),exceptfortheconstructionof muchmorecomplexandorganizedmasscocoonsinallgenera (Fig.9).Oneprobableancestraltraitinthisbehaviourcomplex presentin Sericoceros and Themos (sistergroupoftheremaining taxaincladeXIIc)isthatfemaleslaytheeggsdirectlyon theleafepidermisandincircular/ovaleggclustersinsteadof insidethemesophylloftheleavesandscatteredovertheentire leafasin Dielocerus and Digelasinus (Benson,1938;deSouza Dias,1975;deSouzaDias,1976;Smith,1992;Ciesla,2002; Smith&Janzen,2003;Boraschi etal.,2005).Larvaeofclade XIIsharethepresenceoftwoormorenon-eversiblecoxal glands(Malagón-Aldana etal.,2021b);however,itisunknown ifthisfeatureisrelatedtogregariousorsubsocialbehaviour. Amorphologicalconvergencebetweenadultfemalesthatshow maternalcareinPergidae(e.g., Pseudoperga)andcladeXII
arethelongplantulaeonthetarsi,mainlyonthebasitarsomere (Malagón-Aldana etal.,2021a,char.59).
Conclusions
Ourstudyisthemostcomprehensivephylogeneticanalysisfor theArgidaetodate,althoughthelimitedamountoffreshmaterialandtheprevalenceofmonotypicorsmallgenerarepresented onlybytypematerialmadeitimpossibletoreconstructabetter resolvedphylogeny.Weprovidethefirstmolecularsequences ofsomegenesforthegenera Athermantus, Triarge, Pampsilota, Dielocerus, Sphacophilus, Tanymeles and Manaos,as wellassomeoutgroupspecies,e.g., Perreyiatropica (Pergidae), Andeanafarcta and Waldheimiaamazonica (Tenthredinidae). Wehopethatnewtechnologiesandcollectingeffortswillhelp toincreasetheamountofdataandevidenceforunderstanding theevolutionofthefamily.
Theplacementof Zenarge inaseparatefamilyassuggested byMalagón-Aldana etal. (2021a,2021b),asindicatedbythe relationshipZenargidae + (Argidae + Pergidae),isbettersupportedwhencombiningmolecularandmorphologicalcharactersorwhenexcludingthesaturatedthirdcodonpositioninthe molecularanalysis.Furthermore,themorphological(adultand larva)andecologicalfeatures(e.g.,gymnospermhostplant)of Zenarge clearlymakesitunique.Accordingtothecalibrated phylogenypresentedhere,themorebasalpositionof Zenarge turneri,whichcurrentlyistheonlyspeciesoftheZAPclade knowntofeedongymnosperms,mightchallengethehypothetic scenarioofanancestralangiospermhostforTenthredinoidea proposedbyNyman etal. (2019).Alternatively,thehostplant choiceof Zenarge isasecondaryreversaltofeedingongymnosperms,asindicatedbytheestimatedCenozoicageofits currentnativehostplant,seeabove.
Ourresultsagreemostlywithpreviousphylogeneticresults forthefamilyArgidaes.s.showingtwomainclades,i.e., thesubfamiliesArginaeandSterictiphorinae(Malagón-Aldana etal.,2021a,2021b),bothofthemwithancestraldistribution inGondwana,andwithanorigincoincidingwiththebreakup betweeneastandwestGondwana.ThebiogeographicreconstructionsuggestsArginaedispersedtoEurasiamostlikely throughnorthernAfrica(andtheArabianPeninsula);however, forSterictiphorinaesimilarevidenceisabsent.Weconsiderthat theriseoftheHimalayasplayedasubstantialroleinthediversificationofArginae,andperhapsSterictiphorinae,inthePalearcticregion,ashasbeensuggestedforotherorganismsinconjunctionwithupliftofmountainranges(Wen etal.,2014;Luebert &Muller,2015;Klaus etal.,2016)(Fig.9A).Regardingsubsocialbehaviour,wefindthatmaternalcareevolvedonlyonce withinSterictiphorinae;however,furtherresearchisnecessary tounderstandevolutionofspecifictraitsofthisbehaviourcomplex.
MuchmoreworkstillneedstobedonetoresolvetherelationshipsofNeotropicalSterictiphorinaeandthemegadiverse genus Arge,forwhichweonlyobtainedlowresolution.Besides, Arge inparticularneedstobesampledmuchmoredensely,and thegenuswillprobablyhavetobesubdivided,asindicatedby
ourresultsforAfrotropicalArginae.Atthesametime,anumberofsmallnorthernhemispheregeneranestedinside Arge (cladeV,Fig.4),e.g.,the‘prefix’genera Asiarge, Pseudarge and Spinarge,mighteventuallyhavetobesynonymizedwiththe nominalgenus.Thegenus Sphacophilus isretrievedasmonophyletic,contrarytoprevioushypotheses.Molecularinformationisalsoneededtoconfirmtheplacementofanumberof genera,e.g., Trichorhachus, Tanyphatnidea, Sjoestedtia, Pseudaprosthema, Yasumatsua, Ortasiceros,mostoftheAfrotropical speciesof Arge andNeotropicalSterictiphorinae,includingthe statusoftheNewWorldgenera Ptenos and Didymia currently retrievedasparaphyletic.Similarly,arevisionofthetribalclassificationofArgidaemustawaitadensersamplingthantheone achievedhere.
SupportingInformation
Additionalsupportinginformationmaybefoundonlinein theSupportingInformationsectionattheendofthearticle.
FigureS1. Substitutionsaturationpercodonposition(first andsecondcodontogetherandthirdcodonseparately)per protein-codinggene.Observedtransitionsandtransversion vs.correctedgeneticdivergence.
FigureS2. MaximumlikelihoodIndividualGeneTrees.
FigureS3. Phylogeneticpositionof Zenargeturneri Rohwer whenusingdifferentgenedatasets.Firstandsecondcodon positiontogether(first-second)andthirdcodonposition (third)separately.MaximumlikelihoodMLandBayesian inference(BI)analyses.
FigureS4. CurrentoccurrenceofOrientalgeneraofclade IV.
FileS1. Dispersalmatrix,DispersalExtinctionCladogenesis (DEC)analysis.
FileS2. TotalEvidenceMatricesforNodedating.
TableS1. Listoftaxausedformolecularinformationand GenBankaccessionnumberpergene(Sequencesstarting withOKwereproducedinthiswork).
Acknowledgements
WethankCOLCIENCIASInstitutionofColombiaandthe NaturalHistoryMuseumofDenmark(NHMD),whichfunded thefirstauthor’sPhDprogrammeattheUniversityofCopenhagen.Weareespeciallygratefultothecuratorsofthedifferententomologicalcollectionsthatprovidedspecimenstothis work:StephanBlank(SDEI),FernandoFernández(ICN-MHN), DimitriForero(MPUJ),AndrewListon(SDEI),ClaudiaPeña (ICN-MHN),AndreasTaeger(SDEI),andValentinaVergara (UNAB).ThankstoTobiasMalm,TommiNyman,andSaskia WutkewhosharedwithusalltheirownoriginaldataofArgidae,includingthe16Sand28SrDNAsequencesof Cibdela
janthina.Also,thankstoSerainaKlopfstein,TamaraSpasojevic andLipingYan,whoprovidedseveralsuggestionsandconstructivecriticismofthemaximumlikelihoodandBayesianinference methodsapplied,aswellastoKatjaKrampforherrelevant suggestionsontheDNAextractiontechniques.Finally,wethank SerainaKlopfsteinandGavinBroadwhoreviewedanearlyversionofthemanuscript.AdditionalinputwasprovidedbyBonnie Blaimerandtwoanonymousreviewers.Mentionoftradenames orcommercialproductsinthispublicationissolelyforthepurposeofprovidingspecificinformationanddoesnotimplyrecommendationorendorsementbytheUSDA.USDAisanequal opportunityproviderandemployer.Theauthorsdeclarenoconflictofinterest.
Authorcontributions
LeonardoA.Malagón-Aldana(LAMA)andLarsVilhelmsen (LV)conceivedanddesignedthestudy.DavidR.SmithandAkihikoShinoharaprovidedcrucialmaterialforDNAextraction. LAMAandArnR.Jensengeneratedthemoleculardata.LAMA conductedallanalyses.LAMAwrotethefirstdraftofthepaper, whichwasthenreviewedbyLV.Allauthorscontributedtothe finalversionofthepaper.
Dataavailabilitystatement
NCBISequenceaccessionnumbersareprovidedinTableS1. Thefinaltotalevidencematrix,includingtheusedconcatenated nucleotidealignementandthemorphologicaldatasetareprovidedinFileS2.Therestofdatathatsupportthefindingsofthis studyareavailablefromthecorrespondingauthoruponreasonablerequest.
References
Areces-Berazain,F.&Ackerman,J.D.(2017)Diversificationand fruitevolutionineumalvoids(Malvaceae). BotanicalJournalofthe LinneanSociety, 184(4),401–417.
Benson,D.A.,Cavanaugh,M.,Clark,K.,Karsch-Mizrachi,I.,Lipman, D.J.,Ostell,J.&Sayers,E.W.(2017)GenBank. NucleicAcids Research, 41 (Databaseissue)(D1),D41–D47.
Benson,R.B.(1938)Ontheclassificationofsawflies(Hymenoptera Symphyta). TransactionsoftheRoyalEntomologicalSocietyof London, 87(15),353–384.
Benson,R.B.(1963)TheaffinitiesoftheAustralianArgidae (Hymenoptera). AnnalsandMagazineofNaturalHistory, 13, 631–635.
Bibi,F.(2011)Mio-PliocenefaunalexchangesandAfricanbiogeography:therecordoffossilbovids. PLoSOne, 6(2),e16688.https://doi .org/10.1371/journal.pone.0016688.
Blank,S.M.,Köhler,T.,Pfannenstill,T. etal. (2014)Zig-zaggingacross CentralEurope:recentrangeextension,dispersalspeedandlarval hostsof Aprocerosleucopoda (Hymenoptera,Argidae)inGermany. JournalofHymenopteraResearch, 41,57–74.
Boevé,J.L.,Blank,S.M.,Meijer,G.&Nyman,T.(2013)Invertebrate andavianpredatorsasdriversofchemicaldefensivestrategiesin tenthredinidsawflies. BMCEvolutionaryBiology, 13(1),1–14.
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
Boevé,J.-L.,Nyman,T.,Shinohara,A.&Schmidt,S.(2018)Endogenoustoxinsandthecouplingofgregariousnesstoconspicuousnessin ArgidaeandPergidaesawflies. ScientificReports, 8(1),1–11.
Boraschi,D.,Peruquetti,R.C.&DelLama,M.A.(2005)Biologia, comportamentosocialealocaçãosexualde Digelasinusdiversipes (Kirby,1882)(Hymenoptera,Argidae). RevistaBrasileiradeEntomologia, 49(2),253–263.
Breinholt,J.W.&Kawahara,A.Y.(2013)Phylotranscriptomics:saturatedthirdcodonpositionsradicallyinfluencetheestimationof treesbasedonnext-gendata. GenomeBiologyandEvolution, 5(11), 2082–2092.
Brown,J.H.&Lomolino,M.V.(1998) Biogeography,2ndedn,p.691. SinauerAssociate;distributedintheUKbyMacmillan,Sunderland, Massachusetts.
Chatterjee,S.&Bajpai,S.(2016)India’snorthwarddriftfromGondwanatoAsiaduringtheLateCretaceous-Eocene. Proceedingsofthe IndianNationalScienceAcademy, 82(3),479–487.
Chatterjee,S.,Christopher,R.S.&Sunil,B.(2017) IndianPlateand ItsEpicVoyagefromGondwanatoAsia:ItsTectonic,Paleoclimatic, andPaleobiogeographicEvolution,Vol. 529.GeologicalSocietyof America,Boulder,Colorado.https://doi.org/10.1130/SPE529
Chernomor,O.,vonHaeseler,A.&Minh,B.Q.(2016)Terraceaware datastructureforphylogenomicinferencefromsupermatrices. SystematicBiology, 65(6),997–1008.
Ciesla,W.M.(2002)Observationsonthelifehistoryandhabitsof atropicalsawfly, Sericocerosmexicanus (Kirby)(Hymenoptera: Argidae)onRoatánIsland,Honduras. TheForestryChronicle, 78(4), 515–521.
deSouzaDias,B.F.(1975)Comportamentopre-socialdesinfitasdo BrasilCentral.I. Themosolfersii (Klug)(Hymenoptera:Argidae). Studiaentomologica, 18,401–432.
deSouzaDias,B.F.(1976)Comportamentopre-socialdesinfitas doBrasilcentral.II. Dielocerusdiasi Smith,1975(hymenoptera: Argidae). Studiaentomologica, 19,461–501.
Eiseman,C.S.(2015)Onthedistinctivefeedingpatternof Sterictiphora Billberg(Hymenoptera:Argidae)sawflylarvae. Proceedingsofthe EntomologicalSocietyofWashington, 117(1),65–67.
Endara,M.J.,Nicholls,J.A.,Coley,P.D. etal. (2018)Trackingof hostdefensesandphylogenyduringtheradiationofneotropical Inga-feedingsawflies(Hymenoptera;Argidae). FrontiersinPlant Science, 9,1237.
Farrell,B.D.(2001)Evolutionaryassemblyofthemilkweedfauna: cytochromeoxidaseIandtheageof Tetraopes beetles. Molecular PhylogeneticsandEvolution, 18(3),467–478.
Florens,F.B.V.,Bissessur,P.,Bunsy,Y.&Ramdonee,D.(2017) Cibdela janthina (Klug1834)(Hymenoptera:Tenthredinidae),Réunion’s biocontrolagentof Rubusalceifolius Poir.,recordedonMauritius. AfricanEntomology, 25(1),271–274. Fu,Q.,Diez,J.B.,Pole,M. etal. (2018)Anunexpectednoncarpellate epigynousflowerfromtheJurassicofChina. eLife, 7,1–24. Gao,T.,Ren,D.&Shih,C.(2009)ThefirstXyelotomidae (Hymenoptera)fromthemiddleJurassicinChina. Annalsof theEntomologicalSocietyofAmerica, 102(4),588–596.
Haack,R.A.&Mattson,W.J.(1993)LifehistorypatternsofNorth Americantree-feedingsawflies. SawflyLifeHistoryAdaptationsto WoodyPlants.NorthAmericaTree-FeedingSawflies (ed.byM.R. WagnerandK.F.Raffa),pp.504–545.AcademicPress,Inc.,San Diego,CA.
Hall,R.&Sevastjanova,I.(2012)AustraliancrustinIndonesia. AustralianJournalofEarthSciences, 59(6),827–844. Hara,H.&Shinohara,A.(2018)Taxonomicnotesandnewdistribution andhostplantrecordsforsawfliesandwoodwasps(Hymenoptera, Symphyta)ofJapanIV. BulletinoftheNationalMuseumofNature andScience,Tokyo,SeriesA, 44,93–113.
Heraty,J.,Ronquist,F.,Carpenter,J.M. etal. (2011)Evolutionofthe hymenopteranmegaradiation. MolecularPhylogeneticsandEvolution, 60(1),73–88.
Herendeen,P.S.,Friis,E.M.,Pedersen,K.R.&Crane,P.R.(2017) Palaeobotanicalredux:revisitingtheageoftheangiosperms. Nature Plants, 3(3),1–8.
Hernandez-Gutierrez,R.&Magallon,S.(2019)ThetimingofMalvales evolution:incorporatingitsextensivefossilrecordtoinformabout lineagediversification. MolecularPhylogeneticsandEvolution, 140, 106606.https://doi.org/10.1016/j.ympev.2019.106606.
Hochuli,P.A.,&Feist-Burkhardt,S.(2013)Angiosperm-likepollenand AfropollisfromtheMiddleTriassic(Anisian)oftheGermanicBasin (NorthernSwitzerland). FrontiersinPlantScience, 4,344.https://doi .org/10.3389/fpls.2013.00344
Holt,B.G.,Lessard,J.P.,Borregaard,M.K. etal. (2013)Anupdateof Wallace’szoogeographicregionsoftheworld. Science, 339(6115), 74–78.
Hopkins,D.M.(1959)CenozoichistoryoftheBeringlandbridge. Science, 129(3362),1519–1528.
Houston,T.F.(1989) Leioproctus beesassociatedwithWesternAustraliansmokebushes(Conospermum spp.)andtheiradaptationsfor foragingandconcealment(Hymenoptera:Colletidae:Paracolletini). RecordsoftheWesternAustralianMuseum, 14,275–292.
Isaka,Y.&Sato,T.(2014)Molecularphylogeneticanddivergence timeestimationanalysesofthesawflysubfamilySelandriinae (Hymenoptera:Tenthredinidae). EntomologicalScience, 17(4), 435–439.
Johanson,K.A.&Malm,T.(2010)TestingthemonophylyofCalocidae (Insecta:Trichoptera)basedonmultiplemoleculardata. Molecular PhylogeneticsandEvolution, 54(2),535–541.
Källersjö,M.,Albert,V.A.&Farris,J.S.(1999)Homoplasyincreases phylogeneticstructure. Cladistics, 15,91–93.
Kalyaanamoorthy,S.,Minh,B.Q.,Wong,T.K.,vonHaeseler,A.& Jermiin,L.S.(2017)ModelFinder:fastmodelselectionforaccurate phylogeneticestimates. NatureMethods, 14(6),587–589.
Katoh,K.,Rozewicki,J.&Yamada,K.D.(2019)MAFFTonline service:multiplesequencealignment,interactivesequencechoiceand visualization. BriefingsinBioinformatics, 20(4),1160–1166.
Kimsey,L.S.&Smith,D.R.(1985)Twonewspecies,larvaldescriptions andlifehistorynotesofsomePanamaniansawflies(Hymenoptera: Argidae,Tenthredinidae). ProceedingsoftheEntomologicalSociety ofWashington, 87,191–201.
Kirby,W.F.(1882)ListofHymenopterawithDescriptionsandFigures oftheTypicalSpecimensintheBritishMuseum. Tenthredinidaeand Siricidae,Vol.1,pp.1–450.OrderoftheTrustees,London.
Klaus,S.,Morley,R.J.,Plath,M.,Zhang,Y.P.&Li,J.T.(2016)Biotic interchangebetweentheIndiansubcontinentandmainlandAsia throughtime. NatureCommunications, 7(1),12132.
Klopfstein,S.&Ronquist,F.(2013)Convergentintrongainsin hymenopteranelongationfactor-1α MolecularPhylogeneticsand Evolution, 67(1),266–276.
Klopfstein,S.,Vilhelmsen,L.,Heraty,J.M.,Sharkey,M.&Ronquist,F. (2013)Thehymenopterantreeoflife:evidencefromprotein-coding genesandobjectivelyalignedribosomaldata. PLoSOne, 8(8), e69344.
Koch,F.,Goergen,G.&Noort,S.(2015)ThesawfliesofNamibia andwesternSouthAfrica(Symphyta,Hymenoptera). ABCtaxa The JournalDedicatedtoCapacityBuildinginTaxonomyandCollection Management , 15,1–262.
Lamont,B.B.&He,T.(2012)Fire-adaptedGondwananangiosperm florasevolvedintheCretaceous. BMCEvolutionaryBiology, 12(1), 223.
Lanfear,R.,Calcott,B.,Ho,S.Y.&Guindon,S.(2012)PartitionFinder: combinedselectionofpartitioningschemesandsubstitutionmodels
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
L.A.Malagon-Aldana etal. forphylogeneticanalyses. MolecularBiologyandEvolution, 29(6), 1695–1701.
Lanfear,R.,Frandsen,P.B.,Wright,A.M.,Senfeld,T.&Calcott, B.(2017)PartitionFinder2:newmethodsforselectingpartitioned modelsofevolutionformolecularandmorphologicalphylogenetic analyses. MolecularBiologyandEvolution, 34(3),772–773.
Larkin,M.A.,Blackshields,G.,Brown,N.P. etal. (2007)ClustalWand ClustalXversion2.0. Bioinformatics, 23(21),2947–2948.
Larter,M.,Pfautsch,S.,Domec,J.C.,Trueba,S.,Nagalingum,N.& Delzon,S.(2017)Ariditydrovetheevolutionofextremeembolism resistanceandtheradiationofconifergenus Callitris NewPhytologist , 215(1),97–112.
Leppänen,S.A.,Altenhofer,E.,Liston,A.D.&Nyman,T.(2012)Phylogeneticsandevolutionofhost-plantuseinleaf-miningsawflies (Hymenoptera:Tenthredinidae:Heterarthrinae). MolecularPhylogeneticsandEvolution, 64(2),331–341.
Letunic,I.&Bork,P.(2019)Interactivetreeoflife(iTOL)v4:recent updatesandnewdevelopments. NucleicAcidsResearch, 47(W1), W256–W259.
Linder,H.P.(2014)TheevolutionofAfricanplantdiversity. Frontiers inEcologyandEvolution, 2,1–14.
Linnen,C.R.&Farrell,B.D.(2008)Comparisonofmethodsfor species-treeinferenceinthesawflygenus Neodiprion (Hymenoptera: Diprionidae). SystematicBiology, 57(6),876–890.
Liston,A.D.,Goergen,G.&Koch,F.(2017)Revisionsofthe AfrotropicalgeneraofArgidaeandspeciesof Pampsilota Konow, 1899(Hymenoptera,Tenthredinoidea). DeutscheEntomologische Zeitschrift , 64(1),1–25.
Liston,A.D.,Kan,P.&Kan-VanLimburgStirum,B.(2018)The life-historyof Aprosthematardum (Klug,1814)(Hymenoptera,Tenthredinoidea,Argidae). ContributionstoEntomology:Beiträgezur Entomologie, 68(1),97–106.
Liston,A.D.,Savina,H.,Nagy,Z.T.,Sonet,G.&Boeve,J.L.(2014) Taxonomy,phylogenyandhostplantsofsome Abia sawflies (Hymenoptera,Cimbicidae). Zootaxa, 3821(1),125–132. Luebert,F.&Muller,L.A.(2015)Effectsofmountainformationand upliftonbiologicaldiversity. FrontiersinGenetics, 6,1–2. Maddison,W.P.,&Maddison,D.R.(2019) Mesquite:AModular SystemforEvolutionaryAnalysis.Version3.6 2018.http://www .mesquiteproject.org
Malagón-Aldana,L.A.,Shinohara,A.,Smith,D.R.&Vilhelmsen,L. (2021b)Larvalanatomyofargidsawflies(Hymenoptera:Argidae): aphylogeneticapproach. OrganismsDiversity&Evolution, 1-31(2), 361–392.https://doi.org/10.1007/s13127-021-00485-0.
Malagón-Aldana,L.A.,Smith,D.R.,Shinohara,A.&Vilhelmsen,L. (2021a)From Arge to Zenarge:adultmorphologyandphylogenetics ofargidsawflies(Hymenoptera:Argidae). ZoologicalJournalofthe LinneanSociety, 193,880–938.https://doi.org/10.1093/zoolinnean/ zlaa170.
Malagon-Aldana,L.A.,Smith,D.R.,Vilhelmsen,L.&Serna,F.(2019) Thesawfliesandwoodwasps(Hymenoptera:‘Symphyta’)ofColombia. Zootaxa, 4633(1),1–88.
Malm,T.&Nyman,T.(2015)Phylogenyofthesymphytangradeof Hymenoptera:newpiecesintotheoldjigsaw(fly)puzzle. Cladistics, 31(1),1–17.
Matzke,N.J.(2013a)Probabilistichistoricalbiogeography:newmodels forfounder-eventspeciation,imperfectdetection,andfossilsallow improvedaccuracyandmodel-testing. FrontiersofBiogeography, 5(4),242–248.
Matzke,N.J.(2013b) BioGeoBEARS:BioGeographywithBayesian (andLikelihood)EvolutionaryAnalysisinRScripts.Rpackage, version 0.2.1,publishedJuly27,2013URLhttp://CRAN.R-project .org/package=BioGeoBEARS
Matzke,N.J.(2014)Modelselectioninhistoricalbiogeographyreveals thatfounder-eventspeciationisacrucialprocessinIslandclades. SystematicBiology, 63(6),951–970.
Miller,M.A.,Pfeiffer,W.&Schwartz,T.(2012)TheCIPRESscience gateway:enablinghigh-impactscienceforphylogeneticsresearchers withlimitedresources. Proceedingsofthe1stConferenceofthe ExtremeScienceandEngineeringDiscoveryEnvironment:Bridging fromtheExtremetotheCampusandbeyond ,pp.1–8.Associationfor ComputingMachinery,NewYork. Montes,C.,Cardona,A.,Jaramillo,C. etal. (2015)MiddleMiocene closureofthecentralAmericanseaway. Science, 348(6231), 226–229.
Moore,K.M.(1963a)ObservationsonsomeAustralianforestinsects. 12.Thetaxonomyof Zenargeturneri Rohwer(1918)(Hymenoptera: Argidae),thecypresspinesawfly. ProceedingsoftheLinneanSociety ofNSW , 87,116–124.
Moore,K.M.(1963b)ObservationsonsomeAustralianforestinsects. 13.Acomparisonofthebiologyofthecypresspinesawflysubspecies. ProceedingsoftheLinneanSocietyofNSW , 87,125–136. Morrone,J.J.(2015)Biogeographicalregionalizationoftheworld:a reappraisal. AustralianSystematicBotany, 28(3),81–90.
Muellner-Riehl,A.N.,Weeks,A.,Clayton,J.W. etal. (2016)Molecular phylogeneticsandmolecularclockdatingofSapindalesbasedon plastidrbcL,atpBandtrnL-trnFDNAsequences. Taxon, 65(5), 1019–1036.
Müller,R.D.,Zahirovic,S.,Williams,S.E. etal. (2019)Aglobal platemodelincludinglithosphericdeformationalongmajorriftsand orogenssincetheTriassic. Tectonics, 38(6),1884–1907. Nel,A.(2004)NewandpoorlyknownCenozoicsawfliesofFrance (Hymenoptera,Tenthredinoidea,Pamphilioidea). DeutscheEntomologischeZeitschrift , 2,253–269.
Nguyen,L.T.,Schmidt,H.A.,vonHaeseler,A.&Minh,B.Q.(2015) IQ-TREE:afastandeffectivestochasticalgorithmforestimating maximum-likelihoodphylogenies. MolecularBiologyandEvolution, 32(1),268–274.
Normark,B.B.,Jordal,B.H.&Farrell,B.D.(1999)Originofa haplodiploidbeetlelineage. ProceedingsoftheRoyalSocietyof London.SeriesB:BiologicalSciences, 266(1435),2253–2259. Notredame,C.,Higgins,D.G.&Heringa,J.(2000)T-coffee:anovel methodforfastandaccuratemultiplesequencealignment. Journalof MolecularBiology, 302(1),205–217.
Nyman,T.,Onstein,R.E.,Silvestro,D. etal. (2019)Theearlywasp pluckstheflower:disparateextantdiversityofsawflysuperfamilies (Hymenoptera:“Symphyta”)mayreflectasynchronousswitchingto angiospermhosts. BiologicalJournaloftheLinneanSociety, 128(1), 1–19.
Nyman,T.,Zinovjev,A.G.,Vikberg,V.&Farrell,B.D.(2006)Molecular phylogenyofthesawflysubfamilyNematinae(Hymenoptera:Tenthredinidae). SystematicEntomology, 31(4),569–583. Paleobiologydatabase(Website).Fossilworks.org.[accessedonAugust 192020].
Pasteels,J.(1963)Prodromed’unefaunedesTenthredinoideade l’Afriquenoire.IV.-2esupplementauxArgidae. Bulletinetannales delaSociétéRoyaled’EntomologiedeBelgique, 99,540–560. Pittermann,J.,Stuart,S.A.,Dawson,T.E.&Moreau,A.(2012)Cenozoicclimatechangeshapedtheevolutionaryecophysiologyofthe Cupressaceaeconifers. ProceedingsoftheNationalAcademyofSciences, 109(24),9647–9652.
Portik,D.M.&Papenfuss,T.J.(2015)Historicalbiogeography resolvestheoriginsofendemicArabiantoadlineages(Anura: Bufonidae):evidenceforancientvicarianceanddispersalevents withtheHornofAfricaandSouthAsia. BMCEvolutionaryBiology, 15(1),152.
©2021TheRoyalEntomologicalSociety, SystematicEntomology,doi:10.1111/syen.12527
Prous,M.,Liston,A.,Kramp,K.,Savina,H.,Vårdal,H.&Taeger, A.(2019)ThewestPalaearcticgeneraofNematinae(Hymenoptera, Tenthredinidae). ZooKeys, 875,63–127.
Ratnasingham,S.&Hebert,P.D.(2007)BOLD:thebarcodeoflifedata system(http://www.barcodinglife.org). MolecularEcologyNotes, 7(3),355–364.
Ree,R.H.&Smith,S.A.(2008)Maximumlikelihoodinferenceofgeographicrangeevolutionbydispersal,localextinction,andcladogenesis. SystematicBiology, 57(1),4–14.
Regier,J.C.,Shultz,J.W.,Ganley,A.R.D. etal. (2008)Resolving arthropodphylogeny:exploringphylogeneticsignalwithin41kb ofprotein-codingnucleargenesequence. SystematicBiology, 57(6), 920–938.
Reinert,J.F.(1970)Zoogeographyof Aedes (Diceromyia)Theobald (Diptera:Culicidae). JournaloftheentomologicalSocietyofSouth Africa, 33,129–141.
Rohwer,S.A.,Smith,M.,Rohwer,G.&Cockerell,W.P.(1908)Tertiary Tenthredinoideaoftheexpeditionof1908toFlorissant,Colorado. BulletinoftheAmericanMuseumofNaturalHistory, 24,591–595.
Ronquist,F.&Huelsenbeck,J.P.(2003)MrBayes3:Bayesianphylogeneticinferenceundermixedmodels. Bioinformatics, 19(12), 1572–1574.
Ronquist,F.,Klopfstein,S.,Vilhelmsen,L.,Schulmeister,S.,Murray, D.L.&Rasnitsyn,A.P.(2012a)Atotal-evidenceapproachtodating withfossils,appliedtotheearlyradiationoftheHymenoptera. SystematicBiology, 61(6),973–999.
Ronquist,F.,Teslenko,M.,vanderMark,P. etal. (2012b)MrBayes3.2: efficientBayesianphylogeneticinferenceandmodelchoiceacrossa largemodelspace. SystematicBiology, 61(3),539–542.
Saini,M.S.(2009)FamiliesArgidae,Cimbicidae,Diprionidae,Pamphiliidae,Siricidae,Xiphydriidae,Orussidae. Indiansawfliesbiodiversity.Keys,Catalogue&Illustrations,Vol. 6 (ed.byB.Singhand M.P.Singh),pp.1–168.BishenSinghMahendraPalSinghBSMPS, DehraDun.
Salemi,M.(2009)Geneticdistancesandnucleotidesubstitution models–practice. ThePhylogeneticHandbook:APracticalApproach toPhylogeneticAnalysisandHypothesisTesting (ed.byP.Lemey, M.SalemiandA.M.Vandamme),pp.126–141.CambridgeUniversityPress,NewYork.
SanMartin,I.,Enghoff,H.&Ronquist,F.(2001)Patternsofanimal dispersal,vicarianceanddiversificationintheHolarctic. Biological JournaloftheLinneanSocietyofLondon, 73(4),345–390. SanMartin,I.&Ronquist,F.(2004)Southernhemispherebiogeography inferredbyevent-basedmodels:plantversusanimalpatterns. SystematicBiology, 53(2),216–243.
Schiff,N.(2004) Pseudopergaguerinii (Hymenoptera:Pergidae).A femaleprotectingheryoungonaeucalyptleaf[photographof featuredinsect]. AmericanEntomologiste, 50(3),129.
Schmidt,S.,Taeger,A.,Morinière,J. etal. (2017)Identificationof sawfliesandhorntails(Hymenoptera,‘Symphyta’)throughDNA barcodes:successesandcaveats. MolecularEcologyResources, 17(4),670–685.
Schmidt,S.&Walter,G.H.(2014)Youngcladesinanoldfamily:major evolutionarytransitionsanddiversificationoftheeucalypt-feeding pergidsawfliesinAustralia(insect,Hymenoptera,Pergidae). MolecularPhylogeneticsandEvolution, 74,11–121.
Schmidt,S.,Walter,G.H.,Grigg,J.&Moore,C.J.(2006)SexualcommunicationandhostplantassociationsofAustralianpergidsawflies (Hymenoptera:Symphyta:Pergidae). RecentSawflyResearch:SynthesisandProspects (ed.byS.M.Blank,S.SchmidtandA.Taeger), pp.173–193.Goecke&Evers,Keltern.
Schmidt,S.,Walter,G.H.&Moore,C.J.(2000)Hostplantrelationships inmyrtaceous-feedingpergidsawflies:essentialoilsandthemorphologyandbehaviourof Pergagrapta larvae(Hymenoptera,Symphyta,
Pergidae). BiologicalJournaloftheLinneanSocietyofLondon, 70, 15–26.
Schulmeister,S.(2003a)Reviewofmorphologicalevidenceonthe phylogenyofbasalHymenoptera(Insecta),withadiscussionofthe orderingofcharacters. BiologicalJournaloftheLinneanSociety, 79(2),209–243.
Schulmeister,S.(2003b)SimultaneousanalysisofbasalHymenoptera (Insecta):introducingrobust-choicesensitivityanalysis. Biological JournaloftheLinneanSociety, 79(2),245–275.
Schulmeister,S.,Wheeler,W.C.&Carpenter,J.M.(2002)Simultaneous analysisofthebasallineagesofHymenoptera(Insecta)usingsensitivityanalysis. Cladistics, 18(5),455–484.
Schwarz,G.(1978)Estimatingthedimensionofamodel. Annalsof Statistics, 6,461–464.
Shinohara,A.(1986)AnewapteroussawflyfromSulawesi,Indonesia (Hymenoptera:Pergidae:Perreyiinae),andthepleuraloriginofthe ventralregionofthesawflymesothorax. SystematicEntomology, 11(2),247–253.
Shinohara,A.,Kiyoshi,T.&Kameda,Y.(2016)Newdistribution recordsoftwospeciesof Arge and Spinarge (Hymenoptera,Argidae) fromKyushu,Japan. BulletinoftheNationalMuseumofNatureand Science,Tokyo,SeriesA, 42,81–85.
Silvestro,D.,Cascales-Miñana,B.,Bacon,C.D.&Antonelli,A. (2015)Revisitingtheoriginanddiversificationofvascularplants throughacomprehensiveBayesiananalysisofthefossilrecord. New Phytologist , 207(2),425–436.
Smith,D.R.(1970)Nearcticspeciesofthegenus Ptenus Kirby (Hymenoptera:Argidae). TransactionsoftheAmericanEntomologicalSociety, 96,79–100.
Smith,D.R.(1989)Thesawflygenus Arge (Hymenoptera:Argidae)in thewesternhemisphere. TransactionsoftheAmericanEntomological Society, 115,83–205.
Smith,D.R.(1990)Asynopsisofthesawflies(Hymenoptera:Symphyta)ofAmericasouthoftheUnitedStates:Pergidae. Revista BrasileiradeEntomologia, 34,7–200.
Smith,D.R.(1992)Asynopsisofthesawflies(Hymenoptera:Symphyta)ofAmericasouthoftheUnitedStates:Argidae. Memoirsof theAmericanEntomologicalSocietyNo., 39,201.
Smith,D.R.&Janzen,D.H.(2003)Foodplantsandlifehistoriesof sawfliesofthefamilyArgidae(Hymenoptera)inCostaRica,with descriptionsoftwonewspecies. JournalofHymenopteraResearch, 12,193–208.
Smith,D.R.,Janzen,D.H.&Hallwachs,W.(2013)Foodplantsand lifehistoriesofsawfliesofthefamiliesArgidaeandTenthredinidae (Hymenoptera)inCostaRica,asupplement. JournalofHymenoptera Research, 35,17–31.
Smith,D.R.&Poinar,G.O.(1992)Sawflies(Hymenoptera:Argidae) fromDominicanamber. EntomologicalNews, 103,117–124.
Smith,D.R.,Wheeler,G.S.,Sánchez-Restrepo,A.F.,McKay,F., Guala,M.&Coria,C.(2019)Threespeciesof Heteroperreyia (Hymenoptera:Pergidae)feedingonBrazilianpeppertrees, Schinus spp.(Anacardiaceae),includinganewspecies. Proceedingsofthe EntomologicalSocietyofWashington, 12(4),704–719.
Soltis,D.,Bell,C.,Kim,S.&Soltis,P.(2008)Originandearlyevolution ofangiosperms. AnnalsoftheNewYorkAcademyofSciences, 1133(1),3–25.
Stecher,G.,Tamura,K.&Kumar,S.(2020)Molecularevolutionary geneticsanalysis(MEGA)formacOS. MolecularBiologyandEvolution, 37(4),1237–1239.
Strimmer,K.&vonHaeseler,A.(2009)Geneticdistancesand nucleotidesubstitutionmodels. ThePhylogeneticHandbook:APracticalApproachtoPhylogeneticAnalysisandHypothesisTesting (ed. byP.Lamey,M.SalemiandA.-M.Vandamme),pp.111–141.CambridgeUniversityPress,NewYork.
L.A.Malagon-Aldana etal.
Sun,G.,Dilcher,D.L.,Zheng,S.&Zhou,Z.(1998)Insearchofthefirst flower:aJurassicangiosperm, Archaefructus,fromNortheastChina. Science, 282(5394),1692–1695.
Taeger,A.,Blank,S.M.&Liston,A.D.(2010)Worldcatalogof Symphyta(Hymenoptera). Zootaxa, 2580(1),1–1064.
Taeger,A.,Liston,A.D.,Prous,M.,Groll,E.K.,Gehroldt,T.&Blank S.M.(2018) ECatSym–ElectronicWorldCatalogofSymphyta (Insecta, Hymenoptera).Programversion 5.0(19Dec2018),data version40(23Sep2018).–SenckenbergDeutschesEntomologisches Institut(SDEI),Müncheberg.URLhttps://sdei.de/ecatsym/[accessed on4June2020].
Togashi,I.(1975)Onthespeciesofthegenus Pampsilota Konowin Thailand(Hymenoptera,Argidae). Mushi., 48,87–93. Togashi,I.(1980)NotesonTaiwanesesawflies.I.Hymenoptera, Tenthredinidae. Kontyû, 48,30–34.
Trifinopoulos,J.,Nguyen,L.T.,vonHaeseler,A.&Minh,B.Q.(2016) W-IQ-TREE:afastonlinephylogenetictoolformaximumlikelihood analysis. NucleicAcidsResearch, 44(W1),W232–W235. Vaidya,G.,Lohman,D.J.&Meier,R.(2011)SequenceMatrix: concatenationsoftwareforthefastassemblyofmulti-gene datasetswithcharactersetandcodoninformation. Cladistics, 27(2),171–180.
vanNoort,S.(2020) WaspWeb:HymenopteraoftheAfrotropicalRegion URLwww.waspweb.org[accessedon25November2019].
Vilhelmsen,L.(1997)ThephylogenyoflowerHymenoptera(Insecta), withasummaryoftheearlyevolutionaryhistoryoftheorder. JournalofZoologicalSystematicsandEvolutionaryResearch, 35(2), 49–70.
Vilhelmsen,L.(2001)Phylogenyandclassificationoftheextantbasal lineagesoftheHymenoptera(Insecta). ZoologicalJournalofthe LinneanSociety, 131(4),393–442.
Vilhelmsen,L.&Turrisi,G.F.(2011)Perarboremadastra:morphologicaladaptationstoexploitingthewoodyhabitatintheearlyevolutionofHymenoptera. ArthropodStructure&Development , 40(1), 2–20.
Wei,M.(1997a)TaxonomicalstudiesonArgidae(Hymenoptera)of ChinaIV.RevisionofTanyphatnideinifromChinawithdescriptions
oftwonewspecies. Entomotaxonomia 19 [supplement]:35–42.[In Chinese,abstractinEnglish]
Wei,M.(1997b)TaxonomicalstudiesonArgidae(Hymenoptera)of China–anewgenusandfivenewspeciesofSterictiphorinae. Entomologiasinica, 4(4),295–305.
Wei,M.,Nie,H.&Taeger,A.(2006)Sawflies(Hymenoptera:Symphyta)ofChina–checklistandreviewofresearch. RecentSawfly Research:SynthesisandProspects,Vol. 16 (ed.byS.Blank, S.SchmidtandA.Taeger),pp.505–574.Goecke&Evers,Keltern. Wen,J.,Zhang,J.,Nie,Z.L.,Zhong,Y.&Sun,H.(2014)Evolutionary diversificationsofplantsontheQinghai-Tibetanplateau. Frontiersin Genetics, 5,4.
Wilford,G.E.&Brown,P.J.(1994)MapsoflateMesozoic-Cenozoic Gondwanabreak-up:somepalaeogeographicalimplications. History oftheAustralianVegetation:CretaceoustoRecent (ed.byR.S.Hill), pp.5–13.UniversityofAdelaidePress,SouthAustralia.
Xia,X.&Xie,Z.(2001)DAMBE:softwarepackagefordataanalysis inmolecularbiologyandevolution. JournalofHeredity, 92(4), 371–373.
Xia,X.,Xie,Z.,Salemi,M.,Chen,L.&Wang,Y.(2003)Anindexof substitutionsaturationanditsapplication. MolecularPhylogenetics andEvolution, 26(1),1–7.
Yan,Y.,Wei,M.&He,Y.(2009)Anewspeciesofthegenus Arge SchrankfromChina(Hymenoptera,Argidae)withakeytospeciesof Argexanthogaster groupfromEastAsia. ActaZootaxonomicaSinica, 34,58–61.
Yu,Y.,Harris,A.J.,Blair,C.&He,X.(2015)RASP(reconstruct ancestralstateinphylogenies):atoolforhistoricalbiogeography. MolecularPhylogeneticsandEvolution, 87,46–49.
Zhang,C.(2017) MolecularclockdatingusingMrBayes arXiv:1603.05707[q-bio.PE],1-19.
Zhelochovtzev,A.N.&Rasnitsyn,A.P.(1972)Onsometertiarysawflies (Hymenoptera,Symphyta)fromColorado. Psyche:AJournalof Entomology, 79(4),315–327.
Accepted8October2021
First published online 5 November 2021
Blasticotoma filicetiGB
Runaria reductaGB
Waldheimia amazonica
Abia sppGB
Pterygophorus insignisGB
Dracogymnia fernandezi
Sericoceros nrtannuusGB
Styracotechys dicelysmaGB
Sphacophilus sp1
Dielocerus sp
Dielocerus larva
Ptenos texanus
Aprosthema tardum
Aprosthema intermedium
Aprosthema fusicorne
Sterictiphorinae males
Sterectiphorinae males
Sphacophilus sp3
Sterictiphora geminata
Atomacera pubicornis
Aproceros leucopoda
Cladius pectinicornisGB
Corynis crassicornisGB
Siobla sppGB
Sterictiphora angelicae
Phylacteophaga froggattiGB
Pergagrapta sppGB
Pseudoperga sppGB
Aulacomerus atriventris
Andeana farcta
Monoctenus juniperiGB
Neodiprion sertiferGB
Polyclonus atratusGB
Decameria sppGB
Perreyia picea
Perreyia tropica
Triarge sp
Arge berberidisGB
Arge berberidis
Arge suzuki
Cibdela janthina
Arge melanochra
Arge pagana
Athermantus imperialis
Arge thoracica
Arge gracilicornis
Arge rejecta
Arge rustica
Arge obesa
Scobina bolivariGB
Scobina strophosaGB
Scobina thoracicaGB
Argemali
Pampsilota dahomeyanus
Antargidium allucenteGB
Antargidium atricepsGB
Spinarge metallica
Spinarge fulvicornisP
Spinarge fulvicornisA
Spinargeprunivora
Spinarge affinis
Arge ustulata
Arge nigripes
Arge aruncus
Arge similis
Arge similislarva
Arge similis2