TotalChemicalSynthesisofProteins
Editedby AshrafBrik,PhilipDawson,andLeiLiu
Editors
AshrafBrik
Technion-IsraelInst.ofTechnology
SchulichFacultyofChemistry TechnionCity 32000Haifa
Israel
PhilipDawson DepartmentofChemistry
TheScrippsResearchInstitute 10550N.TorreyPinesRoad UnitedStates
LeiLiu
TsinghuaUniversity DepartmentofChemistry HetianBuilding 100084Beijing China
AllbookspublishedbyWiley-VCHarecarefully produced.Nevertheless,authors,editors,and publisherdonotwarranttheinformationcontained inthesebooks,includingthisbook,tobefreeof errors.Readersareadvisedtokeepinmindthat statements,data,illustrations,proceduraldetailsor otheritemsmayinadvertentlybeinaccurate.
LibraryofCongressCardNo.: appliedfor
BritishLibraryCataloguing-in-PublicationData: Acataloguerecordforthisbookisavailablefromthe BritishLibrary.
BibliographicinformationpublishedbytheDeutsche Nationalbibliothek
TheDeutscheNationalbibliothekliststhis publicationintheDeutscheNationalbibliografie; detailedbibliographicdataareavailableonthe Internetat<http://dnb.d-nb.de>.
©2021WILEY-VCHGmbH,Boschstr.12,69469 Weinheim,Germany
Allrightsreserved(includingthoseoftranslation intootherlanguages).Nopartofthisbookmaybe reproducedinanyform–byphotoprinting, microfilm,oranyothermeans–nortransmittedor translatedintoamachinelanguagewithoutwritten permissionfromthepublishers.Registerednames, trademarks,etc.usedinthisbook,evenwhennot specificallymarkedassuch,arenottobeconsidered unprotectedbylaw.
PrintISBN: 978-3-527-34660-8
ePDFISBN: 978-3-527-82357-4
ePubISBN: 978-3-527-82358-1
oBookISBN: 978-3-527-82356-7
Typesetting SPiGlobal,Chennai,India
Printedonacid-freepaper 10987654321
Contents
Preface xvii
1CharacterizationofProteinMoleculesPreparedbyTotal ChemicalSynthesis 1
StephenB.H.Kent
1.1Introduction 1
1.2ChemicalProteinSynthesis 2
1.3CommentsonCharacterizationofSyntheticProteinMolecules 8
1.3.1Homogeneity 8
1.3.2AminoAcidSequence 9
1.3.3ChemicalAnalogues 10
1.3.4LimitationsofSPPS 10
1.3.5FoldingasaPurificationStep 10
1.4Summary 12 References 12
2AutomatedFastFlowPeptideSynthesis 17
MarkD.Simon,AlexanderJ.Mijalis,KyleA.Totaro,DanielDunkelmann, AlexanderA.Vinogradov,ChiZhang,YutaMaki,JustinM.Wolfe,Jessica Wilson,AndreiLoas,andBradleyL.Pentelute
2.1Introduction 17
2.2Results 19
2.2.1Summary 19
2.2.1.1MechanicalPrinciples 20
2.2.1.2ChemicalPrinciples 20
2.2.1.3UserInterfacePrinciples 20
2.2.1.4DataAnalysisMethod 20
2.2.1.5Outcome 21
2.2.2First-generationAutomatedFastFlowPeptideSynthesis 21
2.2.2.1KeyFindings 21
2.2.2.2DesignofFirst-generationAFPS 21
2.2.2.3CharacterizationofFirst-generationAFPS 23
2.2.3Second-generationAutomatedFastFlowPeptideSynthesis 24
2.2.3.1KeyFindings 24
2.2.3.2DesignofSecond-generationAFPS 24
2.2.3.3CharacterizationandUseofSecond-generationAFPS 26
2.2.4Third-generationAutomatedFastFlowPeptideSynthesis 32
2.2.4.1KeyFindings 32
2.2.4.2DesignofThird-generationAFPS 34
2.2.4.3CharacterizationofThird-generationAFPS 39
2.2.4.4ReagentStabilityStudy 43
2.2.5Fourth-generationAutomatedFastFlowPeptideSynthesis 45
2.2.5.1KeyFindings 45
2.2.5.2EffectofSolventonFastFlowSynthesis 45
2.2.5.3DesignandCharacterizationofFourth-generationAFPS 45
2.3Conclusions 49 Acknowledgments 53 References 53
3 N,S -and N,Se-AcylTransferDevicesinProteinSynthesis 59 VincentDiemer,JenniferBouchenna,FlorentKerdraon,VangelisAgouridas, andOlegMelnyk
3.1Introduction 59
3.2 N,S- and N,Se-AcylTransferDevices:GeneralPresentation,Reactivity andStatisticalOverviewofTheirUtilization 61
3.2.1GeneralPresentationof N,S-and N,Se-AcylTransferDevices 61
3.2.2RelativeReactivityof N,S-and N,Se-AcylTransferDevices 63
3.2.3AStatisticalOverviewoftheSyntheticUseof N,S-and N,Se-Acyl TransferDevicesforProteinTotalChemicalSynthesis 64
3.3PreparationofSEA/SeEAoff andSEAlidePeptides 68
3.3.1PreparationofSEAandSeEAPeptides 68
3.3.2PreparationofSEAlidePeptides 70
3.4Redox-controlledAssemblyofBiotinylatedNK1Domainofthe HepatocyteGrowthFactor(HGF)UsingSEAandSeEAChemistries 71
3.5TheTotalChemicalSynthesisofGM2-APUsingSEAlide-based Chemistry 75
3.6Conclusion 79 References 80
4ChemicalSynthesisofProteinsThroughNativeChemical LigationofPeptideHydrazides 87 ChaoZuo,XiaodanTan,XianglongTan,andLeiLiu
4.1Introduction 87
4.2DevelopmentofPeptideHydrazide-basedNativeChemicalLigation 88
4.2.1ConversionofPeptideHydrazidetoPeptideAzide 88
4.2.2AcylAzide-basedSolid-phasePeptideSynthesis 88
4.2.3AcylAzide-basedSolution-phasePeptideSynthesis 89
4.2.4TheTransesterificationofAcylAzide 90
4.2.5DevelopmentofPeptideHydrazide-basedNativeChemicalLigation 90
4.3OptimizationofPeptideHydrazide-basedNativeChemicalLigation 91
4.3.1PreparationofPeptideHydrazides 91
4.3.1.12-Cl-Trt-ClResin 91
4.3.1.2PeptideHydrazidesfromExpressedProteins 92
4.3.1.3Sortase-mediatedHydrazideGeneration 93
4.3.2ActivationMethodsofPeptideHydrazide 94
4.3.2.1KnorrPyrazoleSynthesis 94
4.3.2.2ActivationinTFA 94
4.3.3LigationSitesofPeptideHydrazide 95
4.3.4MultipleFragmentLigationBasedonPeptideHydrazide 96
4.3.4.1N-to-CSequentialLigation 96
4.3.4.2ConvergentLigation 96
4.3.4.3One-potLigation 96
4.4ApplicationofPeptideHydrazide-basedNativeChemicalLigation 99
4.4.1PeptideDrugsandDiagnosticTools 99
4.4.1.1PeptideHydrazidesforCyclicPeptideSynthesis 99
4.4.1.2Screeningfor D PeptideInhibitorsTargetingPD-L1 99
4.4.1.3ChemicalSynthesisofDCAFforTargetedAntibodyBlocking 101
4.4.1.4PeptideToxins 101
4.4.2SynthesisandApplicationofTwo-photonActivatableChemokine CCL5 102
4.4.3ProteinswithPosttranslationalModification 103
4.4.3.1TheSynthesisofGlycosylation-modifiedFull-lengthIL-6 103
4.4.3.2TheChemicalSynthesisofEPO 105
4.4.3.3ChemicalSynthesisofHomogeneousPhosphorylatedp62 105
4.4.3.4ChemicalSynthesisofK19,K48Bi-acetylatedAtg3Protein 105
4.4.4UbiquitinChains 108
4.4.4.1SynthesisofK27-linkedUbiquitinChains 108
4.4.4.2SynthesisofAtypicalUbiquitinChainsbyUsinganIsopeptide-linkedUb Isomer 109
4.4.4.3SynthesisofAtypicalUbiquitinChainsUsinganIsopeptide-linkedUb Isomer 109
4.4.5ModifiedNucleosomes 110
4.4.5.1SynthesisofDNA-barcodedModifiedNucleosomeLibrary 110
4.4.5.2SynthesisofModifiedHistoneAnalogswithaCysteine Aminoethylation-assistedChemicalUbiquitinationStrategy 111
4.4.5.3SynthesisofUbiquitylatedHistonesforExaminationofthe DeubiquitinationSpecificityofUSP51 111
4.4.6MembraneProteins 112
4.4.7Mirror-imageBiologicalSystems 112
4.5SummaryandOutlook 113 References 114
5ExpandingNativeChemicalLigationMethodologywith SyntheticAminoAcidDerivatives 119
EmmaE.Watson,LaraR.Malins,andRichardJ.Payne
5.1NativeChemicalLigation 120
5.2DesulfurizationChemistries 120
5.3AsparticAcid(Asp,D) 122
5.4GlutamicAcid(Glu,E) 124
5.5Phenylalanine(Phe,F) 125
5.6Isoleucine(Ile,I) 127
5.7Lysine(Lys,K) 130
5.8Leucine(Leu,L) 133
5.9Asparagine(Asn,N) 135
5.10Proline(Pro,P) 138
5.11Glutamine(Gln,Q) 139
5.12Arginine(Arg,R) 139
5.13Threonine(Thr,T) 140
5.14Valine(Val,V) 142
5.15Tryptophan(Trp,W) 144
5.16ApplicationofSelenocysteine(Sec)toLigationChemistry 146
5.17AsparticAcid(Asp,D) 147
5.18GlutamicAcid(Glu,E) 148
5.19Phenylalanine(Phe,F) 149
5.20Leucine(Leu,L) 151
5.21Proline(Pro,P) 151
5.22Serine(Ser,S) 153 References 155
6PeptideLigationsatStericallyDemandingSites 161 YingluWangandSuweiDong
6.1Introduction 161
6.2LigationsUsingThioesters 162
6.2.1ExogenousAdditive-promotedLigations 162
6.2.2LigationsUsingReactiveThioesters 167
6.2.3InternalActivationStrategyinPeptideLigations 169
6.3LigationsUsingOxo-esters 170
6.4PeptideLigationsBasedonSelenoesters 170
6.5Microfluidics-promotedNCL 175
6.6RepresentativeApplicationsinProteinSynthesis 178
6.7SummaryandOutlook 181 References 181
7ControllingSegmentSolubilityinLargeProtein Synthesis 185 RileyJ.Giesler,JamesM.Fulcher,MichaelT.Jacobsen,andMichaelS.Kay
7.1SolventManipulation 185
7.2IsoacylStrategy 187
7.3SemipermanentSolubilizingTags 191
7.3.1N-orC-TerminalSolubilizing“Tails” 192
7.3.2ReversibleBackboneModificationsasSolubilizingTags 194
7.3.3BuildingBlockSolubilizingTags 195
7.3.4ExtendableSide-chain-basedSolubilizingTags 195 References 198
8TowardHPLC-freeTotalChemicalSynthesisofProteins 211 PhucUngandOliverSeitz
8.1Introduction 211
8.1.1CaptureandReleasePurification 212
8.1.2Solid-phaseChemicalLigations(SPCL) 212
8.2SynthesisofPeptideSegmentsforNativeChemicalLigation 213
8.2.1HPLC-freePreparationofN-terminalPeptideSegmentsforNCL 213
8.2.2HPLC-freePreparationofC-terminalPeptideSegmentsforNCL 217
8.3SynthesisofProteinsUsingtheHis6 Tag 220
8.3.1ReversibleHis6 -basedCaptureTags 220
8.3.2His6 -basedImmobilizationfor C-to-N AssemblyofCrambin 221
8.3.3His6 -basedImmobilizationforAssemblyofProteinsonMicrotiter Plates 222
8.3.4His6 andHydrazideTagsforSequential N -to-C Captureand Release 225
8.4SynthesisofProteinsviaOximeFormation 227
8.4.1ReversibleOxime-basedCaptureTags 227
8.4.2Oxime-basedImmobilizationfor N -to-C Solid-phaseChemical Ligations 227
8.4.3Oxime-basedImmobilizationfor C-to-N Solid-phaseChemical Ligations 233
8.4.4Oxime-based C-to-N Solid-phaseChemicalLigations 237
8.5SynthesisofProteinsviaHydrazoneFormation 238
8.5.1ReversibleHydrazone-basedCaptureTags 238
8.5.2Hydrazone-basedImmobilizationforAssemblyofProteinsonMicrotiter Plates 239
8.6SynthesisofProteinsUsingClickChemistry 242
8.6.1Click-basedImmobilizationfor N -to-C Solid-phasePeptideLigations UsingaProtectedAlkyne 242
8.6.2Click-basedImmobilizationfor N -to-C Solid-phasePeptideLigations UsingaSeaGroup 243
8.7SynthesisofProteinsUsingtheKAHALigation 244
8.7.1TheKAHALigation 244
8.7.2HPLC-freeSynthesisofProteinsUsingtheKAHALigation 245
8.8SynthesisofProteinsUsingPhotocleavableTags 246
8.8.1SynthesisofProteinsUsingaPhotocleavableBiotin-basedPurification Tag 246
x Contents
8.8.2SynthesisofProteinsUsingaPhotocleavableHis6 -basedPurification Tag 247
8.9Conclusion 249 References 251
9Solid-phaseChemicalLigation 259
SkanderA.Abboud,AgnèsF.Delmas,andVincentAucagne
9.1Introduction 259
9.1.1ThePromisesofSolidPhaseChemicalLigation(SPCL) 259
9.1.2ChemicalLigationReactionsUsedforSPCL 260
9.1.3KeyRequirementsforaSPCLStrategy 261
9.2SPCLinthe C-to-N Direction 262
9.2.1TemporaryMaskingGroupstoEnableIterativeLigations 262
9.2.2Linkersfor C-to-N SPCL 264
9.2.2.1UseofSameLinkerandSolidSupportforSPPSandSPCL 265
9.2.2.2Re-immobilizationoftheC-TerminalSegment 266
9.3SPCLinthe N -to-C Direction 268
9.3.1TemporaryMaskingGroupstoEnableIterativeLigations 268
9.3.2Linkersfor N -to-C SPCL 270
9.3.3CaseStudy 272
9.3.4SPCLwithConcomitantPurifications 274
9.4Post-LigationSolid-SupportedTransformations 274
9.4.1ChemicalTransformations 274
9.4.2BiochemicalTransformations 275
9.5SolidSupport 275
9.6ConclusionandPerspectives 278
Acknowledgment 278
9.AAppendix 278 References 280
10Ser/ThrLigationforProteinChemicalSynthesis 285 CarinaHeyPuiCheungandXuechenLi
10.1Serine/ThreonineLigation 287
10.2EpimerizationIssue 289
10.3OtherArylAldehydeEsters 289
10.4PreparationofPeptideSalicylaldehydeEsters 289
10.5ScopeandLimitations 294
10.6StrategiesofSer/ThrLigationforProteinChemicalSynthesis 294
10.7 C-to-N Ser/ThrLigation 294
10.8 N -to-C Ser/ThrLigation 296
10.9One-potSer/ThrLigationandNCL 296
10.10Bioconjugation 296
10.11SolubilityIssues 298
10.12ExtensionofSer/ThrLigation 298
10.13Conclusion 302 References 303
11ProteinSemisynthesis 307
NamChuandPhilipA.Cole
11.1Background 307
11.2ExpressedProteinLigation(EPL) 308
11.2.1MethodDevelopment 308
11.2.2ApplicationsofEPLforStudyingProteinPosttranslational Modifications 309
11.2.3Site-specificProteinLabelingwith N -HydroxysuccinimideEsters 311
11.3CysteineModifications 311
11.3.1DehydroalanineGenerationandApplicationsinSemisynthesis 312
11.3.2CysteineAlkylation-relatedMethodstoIntroduceLysMimics 313
11.4Enzyme-catalyzedProtein/PeptideLigations 314
11.4.1Sortase 314
11.4.2Butelase-1 316
11.4.3Subtiligase 317
11.4.4Trypsiligase 318
11.5Enzyme-catalyzedExpressedProteinLigation 318
11.6SummaryandOutlook 319
Acknowledgments 320 References 320
12Bio-orthogonalImineChemistryinChemicalProtein Synthesis 327
StijnM.Agten,IngridDijkgraaf,StanH.E.vanderBeelen,andTilmanM. Hackeng
12.1Introduction 327
12.2CarbonylFunctionalization 328
12.3Aminooxy,Hydrazine,andHydrazideFunctionalization 335
12.4OximeLigation 337
12.5HydrazoneLigation 342
12.6Pictet–SpenglerReaction 344
12.7CatalysisofOximeandHydrazoneLigations 346 References 348
13DecipheringProteinFoldingUsingChemicalProtein Synthesis 357 VladimirTorbeev
13.1Introduction 357
13.2ModificationofProteinBackboneAmides 358
13.3Insertionof β-turnMimetics 361
13.4InversionofChiralCentersinProteinBackboneandSideChains 362
13.5Modulating cis–trans ProlineIsomerization 366
13.6SteeringOxidativeProteinFolding 368
13.7CovalentTetheringtoFacilitateFoldingofDesignedProteins 371
13.8DiscoveryofPreviouslyUnknownProteinFolds 373
13.9Site-specificLabelingwithFluorophores 373
13.10FoldamersandFoldamer–PeptideHybrids 375
13.11ConclusionsandOutlook 377 Acknowledgement 378 References 378
14ChemicalSynthesisofUbiquitinatedProteinsforBiochemical Studies 383 GandhesiriSatish,GangaB.Vamisetti,andAshrafBrik
14.1TheUbiquitinSystem 383
14.2Non-enzymaticUbiquitination:ChallengesandOpportunities 386
14.2.1ChemicalSynthesisofUbBuildingBlocks 387
14.2.2IsopeptideLigation 387
14.2.3TotalChemicalSynthesisofTetra-UbChains 390
14.3SynthesisandSemisynthesisofUbiquitinatedProteins 393
14.3.1MonoubiquitinatedProteins 393
14.3.2Tetra-ubiquitinatedProteins 395
14.3.3ModificationofExpressedProteinswithTetra-Ub 400
14.4SynthesisofUniqueUbConjugatestoStudyandTargetDUBs 401
14.5Activity-basedProbes 403
14.6Perspective 405
ListofAbbreviations 406 References 407
15GlycoproteinSynthesis 411
ChaitraChandrashekar,KentoIritani,TatsuyaMoriguchi,andYasuhiro Kajihara
15.1Introduction 411
15.2TotalChemicalSynthesisofGlycoproteins 411
15.3SemisynthesisofGlycoproteins 413
15.4ChemoenzymaticSynthesis 413
15.5 α-Synuclein 414
15.6HirudinP6 415
15.7SaposinD 416
15.8Interleukin2 417
15.9Interleukin25 417
15.10Mucin1 419
15.11Crambin 421
15.12TauProtein 422
15.13ChemicalDomainofFractalkine 423
15.14CCL1 424
15.15Interleukin6 424
15.16Interleukin8 425
15.17Erythropoietin 426
15.18Trastuzumab 430
15.19AntifreezeGlycoprotein 432
15.20Conclusion 434
References 434
16ChemicalSynthesisofMembraneProteins 437
AlancaSchmidandChristianF.W.Becker
16.1Introduction 437
16.2SolidPhaseSynthesisofTMPeptides 438
16.3PurificationandHandlingStrategiesofTMPeptides 442
16.4SolubilityTags 443
16.4.1TerminalTags 443
16.4.2SideChainTags 445
16.5RemovableSolubilizingBackboneTags 445
16.6ChemicalSynthesisofMembraneProteins 449
16.6.1ProteinsWith1TMDomain 449
16.6.2Proteinswith2TMDomains 450
16.6.3Proteinswith3andMoreTMDomains 454
16.7Outlook 456
References 457
17ChemicalSynthesisofSelenoproteins 463
RebeccaN.Dardashti,ReemGhadir,HibaGhareeb,OritWeil-Ktorza,and NormanMetanis
17.1WhatareSelenoproteins? 463
17.2ExpressionofSelenoproteins 466
17.3SecasaReactiveHandle 469
17.4SynthesisandSemisynthesisofNaturalSelenoproteins 473
17.5SeleniumasaToolforProteinFolding 475
17.6Conclusions 478
References 478
18HistoneSynthesis 489
ChampakChatterjee
18.1TheHistonesandTheirChemicalModifications 489
18.1.1HistoneProteins 489
18.1.2HistonePosttranslationalModifications 490
18.2ChemicalLigationforHistoneSynthesis 492
18.2.1NativeChemicalLigation 492
18.2.2ExpandingtheScopeofNativeChemicalLigationWithInteins 494
18.3HistoneOctamerandNucleosomeCoreParticleAssembly 494
18.4StudyingtheHistoneCodeWithSyntheticHistones 496
18.4.1SynthesisofHistonesModifiedbySmallerFunctionalGroups 497
18.4.1.1HistonePhosphorylation 497
18.4.1.2HistoneAcetylation 499
18.4.1.3HistoneMethylation 502
18.4.2SynthesisofSumoylatedHistones 505
18.5Conclusions 506
Acknowledgments 506
References 506
19ApplicationofChemicalSynthesistoEngineerProtein BackboneConnectivity 515
ChinoC.CabaltejaandW.SethHorne
19.1Introduction 515
19.2BackboneEngineeringtoFacilitateSynthesis 516
19.3BackboneEngineeringtoExploretheConsequencesofChirality 517
19.4BackboneEngineeringtoUnderstandandControlFolding 520
19.5BackboneEngineeringtoCreateProteinMimetics 522
19.6Conclusions 525
References 526
20BeyondPhosphateEsters:SynthesisofUnusually PhosphorylatedPeptidesandProteinsforProteomic Research 533
AnettHauser,ChristianE.Stieger,andChristianP.R.Hackenberger
20.1Introduction 533
20.2GeneralMethodsfortheIncorporationofHydroxy-phosphorylated AminoAcidsintoPeptidesandProteins 534
20.3IncorporationofOtherPhosphorylatedNucleophilicAminoAcidsinto PeptidesandProteins 537
20.3.1 Phosphoarginine(pArg) 537
20.3.2Phosphohistidine(pHis) 538
20.3.3Phospholysine(pLys) 539
20.3.4Phosphocysteine(pCys) 539
20.3.5PyrophosphorylationofSerineandThreonine(ppSer,ppThr) 541
20.4DevelopmentofPhospho-analoguesasMimicsforEndogenous Phospho-AminoAcids 541
20.4.1AnaloguesofPhosphoserine,Phosphothreonine,and Phosphotyrosine 541
20.4.2StableAnaloguesofPhosphoaspartateandPhosphoglutamate 543
20.4.3StableAnaloguesofPhosphoarginine 544
20.4.4StableAnaloguesofPhosphohistidine 545
20.4.5StableAnaloguesofPyrophosphorylatedSerine 547
20.5Conclusion 547
References 547
21CyclicPeptidesviaLigationMethods 553
TristanJ.TylerandDavidJ.Craik
21.1Introduction 553
21.2CyclicPeptideSynthesis 554
21.3Orbitides 557
21.4Paws-derivedPeptides(PDPs) 559
21.5CyclicConotoxins 561
21.6 θ-Defensins 563
21.7Cyclotides 563
21.8Outlook 568
Acknowledgements 568
Funding 568
References 569
Index 579
Preface
Chemicalproteinsynthesisenablesthegenerationofproteinsofanyarchitecture andfacilitatesthesite-selectiveintroductionofunnaturalaminoacids,biophysical probes,posttranslationalmodifications,andotherfunctionalities.Thepreparation oftheseproteinanaloguesenablesimportantbiochemicalandbiophysicalstudies andcontributestothefundamentalunderstandingofproteins.Inthisbook,we presentasalientcollectionofarticlesthatcoverthemodernmethodsandapplicationsofthefield,alongwithdescriptionsofvariouscasestudiesinwhichchemical proteinsynthesisisleveragedtoshedlightonfundamentalquestionsinprotein science.
InChapter1,StephenKent,thepioneerandleadingfigureinmodernchemicalproteinsynthesis,introducestheessentialroleofchemicalproteinsynthesis indevelopingafundamentalunderstandingoftheprinciplesthatgiverisetothe structuresandbiologicalfunctionsofproteinmolecules.Strongemphasiswasgiven toanalyticalcontrolanddocumentationofthesyntheticprocessinordertomeet standards,similartothoseusedtocharacterizesmallorganicmolecules.Characterizationofasyntheticproteinmoleculeshouldrigorouslycharacterizethemolecular homogeneity,correctcovalentstructure,anddefinedfoldedstructure.
Newexcitingadvancesinsyntheticmethodologyhavegreatlyreshapedthe landscapeofchemicalproteinsynthesisoverthepast10years.InChapter2, BradleyL.Penteluteetal.describethenewtechnologyofautomatedflow-based solid-phasepeptidesynthesizer(AFPS)thatcanincorporateeachaminoacid residueinaslittleas40seconds.Thistechnologyoffersatemplateforaccelerating synthesesoflongpolypeptidesequencesusinganautomatedflowinstrument. Thisapproachmayopenthechemicalbiologyfieldtoroutinemanufactureof custom-made,fullysyntheticproteins.InChapter3,VangelisAgouridas,Oleg Melnyk,andcoworkersdescribethedevelopmentof N,S(Se)-acylshiftsystemsfor linkingpeptidesegmentstogetherthroughthemostwidelyusedmethodofchemicalproteinsynthesis,namely,nativechemicalligation.Thisstrategyisespecially usefulforsystemsthatrequirelatentthioesters.Manybiologicallyactiveproteins havebeensuccessfullysynthesizedwiththeSEA(bis(2-sulfanylethyl)amido)and SEAlide(N -sulfanylethylanilide)chemistries.InChapter4,LeiLiuetal.describe thedevelopmentandoptimizationofhydrazide-basednativechemicalligation, aswellastheoptimizationofthepreparation,activation,andligationofpeptide
hydrazides.Thehydrazide-basedmethodexhibitsseveraladvantagesincludingeasy preparation,highstability,goodhandlingproperties,andcontrollableactivation.It hasbeensuccessfullyusedinthesynthesisofpeptidepharmaceutics,posttranslationallymodifiedproteins,photo-activatableproteintools,membraneproteins,and mirror-imageproteins.
Tofurtherexpandthescopeandutilityofnativechemicalligation,RichardJ. Payneetal.describeinChapter5theeffortsmadetodevelopvariousthiolatedamino acids,which,withthehelpofmild,radical-mediateddesulfurizationconditions, canenableligationat16ofthe20proteinogenicaminoacids.Theyalsodescribe theirseminalstudiesonselenocysteine(Sec),whichhaveenabledrapidchemical proteinsynthesisthroughthediselenide–selenoesterligation(DSL)andnovelstrategiessuchas“selenolligationauxiliary.”InChapter6,SuweiDongetal.describe therecentdevelopmentonnewstrategiesthataimtofacilitatepeptideligationsat stericallydemandingsites.Theseligationreactionsareusuallychallenging,asthe strongactivatingconditionsmayresultincompetitivehydrolysis.Furthermore,in Chapter7,MichaelS.Kayetal.describetherecentstudiesonhowtocontrolsegment solubilityinlargeproteinsynthesis.Variousstrategiesthatutilizesolventchoice, isoacyldipeptides,andsemipermanentsolubilizingtagsaresurveyedandanalyzed indepth.Thischapterprovidesveryusefulguidesforresearcherstoovercomethe often-encounteredsolubilityproblemsinchemicalproteinsynthesis.
Withtheadventoffastpeptidesynthesisandligation,otherlimitingfactorshave receivedattentionsforchemicalproteinsynthesis.InChapter8,OliverSeitzetal. describethecurrentstateoftheartinthedevelopmentofmethodsthatallowminimizingthenumberofhigh-performanceliquidchromatograpohy(HPLC)purificationsteps.ThesemethodstypicallyrelyonrapidandHPLC-freeassemblyofproteinsusingcaptureandreleasepurificationhandles,openingopportunitiesforthe chemicalsynthesisofproteinarraysunderparallelconditions.InChapter9,Vincent Aucagneetal.describesolid-phasepeptidechemicalligation(SPCL),whichentails aligation-basedassemblyonasolidsupporttoavoidthelaboriousintermediate chromatographicpurifications.Thedevelopmentofnewpolymers,newlinkers,new ligationsreactionswithimprovedkinetics,andnewmasking/unmaskingstrategies hasbroadenedthescopeofSPCLtomoreambitioustargets.Onadifferentfront,to expandtheflexibilityofpeptidecondensation,ligationstrategiesotherthannative chemicalligationneedtobedeveloped.InChapter10,XuechenLietal.describe theapproachofserine/threonineligation(STL)betweenpeptideC-terminalsalicylaldehydeestersandN-terminalSer/Thrresidues.Thisapproachtakesadvantageof thehighabundanceofSer/Thrresiduesintheproteinsequenceandhaspavedthe wayforbroadapplicationsinproteinssynthesisandrelatedconstructs.
Moderntotalchemicalsynthesisofproteinscannowreachapproximately 200–400aminoacids.Toamplifytheabilitytomakeproteinscontainingunnatural aminoacidsandposttranslationalmodifications,animportantstrategyisprotein semisynthesiswhereabiologicallyproducedproteinorproteinfragmentisselectivelymodifiedthroughcovalentchemicalreactions.InChapter11,PhilipA.Cole etal.describethedevelopmentofmethodsforproteinsemisynthesisincluding expressedproteinligation,cysteinemodificationreactions,andenzyme-catalyzed
Preface xix protein/peptideligations.Thesemethodshavebeensuccessfullyemployedto augmentourunderstandingonproteinposttranslationalmodification.Asaunique exampleofproteinsemisynthesismethods,inChapter12,TilmanM.Hackengetal. describebio-orthogonaliminechemistryforproteinmodification.Detailswere providedforhowtoincorporatecarbonylor α-nucleophilesmoietiesintoproteins, andhowtocarryoutdifferentiminechemistriessuchasoximeandhydrazone ligation.
Nextaretheapplicationsofchemicalproteinsynthesistosolvevariousbiochemical,biophysical,andbiomedicalproblems.InChapter13,VladimirTorbeev describeshowtousechemicalproteinsynthesistodecipherthemechanismof proteinfolding.Manyinterestingmethodshavebeendevelopedincludingmodificationofproteinbackboneamides,insertionof β-turnmimetics,inversionof chiralcentersinproteinbackboneandsidechains,modulatingcis–transproline isomerization,covalenttetheringtofacilitatefoldingofdesignedproteins,and foldamersandfoldamer–peptidehybrids.Thesemethodsfacilitatebiophysical studiesonintrinsicallydisorderedproteins(IDPs)thatperformimportantfunctions suchasgenetranscriptionandchromatinremodeling.Theyarealsohelpfulto studiesonimproperproteinfoldingrelatedtomanydiseasessuchasAlzheimer’s andParkinson’sdisorders.
Oneofthemostexcitingapplicationsofchemicalproteinsynthesisisthedevelopmentofchemicaltoolstostudythebiologicalfunctionsandmechanismsofproteins bearingposttranslationalmodifications.InChapter14,AshrafBriketal.described theircontributionsindevelopinginnovativemethodstoconstructavarietyofubiquitinconjugatesofhighpurity,insufficientquantitytofacilitatedetailedbiophysical andbiochemicalanalysis.Asa“gamechanger”toovercometheinheritlimitations oftheenzymaticapproaches,chemicalproteinsynthesishasenabledreadilyconstructionofversatileubiquitinconjugatestostudyandtargetubiquitin-processing enzymes.InChapter15YasuhiroKajiharaetal.describethecutting-edgeexamples ofglycoproteinsynthesisemployingvarioussyntheticmethodologiesandligation strategies.Thecombinationofbothchemicalandenzymaticmethodologiesgives insightintohowglycansfunctionintheirbiologicalenvironmentandmayeventuallyleadtohomogeneousglycoproteinpharmaceuticals.
Inadditiontoproteinposttranslationalmodification,chemicalproteinsynthesisalsoplaysimportantrolesinthebiochemicalstudiesonsomeuniqueprotein families.InChapter16,ChristianBeckeretal.delineatethechemicalsynthesisof membraneproteins,auniqueproteinfamilycomprisingabout30%oftheproteins encodedbythehumangenome.Accesstomembraneproteinsbychemicalsynthesishashelpedtodecipherimportantaspectsofmembraneproteinfunctionsdue totheuniqueopportunitiesthatchemicalsynthesisprovidesintermsofmanipulationsofthebackboneandsidechainsofproteins.InChapter17,NormanMetanis etal.describechemicalsynthesisofselenoproteins,whichcontaintherarecoded aminoacidselenocysteine.Studiesonsyntheticselenoproteinsfacilitateelucidation oftheroleofseleniuminbothnaturalandunnaturalprocesses,providingincreasing insightsintothispreviouslymysteriousmicronutrient’sroleinhumanhealthand disease.Furthermore,inChapter18,ChampakChatterjeedescribethechemical
xx Preface synthesisofhistonesbearingvariousmodifications.Dueinlargeparttotheadditionofsemisyntheticandfullysyntheticsite-specificallymodifiedhistonestothe repertoireoftoolsavailabletoinvestigatethehistonecodehypothesis,ourunderstandingofthemanymechanisticrolesforhistoneshasrapidlyexpandedinthe past10years.
Inthelastchapters,W.SethHorneetal.describeinChapter19howtoapply chemicalsynthesistoengineerproteinbackboneconnectivity.Studiesinthisarea enableexplorationofthestructuralandfunctionalconsequencesofproteinbackbonealterationandopenthedoortonewbio-inspiredentitieswithmyriadpotential applications.InChapter20,ChristianHackenbergeretal.describehowtosynthesizeunusuallyphosphorylatedpeptidesandproteinsforproteomicresearch.Details areprovidedonmimicsofendogenousphosphateestersaswellasrare,naturally occurringphosphorylationsoffunctionalaminoacidssuchascysteine,lysine,histidine,andarginine.Finally,inChapter21,DavidJ.Craiketal.describehowto synthesizecyclicpeptidesvialigationmethods.Targetmoleculesincludeorbitides, paws-derivedpeptides,cyclicconotoxins, θ-defensin,andcyclotides.Thesecyclic peptideshavebeenatopicofconsiderableinterestinrecentyearsduetotheirunique structuralandpharmacologicalfeaturesthatmakethemexcellentstartingpointsin drugdesign.
Webelievethatthe21chaptersexemplifiesthemultidisciplinarynatureof researchinthefieldofchemicalproteinsynthesisinthetwenty-firstcentury.The readersofthebookwillbeexposedtothestate-of-the-artchemistrythatthefield hasbeendevelopingthroughthelastfewdecadesandtheremainingchallenges remainedtobetackled.Thebookwillalsobeveryusefulsourceforstudents andscientistsaswelltolearnaboutthevarioussyntheticaspectsofthepeptide fragmentssynthesis,peptideligationbasedondifferentstrategies.
CharacterizationofProteinMoleculesPreparedbyTotal ChemicalSynthesis
StephenB.H.Kent
DepartmentofChemistry,TheUniversityofChicago,Chicago,IL60637,USA
“Nevertheless,thechemicalenigmaofLifewillnotbesolveduntilorganicchemistryhasmasteredanother,evenmoredifficultsubject,theproteins,inthesame wayasithasmasteredthecarbohydrates.”
Source:EmilFischer(NobelLecture1902)
1.1Introduction
Proteinsarethe“naturalprodicts”ofthetwenty-firstcentury.Proteinmoleculesare ubiquitousinthebiologicalworld,withnumerousdiversefunctionsthatrangefrom actingasstructuralmaterialssuchasthekeratins,tointegralmembraneproteins thatserveasionchannelsorasactivemoleculartransportersincells,toproteins thatactashormonalmessengersinhigheranimalspecies,andtoproteinsthatregulategeneexpression[1].Themostimportantfunctionofproteinmoleculesisas enzymes,thepotentandspecificcatalystsofthechemicalreactionsofbiological metabolism,withoutwhichlifewouldbeimpossible[2].ThankstomodernDNA sequencingmethodsappliedtogenome[3]andmetagenome[4]sequencing,vast numbersofproteinsarebeingdiscoveredatthenucleicacidlevelasopenreading framesthatcodeforaprotein’spolypeptidechain.
Thecentraldogmaofproteinscienceisthattheaminoacidsequenceofthe polypeptidechainencodesthefoldedstructureoftheproteinmoleculeinitsnatural environment,andthatitisthefoldedstructureoftheproteinmoleculethatgives risetoitsbiologicalfunction(s)[5].Proteinsrangeinmassfromlessthan5kDato morethan100kDa.Themediansizeofglobularproteinmoleculesis ∼35–45kDa, comprisingapolypeptidechainof ∼300–400aminoacidresidues.Proteinsare typicallymadeupoftwoormoredomains,autonomousunitsoffolding,eachof ∼120–160aminoacidresidues[6].
Proteinmoleculesarenotsimplyreallybigpeptides.Initsnativeenvironment, eachglobularproteinmoleculehasadefinedfoldedstructurethatgivesrisetothe TotalChemicalSynthesisofProteins, FirstEdition.EditedbyAshrafBrik,PhilipDawson,andLeiLiu. ©2021WILEY-VCHGmbH.Published2021byWILEY-VCHGmbH.
1CharacterizationofProteinMoleculesPreparedbyTotalChemicalSynthesis functionalpropertiesofthatprotein,includingbiochemicalandbiologicalactivities. Syntheticproteinsareorganicmoleculesofhighmolecularmass,comprisedoflinearpolypeptidechains(typicallyof50–300ormoreaminoacidresidues)thatfold toformcomplex,dynamicstructures.Thelargesizeofproteinstogetherwiththe intricacyoftheircovalentandfoldedstructurescreatesspecialchallengesinthe characterizationofthesesyntheticmolecules.Forthisreason,itisimportantfor researcherstorigorouslycharacterizeproteinmoleculespreparedbytotalchemicalsynthesistomeetstandardssimilartothoseusedinsyntheticorganicchemistry. Analyticalmethodsandcriteriafortherigorouscharacterizationofsyntheticproteinshaverecentlybeenenunciated[7].
1.2ChemicalProteinSynthesis
“Thechemicalligationapproach....breakstheconceptualshacklesimposed bythepeptidebond,freesusfromthelinearparadigmofthegeneticcode,and openstheworldofproteinstotheentirerepertoireofchemistry.”
StephenKent(23rdEuropeanPeptideSymposium1994)
Chemistry,enabledbytotalsynthesis,hasanessentialroletoplayindevelopinga fundamentalunderstandingoftheprinciplesthatgiverisetothestructuresandbiologicalfunctionsofproteinmolecules.Thankstomodernchemicalligationmethods [8–10],thetotalsynthesisofproteinmoleculesintheresearchlaboratoryisboth practicalandincreasinglyrobust.
Characterizationofaproteinpreparedbytotalchemicalsynthesisshouldbegin withthesyntheticprocessitself.Analyticalcontrolofthebond-formingsteps,purificationandanalysisofeachsyntheticintermediate,andfoldingofthefull-length syntheticpolypeptidechaintoformadefinedtertiarystructureareintegraltoverifyingthestructureofthefinalsyntheticproteinproduct.Thestepsinvolvedina typicaltotalchemicalsynthesisofaproteinmoleculearelistedbelow.Keyaspects pertinentto characterization ofthesyntheticproteinarein bold:
● Establishaverifiedaminoacidsequenceofthepolypeptidechainofthe targetproteinmolecule.
⚬ UseadatabasesuchasUniProt(www.uniprot.org);includeposttranslational processingwherethatisknowntogivethematureproteinmolecule.
⚬ Resolvingdatabase/literatureambiguitiesinthereportedaminoacidsequence ofaproteincanbeproblematic.Itiseasytomakeamistakeandtoendupmakinganincorrecttargetsequence.
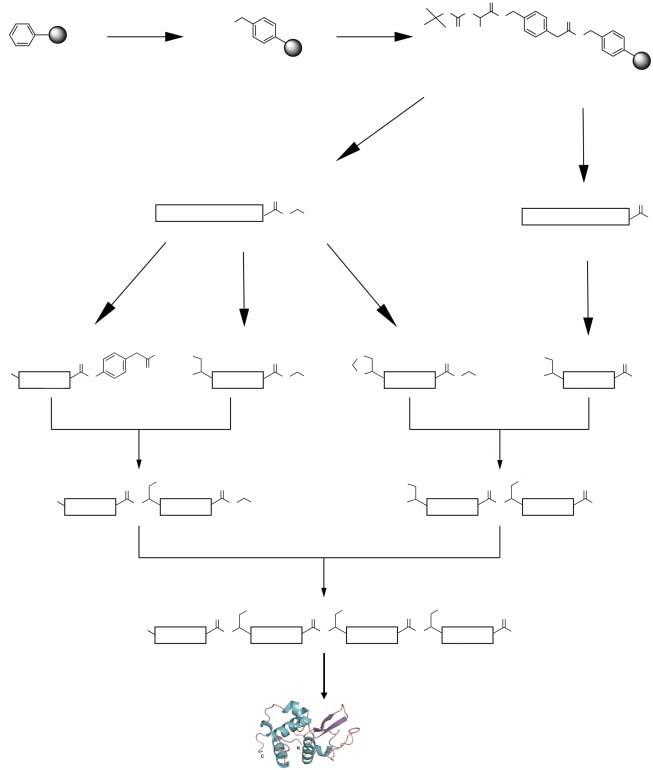
● Designa(convergent)synthesisoftheprotein’spolypeptidechain,starting frompeptidesegmentscontainingfewerthan40–50aminoacidresidues (Scheme1.1).
● Stepwisesolid-phasesynthesis(SPPS)ofpeptidesegmentsequippedwithsuitable functionalitiesforchemicalligation.
● Bocchemistry[11]orFmocchemistrySPPS[12],with documentationofthe aminoacidsequenceactuallymade.
Polystyrene resin Bio-beads S-X1
1
2
1
2
1
2
3
3
4
4
Scheme1.1 Convergentchemicalsynthesisofaproteinmoleculefromfoursynthetic peptidesegmentspreparedbystepwiseSPPS.Key:NCL,nativechemicalligation;KCL, kineticallycontrolledligation;Thz-Cys,conversionofN-terminalthiazolidine-CO–toCys–. Source:AdaptedfromDureketal.[11].
⚬ Purificationbypreparativehigh-performanceliquidchromatography(HPLC), whichshouldbeperformedunder displacementmode [13].
⚬ Combinedfractionsshouldbecheckedbyahighresolutiontechniquebased ona separationprincipledistinctfromthatusedinthepurification, suchascapillaryisoelectricfocusing(CIEF)[14].BecauseHPLCisusedfor thepurificationofthesyntheticpeptidesegment,itisNOTsufficienttouse analyticalHPLCorLCMSrunundersimilarchromatographicconditionsto verifyitshomogeneity.
⚬ Directinfusionelectrosprayionizationmassspectrometry(ESI-MS)forcheckingpurifiedpeptidesegmentsforimpuritiesthathavedifferentmasses.
Peptide
Peptide
Peptide
Peptide
1CharacterizationofProteinMoleculesPreparedbyTotalChemicalSynthesis
⚬ Confirmationofthecovalentstructureofeachsyntheticpeptidesegment.
● Precisemassmeasurement
–fromanalyticalLCMS,themassspectrometricdata must becollectedacross the entireUVabsorbingpeakcorrespondingtothepurifiedpeptide. –acorrectmassisanecessarybutnotsufficientanalyticalcriterion.
● Aminoacidsequence
–byMALDI-TOFMS‘laddersequencing’[15]ofterminatedbyproductsthatare invariablypresentatlow-levelsincrudesyntheticpeptidesmadebySPPS.
–byMS-MSofthepurifiedpeptide.Thisprocessisrenderedmorestraightforwardbecausethetargetsequenceofthepeptidesegmentisknown.
● Convergentcovalentcondensationofthepeptidesegments
⚬ Chemicalconversionofcrypticfunctionalgroupstothereactiveformsuitable forchemicalligation(e.g.Thz–peptidetoCys–peptide;[16]peptide–CONHNH2 topeptide–thioester[17]).
⚬ Nativechemicalligation(NCL);[18]pH7.0;aqueous6MGu.HCLasa near-universalsolvent;ambienttemperature;arylthiolcatalyst[19].
● Analyticalcontrolofsyntheticsteps
⚬ “Hands-on”real-timeanalyticalLCMSduringligationreactionsisanessential toolforfollowingthecourseofeachbond-formingstep.
● Purificationandcharacterizationofeachintermediateproduct
⚬ Usingthesameprotocolsasthoseusedforthesyntheticpeptidesegments.
● Purificationtohomogeneityandrigorouscharacterizationofthe full-lengthsyntheticpolypeptidechain.
⚬ PurificationbypreparativeHPLC,whichshouldbeperformedunder displacementmode [13].
⚬ Combinedfractionsshouldbecheckedbyahigh-resolutiontechniquebasedon a separationprincipledistinctfromthatusedinthepurification, suchas CIEF[14].BecauseHPLChasbeenusedforthepurificationofthesynthetic polypeptide,itisNOTsufficienttouseanalyticalHPLCorLCMSrununder similarconditionstoverifyitshomogeneity.
⚬ DirectinfusionESI-MSisalsoagoodtechniqueforcheckingthefull-lengthsyntheticpolypeptidechainforimpuritiesthathavedifferentmasses.
● Foldingthesyntheticpolypeptidechaintoformthefunctionalproteinmolecule
⚬ Fordisulfide-containingproteins,standardthiolredoxcoupleconditions areusedalongwithmoderateamountsofsolubility-enhancingagents (“denaturants”)tokeepmisfoldedpolypeptidechainsinsolutionsothatthey tooareeventuallyabletofoldcorrectly[20].
⚬ Forproteinsthatdonotcontaindisulfidebonds,itisfrequentlysufficientto simplydilutethepolypeptidechainintonativebufferortoslowlydialyzefrom 6MGu.HClintonativebufferconditions.
● Analyticalcontrolofproteinfolding
⚬ Inthecaseofdisulfide-containingproteins,thefoldedproteinmoleculewill usuallyelute earlier thantheunfoldedpolypeptidechainonanalyticalreverse phaseHPLC,becausehydrophobicsidechainsarelessexposedinthefolded, disulfidecross-linkedproteinmolecule[21].
⚬ Forproteinsthatdonotcontaindisulfidebonds,othertechniquessuchas CD-ORD1 ormultidimensionalnuclearmagneticresonance(NMR)mustbe usedtomonitortheprogressoffolding.
● Purificationofthefoldedsyntheticproteintogiveasingledefinedmolecularspecies.
⚬ Byoneormoreofthetechniquesofreverse-phaseHPLC,ionexchangechromatography,sizeexclusionchromatography.
● Homogeneityofthepurifiedsyntheticproteinmustbeverifiedbya high-resolutiontechniquebasedonaseparationprinciple distinct from thatusedforpurification.
⚬ CIEF[14]isthepreferredtechniqueforrigorouslyestablishingthehomogeneityofthepurifiedsyntheticproteinmolecule.
● Precisemeasurementofthemassofthesyntheticproteinmoleculeby directinfusion ESI-MS(directinfusionESI-MS).
⚬ Directinfusion ESI-MSprovidesdatarepresenting all themolecularspecies presentinthefinalsyntheticproduct.Itthusprovidesbothapreciseexperimentalmeasurementofthemassofthesyntheticproteinandatthesametime canrevealthepresenceofanyotherproductsthathaveamassdistinctfrom thatofthetargetproteinmolecule.Allmeasuredmassdatamustincludean experimentaluncertainty(Figure1.1).
⚬ HPLCwithon-lineESMScanbeused.However,impurityproteinspecies presentinthesyntheticproductmaynotbeelutedfromthereversephase support.MassspectrometricdatafromLCMS must beacquiredoverthe entireUV-absorbingpeak correspondingtothepurifiedsyntheticprotein product.
⚬ **ItisNOTacceptabletoreportamassdeterminedatasingletimepointof theHPLCchromatogram,whileignoringmolecularspecieswithdifferent massesthatarepresentatothertimepointsunderthesameUVabsorbing peak.**
⚬ Whereverpossible,the monoisotopicmass ofasyntheticproteinshouldbe reported(togetherwithexperimentaluncertainty),alongwiththecalculated monoisiopicmass.Anexampleisshownbelow(Figure1.2).
● Verificationofthecysteinepairingfordisulfidecontainingproteins.
⚬ ProteolyticdigestionfollowedbyLCMSpeptidemapping[24].
⚬ CysteinepairingcanalsobeverifiedbyhighresolutionX-raycrystallography (seebelow).
● Conformationalhomogeneityofthesyntheticproteinmolecule.
⚬ MultidimensionalNMR“fingerprinting”canbeusedtoestablishthatthe syntheticproteinhasasinglefoldedstructure[11,25].Notes:(i)thereare examplesofnaturalproteinmoleculesthatformmorethanonedefinedfolded
1Whilealow-resolutiontechniquesuchasCD-ORDisusefulformonitoringtheprogressof folding,itmustbesupplementedwithhigh-resolutioncharacterizationofthefoldedstructureof thesyntheticproteinbytechniquessuchasNMRandX-raycrystallography.
1CharacterizationofProteinMoleculesPreparedbyTotalChemicalSynthesis
Figure1.1 DirectinfusionESI-MSdatafor[Lys24,38,83 ]erythropoietinaglyconepreparedby totalchemicalsynthesis.Impuritiesofmasslessthanthetargetproteinmoleculeare essentiallyabsent,asshownbythelackofpeaksonthelow m/z sideofeachchargestate. Thepeaksonthehigh m/z sideofeachchargestateareNaandCaionadducts.Source: AdaptedfromLiuetal.[22].
Figure1.2 AnalyticalHPLCandMScharacterizationofasyntheticprotein.Withmodern massspectrometricinstrumentation,themonoisotopicmasscanbeexperimentally determinedandcomparedwiththetheoreticalmass.Source:AdaptedfromDureketal.[23].
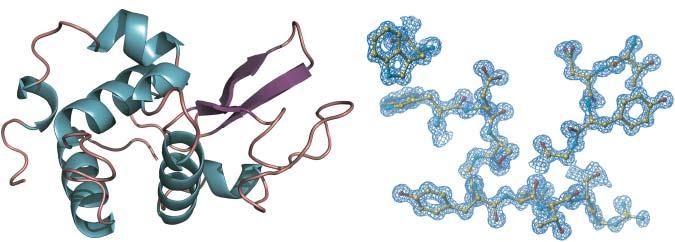
Figure1.3 X-raydiffractionstructureofcrystallinehumanlysozymepreparedbytotal chemicalsynthesis.(a)Cartoonrepresentationofthesecondarystructureofthefolded humanlysozymesyntheticproteinmolecule;(b)2Fo F c electrondensitymap,showingthe qualityofthedataacquiredataresolutionof1.04Å.Source:Dureketal.[11].©2007 NationalAcademyofSciences.
structure[26];(ii)intrinsicallydisorderedproteinsonlyfold,ifatall,inthe presenceoftheirtargetmolecules[27].
● Determinationoftheatomicstructureofthefoldedproteinmolecule.
⚬ X-raycrystallography,includingracemicproteincrystallography,[28]canbe usedtodeterminethestructureofthefolded,homogeneoussyntheticprotein molecule[29,30].
⚬ X-raystructuraldatawillalsoconfirmtheconnectivityofanydisulfidesthatare present,andatsufficientlyhighresolutioncanrevealthepresenceofaberrant chemicalmodificationsofthesyntheticprotein(Figure1.3).
⚬ Notethatbecausefractionalcrystallizationfromproteinmixturesiscommon,determinationofthecrystalstructureofasyntheticproteinbyX-ray diffractiondoesnotinitselfestablishthatallofthesyntheticproteinmolecules havethatstructureandareundamaged.Homogeneityofthesynthetic proteinmoleculemustbeestablished before determinationofthecrystal structure.
⚬ Forsmallproteins,thefoldedstructurecanbedeterminedusingnaturalabundanceproteinNMRtechniques[31].
● Biological/biochemicalassays.
⚬ Quantitativeassays,especiallymeasurementsofthecatalyticactivityofasyntheticenzymemolecule,canbeveryinformativebutareinadequateasproofof homogeneityorcorrectstructureofthesyntheticprotein.
ThetotalsynthesisofthechemokineCCL2(MCP-1)byGrygieletal.[32]isa near-perfectexampleofaproperlydocumentedtotalchemicalsynthesisofaprotein molecule,illustratingessentiallyallofthesyntheticstepsdescribedabovetogether withmeticulousandcompletecharacterizationofthesyntheticproteinasasingle molecularspeciesofdefinedstructure(Figure1.4).
Selectedfurtherexamplesofwell-characterizedsyntheticproteinscanbefound inRefs[24,33–42].
8 1CharacterizationofProteinMoleculesPreparedbyTotalChemicalSynthesis
Figure1.4 AnalyticalcharacterizationoftheproteinCCL2(MCP-1)preparedbytotal chemicalsynthesis.[32]PurityofthesyntheticproteinwasverifiedbyreversephaseHPLC andbycapillaryelectrophoresis,twohighresolutionanalyticalmethodsbasedondifferent separationprinciples(hydrophobicity;charge).Themonoisotopicmasswasmeasuredby LC-ESIQq/TOFmassspectrometry.Inadditiontothedatashownhere,thedisulfidebonds areverifiedbyenzymaticdigestionandLC-(MS-MS)oftheresultingpeptidefragments,and thecrystalstructureofthesyntheticproteinisdeterminedataresolutionof1.9Å.Source: FromGrygieletal.[32].
1.3CommentsonCharacterizationofSyntheticProtein Molecules
“Inthefieldofproteinsynthesisitismyconfidenthopethattomorrow’sdeeds willcatchupwithtoday’stitles,andthatweshalltrulybeabletoobtainenzymaticallyactiveproteinsassyntheticsubstances:asmaterialscomposedofasingle molecularspecies.”
JosefRudinger(3rdAmericanPeptideSymposium1972)
1.3.1Homogeneity
Thesinglemostimportantaspectofthecharacterizationofasyntheticproteinis rigorousverificationofitshomogeneity,i.e.thatthesyntheticproductisasingle
1.3CommentsonCharacterizationofSyntheticProteinMolecules 9 molecularspecies.Inverifyingthehomogeneityofasyntheticproteinitisessential touseanalyticaltechniquesbasedonseparationprinciplesthataredistinctfrom thepurificationmethod(s)usedinthesynthesisoftheproteinmolecule.PreparativeHPLCisalmostinvariablyusedasthefinalpurificationstepandcloselyrelated by-productsfromthesynthesismaybeco-purifiedwiththedesiredproteinproduct. Forthatreason,analyticalHPLCisnotasufficientproofofmolecularhomogeneity ofasyntheticproteinproduct. Directinfusion ESI-MSisagoodtechniqueforcheckingforthepresenceofproteinimpuritiesthathavemassesthatdifferfromthemass ofthetargetproteinmolecule.However,impuritiesthathavethesamemassasthe targetproteinwillnotbedetected.
Characterizationofasyntheticproteinmustalsoruleoutthepresenceofinadvertentmodificationsofthecovalentstructurethatmayhavebeenintroducedin thecourseofchemicalsynthesis.Forexample,chemicalmanipulationsofpeptide chainscanleadtotheformationofisopeptidebonds(i.e.beta-amidelinks)atasparticacidresidues,formationofsuccinimidesatAsnresidues,deamidationsofAsn and/orGlnsidechains, N -to-O acylshiftsatGly-Sersequences,andanyofavariety ofotherpossibleby-products.TheseabnormalmodificationsofthecovalentstructureareoftennotresolvedbyanalyticalHPLCandfrequentlyresultinsmall(1Da) orzerodifferencesintheobservedmassofthesyntheticproteinmolecule,andthus canbedifficultorimpossibletodetectbymassspectrometryalone.
Astate-of-the-arttechniquefordetectionofbyproductsandfortherigorous verificationofthemolecularhomogeneityofasyntheticproteinisCIEF.CIEF isacharge-based,ultrahigh-resolutionanalyticalmethodthatcanrevealthe presenceofimpuritiesthatdonotdirectlyinvolveaunitchargechange,evensubtle differencesasbenignasthereplacementofanunchargedaminoacidresiduein theprotein’spolypeptidechainbyanotherunchargedresidue.Forthatreason, CIEFistobepreferredfordocumentingtheabsenceofunexpectedby-products andfortheverificationofthemolecularhomogeneityofsyntheticpeptides andproteins.Recentdescriptionsofstate-of-the-artCIEFcanbefoundhere [43,44].
Thetimehascomeforcapillaryisoelectricfocusingdatatobemandatoryverificationofhomogeneityinthefinalcharacterizationofasyntheticprotein.
1.3.2AminoAcidSequence
Theaminoacidsequenceofaproteindetermineseveryaspectofitsmolecular structure.Ironically,theaspectofthestructureofachemicallysynthesizedprotein moleculethatismostoftenNOTexperimentallyverifiedistheaminoacidsequence ofaprotein‘spolypeptidechain.Becausethetargetaminoacidsequenceofthe syntheticprotein’spolypeptidechainisknown,itisstraightforwardtoverifythe aminoacidsequenceofasyntheticpolypeptidechainbyenzymaticdigestionand HPLC-(MS-MS)analysisofthepeptidefragments[33].
1CharacterizationofProteinMoleculesPreparedbyTotalChemicalSynthesis
Theall-to-frequentfailuretoverifytheaminoacidsequenceofasyntheticproteinproductshouldberectified
1.3.3ChemicalAnalogues
Animportantobjectiveoftotalproteinsynthesisistomake uniquelychemical analogues ofthenaturalproteinmolecule,includingnon-codedaminoacids [45],stableanaloguesofpost-translationalmodifications[46],fixedelementsof secondarystructure[47],chemicalengineeringofthepolypeptidechainbackbone [48],site-specificlabelingwithisotopes[49]orfluorophores[50],glycoproteins [51,52]andglycoproteinmimetics[53],noveltopologiessuchascircular[54] orconcatenatedpolypeptidechains[55],proteindiastereomers[56–58],and mirror-imageproteinmolecules[59].Thus,inadditiontocharacterizingfeaturesof proteinmolecularstructurefoundinnaturalproteins,characterizationofsynthetic proteinsmustinclude verificationofthenovelfeaturesintroducedbychemical synthesis
1.3.4LimitationsofSPPS
Stepwisesynthesisofpolypeptidechainshasitslimitations.Becauseofthestatistical accumulationofresin-boundby-products,eventhemosthighlyoptimizedstepwise SPPSmethodsshould never beusedforthepreparationofpeptidesegmentslonger than ∼50aminoacidresidues.Dependingonaminoacidcompositionandthe sequenceofthetargetpeptide,thepracticallimitforpreparinghomogeneous syntheticpeptidesfreeofcloselyrelatedcontaminantpeptideco-products(e.g.deletions,terminations,deamidations,isopeptidebonds, N -to-O acylshifts,covalent modifications)canbeasshortas40orevenfeweraminoacidresidues[60].Effective purificationandcarefulcharacterizationofeachsyntheticpeptidesegmentusedin totalproteinsynthesisisessential.
1.3.5FoldingasaPurificationStep
Therealityisthatnomatterhowmeticulouslytheyhavebeenpurified,30–50amino acidresiduepeptidesegmentspreparedbystepwiseSPPSwillalmostalwayscontain somelevelofcloselyrelatedpeptideimpurities.Evensmallamountsofimpurities presentinthepeptidesegmentsusedinproteinsynthesiswillcarryoverineach bond-formingstepandfrequentlywillbefoundinthefull-lengthsyntheticpolypeptidechainafterpurification.Fortunately,particularlyfordisulfide-constrainedglobularproteins,thefoldingstepcanbeaneffectivefurtherpurificationinwhichclosely relatedpolypeptidechainsthatdonotfoldcorrectlyaresubsequentlyremovedduringpurificationofthefoldedsyntheticprotein.ExamplesofLCMSdatabeforeand directinfusionESI-MSdataafterfoldingareshowninFigure1.5.
HPLC on line ESI-MS
Linear 102 aa polypeptide chain
11932.8±0.4 Da
Direct infusion ESI-MS
Folded 204 aa protein molecule
Covalent dimer 23 849.0±1.5 Da calc. mass: 23 849.1Da (av. isotopes)
05101520
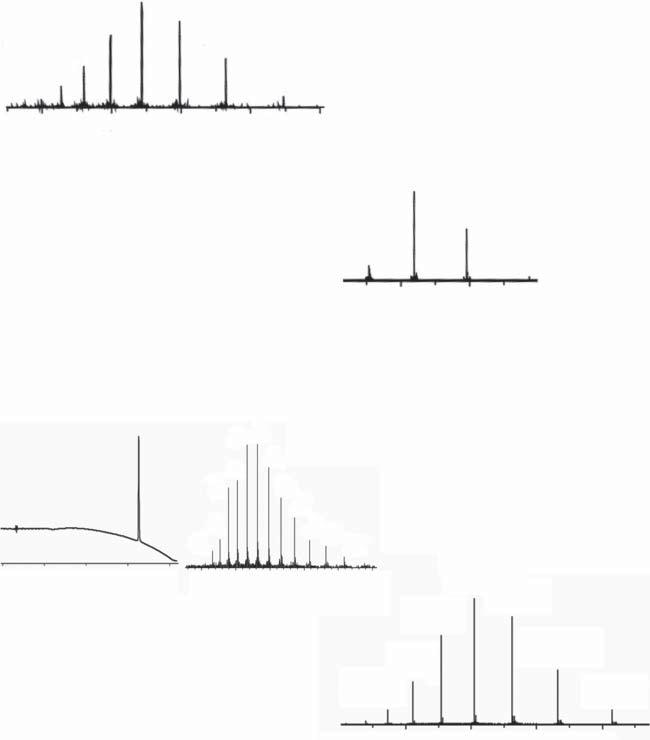
Figure1.5 Thefoldingstepcansignificantlyreducetheamountsofresidualimpurities presentinsyntheticpolypeptidechains.Twoexamplesareshownhere.(a)VEGFA:(upper left)MSdatafromLCMSofpurifiedlinearVEGFApolypeptidechain–ESI-MSdatawere acquiredacrosstheentireUVabsorbingpeakintheLCchromatogram;(lowerright)Direct infusionESI-MSofpurifiedfoldedcovalenthomodimerVEGFAsyntheticproteinmolecule. Notethereducedamountsofimpuritiesonthelow m/z sideofeachchargestateinthe foldedproteindata.Source:AdaptedfromMandalandKent[21].(b)[Lys24,38,83 ]EPO:[upper left]MSdatafromLCMSofpurifiedlinear[Lys24,38,83 ]EPOpolypeptidechain–ESI-MSdata wereacquiredacrosstheentireUVabsorbingpeakshownintheLCchromatogramtothe left;(lowerright)DirectinfusionESI-MSofpurifiedfolded[Lys24,38,83 ]EPOsyntheticprotein molecule.Notetheessentialabsenceofimpuritiesonthelow m/z sideofeachchargestate inthefoldedproteindata.Smallpeakstotherightofeachchargestatearesodiumand calciumionadducts.MSdatarepresentationshavebeendigitallyscaledtogivethesame verticaltotalioncountandhorizontal m/z scales.Adaptedfrom[22].