Population Genetics and Microevolutionary Theory 2nd Edition
Alan R. Templeton
Visit to download the full and correct content document: https://ebookmass.com/product/population-genetics-and-microevolutionary-theory-2n d-edition-alan-r-templeton/
More products digital (pdf, epub, mobi) instant download maybe you interests ...
Molecular population genetics Hahn
https://ebookmass.com/product/molecular-population-genetics-hahn/
Circuit Theory and Transmission Lines 2nd Edition Ravish R. Singh
https://ebookmass.com/product/circuit-theory-and-transmissionlines-2nd-edition-ravish-r-singh/
Population Health 2nd Edition, (Ebook PDF)
https://ebookmass.com/product/population-health-2nd-editionebook-pdf/
Genetics and Evolution of Infectious Diseases 2nd Edition Michel Tibayrenc (Editor)
https://ebookmass.com/product/genetics-and-evolution-ofinfectious-diseases-2nd-edition-michel-tibayrenc-editor/
Circuit Theory and Networks Analysis and Synthesis 2nd Edition Ravish R. Singh
https://ebookmass.com/product/circuit-theory-and-networksanalysis-and-synthesis-2nd-edition-ravish-r-singh/
Genetics and Genomics in Nursing Health Care 2nd Edition
https://ebookmass.com/product/genetics-and-genomics-in-nursinghealth-care-2nd-edition/
Population: An Introduction to Concepts and Issues 13th Edition John R. Weeks
https://ebookmass.com/product/population-an-introduction-toconcepts-and-issues-13th-edition-john-r-weeks/
Diplomacy: Theory and Practice 6th Edition G. R. Berridge
https://ebookmass.com/product/diplomacy-theory-and-practice-6thedition-g-r-berridge/
The Wiley International Handbook on Psychopathic Disorders and the Law 1st Edition Alan R. Felthous
https://ebookmass.com/product/the-wiley-international-handbookon-psychopathic-disorders-and-the-law-1st-edition-alan-rfelthous/
PopulationGeneticsand MicroevolutionaryTheory
PopulationGeneticsandMicroevolutionaryTheory
SecondEdition
AlanR.Templeton DepartmentofBiology
WashingtonUniversity St.Louis,MO,USA
Thissecondeditionfirstpublished2021
©2021JohnWiley&Sons,Inc.
EditionHistory
2006JohnWiley&Sons,Inc.
Allrightsreserved.Nopartofthispublicationmaybereproduced,storedinaretrievalsystem,ortransmitted, inanyformorbyanymeans,electronic,mechanical,photocopying,recording,orotherwise,exceptas permittedbylaw.Adviceonhowtoobtainpermissiontoreusematerialfromthistitleisavailableat http://www.wiley.com/go/permissions.
TherightofAlanR.Templetontobeidentifiedastheauthorofthisworkhasbeenassertedinaccordancewithlaw.
RegisteredOffice
JohnWiley&Sons,Inc.,111RiverStreet,Hoboken,NJ07030,USA
EditorialOffice 9600GarsingtonRoad,Oxford,OX42DQ,UK
Fordetailsofourglobaleditorialoffices,customerservices,andmoreinformationaboutWileyproducts,visitusat www.wiley.com.
Wileyalsopublishesitsbooksinavarietyofelectronicformatsandbyprint-on-demand.Somecontentthatappearsin standardprintversionsofthisbookmaynotbeavailableinotherformats.
LimitofLiability/DisclaimerofWarranty
Thecontentsofthisworkareintendedtofurthergeneralscientificresearch,understanding,anddiscussiononlyand arenotintendedandshouldnotberelieduponasrecommendingorpromotingscientificmethod,diagnosis,or treatmentbyphysiciansforanyparticularpatient.Inviewofongoingresearch,equipmentmodifications,changesin governmentalregulations,andtheconstantflowofinformationrelatingtotheuseofmedicines,equipment,and devices,thereaderisurgedtoreviewandevaluatetheinformationprovidedinthepackageinsertorinstructionsfor eachmedicine,equipment,ordevicefor,amongotherthings,anychangesintheinstructionsorindicationofusage andforaddedwarningsandprecautions.Whilethepublisherandauthorshaveusedtheirbesteffortsinpreparingthis work,theymakenorepresentationsorwarrantieswithrespecttotheaccuracyorcompletenessofthecontentsofthis workandspecificallydisclaimallwarranties,includingwithoutlimitationanyimpliedwarrantiesofmerchantability orfitnessforaparticularpurpose.Nowarrantymaybecreatedorextendedbysalesrepresentatives,writtensales materials,orpromotionalstatementsforthiswork.Thefactthatanorganization,website,orproductisreferredtoin thisworkasacitationand/orpotentialsourceoffurtherinformationdoesnotmeanthatthepublisherandauthors endorsetheinformationorservicestheorganization,website,orproductmayprovideorrecommendationsitmay make.Thisworkissoldwiththeunderstandingthatthepublisherisnotengagedinrenderingprofessionalservices. Theadviceandstrategiescontainedhereinmaynotbesuitableforyoursituation.Youshouldconsultwithaspecialist whereappropriate.Further,readersshouldbeawarethatwebsiteslistedinthisworkmayhavechangedor disappearedbetweenwhenthisworkwaswrittenandwhenitisread.Neitherthepublishernorauthorsshallbeliable foranylossofprofitoranyothercommercialdamages,includingbutnotlimitedtospecial,incidental,consequential, orotherdamages.
LibraryofCongressCataloging-in-PublicationData
Names:Templeton,AlanRobert,author.
Title:Populationgeneticsandmicroevolutionarytheory/AlanR. Templeton.
Description:Secondedition.|Hoboken,NJ:Wiley-Blackwell,2021.| Includesbibliographicalreferencesandindex.
Identifiers:LCCN2021006543(print)|LCCN2021006544(ebook)|ISBN 9781118504239(hardback)|ISBN9781118504369(adobepdf)|ISBN 9781118504345(epub)
Subjects:LCSH:Populationgenetics.|Evolution(Biology)
Classification:LCCQH455.T462021(print)|LCCQH455(ebook)|DDC 576.5/8–dc23
LCrecordavailableathttps://lccn.loc.gov/2021006543
LCebookrecordavailableathttps://lccn.loc.gov/2021006544
CoverDesign:Wiley
CoverImage:CourtesyofAlanR.Templeton
Setin9.5/12.5ptSTIXTwoTextbySPiGlobal,Pondicherry,India
10987654321
ToBonnie andtotheMemoryofLeonBlaustein
Contents
Preface xii
AbouttheCompanionWebsite xiv
1TheScopeandBasicPremisesofPopulationGenetics 1
BasicPremisesofPopulationGenetics 1
DNACanReplicate 1
DNACanMutateandRecombine 3
PhenotypesEmergefromtheInteractionofDNAandEnvironment 7
PopulationGenomics 9
Part1TheScopeandBasicPremisesofPopulationGenetics 17
2ModelingEvolutionandtheHardy–WeinbergLaw 19
HowtoModelMicroevolution 19
TheHardy–WeinbergModel 21
AnExampleoftheHardy–WeinbergLaw 28
ImportanceoftheHardy–WeinbergLaw 30
Hardy–WeinbergforTwoLoci 32
SourcesofLinkageDisequilibrium 39
SomeImplicationsoftheImpactofEvolutionaryHistoryuponDisequilibrium 41
3SystemsofMating 45
Inbreeding 45
DefinitionsofInbreeding 47
AssortativeMating 61
ASimpleModelofAssortativeMating 61
TheCreationofLinkageDisequilibriumbyAssortativeMating 64
AssortativeMatingVersusInbreeding 68
AssortativeMating,LinkageDisequilibrium,andAdmixture 71
DisassortativeMating 73
4GeneticDrift 77
BasicEvolutionaryPropertiesofGeneticDrift 78
FounderandBottleneckEffects 83
GeneticDriftandDisequilibrium 88
GeneticDrift,Disequilibrium,andSystemofMating 89
EffectivePopulationSize 91
InbreedingEffectiveSize 93
VarianceEffectiveSize 97
SomeContrastsBetweenInbreedingandVarianceEffectiveSizes 102
EstimatingEffectivePopulationSizes 110
5GeneticDriftinLargePopulationsandCoalescence 121 NewlyArisenMutations 121
NeutralAlleles 122
TheNeutralTheoryandItsOrigins 122 CritiquesoftheNeutralTheory 129
TheCoalescent 134
TheBasicCoalescentProcess 135
CoalescencewithMutation 143
Genealogies,GeneTrees,andHaplotypeTrees 146 CoalescenceandSpeciesTrees 160
RecombinationandCoalescence 162
6GeneFlowandPopulationSubdivision 169
GeneFlowBetweenTwoLocalPopulations 169
TheBalanceofGeneFlowandDrift 172
AnExampleoftheBalanceofDriftandGeneFlow 183 FactorsInfluencingtheAmountandPatternofGeneFlow 197 Dispersal 197
IsolationbyDistanceandResistance 200
TotalEffectivePopulationSizeinSubdividedPopulations 212 MultipleModesofInheritanceandPopulationStructure 217 Admixture 220
IdentifyingSubpopulationsandPopulationStructure 223 AFinalWarning 235
7PopulationHistory 237
InferringHistoricalEffectivePopulationSizes 239
InferringHistoricalGeneFlowPatternsandAdmixtureEvents 243 UsingHaplotypeTreestoStudyPopulationHistory 249
ExpectedPatternsUnderIsolationbyDistance 257
ExpectedPatternsUnderFragmentation 257
ExpectedPatternsUnderRangeExpansion 259
MultiplePatternsinNested-CladeAnalysis 263
IntegratingHaplotypeTreeInferencesAcrossLociorDNARegions 264
Model-BasedApproachestoPhylogeographicAnalysis 274
DirectStudiesoverSpaceandPastTimes 282
HistoricalPopulationGeneticsandMacroevolution 287
Part2GenotypeandPhenotype 295
8BasicQuantitativeGeneticDefinitionsandTheory 297
“Simple” MendelianPhenotypes 298
ThePhenotypeofElectrophoreticMobility 298
ThePhenotypeofSickling 299
ThePhenotypeofSickleCellAnemia 300
ThePhenotypeofMalarialResistance 301
ThePhenotypeofHealth(Viability) 301
NatureVersusNurture? 302
TheFisherianModelofQuantitativeGenetics 304
QuantitativeGeneticMeasuresRelatedtotheMean 305
QuantitativeGeneticMeasuresRelatedtotheVariance 318
9QuantitativeGenetics:UnmeasuredGenotypes 321
CorrelationBetweenRelatives 322
TheDistinctionBetweenHeritabilityandInheritance 329
ResponsetoSelection 331
TheProblemofBetween-PopulationDifferencesinMeanPhenotype 332 ControlledCrossesfortheAnalysesofBetweenPopulationDifferences 338
TheBalanceBetweenMutation,Drift,andGeneFlowUponPhenotypicVariance 343
10QuantitativeGenetics:MeasuredGenotypes 345
MarkerAssociationStudies 347
AdmixtureMapping 347
MarkersofLinkage 350
Genome-WideAssociationStudies 356
CandidateLoci 366
CandidateLociandGeneticArchitecture 379
Pleiotropy 380
Epistasis 381
PleiotropyandEpistasis 387
GenebyEnvironmentInteractions 389
GeneticArchitecture:IstheWholeGreaterthantheSumoftheParts? 390
Part3NaturalSelectionandAdaptation 393
11NaturalSelection 395
TheFundamentalEquationofNaturalSelection:MeasuredGenotypes 397
Sickle-cellAnemiaasanExampleofNaturalSelection 400
AdaptationasaPolygenicProcess 409
TheFundamentalTheoremofNaturalSelection:UnmeasuredGenotypes 411
SomeImplicationsoftheFundamentalEquationsofNaturalSelection 413
TheCourseofAdaptationandNaturalSelection 422
Contents
12InteractionsofNaturalSelectionwithOtherEvolutionaryForcesandthe DetectionofNaturalSelection 423
TheInteractionofNaturalSelectionwithMutation 424
TheInteractionofNaturalSelectionwithMutationandSystemofMating 426
TheInteractionofNaturalSelectionwithGeneFlow 429
TheInteractionofNaturalSelectionwithGeneticDrift 435
TheInteractionsofNaturalSelection,GeneticDrift,andGeneFlow 441
PopulationSubdivision 445
GeneticArchitecture 446
TheInteractionsofNaturalSelection,GeneticDrift,andMutation 451
TheInteractionsofNaturalSelection,GeneticDrift,Mutation,andRecombination 465 CandidateLoci 468
QuantitativeGeneticApproachestoDetectingSelection 471
TheNeutralist/SelectionistDebate 474
13UnitsandTargetsofSelection 475
TheUnitofSelection 477
TargetsofSelectionBelowtheLeveloftheIndividual 483
TheGenome 483
Gametes 489
SomaticCells 501
OverviewofSelectionBelowtheLevelofanIndividual 502
TargetsofSelectionAbovetheLeveloftheIndividual 503
SexualSelection 503
Fertility/Fecundity 508
CompetitionandCooperation 511
Kin/FamilySelection 516
14SelectioninHeterogeneousEnvironments 523
Coarse-grainedSpatialHeterogeneity 524
Coarse-grainedTemporalHeterogeneity 546
SeasonalandCyclicalVariation 547
RandomorFrequentTemporalVariation 549
Sporadic,RecurrentEnvironments 551
TransitionstoaNewLong-termEnvironment 554
Fine-grainedHeterogeneity 558
Coevolution 567
15SelectioninAge-StructuredPopulations 574
LifeHistoryandFitness 574
TheEvolutionofSenescence 581
AbnormalAbdomen:AnExampleofSelectioninanAge-StructuredPopulation 586
GeneticArchitectureandUnitsandTargetsofSelectionBelowtheLevelof theIndividual 586
GeneticArchitectureandUnitsofSelectionattheLeveloftheIndividual 592
PhenotypesandPotentialTargetsofSelectionattheLeveloftheIndividual 594
NaturalSelectiononthe aa SupergeneinaSpatiallyandTemporallyHeterogeneous
Environment 603
NaturalSelectionontheComponentsofthe aa Supergene 614
Overview 616
ComparativeAnalysis 617
Reductionism 617
Holism 618
MonitoringPopulations 620
AppendixA:GeneticSurveyTechniques 622
AppendixB:ProbabilityandStatistics 636
References 668
Index 723
Preface
Muchhaschangedinpopulationgeneticssincethefirsteditioncameoutin2006,andmuchhas stayedthesame.Thescope,questions,andmethodsofpopulationgeneticshavealwaysbeenconstrainedbythetechniquesavailableforsurveyinggeneticvariation,andtheDNA/genomicsrevolutioncontinuestoaltertheseconstraints.Yet,thebasicpremisesofpopulationgenetictheoryand practicehaveremainedthesame.Thesecorepremisesallowthissecondeditiontogrowsmoothly outofthefirstedition,sothebasicstructureofthiseditionisidenticaltothatofthefirst.However, therevolutioninDNAtechnologiesandgenomicsleadstoasignificantexpansionoftopicsand scope.Theconceptsandmethodsfrompopulationgeneticsarenowwidelyusedinalmostallfields ofbiology,frommolecularbiologytoecology,andinappliedfieldsfromgeneticepidemiologyto conservationbiology.
Thisexpandedapplicabilityofpopulationgeneticsisillustratedwellbythefinalresearchproject ofmylong-termcollaboratorandfriend,LeonBlaustein,whosadlydiedalltooyoungin2020.By trainingandcareer,Leonwasafreshwaterpopulation/communityecologistwithagreatconcern forconservationapplications.Hisfinalprojectfocusedonplasticityinaquaticresourceuseinthe endangeredspecies Salamandrainfraimmaculta atitssouthernmostboundaryinnorthernIsrael. Mostsalamandersspecializeinthetypeofaquatichabitattheyusefortheirlarvalphase,but S. infraimmaculta canusepermanentpondsandsprings,permanentandseasonalstreams,aswell asrockpoolsthatfillwithrainwaterforonlyafewmonthsoftheyear.Leonassembledadiverse teamofgraduatestudents,post-docs,andcollaboratorstoaddressthissystemfrommultipledirections.Leonfirstsoughttodefinetheevolutionaryandecologicalcontextinwhichthisplasticityis manifest.Leonandhisgroupperformedsurveysofmoleculargeneticdiversity,maximumentropy modelingofhabitatdatatodetermineoptimalandsub-optimalareas,andphylogeographicanalysestouncoverhistoricaleffects.Hisgroupperformedmark/recapturestudiestoestimateadult populationsizesinadiversityofhabitats.Thesestudiesrevealedthatthissouthernmostboundary washighlyheterogeneous,varyingfromareasofoptimalhabitatwithhighlevelsofgeneticdiversityandgeneflow,toareasofmarginalhabitatthatwerestronglysubdividedgenetically,andtoan areaofoptimalhabitatthatwasanhistoricalisolatewithaseverereductioningeneticdiversitydue toapastfounderevent.Fieldandlaboratoryexperimentsonlarvaldevelopmentalandmorphologicalplasticityaswellasontheirtranscriptomesrevealedthatplasticityatboththeorganismaland transcriptomelevelsdisplayedbothgeneticallybaseddifferencesamongthesepopulationsand directindividualresponsivenesstoenvironmentalvariation.Couplingclimateprojectionmodels withthemaximumentropymodelsrevealeddiverseconservationchallengesinthesedifferent areasofthesouthernmostboundary.Hence,boththeevolvabilityofplasticityandcurrentindividualenvironmentalplasticitycouldplayanimportantroleinthesurvivalofthisendangeredspecies.
Althoughnotapopulationgeneticist,Leon’sfinalprojectillustrateswelltheimportanceofusing populationgeneticsandgenomicsinotherfieldsofbiology.
Theincreasingscopeandrelevanceofpopulationgeneticstomanyfieldsofbasicandapplied biologyalsomeansanexpandedaudiencethatneedstobeknowledgeableofpopulationgenetic theoryandpractice.Thiseditioniswrittenwiththisexpandedaudienceinmind.Manyexamples aregivenfromconservationbiology,humangenetics,andgeneticepidemiology,yetthefocusof thisbookremainsonthebasicmicroevolutionarymechanismsandhowtheyinteracttocreateevolutionarychange.Thisbookisintendedtoprovideasolidunderstandingofthecoreconceptsin populationgeneticsbothforthosestudentsprimarilyinterestedinevolutionarybiologyandgeneticsandforthosestudentsprimarilyinterestedinapplyingthetoolsofpopulationgeneticsinother areassuchasconservationbiology,geneticepidemiology,andgenomics.Withoutasolidfoundationinpopulationgenetics,theanalyticaltoolsemergingfrompopulationgeneticswillfrequently bemisappliedandincorrectinterpretationscanbemade.Thisbookisdesignedtoprovidethat foundationbothforfuturepopulationandevolutionarygeneticistsandforthosewhowillbeapplyingpopulationgeneticconceptsandtechniquestootherareas.
IthankDavidQuellerforreadingoveradraftofthisedition.Hismanysuggestionsandcorrectionswerehighlyvaluable,andIgreatlyappreciatehisefforts.Davidalsousedsomeofthedraftsof thiseditioninthe2020classonPopulationGeneticsatWashingtonUniversity.Ithankthestudents ofthatclassfortheirsuggestionsandcorrections.Finally,Ithankallthepaststudentsofmycourse onpopulationgeneticsandmyformergraduatestudentsandpost-docs.Theyweretheinspiration forthisbookinthefirstplace,andtheycontributedvaluableinputtothefirstedition,muchof whichhascarriedovertothesecondedition.
AbouttheCompanionWebsite
Thisbookisaccompaniedbyacompanionwebsite.
www.wiley.com/go/templeton/populationgenetics2
Thiswebsiteincludes:
• PowerpointslidesofFiguresfromthebook
• ProblemandAnswersets
TheScopeandBasicPremisesofPopulationGenetics
Populationgeneticsisconcernedwiththeorigin,amount,anddistributionofgeneticvariation presentinpopulationsoforganismsandthefateofthisvariationthroughspaceandtime.Thekinds ofpopulationsthatwillbetheprimaryfocusofthisbookarepopulationsofsexuallyreproducing diploidorganisms,andthefateofgeneticvariationinsuchpopulationswillbeexaminedator belowthespecieslevel.Variationingenesthroughspaceandtimeconstitutesthefundamental basisofevolutionarychange;indeed,initsmostbasicsense, evolution isthegenetictransformationofreproducingpopulationsoverspaceandtime.Populationgeneticsis,therefore,atthevery heartofevolutionarybiologyandcanbethoughtofasthescienceofthemechanismsresponsible for microevolution,evolutionwithinspecies.Manyofthesemechanismshaveagreatimpact ontheoriginofnewspeciesandonevolutionabovethespecieslevel(macroevolution).Afew oftheimpactsofpopulationgeneticsuponspeciesandspeciationwillbediscussed,butthisis notthemainfocusofthisbook.
BasicPremisesofPopulationGenetics
Microevolutionarymechanismsworkupongeneticvariability,soitisnotsurprisingthatthe fundamentalpremisesthatunderliepopulationgenetictheoryandpracticealldealwithvarious propertiesofDNA,themoleculethatencodesgeneticinformationinmostorganisms.(AfeworganismsuseRNAastheirgeneticmaterial,andthesamepropertiesapplytoRNAinthosecases.) Indeed,thetheoryofmicroevolutionarychangestemsfromjustthreepremises:
• DNAcanreplicate
• DNAcanmutateandrecombine
• PhenotypesemergefromtheinteractionofDNAandenvironment
Theimplicationsofeachofthesepremiseswillnowbeexamined.
DNACanReplicate
BecauseDNAcanreplicate,aparticularkindofgene(specificsetofnucleotides)canbepassedon fromonegenerationtothenextandcanalsocometoexistasmultiplecopiesindifferentindividuals.Genes,therefore,haveanexistenceintimeandspacethattranscendstheindividualsthat temporarilybearthem.Thebiologicalexistenceofgenesoverspaceandtimeis thephysicalbasis ofevolution
PopulationGeneticsandMicroevolutionaryTheory,Second Edition.AlanR.Templeton. ©2021JohnWiley&Sons,Inc.Published2021byJohnWiley&Sons,Inc. Companionwebsite:www.wiley.com/go/templeton/populationgenetics2
Thephysicalmanifestationofagene’scontinuityovertimeandthroughspaceisareproducing populationofindividuals.Individualshavenocontinuityoverspaceortime;individualsareunique eventsthatliveandthendieandcannotevolve.ButthegenesthatanindividualbearsarepotentiallyimmortalthroughDNAreplication.Forthispotentialtoberealized,theindividualsmust reproduce.Therefore,toobserveevolution,itisessentialtostudyapopulationofreproducingindividuals.Areproducingpopulationdoeshavecontinuityovertimeasonegenerationofindividuals isreplacedbythenext.Areproducingpopulationgenerallyconsistsofmanyindividuals,andthese individualscollectivelyhaveadistributionoverspace.Hence,areproducingpopulationhascontinuityovertimeandspaceandconstitutesthephysicalrealityofagenes’ continuityovertimeand space.Evolutionisthereforepossibleonlyatthelevelofareproducingpopulationandnotatthe leveloftheindividualscontainedwithinthepopulation.
Thefocusofpopulationgeneticsmustbeuponreproducingpopulationstostudymicroevolution. However,theexactmeaningofwhatismeantbyapopulationisnotfixed,butrathercanvary dependinguponthequestionsbeingaddressed.Thepopulationcouldbealocalbreedinggroup ofindividualsfoundinclosegeographicproximity,oritcouldbeacollectionoflocalbreeding groupsdistributedoveralandscapesuchthatmostindividualsonlyhavecontactwithothermembersoftheirlocalgroupbutthat,onoccasion,thereissomereproductiveinterchangeamonglocal groups.Alternatively,apopulationcouldbeagroupofindividualscontinuouslydistributedovera broadgeographicalareasuchthatindividualsattheextremesoftherangeareunlikelytoevercome intocontact.Sometimes,thepopulationincludestheentirespecies.Withinthishierarchyofpopulationsfoundwithinspecies,muchofpopulationgeneticsfocusesuponthe localpopulation or deme,acollectionofinterbreedingindividualsofthesamespeciesthatliveinsufficientproximity thattheyshareasystemofmating.Systemsofmatingwillbediscussedinmoredetailinsubsequent chapters,but,fornow,the systemofmating referstotherulesbywhichindividualspairforsexual reproduction.Theindividualswithinademeshareacommonsystemofmating.Becauseademeis abreedingpopulation,individualsarecontinuallyturningoverasbirthsanddeathsoccur,butthe localpopulationisadynamicentitythatcanpersistthroughtimefarlongerthantheindividuals thattemporarilycompriseit.Thelocalpopulation,therefore,hastheattributesthatallowthephysicalmanifestationofthegeneticcontinuityoverspaceandtimethatfollowsfromthepremisethat DNAcanreplicate.
Becauseourprimaryinterestisongeneticcontinuity,wewillmakeausefulabstractionfrom thedeme.Associatedwitheverypopulationofindividualsisacorrespondingpopulationofgenes calledthe genepool ,thesetofgenescollectivelysharedbytheindividualsofthepopulation.An alternative,andoftenmoreuseful,wayofdefiningthegenepoolisthatthegenepoolisthepopulationofpotentialgametesproducedbyalltheindividualsofthepopulation.Gametesarethe bridgesbetweenthegenerations,sodefiningagenepoolasapopulationofpotentialgametes emphasizesthegeneticcontinuityovertimethatprovidesthephysicalbasisforevolution. Forempiricalstudies,thefirstdefinitionisprimarilyused;fortheory,theseconddefinitionis preferred.
Thegenepoolassociatedwithapopulationisdescribedbymeasuringthenumbersandfrequenciesofthevarioustypesofgenesorgenecombinationsinthepool. Evolution isdefinedasachange throughtimeofthefrequenciesofvarioustypesofgenesorgenecombinationsinthegenepool. Thisdefinitionisnotintendedtobeanall-encompassingdefinitionofevolution.Rather,itis anarrowandfocuseddefinitionofevolutionthatisusefulinmuchofpopulationgenetics preciselybecauseofitsnarrowness.Thiswillthereforebeourprimarydefinitionofevolutionin thisbook.
Sinceonlyalocalpopulationattheminimumcanhaveagenepool,onlypopulationscanevolve underthisdefinitionofevolution,notindividuals.Therefore,evolutionisanemergentpropertyof reproducingpopulationsofindividualsthatisnotmanifestedintheindividualsthemselves.However,therecanbehigherorderassemblagesoflocalpopulationsthatcanevolve.Inmanycases,we willconsidercollectionsofseverallocalpopulationsthatareinterconnectedbydispersalandreproduction,uptoandincludingtheentirespecies.However,anentirespeciesinsomecasescouldbe justasingledemeoritcouldbeacollectionofmanydemeswithlimitedreproductiveinterchange. Aspeciesisthereforenotaconvenientunitofstudyinpopulationgeneticsbecausespeciesstatus itselfdoesnotdefinethereproductivestatusthatissocriticalinpopulationgenetictheory.Wewill alwaysneedtospecifythetypeandlevelofreproducingpopulationthatisrelevantforthequestions beingaddressed.
DNACanMutateandRecombine
Evolutionrequireschange,andchangecanonlyoccurwhenalternativesexist.IfDNAreplication werealways100%accurate,therewouldbenoevolution.Anecessaryprerequisiteforevolutionis geneticdiversity.Theultimatesourceofthisgeneticdiversityismutation.Therearemanyformsof mutation,suchassinglenucleotidesubstitutions,insertions,deletions,transpositions,andduplications.Fornow,ouronlyconcernisthatthesemutationalprocessescreatediversityinthepopulationofgenespresentinagenepool.Becauseofmutation,alternativecopiesofthesame homologousregionofDNAinagenepoolwillshowdifferentstates.
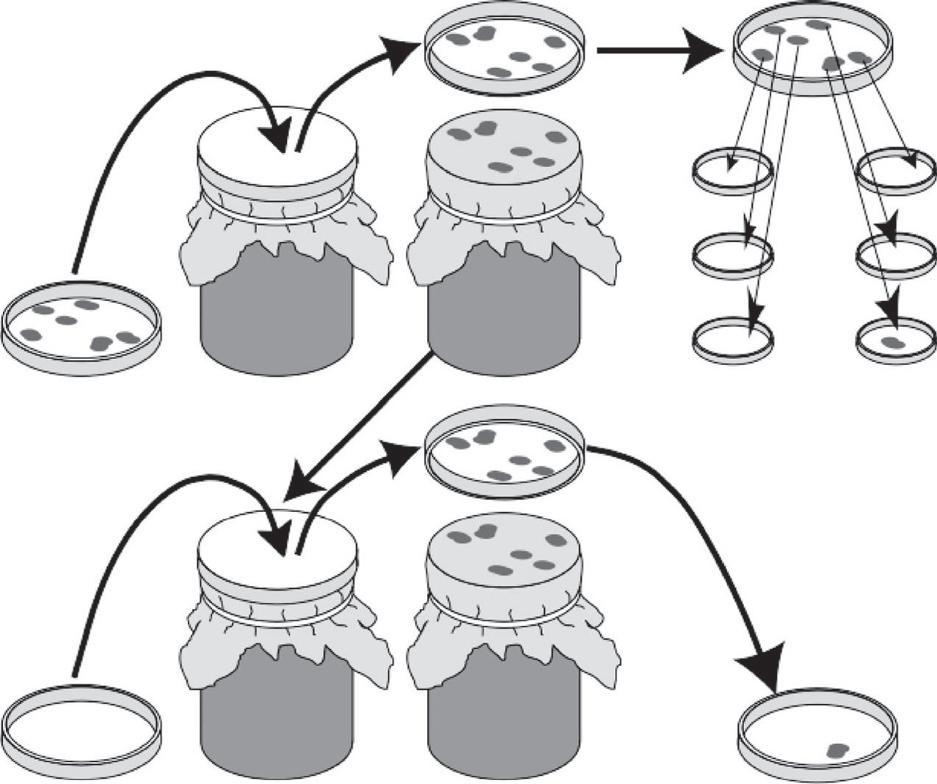
Mutationoccursatthemolecularlevel.Althoughmanyenvironmentalagentscaninfluencethe rateandtypeofmutation,oneofthecentraltenetsofDarwinianevolutionisthatmutationsare randomwithrespecttotheneedsoftheorganismincopingwithitsenvironment.Therehavebeen manyexperimentsaddressingthistenet,butoneofthemoreelegantandconvincingisreplicaplating,firstusedbyJoshuaandEstherLederberg(1952)(Figure1.1).Replicaplatingandotherexperimentsprovideempiricalproofthatmutation,occurringonDNAatthemolecularlevel,isnotbeing directedtoproduceaparticularphenotypicconsequenceatthelevelofanindividualinteracting withitsenvironment.Therefore,wewillregardmutationsasbeingrandomwithrespecttothe organism’sneedsincopingwithitsenvironment(although,aswewillseesoon,mutationishighly nonrandominotherrespectsatthemolecularlevel).
Mutationcreatesallelicdiversity. Alleles arealternativeformsofagene.Insomecases,genetic surveysfocusonaregionofDNAthatmaynotbeageneinaclassicalsense;itmaybeaDNAregion muchlargerorsmallerthanagene,oranoncodingregion.Wewillusetheterm haplotype to refertoanalternativeform(specificnucleotidesequence)amongthehomologouscopiesofa definedDNAregion,whetherageneornot.Theallelicorhaplotypicdiversitycreatedbymutation canbegreatlyamplifiedbythegeneticmechanismsofrecombinationanddiploidy.Inmuchof genetics,recombinationreferstomeioticcrossingover,butweusethetermrecombinationin abroadersenseasanygeneticmechanismthatcancreatenewcombinationsofallelesorhaplotypes.Thisdefinitionofrecombinationencompassesthemeioticeventsofbothindependent assortmentandofcrossingoverandalsoincludesgeneconversionandanynon-meioticeventsthat createnewgenecombinationsthatcanbepassedonthroughagametetothenextgeneration.Sexualreproductionanddiploidycanalsobethoughtofasmechanismsthatcreatenewcombinations ofgenes.
Asanillustrationofthegeneticdiversitythatcanbegeneratedbythejointeffectsofmutationand recombination,considerthe MHC complex(majorhistocompatibilitycomplex,alsoknownin
Plate 1. Bacteria Grown in Absence of Streptomycin
Bacterial Plate Pressed on Velvet
Plate 2. Sterile Plate with Streptomycin
Sterile Plate Pressed on Imprinted Velvet
Bacterial Colonies Imprinted on Velvet
Each Bacterial Colony on Plate 1 is Isolated and Tested for Growth on a Plate with Streptomycin: Only one Colony Grows
Only one Colony Crows on Streptomycin
Figure1.1 Replicaplating.AsuspensionofbacterialcellsisspreaduponaPetridish(plate1)suchthateach individualbacteriumshouldbewellseparatedfromallothers.Eachbacteriumthengrowsintoacolonyof geneticallyidenticalindividuals.Next,acircularblockcoveredwithvelvetispressedontothesurfaceofplate 1.Somebacteriafromeachcolonysticktothevelvet,soaduplicateoftheoriginalplateismadewhenthe velvetispressedontothesurfaceofasecondPetridish(plate2),calledthereplicaplate.Themediumonthe replicaplatecontainsstreptomycin,anantibioticthatkillsmostbacteriafromtheoriginalstrain.Inthe exampleillustrated,onlyonebacterialcolonyonthereplicaplatecangrowonstreptomycin,anditsposition onplate2identifiesitasthedescendantofaparticularcolonyonplate1.Eachbacterialcolonyonplate1is thentestedforgrowthonaplatewiththeantibioticstreptomycin.Ifmutationswererandomandstreptomycin simplyselectedpreexistingmutationsratherthaninducingthem,thenthecoloniesonplate1thatoccupied thepositionsassociatedwithresistantcoloniesonplate2shouldalsoshowresistance,eventhoughthese colonieshadnotyetbeenexposedtostreptomycin.Asshown,thiswasindeedthecase.
humansas HLA,humanleukocyteantigen)ofabout200genesonthesamechromosome.Table1.1 showsthenumberofallelesfoundat26oftheselociasofJuly2018inhumanpopulationsin an HLA database(Robinsonetal.2015).Ascanbeseen,mutationalchangesattheseloci havegeneratedfrom1to5212alleles/locuswithatotalof18690allelesoverall26loci.However, theselocicananddorecombine.Hence,recombinationhasthepotentialofcombiningthese 18690allelesinto7.48×1042 distinctgametetypes(obtainedbymultiplyingtheallelenumbers ateachlocus).Sexualreproductionhasthepotentialofbringingtogetherallpairsofthesegamete typesinadiploidindividual,resultingin2.07×1079 genotypes.Andthisisonlyfrom26lociinone smallregionofonechromosomeofthehumangenome!Giventhatthereareonlyabout7.7×109 humansintheworldatthebeginningof2019,everyoneintheworld(withtheexceptionofidentical twins)willhaveaunique HLA genotypewhenthese26lociareconsideredsimultaneouslyasthere aremanymorepossiblegenotypesthanhumanindividuals.But,ofcourse,humansdifferatmany
Table1.1 Numbersofallelesknownin2018at26loci withinthe human MHC (HLA)region.
Source: DatabasedescribedinRobinsonetal.(2015)asupdated onJuly2018.
morelocithanjustthese26.Asof2015,88milliongeneticvariantswereknowninthehuman genome(The1000GenomesProjectConsortium2015).Assumingthatmostofthesearebiallelic, eachpolymorphicnucleotidedefinesthreegenotypes,socollectivelythenumberofpossible genotypesdefinedbytheseknownpolymorphicsitesis388,000,000 =1041,986670 genotypes.Toput thisnumberintoperspective,thenumberofelectronsintheknownuniverseis1081 (https://www.quora.com/Approximately-how-many-electrons-are-there-in-the-known-Universe), anumberfarsmallerthanthenumberofpotentialgenotypesthatarepossibleinhumanityjust withknowngeneticvariation.Hence,mutationandrecombinationcangeneratetrulyastronomical levelsofgeneticvariation.
Thedistinctionbetweenmutationandrecombinationisoftenblurredbecauserecombination canoccurwithinageneandtherebycreatenewallelesorhaplotypes.Forexample,71individualsfromthreehumanpopulationsweresequencedfora9.7kbregionwithinthe lipoprotein lipase (LPL)locus(Nickersonetal.1998).Thisrepresentsjustaboutathirdofthisonelocus.In all,88variablesiteswerediscovered,and69ofthesesiteswereusedtodefine88distincthaplotypesoralleles.These88haplotypesarosefromatleast69mutationalevents(aminimumof onemutationforeachofthe69variablenucleotidesites)coupledwithabout30recombination andgeneconversionevents(Templetonetal.2000a).Thus,intragenicrecombinationandmutationhavetogethergenerated88haplotypesasinferredusingonlyasubsetoftheknownvariable sitesinjustathirdofasinglegeneinasampleof142chromosomes.These88haplotypes inturndefine3916possiblegenotypes – anumberconsiderablylargerthanthesamplesize of71people.
Studiessuchasthosementionedabovemakeitclearthatmutationandrecombinationcan generatelargeamountsofgeneticdiversityatparticularlociorchromosomalregions,butthey donotaddressthequestionofhowmuchgeneticvariationispresentwithinspeciesingeneral. Howmuchgeneticvariationispresentinnaturalpopulationswasoneofthedefiningquestions ofpopulationgeneticsupuntilthemid-1960s.Beforethen,mostofthetechniquesusedtodefine genesrequiredgeneticvariationtoexist.Forexample,manyoftheearlyimportantdiscoveriesin MendeliangeneticsweremadeinthelaboratoryofThomasHuntMorganduringthefirstfew decadesofthetwentiethcentury.Thislaboratoryusedmorphologicalvariationinthefruitfly Drosophilamelanogaster asitssourceofmaterialtostudy.Amongthegenesidentifiedinthis laboratorywasthelocusthatcodesforanenzymeineyepigmentbiosynthesisknownas vermillion in Drosophila.Morganandhisstudentscouldonlyidentify vermillion asageneticlocus becausetheyfoundamutantthatcodedforadefectiveenzyme,therebyproducingaflywith brightredeyes.Ifageneexistedwithnoallelicdiversityatall,itcouldnotevenbeidentified asalocuswiththetechniquesusedinMorgan’slaboratory.Hence, all observablelocihadatleast twoallelesinthesestudies(the “wildtype” and “mutant ” allelesinMorgan ’sterminology).Asa result,eventhesimplequestionofhowmanylocihavemorethanoneallelecouldnotbe answereddirectly.Thissituationchangeddramaticallyinthemid-1960swiththefirstapplicationsofmoleculargeneticsurveys(firstonproteins,laterontheDNAdirectly;seeAppendix A,whichgivesabriefsurveyofthemoleculartechniquesusedtomeasuregeneticvariation). Thesenewmoleculartechniquesallowedgenestobedefinedbiochemicallyandirrespective ofwhetherornottheyhadallelicvariation.Theinitialstudies(Harris1966;Johnsonetal. 1966;LewontinandHubby1966),usingtechniquesthatcouldonlydetectmutationscausing aminoacidchangesinproteincodingloci(andonlyasubsetofallaminoacidchangesatthat), revealedthataboutathirdofallproteincodinglociwerepolymorphic(i.e.alocuswithtwo ormoreallelessuchthatthemostcommonallelehasafrequencyoflessthan0.95inthegene pool)inavarietyofspecies.Asourgeneticsurveytechniquesacquiredgreaterresolution (AppendixA),thisfigurehasonlygoneup.
Thesegeneticsurveyshavemadeitclearthatmanyspecies,includingourown,haveliterally astronomicallylargeamountsofgeneticvariation.ThechaptersinSection1ofthisbookwillexaminehowpremises1and2combinetoexplaingreatcomplexityatthepopulationlevelintermsof theamountofgeneticvariationanditsdistributioninindividuals,withindemes,amongdemes, andoverspaceandtime.Becauseitisnowclearthatmanyspecieshavevastamountsofgenetic variation,thefieldofpopulationgeneticshasbecomelessconcernedwiththeamountofgenetic variationandmoreconcernedwithitsphenotypicandevolutionarysignificance.Thisshiftin emphasisleadsdirectlyintoourthirdandfinalpremise.
PhenotypesEmergefromtheInteractionofDNAandEnvironment
A phenotype isameasurabletraitofanindividual(oraswewillseelater,itcanbegeneralized tootherunitsofbiologicalorganization).InMorgan’sday,genescouldonlybeidentifiedthrough theirphenotypiceffects.Thegenewasoftennamedforitsphenotypiceffectinahighlyinbred laboratorystrainmaintainedundercontrolledenvironmentalconditions.Thismethodofidentifyinggenesledtoasimple-mindedequatingofgeneswithphenotypesthatstillplaguesustoday. Almostdaily,onereadsabout “thegeneforcoronaryarterydisease,”“thegeneforthrill seeking,” etc.Equatinggeneswithphenotypesisreinforcedbymetaphorsappearinginmany textbooksandsciencemuseumstotheeffectthatDNAisthe “blueprint” oflife.However, DNAisnotablueprintforanything;thatisnothowgeneticinformationisencodedorprocessed. Forexample,thehumanbraincontainsabout1011 neuronsand1015 neuronalconnections (CoveneyandHighfield1995).DoestheDNAprovideablueprintforthese1015 connections? Theanswerisanobvious “NO.” Thereareonlyabout3billionbasepairsinthehumangenome. Evenifeverybasepaircodedforabitofinformation,thereisinsufficientinformationstorage capacityinthehumangenomebyseveralordersofmagnitudetoprovideablueprintfortheneuronalconnectionsofthehumanbrain.DNAdoesnotprovidephenotypicblueprints;instead,the informationencodedinDNAcontrolsdynamicprocesses(suchasaxonalgrowthpatternsand signalresponses)thatalwaysoccurinanenvironmentalcontext.Thereisnodoubtthatenvironmentalinfluenceshaveanimpactonthenumberandpatternofneuronalconnectionsthat developinmammalianbrainsingeneral.Itisthisinteractionofgeneticinformationwith environmentalvariablesthroughdevelopmentalprocessesthatyieldsphenotypes(suchasthe precisepatternofneuronalconnectionsofaperson’sbrain).Genesshouldneverbeequated tophenotypes.Phenotypesemergefromgeneticallyinfluenceddynamicprocesseswhoseoutcome dependsuponenvironmentalcontext.
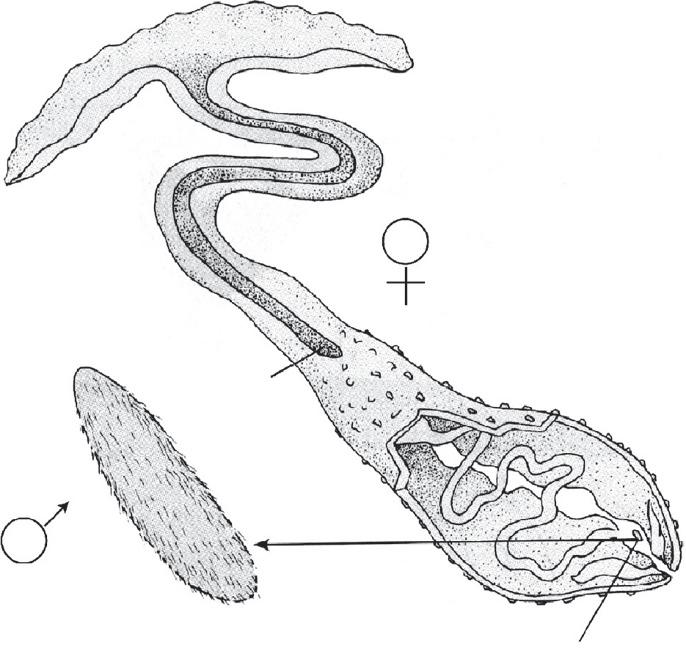
Inthisbook,phenotypesarealwaysregardedasarisingfromaninteractionofgenotypewith environment.Themarineworm Bonellia (Figure1.2)providesanexampleofthisinteraction (GilbertandSarkar2000).Thefree-swimminglarvalformsofthesewormsaresexuallyundifferentiated.Ifalarvasettlesaloneonthenormalmudsubstrate,itbecomesafemalewithalong (about15cm)tubeconnectingaproboscistoamoreroundedpartofthebodythatcontainsthe uterus.Ontheotherhand,thelarvaeareattractedtofemales,andifitcanfindafemale,itdifferentiatesintoamalethatexistsasaciliatedmicroparasiteinsidethefemale.Thebodyforms aresodifferenttheywereinitiallythoughttobetotallydifferentcreatures.Hence,thesame genotype,dependinguponenvironmentalcontext,canyieldtwodrasticallydifferentbodytypes. Theinteractionbetweengenotypeandenvironmentinproducingphenotypeiscriticalforunderstandingtheevolutionarysignificanceofgeneticvariability,sothechaptersinSection2will bedevotedtoanexplorationofthepremisethatphenotypesemergefromagenotype-byenvironmentinteraction.
Asapreludetowhytheinteractionofgenotypeandenvironmentissocriticaltoevolution, considerthefollowingphenotypesthatanorganismcandisplay:
• Beingaliveversusbeingdead:thephenotypeof viability (theabilityoftheindividualtosurvive intheenvironment)
• Givenbeingalive,havingmatedversusnothavingmated:thephenotypeof matingsuccess (theabilityofalivingindividualtofindamateintheenvironment)
• Givenbeingaliveandmated,thenumberofoffspringproduced:thephenotypeof fertility or fecundity (thenumberofoffspringthemated,livingindividualcanproduceinthe environment).
Figure1.2 Sexesin Bonellia.Thefemalehasawalnut-sizedbodythatisusuallyburiedinthemudwitha protrudingproboscis.Themaleisaciliatedmicroorganismthatlivesinsidethefemale. Source: Adapted fromFigure3.18from Genetics,3rdEdition,byPeterJ.Russell.Copyright©1992byPeterJ.Russell. ReprintedbypermissionofPearsonEducation,Inc.
Thethreephenotypesgivenaboveplayanimportantroleinmicroevolutionarytheorybecause, collectively,thesephenotypesdeterminethechancesofanindividualpassingonitsDNAinthe contextoftheenvironment. Reproductivefitness isthecollectivephenotypeproducedbycombiningthesethreecomponentsrequiredforpassingonDNAintoasinglemeasure.Fitnesswillbe discussedindetailinSection3ofthisbook.Reproductivefitnessturnspremise1(DNAcanreplicate)intoreality.DNAisnottrulyself-replicating.DNAcanonlyreplicateinthecontextofan individualsurvivinginanenvironment,matinginthatenvironment,andproducingoffspring inthatenvironment.Hence,thephenotypeofreproductivefitnessunitespremise3(phenotypes aregene-by-environmentinteractions)withpremise1(DNAcanreplicate).Thisunificationof premisesimpliesthattheprobabilityofDNAreplicationisdeterminedbyhowthegenotypeinteractswiththeenvironment.Inapopulationofgeneticallydiverseindividuals(arisingfrompremise 2thatDNAcanmutateandrecombine),itispossiblethatsomegenotypeswillinteractwiththe environmenttoproducemoreorfeweractsofDNAreplicationthanothergenotypes.Hence, theenvironmentinfluencestherelativechancesforvariousgenotypesofreplicatingtheirDNA. AswewillseeinSection3ofthisbook,thisinfluenceoftheenvironment(premise3)upon DNAreplication(premise1)ingeneticallyvariablepopulations(premise2)isthebasisfornatural selectionandoneofthemajoremergentfeaturesofmicroevolution: adaptation totheenvironment.Adaptationreferstoattributesandtraitsdisplayedbyorganismsthataidtheminliving andreproducinginspecificenvironments.Adaptationisoneofthemoredramaticfeaturesof evolution,and,indeed,itwasthemainfocusofthetheoriesofDarwinandWallace.Adaptation canonlybeunderstoodintermsofathree-wayinteractionamongallofthecentralpremisesof populationgenetics(Figure1.3).
Thisbookusesthesethreepremisesinaprogressivefashion:Section1utilizespremises1 (DNAcanreplicate)and2(DNAcanmutateandrecombine),whicharemolecularinfocus,
Proboscis
Mouth
Uterus
Premise I : DNA Can Replicate
Premise II: DNA Can Mutate & Recombine
Heritable Variation in the Phenotype of Reproductive Fitness
Premise III: Phenotypes Emerge from the Interaction of DNA and Environment
Genetically Variable Population of Individuals
Figure1.3 Theintegrationofthethreefundamentalpremisesofpopulationgenetictheorythroughthe phenotypeofreproductivefitness.
toexplaintheamountandpatternofgeneticvariationundertheassumptionthatthevariationhas nophenotypicsignificanceonreproductivefitness.Section2focusesuponpremise3(phenotypes emergefromtheinteractionofDNAandenvironment)andconsiderswhathappenswhen geneticvariationdoesinfluencephenotype.Finally,Section3considerstheemergentevolutionary propertiesthatarisefromtheinteractionsofallthreepremises(Figure1.3)andspecificallyfocuses uponadaptationthroughnaturalselection.Inthismanner,wehopetoachieveathoroughand integratedtheoryofmicroevolutionaryprocesses.
PopulationGenomics
Microevolutionaryprocessesdependupontheexistenceofgeneticvariationinthegenepool,soitis notsurprisingthatthequestionsaskedandapproachesusedinpopulationgeneticresearchare heavilyinfluencedbythetechniquesavailableforsurveyinggeneticvariationinapopulation. Theimportanceofthetechniqueusedtosurveyvariationwasalreadymentionedabovewith respecttooneoftheearliestandmostfundamentalquestionsofpopulationgenetics – howmuch variationexistsinagenepool?Asthefieldofmoleculargeneticsmaturedintogenomics,therehas beenexplosivegrowthintheabilitytoaddressevermorefundamentalquestionsinunprecedented detailinpopulationgenetics.Theapplicationofgenomictechniquestoaddresspopulationgenetic questionsisreferredtoas populationgenomics . Thefocusofthisbookisuponeukaryotic,sexuallyreproducingdiploidspecies.Suchspecieshave multiplegenomes. Agenome isacompletesetofthechromosomesthatarenormallypassedon throughagamete.BecauseeukaryotesarosefromasymbioticfusionoftwoorthreedifferentPrecambrianlineagesofprokaryotes,eukaryoticcellstypicallycarrytwoorthreedifferentgenomes withseparateprokaryoticorigins(Koonin2010).Thefirstisthenucleargenome,whichevolved

Figure1.4 Aphotomicrographofthenucleargenomeof Drosophilabusckii asvisualizedthroughpolytene chromosomes.Polytenechromosomesareformedinsometissuesfromendoreduplication – repeatedrounds ofDNA(deoxyribonucleicacid)replicationwithoutnucleardivision.Homologousstrandssitsidebyside, allowingahigh-resolutionvisualizationofthenuclearchromosomes. Source: AndrewSyred/ScienceSource.
fromanArchaebacteriaancestor.Thisgenomeinmoderneukaryotesgenerallyconsistsofseveral linearanddistinctDNAmoleculescomplexedwithanumberofproteinsandotherchemicalsto formdistinctchromosomes.Thenucleargenomereferstoahaploidsetofthesechromosomes.
Figure1.4showsaphotomicrographofanucleargenomeofthefruitfly Drosophilabusckii.Fruit flies,likehumans,haveanXYsexchromosomesystem,sosomegenomesbearanXchromosome, andanalternativenucleargenomebearsaYchromosomeandnoXchromosome.Ychromosomes areoftenmuchsmallerinmanyspecies,including Drosophila andhumans.Insomespecies,sexis determinedbyfactorsotherthanchromosomes,suchasenvironmentalfactorsasillustratedin Figure1.2,inwhichcasethereisonlyonetypeofnucleargenome.Indiploidspecies,cellscarry twonucleargenomes:onederivedfromthefemalegameteandonederivedfromthemalegamete. Themodeofinheritanceoftheautosomalportionofthegenomeisbisexualandsexuallybalanced, butthesexchromosomesdisplayadifferentpatternofinheritance.Forexample,inanXYsystemof sexchromosomes,suchasthosefoundinhumansand Drosophila,theXchromosomeisinherited bisexually,butfemalesarediploidfortheXchromosomewhereasmalesarehaploid,resultingin haplo–diploidinheritance.TheYchromosomeisunisexualininheritance,beingpassedonfrom fathertoson,andishaploidinmales.
DuringthePrecambrian,certaineubacteriallineagesevolvedthecapacitytoutilizeoxygenfor aerobicmetabolismaftertheearlierevolutionofprokaryoticphotosynthesis.Photosynthesis resultedinthereleaseoflargeamountsofoxygenintoEarth’sancientreducingatmosphere.
Oxygenwasinitiallyadeadlypollutantformostlifeforms,butsomeprokaryoticlineagesadapted tothisnewenvironmentalfactorthroughtheevolutionofaerobicmetabolismthatallowedoxygen tobeusedasanefficientmeansofextractingenergyfromsugars.Someoftheseaerobicbacteria wereingestedbyArchaebacteriabutnotdigested,resultingultimatelyinasymbioticrelationshipin whichtheaerobicsymbiotsevolvedintotheorganellenowcalledmitochondria.Thishorizontal transferbetweenthesetwobacteriallineagesresultedintheeukaryoticcellwithbothnuclear andmitochondrialgenomes.Themitochondriaretainedamorebacteria-likegenomeconsisting ofasingle,circularpieceofDNA(Figure1.5).Overthecourseofevolution,manyofthegenes neededforaerobicmetabolismweretransferredfromthemitochondrialgenomeintothenuclear genome,butothergenesessentialforaerobicmetabolismareretainedinthemitochondrial genome.MitochondrialDNAisofteninheritedonlythroughfemalesinbisexualanimalsand displaysaclonal,non-recombininghaploidpatternofinheritanceratherthanabisexual,recombiningmode.
tRNA Leu
tRNA Ile tRNA Gln tRNA Met
tRNA Trp tRNA Ala tRNA Asn tRNA Cys tRNA Tyr
tRNA Leu tRNA Ser tRNA His
tRNA Ser
Figure1.5 Thehumanmitochondrialgenome.Thenamesandpositionsofthevariousgenesandcontrol elementsareindicatedonthecirculargenome.
Eukaryoticplantsacquiredanothergenome,thechloroplastgenome,throughendosymbiosis withaphotosyntheticcyanobacterialineage.Thechloroplastgenomeconfersuponplantstheabilityofphotosynthesis.Thechloroplastgenome,likethemitochondrialgenome,consistsofcircular DNA,althoughitistypicallylargerandcontainsmoregenesthanthemitochondrialgenome.Like themitochondrialgenome,thechloroplastgenometypicallydisplaysunisexualinheritancethatis effectivelyhaploid.Maternalinheritanceisthemostcommonpattern,butsomegroupsofplants displaybiparentalandpaternalinheritance(Birky2008).
Thefactthatautosomes,sexchromosomes,andorganelleDNAsoftendisplaydistinctpatterns ofinheritancehasmadegeneticsurveysoftwoormoreofthesetypesofDNAapowerfultoolin populationgeneticsandgenomics.Aswewillseeinlaterchapters,suchjointsurveysallowthe separationoftheroleofthesexes,coverageoflongerperiodsoftimeinreconstructingthepast, andtheexplorationofthebalanceofmanydifferentevolutionaryforcesactinginareproducing population.Thediversityinmodesofinheritancehastherebymadepremise1,DNAcanreplicate, morepowerfulthaneverinthegenomicera.
Genomesarethephysical–chemicalstructuresinwhichtheprocessesofmutationandrecombinationarecarriedout.Althoughmutationisrandomwithrespecttotheneedsoftheorganism, aspointedoutaboveandinFigure1.1,mutationishighlynonrandomatthegenomiclevel,afact oftenignoredinthepopulationgeneticliterature.Onecommonclassofmutationsiswhenasingle nucleotidemutatestoanothernucleotidestate.Itiscriticaltonotethatsuchsinglenucleotide substitutionsarechemicalprocesses,and,assuch,theprobabilityandtypeofsubstitutionagiven singlenucleotideislikelytoexperienceareinfluencedbythephysical–chemicalenvironment createdbythenucleotidessurroundingthetargetnucleotide(Sungetal.2015;Zhuetal.2017; Suárez-Villagránetal.2018).Inotherwords,singlesitemutationscanberegardedasphenotypes thatdependinpartupontheenvironmentalcontextasdeterminedbytheirnucleotideneighbors. Hence,singlenucleotidemutagenesisisactuallyamulti-nucleotideprocessthatcreatesmutational hotspotsinthegenome,sometimesforjustasinglenucleotidewithinamulti-nucleotidemutagenic motif,butsometimesforseveralnucleotidesinthemotifleadingtonotonlyahotspotbutclosely spacedmutationalclusters(Schrideretal.2011;Besenbacheretal.2016).Templetonetal.(2000a) exploredtheimpactofmutagenicmotifsuponthenucleotidevariationina9.7kbsegmentofthe LPL genein71individuals.Theydiscoveredthatabouthalfofthenucleotidesitesshowingvariationwereassociatedwithjustthreemulti-nucleotidemotifs,withCGdinucleotidesbeingthemost likelytoshowvariation(Table1.2).CGdinucleotidesarehypermutagenicwhenthecytosineis methylated,makingaCtoTtransitionhighlylikely,andmutabilitycanbefurtherenhanced bythe5 nucleotideadjacenttoaCGpair(Baeleetal.2008).Mutationalhotspotsassociatedwith
Table1.2 Thedistributionofpolymorphicnucleotidesitesina9.7kbregionofthehuman LipoproteinLipase geneovernucleotidesassociatedwiththreeknownmutagenicmotifsandallremainingnucleotidepositions.
Source: ModifiedfromTempletonetal.(2000a).
DNAsequencemotifsarenotlimitedjusttosinglenucleotidesubstitutionsbutarealsofoundfor indel(insertion/deletion)andcopy-numbermutations(KvikstadandDuret2014;Pressetal.2019).
MethylatedCGdinucleotidemutationsillustratethatnonrandommutagenesisnotonlycreates mutationalhotspotsinthegenomebutalsoincreasestheprobabilityof mutationalhomoplasy in whichtwoindependentmutationaleventsatthesamenucleotidesitecreatethesamenucleotide substitution.Suchhomoplasystrikesdirectlyatafundamentalconceptinpopulationgenetics –thedistinctionbetweenidentity-by-descentversusidentity-by-state. Identity-by-descent occurs whentwohomologouscopiesofaDNAregionareidenticalinnucleotidestatebecausenomutationsoccurredineitherDNAlineage,tracingbacktotheircommonancestralDNAmolecule(by definition,homologouscopiesaredescendedfromacommonancestralmolecule).Incontrast, identity-by-state occurswhentwohomologouscopiesofaDNAregionareidenticalinnucleotide stateregardlessoftheirmutationalhistory.Aswillbeseeninlaterchapters,muchofpopulation genetictheoryhasbeenformulatedintermsofidentity-by-descent,andthistheorycanbemost easilyappliedtodatawhenidentity-by-stateisequatedtoidentity-by-descent.HomoplasyunderminesthisassumptionbyallowingtwoidenticalDNAcopiestohavehadmutationsinoneorboth lineagesfromtheancestralmolecule.
Despitetheoverwhelmingevidencethatsinglenucleotidemutagenesisisamulti-nucleotide process,manypopulationgeneticmodelsandcomputersimulationstreateachnucleotideorindel asindependentmutationalunitsthatdonotdependuponthesequencecontext.Forexample,one ofthemostcommonlyusedmodelsofmutationinpopulationgeneticsisthe infinitesitesmodel thatassumesthateverymutationoccursatadifferentmutationalsitebecausetheprobabilityof mutationatanygivensiteissmallandtherearealargenumberofpotentialsites.Theinfinitesites modeleliminatesthepossibilityofmultiplehitsandhomoplasyandalwaysresultsinidentity-bystatebeingthesameasidentity-by-descent.Morecomplicatedmodelsofmutationarepossiblethat allowmultiplehitsandhomoplasy,aswillbegivenlaterinthisbook.Computerprogramssuchas ModelTestandjModelTest(Posada2008)assessthebestfittingmutationalmodelfromDNA sequencedataoveralargenumberofmutationalmodels.Mutationalhotspotscanbeincorporated intosomeofthesemodelsbyallowingratevariationacrosssites.However,noneofthemodelsin ModelTestincorporatetheimpactofmulti-nucleotidesequencecontextintothemutationalmodel. However,justbecauseamodelmakesunrealisticbiologicalassumptionsdoesnotmeanthatitis useless.Inthenextchapter,wewillpresenttheHardy–Weinbergmodel,abasicmodelofevolution thatmakesmanyunrealisticassumptionsyethasturnedouttobeextremelyusefulandyields resultsthataccuratelydescribemanypopulations.Thequestionis,therefore,notiftheassumptions ofamodelareunrealisticbutratherhowrobustarethemodel’spredictionstoviolationsofits assumptions.Unfortunately,becauseoftheanalyticaldifficultyandcomputationalcomplexity ofmultisitemutationalmodels,therehaveonlybeenafewexaminationsofrobustnesstodeviations fromthesimplermutationalmodels.Thefewstudiesthathavebeenperformedindicatethat directlymodelingmultisitecontextresultsinmuchbetterfitstosequencedatathanindependent-sitemodels,andthefailuretoincorporatemultisitecontextproducesbiasesandfalsepositives inphylogeneticinferenceanddetectingnaturalselection(SiepelandHaussler2004;Baeleetal. 2008;Lawrieetal.2011;BérardandGuéguen2012;ChachickandTanay2012;Bloom2014;KvikstadandDuret2014).
Manyofthestudiesmentionedaboveinvolvelongevolutionarytimescales,andafrequentjustificationfortheinfinitesitesmodelisthatmultiplehitsandhomoplasyarenotlikelyontheshorter timescalesrelevantformostpopulationgeneticstudies.However,thisargumentdoesnotseemto beborneoutbygenomicsurveys.Forexample,multiplehitsandparallelchangesminimally occurredat51%ofthe88singlenucleotidepolymorphisms(SNPs)inthe LPL geneinhumans
(Templetonetal.2000a),andextensivehomoplasyhasbeenobservedinotherregionsofthehuman genome(e.g.Fullertonetal.2000).Hence,theverygeneticvariationthatisthemainfocusofpopulationgeneticsisgreatlyinfluencedbymutationalprocessesthatviolatetheinfinitesitesmodel. Theseresultsareworrisomebecausemanyoftheanalysesandcomputersimulationsusedinpopulationgeneticsdependupontheinfinitesitesmodeloratleastindependentsitesmodelsofmutation;yet,therobustnessofthepredictionshasrarelybeenexamined.Afewstudieshaveexamined thequestionofrobustnesstothemutationalmodel,andmanypredictionsarenotrobust.Forexample,inferencesconcerningrecombination(Templetonetal.2000a),geneflow(StrasburgandRieseberg2010),populationstructure/demographichistory(Cutteretal.2012),andnaturalselection (Baeleetal.2008;Cutteretal.2012)canallbegreatlyaffectedbyviolationsofcommonlyused simplemutationalmodels.Giventheseresults,amorecompleteexaminationofrobustnesstosimplemutationalmodelsisgreatlyneededinpopulationgenetics.
Recombinationisanothermajorsourceofgeneticvariation,andittooisoftenconcentratedinto recombinationalhotspotsthatareassociatedwithspecificDNAsequencemotifs(Singhetal.2013; deMassy2014;Singhaletal.2015).Moreover,recombinationhotspotsarefrequentlyhotspotsfor mutationand geneconversion (Prattoetal.2014;Arbel-EdenandSimchen2019;Halldorsson etal.2019),aprocessrelatedtorecombinationthatplacesasmallsegmentfromonechromosome ontoahomologouschromosomeandtherebycancreateanewhaplotype.Forexample,30statisticallysignificantrecombinationandgeneconversionevents(aftercorrectionforfalsepositives) weredetectedinthe9.7kbregioninthehuman LPL gene.Alltheseeventsmappedtoasmall regionassociatedwithamicrosatellitesequenceinthe6thintron(Templetonetal.2000a),as showninFigure1.6,withnotasinglesignificantrecombinationorgeneconversioneventdetected anywhereelseinthis9.7kbregion.Justasmutationalhotspotscancreatefalseormisleadingsignalsforevolutionaryforcessuchasnaturalselection,socanrecombinationandgeneconversion hotspots(Bolívaretal.2019).
ThefactthatDNAsequenceinfluencesboththelocalratesofmutationandrecombinationatthe genomiclevelcreatesaninterestingtwisttopremise2inpopulationgenetics,namely,localmutationandrecombinationcanberegardedasagenomiclevel,inheritedphenotypethatispotentially responsivetonaturalselection.MartincorenaandLuscombe(2013)investigatedthepossibilitythat populationsmightevolvelowermutationratesatgenomicpositionswheremostmutationsare
Figure1.6 Recombinationandgeneconversioneventsina9.7kbintervalofthe LPL gene.Thehorizontalline ismapoftheregion,showingthepositionsofexons(E4throughE9)inthicklinesandintronsinthinlines.The numberofstatisticallysignificantrecombinationorgeneconversioneventsthatcouldhaveoccurredinan intervaldefinedbyadjacentsinglenucleotidepolymorphismpairsisplotted. Source: Basedondatain Templetonetal.(2000a).
deleteriousandincreasedmutationrateswheremanymutationsareadvantageous.Theyfound thatnaturalselectioncouldresultintargetedhyper-andhypomutation,withtheconditionsfavoringhypermutationbeingmorerestrictive.Livnat(2013,2017)hasalsoproposedmodelsofnatural selectioninteractingwithnonrandommutationandrecombinationatthegenomeleveltoproduce adaptiveevolutionarychange.Atfirstglance,theabilityofnaturalselectiontoadjustlocalmutationandrecombinationratesmayseemtocontradictourearlierconclusionthatmutationsarerandomwithrespecttotheenvironmentalneedsoftheorganism,asillustratedbythereplicaplating experimentshowninFigure1.1.However,thereisnothingLamarckianabouttheselectivemodels ofMartincorenaandLuscombe(2013)andLivnat(2013,2017)thatdealwiththeadjustmentof mutation rates throughordinarynaturalselectiononDNAsequencesandnotwiththeproduction ofa specific adaptivemutationinducedbyaspecificenvironmentalneed(theLamarckianmodel). Thesemodelsdoshowthatnonrandommutationandrecombinationatthegenomiclevelcanplay animportantroleinadaptiveresponse(Figure1.3)andthatthedetailsunderlyingpremise2are themselvessubjecttoadaptiveevolutionarychange.
Genomicsciencehasalsohadaprofoundimpactonourunderstandingofpremise3thatphenotypesariseoutoftheinteractionofgenotypesrespondingtotheenvironment.Thegenesinanyof thegenomesconstitutebiologicalinformationbutnotfunctionunlesstheyareexpressed.Whether agenecodesforaproteinorforafunctionalRNAmolecule(BonasioandShiekhattar2014;Lyu etal.2014),thefirststepingoingfromstoredinformationtobiologicalfunctionalityisthetranscriptionoftheDNAintoRNAbyRNApolymerases.The transcriptome referstothefraction ofthegenomethatisactuallybeingtranscribed,andthetranscriptomevariesoverdevelopmental stages,acrosscelltypes,acrossindividuals,andacrossmanyenvironmentalfactors.Thereisnota singletranscriptomeforaspeciesorevenanindividual,butmany,andall,arehighlycontext dependent.Theportionsofagenomethatarebeingtranscribedatagiventimeandcelltype areinfluencedbymanyfactors,includingthethree-dimensionalstructureofthegenomeinterms oftherelativepositioningofsequencesonthesamechromosomeandbetweenchromosomes (BouwmananddeLaat2015;BonevandCavalli2016),chromatinremodeling(Chambersetal. 2013),andDNAsequencessuchaspromoters,enhancers,silencers,responseelements,andinsulators.TheseDNAsequencesoftenhavebindingdomainsthatwhenboundtoaproteinorsome othermolecule(calledtranscriptionfactors)canactivateordeactivatethetranscriptionalfunction ofthepromoters,enhancers,silencers,etc.Thesebindingmoleculesthemselvesareofteninduced byvariousenvironmentalfactors.Environmentalfactorscanalsoinfluencehowthetranscribed RNAisprocessed,edited,andtranslatedintoproteinsandhowtheseproteinsaresometimesmodifiedpost-translationally.Hence,theenvironmentcanplayadirectroleinhowtheinformationin theDNAinfluencesphenotypes.
Manybindingdomainsarefoundin transposableelements (e.g.Cuietal.2011),segmentsof DNAthathavetheabilitytomoveormakecopiesofthemselvesatdifferentlocationsinthe genome.Transposableelements,alsoknownastransposons,oftenconstituteasizableportion ofthenucleargenomeineukaryotes,goingupto90%ofthemaizegenome(SanMigueletal. 1996).Whentransposonsbearingbindingdomainstransposetonewlocationswithinagenome, thereisthepotentialforbringinganewgeneintoanestablishedregulatorynetwork,therebyinducinggeneticvariationinthecoordinationofgene-networktranscriptionregulation.Moreover, manybindingdomainscanbindseveraldifferenttranscriptionfactors,andevenasinglemutation inabindingdomaincangreatlyalterthetranscriptionfactorsthatcaninfluencetranscriptionin theregioninfluencedbythedomain(PayneandWagner2014).Thisabilityofmutationtochange thearrayoftranscriptionfactorsthatwillbindaspecificdomainandthemovementofsuch domainsbytransposonscangeneratemuchvariationintranscriptionalregulatorypatterns.This
variationinturnconfersahighdegreeof evolvability (theabilitytobringforthnoveladaptations) uponthepopulation.Evolvabilityisalsoenhancedbythenonrandomnatureofmutationand recombinationinthemodelsofLivnat(2013,2017).
Anotherimportantaspectofthetranscriptomeis epigenetics,thedevelopmentandmaintenanceofheritablestatesofgeneexpressionpatternsthatdonotdirectlydependontheDNA sequenceandthataretypicallyreversible,ofteninresponsetoenvironmentalcues(Bonasio etal.2010).Epigeneticstatesarefrequentlycontrolledbychemicalmodificationsofhistonesin thechromatinand/orthemethylationofcytosinesatCGdinucleotides(RiveraandRen2013; Wonetal.2013).Thesechemicallymodifiedepigeneticstatesarestableyetreversibleandcan becopiedduringtheprocessofmitosisandtherebypersistovermultiplecellgenerationsand cansometimesbepassedonacrossgenerations(BoskovicandRando2018;Zhangetal.2018). ThesechemicalmodificationsarecontrolledbyenzymesandnoncodingRNAs(FaticaandBozzoni 2014),whichinturnarecodedforingenes.Geneticvariationinthesegenesinfluencesepigenetic phenomena(Klironomosetal.2013;Kilpinenetal.2013),sotheepigenome,includingitssensitivitytoenvironmentalcues,isitselfaphenotypethatisinfluencedbyunderlyinggeneticvariation, andhencesubjecttochangebynaturalselectionasshowninFigure1.3.
Asillustratedbythediscussionabove,theDNAinthegenomeis not astaticblueprintoflifebut ratherisadynamicentitythatisconstantlychanginginresponsetotheenvironmenttoproduce phenotypesandwhosedynamicattributesaresubjecttoevolutionarychange,includingadaptation.Hence,genomicstudiesstronglyreinforcethevalidityofpremise3inpopulationgenetics anditsintegrationwiththeotherpremises,asshowninFigure1.3.
ModelingEvolutionandtheHardy–WeinbergLaw
Throughoutthisbook,wewillconstructmodelsofreproducingpopulationstoinvestigatehowvariousfactorscancauseevolutionarychanges.Inthischapter,wewillconstructsomesimplemodels ofanisolatedlocalpopulation.Thesemodelseliminatemanypossiblefeaturesinordertofocusour inferenceupononeorafewpotentialmicroevolutionaryfactors.Themodelswillalsoprovide insightsthathavebeenhistoricallyimportanttotheacceptanceoftheneoDarwiniantheoryofevolutionatthebeginningofthetwentiethcenturyandareofincreasingimportancetotheapplication ofgeneticstohumanhealthandothercontemporaryproblemsinthetwenty-firstcentury.
HowtoModelMicroevolution
Givenourdefinitionthatevolutionisachangeovertimeinthefrequencyofallelesorallelecombinationsinthegenepool,anymodelofevolutionmustincludeattheminimumthepassingof geneticmaterialfromonegenerationtothenext.Hence,ourfundamentaltimeunitwillbethe transitionbetweentwoconsecutivegenerationsatcomparablestages.Wecanthenexaminethe frequenciesofallelesoralleliccombinationsintheparentalversusoffspringgenerationtoinfer whetherornotevolutionhasoccurred.Allsuchtrans-generationalmodelsofmicroevolutionhave tomakeassumptionsaboutthreemajormechanisms:
• Mechanismsofproducinggametes
• Mechanismsofunitinggametes
• Mechanismsofdevelopingphenotypes
Inordertospecifyhowgametesareproduced,wehavetospecifythegeneticarchitecture. Geneticarchitecture isthenumberoflociandtheirgenomicpositions,thenumberofalleles perlocus,themutationrates,andthemodeandrulesofinheritanceofthegeneticelements. Forexample,thefirstmodelwewilldevelopassumesageneticarchitectureofasingleautosomal locuswithtwoalleleswithnomutation.Thegeneticarchitectureprovidestheinformationneeded tospecifyhowgametesareproduced.Forasingle-locus,two-alleleautosomalmodelwithnomutation,weneedonlytouseMendel’sfirstlawofinheritance(thelawofequalsegregationofthetwo allelesinanindividualheterozygousatanautosomallocus)tospecifyhowgenotypesproduce gametes.Othersingle-locusgeneticarchitecturescandisplaydifferentmodesofinheritance, includingX-linkedloci(withahaplo–diploid,sex-linkedmodeofinheritanceinorganismswith anXYsex-determiningsystem),Ychromosomalloci(withahaploid,unisexualpaternalmode ofinheritanceinorganismswithanXYsex-determiningsystem),ormitochondrialDNAand
PopulationGeneticsandMicroevolutionaryTheory,Second Edition.AlanR.Templeton. ©2021JohnWiley&Sons,Inc.Published2021byJohnWiley&Sons,Inc. Companionwebsite:www.wiley.com/go/templeton/populationgenetics2