PatterningandCellTypeSpecification
intheDevelopingCNSandPNS
ComprehensiveDevelopmentalNeuroscience
SecondEdition
SeniorEditors-in-Chief
JohnRubenstein
DepartmentofPsychiatry & WeillInstituteforNeurosciences UniversityofCalifornia,SanFrancisco,SanFrancisco,CA,UnitedStates
PaskoRakic
DepartmentofNeuroscience & KavliInstituteforNeuroscience YaleSchoolofMedicine,NewHaven,CT,UnitedStates
Editors-in-Chief
BinChen
DepartmentofMolecular,Cell & DevelopmentalBiology UniversityofCalifornia,SantaCruz,SantaCruz,CA,UnitedStates
KennethY.Kwan
MichiganNeuroscienceInstitute & DepartmentofHumanGenetics UniversityofMichigan,AnnArbor,MI,UnitedStates
SectionEditors:
ElizabethA.Grove
DepartmentofNeurobiology & GrossmanInstituteforNeuroscience UniversityofChicago,Chicago,IL,UnitedStates
ShubhaTole
DepartmentofBiologicalSciences
TataInstituteofFundamentalResearch,Mumbai,India
FrancoisGuillemot
TheFrancisCrickInstitute,London,UnitedKingdom
KennethCampbell
DivisionofDevelopmentalBiology,CincinnatiChildren’sHospitalMedicalCenter UniversityofCincinnatiCollegeofMedicine,Cincinnati,OH,UnitedStates
ArturoAlvarez-Buylla
EliandEdytheBroadCenterofRegenerationMedicineandStemCellResearch UniversityofCalifornia,SanFrancisco,SanFrancisco,CA,UnitedStates
DavidRowitch
DepartmentofPaediatrics,UniversityofCambridgeandWellcome-MRCCambridgeStemCellInstitute, Cambridge,UnitedKingdom
AdjunctProfessorofPediatrics,UCSF
AcademicPressisanimprintofElsevier 125LondonWall,LondonEC2Y5AS,UnitedKingdom 525BStreet,Suite1650,SanDiego,CA92101,UnitedStates 50HampshireStreet,5thFloor,Cambridge,MA02139,UnitedStates TheBoulevard,LangfordLane,Kidlington,OxfordOX51GB,UnitedKingdom
Copyright©2020ElsevierInc.Allrightsreserved.
Nopartofthispublicationmaybereproducedortransmittedinanyformorbyanymeans,electronicormechanical,including photocopying,recording,oranyinformationstorageandretrievalsystem,withoutpermissioninwritingfromthepublisher. Detailsonhowtoseekpermission,furtherinformationaboutthePublisher’spermissionspoliciesandourarrangementswith organizationssuchastheCopyrightClearanceCenterandtheCopyrightLicensingAgency,canbefoundatourwebsite: www. elsevier.com/permissions
ThisbookandtheindividualcontributionscontainedinitareprotectedundercopyrightbythePublisher(otherthanasmay benotedherein).
Notices
Knowledgeandbestpracticeinthis fieldareconstantlychanging.Asnewresearchandexperiencebroadenourunderstanding, changesinresearchmethods,professionalpractices,ormedicaltreatmentmaybecomenecessary.
Practitionersandresearchersmustalwaysrelyontheirownexperienceandknowledgeinevaluatingandusingany information,methods,compounds,orexperimentsdescribedherein.Inusingsuchinformationormethodstheyshouldbe mindfuloftheirownsafetyandthesafetyofothers,includingpartiesforwhomtheyhaveaprofessionalresponsibility.
Tothefullestextentofthelaw,neitherthePublishernortheauthors,contributors,oreditors,assumeanyliabilityforany injuryand/ordamagetopersonsorpropertyasamatterofproductsliability,negligenceorotherwise,orfromanyuseor operationofanymethods,products,instructions,orideascontainedinthematerialherein.
LibraryofCongressCataloging-in-PublicationData
AcatalogrecordforthisbookisavailablefromtheLibraryofCongress
BritishLibraryCataloguing-in-PublicationData
AcataloguerecordforthisbookisavailablefromtheBritishLibrary
ISBN:978-0-12-814405-3
ForinformationonallAcademicPresspublicationsvisitour websiteat https://www.elsevier.com/books-and-journals
Publisher: NikkiLevy
AcquisitionsEditor: NatalieFarra
EditorialProjectManager: AndraeAkeh
ProductionProjectManager: SuryaNarayananJayachandran
CoverDesigner: DavidTastad
TypesetbyTNQTechnologies
Contributors
KaterinaAkassoglou,GladstoneInstituteofNeurological DiseaseandDepartmentofNeurology,Universityof California,SanFrancisco,CA,UnitedStates
NicolaJ.Allen,MolecularNeurobiologyLaboratory,Salk InstituteforBiologicalStudies,LaJolla,CA,United States
FernandoC.Alsina,DepartmentofMolecularGenetics andMicrobiology
AlessandroAlunni,ZebrafishNeurogeneticsUnit, Developmental & StemCellBiologyDepartment, InstitutPasteur,UMR3738,CNRS,Paris,France
A.Alvarez-Buylla,UniversityofCalifornia,SanFrancisco,CA,UnitedStates
MadelineG.Andrews,UniversityofCalifornia,San Francisco,CA,UnitedStates
S.-L.Ang,FrancisCrickInstitute,London,United Kingdom
B.Appel,UniversityofColoradoSchoolofMedicine, Aurora,CO,UnitedStates
BadrulAre fin,DepartmentofClinicalandExperimental Medicine,LinköpingUniversity,Linköping,Sweden
ShahrzadBahrampour ,DepartmentofClinicaland ExperimentalMedicine,LinköpingUniversity,Linköping,Sweden
Q.-R.Bai,TongjiUniversity,Shanghai,China
LaureBally-Cuif,ZebrafishNeurogeneticsUnit,Developmental & StemCellBiologyDepartment,Institut Pasteur,UMR3738,CNRS,Paris,France
RenataBatista-Brito,DominickP.PurpuraDepartmentof Neuroscience,AlbertEinsteinCollegeofMedicine, Bronx,NY,UnitedStates
MagnusBaumgardt,DepartmentofClinicalandExperimentalMedicine,LinköpingUniversity,Linköping, Sweden
JonathanBenito-Sipos,DepartamentodeBiología,UniversidadAutónomadeMadrid,Madrid,Spain
D.E.Bergles,JohnsHopkinsSchoolofMedicine,Baltimore,MD,UnitedStates
AparnaBhaduri,UniversityofCalifornia,SanFrancisco, CA,UnitedStates
S.Blaess,InstituteofReconstructiveNeurobiology,LIFE & BRAINCenter,UniversityofBonn,MedicalFaculty andUniversityHospitalBonn,Bonn,Germany
StephanieBonney,DepartmentofPediatrics,Sectionof DevelopmentalBiology,UniversityofColorado, AnschutzMedicalCampus,Aurora,CA,UnitedStates
BernadettBosze,DepartmentofCellBiologyandHuman Anatomy,UniversityofCaliforniaDavis,Davis,CA, UnitedStates
JoshuaJ.Breunig,BoardofGovernorsRegenerative MedicineInstitute,LosAngeles,CA,UnitedStates; DepartmentofBiomedicalSciences,LosAngeles,CA, UnitedStates;SamuelOschinComprehensiveCancer Institute,LosAngeles,CA,UnitedStates;Department ofMedicine,DavidGeffenSchoolofMedicine,UCLA, LosAngeles,CA,UnitedStates
NadeanL.Brown,DepartmentofCellBiologyandHumanAnatomy,UniversityofCaliforniaDavis,Davis, CA,UnitedStates
S.A.Buffi ngton,BaylorCollegeofMedicine,Houston, TX,UnitedStates
C.L.Call,JohnsHopkinsSchoolofMedicine,Baltimore, MD,UnitedStates
K.Campbell,CincinnatiChildren’sHospitalMedical Center,UniversityofCincinnatiCollegeofMedicine, Cincinnati,OH,UnitedStates
AstridE.Cardona,UTSABrainHealthConsortiumand SouthTexasCenterforEmergingInfectiousDiseases, DepartmentofBiology,TheUniversityofTexasatSan Antonio,SanAntonio,TX,UnitedStates
CatarinaCatela,DepartmentofNeurobiology,University ofChicago,Chicago,IL,UnitedStates
A.Cebrián-Silla,UniversityofCalifornia,SanFrancisco, CA,UnitedStates;UniveristatdeValència, CIBERNED,Valencia,Spain
Yi-TingCheng,CenterforCellandGeneTherapy,Baylor CollegeofMedicine,OneBaylorPlaza,Houston,TX, UnitedStates
VictorV.Chizhikov,UniversityofTennesseeHealth ScienceCenter,DepartmentofAnatomyandNeurobiology,Memphis,TN,UnitedStates
MarionCoolen ,ZebrafishNeurogeneticsUnit,Developmental & StemCellBiologyDepartment,Institut Pasteur,UMR3738,CNRS,Paris,France
JesúsRodriguezCurt,DepartmentofClinicaland ExperimentalMedicine,LinköpingUniversity, Linköping,Sweden
DimitriosDavalos,NeuroinflammationResearchCenter, DepartmentofNeurosciences,LernerResearchInstitute,ClevelandClinic,Cleveland,OH,UnitedStates
L.M.DeBiase,JohnsHopkinsSchoolofMedicine,Baltimore,MD,UnitedStates
BenjaminDeneen,CenterforCellandGeneTherapy, DepartmentofNeuroscience,BaylorCollegeofMedicine,OneBaylorPlaza,Houston,TX,UnitedStates
OmerDurak,DepartmentofStemCellandRegenerative Biology,andCenterforBrainScience,HarvardUniversity,Cambridge,MA,UnitedStates
RyannM.Fame,DepartmentofStemCellandRegenerativeBiology,andCenterforBrainScience,Harvard University,Cambridge,MA,UnitedStates;Department ofPathology,BostonChildren’sHospital,Boston,MA, UnitedStates
StephenP.J.Fancy,NeurologyandPediatrics,University ofCalifornia,SanFrancisco,SanFrancisco,CA,United States
GordFishell,DepartmentofNeurobiology,Blavatnik Institute,HarvardMedicalSchool,Boston,MA,United States;StanleyCenterforPsychiatricResearch,Broad Institute,Cambridge,MA,UnitedStates
IsabelleFoucher,ZebrafishNeurogeneticsUnit,Developmental & StemCellBiologyDepartment,Institut Pasteur,UMR3738,CNRS,Paris,France
L.Fuentealba,UniversityofCalifornia,SanFrancisco, CA,UnitedStates
FredH.Gage,SalkInstituteforBiologicalStudies,La Jolla,CA,UnitedStates
LudovicGalas,NormandieUniversity,UNIROUEN, INSERM,PRIMACEN,Mont-Saint-Aignan,France
AndrewW.Grande,DepartmentofNeurosurgery, UniversityofMinnesota,Minneapolis,MN,United States
ElizabethA.Grove,DepartmentofNeurobiology,The GrossmanInstituteforNeuroscience,Universityof Chicago,Chicago,IL,UnitedStates
J.L.Haigh,UniversityofCalifornia,Davis,CA,United States
JeanHébert,Neuroscience,Genetics,StemCells,Albert EinsteinCollegeofMedicine,Bronx,NY,UnitedStates
OliverHobert,DepartmentofBiologicalSciences, HowardHughesMedicalInstitute,ColumbiaUniversity,NewYork,NY,UnitedStates
RobertB.Hufnagel,MedicalGeneticsandOphthalmic GenomicsUnit,NationalEyeInstitute,Bethesda,MD, UnitedStates
WielandB.Huttner ,MaxPlanckInstituteofMolecular CellBiologyandGenetics,Dresden,Germany
YasuhiroItoh,DepartmentofStemCellandRegenerative Biology,andCenterforBrainScience,HarvardUniversity,Cambridge,MA,UnitedStates
K.R.Jessen,UniversityCollegeLondon,London,United Kingdom
JaneE.Johnson,DepartmentofNeuroscience,University ofTexasSouthwesternMedicalCenter,Dallas,TX, UnitedStates
EyalKarzbrun,KavliInstituteofTheoreticalPhysicsand DepartmentofPhysics,UniversityofCalifornia,Santa Barbara,CA,UnitedStates
YutaroKomuro,DepartmentofNeurology,DavidGeffen SchoolofMedicine,UniversityofCalifornia,Los Angeles,LosAngeles,CA,UnitedStates
HitoshiKomuro,DepartmentofNeuroscience,Yale UniversitySchoolofMedicine,NewHaven,CT, UnitedStates
ArnoldR.Kriegstein,UniversityofCalifornia,San Francisco,CA,UnitedStates
J.T.Lambert,UniversityofCalifornia,Davis,CA,United States
KatherineR.Long,MaxPlanckInstituteofMolecular CellBiologyandGenetics,Dresden,Germany
GuillerminaLópez-Bendito ,InstitutodeNeurocienciasde Alicante,UniversidadMiguelHernández-ConsejoSuperiordeInvestigacionesCientí fi cas(UMH-CSIC), SantJoand’Alacant,Spain
JessicaL.MacDonald,DepartmentofStemCelland RegenerativeBiology,andCenterforBrainScience, HarvardUniversity,Cambridge,MA,UnitedStates; DepartmentofBiology,SyracuseUniversity,Syracuse, NY,UnitedStates
JeffreyD.Macklis,DepartmentofStemCellandRegenerativeBiology,andCenterforBrainScience,Harvard University,Cambridge,MA,UnitedStates;Bauer Laboratory,Cambridge,MA,UnitedStates
MariaCarolinaMarchetto,SalkInstituteforBiological Studies,LaJolla,CA,UnitedStates
FranciscoJ.Martini,InstitutodeNeurocienciasdeAlicante,UniversidadMiguelHernández-ConsejoSuperior deInvestigacionesCientí ficas(UMH-CSIC),SantJoan d’Alacant,Spain
MichaelP.Matise,DepartmentofNeuroscience & Cell Biology,Rutgers-RobertWoodJohnsonMedical School,Piscataway,NJ,UnitedStates
F.T.Merkle,MetabolicResearchLaboratoriesand MedicalResearchCouncilMetabolicDiseasesUnit, WellcomeTrust-MedicalResearchCouncilInstituteof MetabolicScience,andtheWellcomeTrust-Medical ResearchCouncilCambridgeStemCellInstitute,UniversityofCambridge,Cambridge,UnitedKingdom
A.Meunier,InstitutNationaldelaSantéetdela RechercheMédicale,Paris,France;CentreNationalde laRechercheScienti fique,Paris,France;Institutde Biologiedel’EcoleNormaleSupérieure(IBENS), Paris,France
KathleenJ.Millen,SeattleChildren’sHospitalResearch InstituteCenterforIntegrativeBrainResearch,Seattle, WA,UnitedStates
RobertH.Miller,AnatomyandCellBiology,Schoolof MedicineandHealthSciences,GeorgeWashington University,Washington,DC,UnitedStates
R.Mirsky,UniversityCollegeLondon,London,United Kingdom
SwatiMishra,DepartmentofPediatrics,Sectionof DevelopmentalBiology,UniversityofColorado, AnschutzMedicalCampus,Aurora,CA,UnitedStates; DepartmentofPathology,InstituteforStemCell & RegenerativeMedicine,UniversityofWashington, Seattle,WA,UnitedStates
AnnaVictoriaMolofsky,LaboratoryofMolecular Neurobiology,CentreofNewTechnologies,University ofWarsaw,Warsaw,Poland
IgnacioMonederoCobeta,DepartmentofClinicaland ExperimentalMedicine,LinköpingUniversity, Linköping,Sweden
K.Monk,VollumInstitute,OregonHealthScienceCenter, Portland,OR,UnitedStates
EdwinS.Monuki,Pathology & LaboratoryMedicine, Developmental & CellBiology,UniversityofCaliforniaIrvine,Irvine,CA,UnitedStates
MasatoNakafuku,DivisionofDevelopmentalBiology, CincinnatiChildren’sHospitalMedicalCenter,DepartmentsofPediatricsandNeurosurgery,Universityof CincinnatiCollegeofMedicine,Cincinnati,OH,United States
HarukazuNakamura,LaboratoryofOrganMorphogenesis,GraduateSchoolofLifeSciences,Tohoku University,Aoba-ku,Sendai,Japan
BrandenR.Nelson,CenterforIntegrativeBrainResearch, SeattleChildren’sResearchInstitute,Seattle,WA, UnitedStates
A.S.Nord,UniversityofCalifornia,Davis,CA,United States
K.Obernier,UniversityofCalifornia,SanFrancisco,CA, UnitedStates
NobuhikoOhno,DepartmentofAnatomy,Divisionof HistologyandCellBiology,JichiMedicalUniversity, Shimotsuke-Shi,Tochigi,Japan;DivisionofUltrastructuralResearch,NationalInstituteforPhysiological Sciences,Okazaki,Aichi,Japan
AbdulkadirOzkan,DepartmentofStemCelland RegenerativeBiology,andCenterforBrainScience, HarvardUniversity,Cambridge,MA,UnitedStates
DavidB.Parkinson,MedicineandDentistry,Plymouth University,Plymouth,Devon,UnitedKingdom
ManuelPeter ,DepartmentofStemCellandRegenerative Biology,andCenterforBrainScience,Harvard University,Cambridge,MA,UnitedStates
SamuelJ.Pleasure,DepartmentofNeurology,Programs inNeuroscienceandDevelopmentalBiology,Institute forRegenerativeMedicine,UniversityofCalifornia, SanFrancisco,CA,UnitedStates
M.N.Rasband,BaylorCollegeofMedicine,Houston,TX, UnitedStates
OrlyReiner,DepartmentofMolecularGenetics,The WeizmannInstituteofScience,Rehovot,Israel
D.H.Rowitch,UniversityofCalifornia,SanFrancisco, CA,UnitedStates
J.L.R.Rubenstein,UniversityofCaliforniaatSanFrancisco,SanFrancisco,CA,UnitedStates
DebosmitaSardar,CenterforCellandGeneTherapy, BaylorCollegeofMedicine,OneBaylorPlaza,Houston,TX,UnitedStates
AninditaSarkar,SalkInstituteforBiologicalStudies,La Jolla,CA,UnitedStates
K.Sawamoto,NagoyaCityUniversityGraduateSchoolof MedicalSciences,Nagoya,Japan;NationalInstitutefor PhysiologicalSciences,Okazaki,Japan
KamalSharma,DepartmentofAnatomy & CellBiology, UniversityofIllinoisatChicago,Chicago,IL,United States
Q.Shen,TongjiUniversity,Shanghai,China
JulieA.Siegenthaler,DepartmentofPediatrics,Sectionof DevelopmentalBiology,UniversityofColorado, AnschutzMedicalCampus,Aurora,CA,UnitedStates
DebraL.Silver,DepartmentofMolecularGeneticsand Microbiology;DepartmentofCellBiology;Department ofNeurobiology;DukeInstituteforBrainSciences, DukeUniversityMedicalCenter,Durham,NC,United States
N.Spassky,InstitutNationaldelaSantéetdelaRecherche Médicale,Paris,France;CentreNationaldelaRechercheScienti fique,Paris,France;InstitutdeBiologiede l’EcoleNormaleSupérieure(IBENS),Paris,France
S.R.W.Stott,TheCureParkinson’sTrust,London,United Kingdom
JohannesStratmann,DepartmentofClinicalandExperimentalMedicine,LinköpingUniversity,Linköping, Sweden
L.Subramanian,UniversityofCalifornia,SanFrancisco, CA,UnitedStates
JohnSvaren,DepartmentofComparativeBiosciencesand WaismanCenter,UniversityofWisconsin,Madison, WI,UnitedStates
LukaszMateuszSzewczyk,DepartmentofPsychiatryand WeillInstituteforNeurosciences,UniversityofCalifornia,SanFrancisco,SanFrancisco,CA,UnitedStates; LaboratoryofMolecularNeurobiology,CentreofNew Technologies,UniversityofWarsaw,Warsaw,Poland
S.Temple,NeuralStemCellInstitute,Rensselaer,NY, UnitedStates
StefanThor,DepartmentofClinicalandExperimental Medicine,LinköpingUniversity,Linköping,Sweden; SchoolofBiomedicalSciences,UniversityofQueensland,StLucia,QLD,Australia
ShubhaTole,DepartmentofBiologicalSciences,Tata InstituteofFundamentalResearch,Mumbai,Maharashtra,India
GregorioValdez,BrownUniversity,Providence,RI, UnitedStates
DavidVaudry,NormandieUniversity,UNIROUEN, INSERM,PRIMACEN,Mont-Saint-Aignan,France; NormandieUniversity,UNIROUEN,INSERM,U1239, DC2N,Mont-Saint-Aignan,France
ClaireWard,DominickP.PurpuraDepartmentofNeuroscience,AlbertEinsteinCollegeofMedicine,Bronx, NY,UnitedStates
MichaelWegner,InstitutfürBiochemie,Emil-FischerZentrum,UniversitätErlangen-Nürnberg,Erlangen, Germany
BehzadYaghmaeianSalmani,DepartmentofClinicaland ExperimentalMedicine,LinköpingUniversity, Linköping,Sweden
Morphogens,patterningcenters,and theirmechanismsofaction
ElizabethA.Grove1 andEdwinS.Monuki2
1DepartmentofNeurobiology,UniversityofChicago,Chicago,IL,UnitedStates; 2Pathology & LaboratoryMedicine,Developmental & Cell Biology,UniversityofCaliforniaIrvine,Irvine,CA,UnitedStates
Chapteroutline
1.1.Generalprinciplesofmorphogengradients3
1.1.1.Historyofthemorphogenandmorphogeneticfield3
1.1.2.Howmorphogengradientspatterntissues4
1.1.3.Howmorphogensaredistributed5
1.1.4.Howmorphogensignalingistransducedand interpreted6
1.1.5.Howmorphogengradientsareconvertedintosharp boundaries6
1.1.6.Summary generalprinciplesofmorphogen gradients7
1.2.Localsignalingcentersandprobablemorphogensinthe telencephalon7
1.2.1.Earlyforebrainpatterning8
1.2.2.TheRPC8
1.2.3.Thetelencephalicroofplateandcorticalhem8
1.2.4.Theantihem9
1.3.BMPsasmorphogensintelencephalicpatterning9
1.3.1.PerformanceobjectivesforaBMPgradientinthe dorsaltelencephalon9
1.3.2.Midlineexpressionandhomeogeneticexpansionof BMPproduction10
1.3.3.BMPsignalinggradientinthedorsaltelencephalon11
1.3.4.BMPsasdorsaltelencephalicmorphogens11
1.3.5.LinearconversionofBMPsignalingbycorticalcells12 1.3.6.NonlinearconversionofBMPsignalingbyDTM cells12
1.3.7.Summary theBMPsignalinggradient13 1.4.FGF8asamorphogenintelencephalicpatterning13 1.5.Interactionsamongsignalingcentersintelencephalic patterning14
1.5.1.FGF8,Shh,andBMPsignaling15 1.5.2.Cross-regulationofBMP,FGF,andWNTsignaling15 1.5.3.InteractionsofShh,FGFs,andGli315 1.6.Morphogensinhumanbraindisease15 1.6.1.HoloprosencephalyandKallmannsyndrome15 1.6.2.Gradientsinholoprosencephalyneuropathology17 1.6.3.Gradientsinotherhumanbraindisorders17 References18
1.1Generalprinciplesofmorphogengradients
1.1.1Historyofthemorphogenandmorphogeneticfield
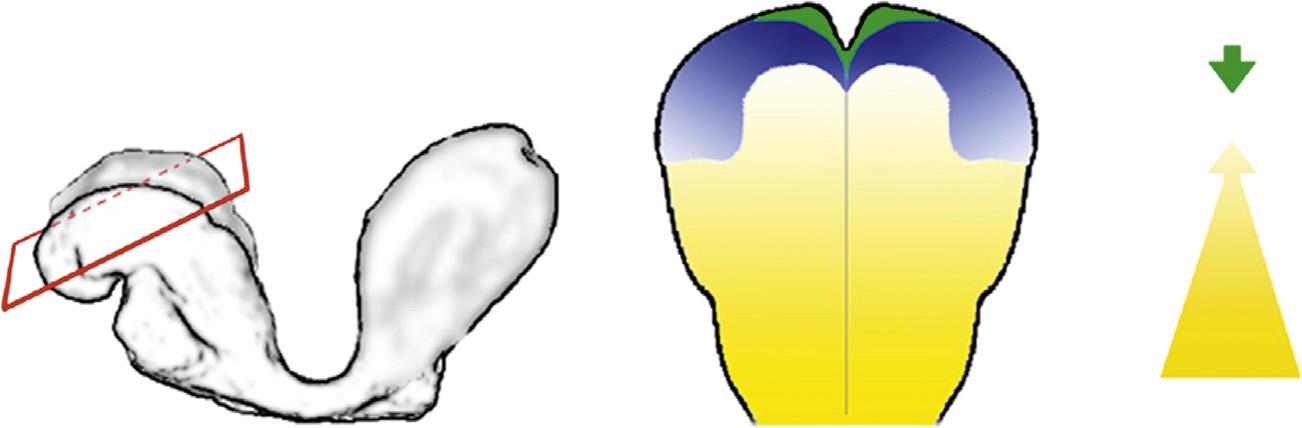
Theconceptofamorphogencanbetracedtotheturnofthe20thcentury,whenMorganpostulatedthepresenceof “formativesubstances” asthebasisfordifferentregenerationratesinworms(Morgan,1901).Verysoonthereafter,Boveri entertainedthisideafornormaldevelopment(Boveri,1901).Aseminaleventforthis fieldwasthediscoveryofalocalized sourceformorphogensknownastheSpemannorganizer(SpemannandMangold,1924).Theterm “morphogen” was coinedbyTuring,whodescribedhowuniformlydistributedsignalsmadebycellscanspread,self-organize,andgenerate pattern(Turing,1952).Turingpatternsremainhighlyrelevantindevelopment,butforthischapterandthedeveloping forebrain,themorerelevantconceptisthatofnonuniformgradeddistributionsofmorphogens,anideaformalizedinthe famous “French flag” modelofWolpert(Fig.1.1)(Wolpert,1969).



FIGURE1.1 TheFrench flagmodel. Schematicofhowadiffusiblemorphogencanassignpositionalvaluesandinstructcellsfates.Morphogen (green) secretedfromasourcecellformsaconcentrationgradientwithinatissue.Atintermediateconcentrationsabovethreshold1,respondingcellsadopt “white” fate.Athighconcentrationsabovethreshold2,cellsadopt “blue” fate. BasedonKicheva,A.,Gonzalez-Gaitan,M.,2008.TheDecapentaplegic morphogengradient:aprecisedefinition.Curr.Opin.CellBiol.20,137 143;Rogers,K.W.,Schier,A.F.,2011.Morphogengradients:fromgeneration tointerpretation.Annu.Rev.CellDev.Biol.27,377 407.
Inthismodel,Wolpertdescribedsmoothlydeclininggradientsofmorphogenconcentrationwithina “morphogenetic field” ofcells.Thesegradientswereimaginedtoariseviadiffusionfromalocalizedsourcetowardasink,thusgivingcells withinthemorphogenetic fielddifferentpositionalvaluesbasedonmorphogenconcentration.Thepositionalvaluesthen determinedthefatesadoptedbycellsinthe field(Fig.1.1).Itwasnotuntilthe1980sthatthemolecularidentityofa morphogenwasdefined(bicoid)(DrieverandNusslein-Volhard,1988a,1988b).The firstsecretedmorphogenwas identi fiedsoonthereafter(decapentaplegicordpp)(FergusonandAnderson,1992).Sincethen,manymoremorphogens havebeendiscovered.Most,butnotall,aresecretedproteins;examplesofothermolecularclassesincludetranscription factors(bicoidanddorsal)andavitaminderivative(retinoicacid).
1.1.2Howmorphogengradientspatterntissues
Theconceptofpositionalidentityisimportantforunderstandinghowmorphogengradientsworkbecauseitiserroneousto considermorphogensasthesoledeterminantsofcellfate.Asitturnsout,thesamelimitedrepertoireofmorphogensis usedoverandoveragainacrossontogenyandphylogenytogeneratethedizzyingarrayofcelltypesfoundintheanimal kingdom.Thus,morphogenscouldnotpossiblybeinstructiveofcellfateontheirown.Rather,morphogensactupon tissueswithdifferentprepatternsandcompetencies,andthesecompetenciesincombinationwiththepositionalinformation providedbymorphogensdeterminecellfate.Forexample,inthischapter,wediscussindetailhowbonemorphogenetic proteins(BMPs,theorthologuesofdpp)and fibroblastgrowthfactors(FGFs)providepositionalinformationtodorsal telencephaliccellswithrestrictedneuralpotential.
Thedefi ningpropertyofamorphogenistheabilitytospecifytwoormorecellfatesinaconcentration-dependent manner.Forsome,atleastthreefatesarenecessarytoensurethatamorphogenistrulyinstructive(Freemanand Gurdon,2002).Morphogensoftenspecifybetweenthreetosevenfateswithinatissue(AsheandBriscoe,2006),whichare separatedbysharp,discreteboundaries.Theacquisitionofmaturecellfatesandboundariesisprecededbycell-intrinsic differencesintheexpressionof “selector” genes(mostoftentranscriptionfactors)thatspecifycellfatesinparticularways (Garcia-Bellido,1975).Understandinghowgradedmorphogenicinformationisconvertedintosharp(switch-likeor ultrasensitive)changesindownstreamgeneexpressionremainsacentralproblemfordevelopmentalbiologists,although, aswewillseebelow,severalmechanismsunderlyingsuch “switches” havebeendefined.
Oneimportantobjectiveformanytissuespatternedbymorphogensistheestablishmentofsecondaryorganizersor signalingcenters(Meinhardt,2009).Thesesecondarysitesofmorphogenproductionexpandtherangesoverwhich morphogenscanact,providefor fi nersubdivisionsofpattern,orboth,andarelocatedatboundariesestablishedbythe primarymorphogengradientandselectorgenes.Otherconsequencesofprimarypatterningincludeapoptosis,cellsorting tofurtherrefi neborders,andotherformsofcell-to-cellsignaling(Landeretal.,2009a).Theseeventscanberegulatedby morphogensormaybecomelargelycell-autonomousandimmunetoextrinsiccontrol.Aninterestingpotentialuseof
morphogengradientsisthecontrolofproliferationandgrowth,whichisoftenrelativelyuniformwithintissues,butthe juryremainsoutonthisissue(SchwankandBasler,2010;DekantyandMilan,2011).(Note:Interestingly,generating uniformgrowthfromgradedinformationwouldbeexactlytheoppositeproblemofmakingsharpborders!)
Overthelastdecadeorso,theapplicationofmathematic almodelingandcomputersimulationshasprovideddeep insightsnotonlyintothe “what’ s” and “how ’s ” ofmorphogengradientsbutalsointothemanyinteresting “why ’s” . Theseapproacheshaveprovidedinsightsintosystems-levelfeaturessuchasrobustness(insensitivitytoperturbations), adaptabilityorresilience(theabilitytoadapttoperturbations),precision,noisebuffering,andscalingofpatterntotissue size,whichwouldbeimpossibleorimpracticaltoaddressviawetlabexperimentationalone.(Considerthemany advantagesofacomputer,ratherthanabenchscientist,testing1000pointsinparameterspace.)Severalgeneral principleshaveemergedfromthiswork,whichhavebeenreviewedbyseveralothers(BarkaiandShilo,2009;Lander etal.,2009a;Wartlicketal.,2009;BriscoeandSmall,2015).Amongitsmanylessons,morphogensystemsbiologyhas taughtus(1)thatde fi ningindividualoperationswithinamorphogensystem(e.g.,whethergene x isnecessaryand suf fi cientforfunction y )provideslittleunderstandingofthesystem itself;(2)thattheperfo rmanceobjectivesof morphogensystemsdifferasaresultoftheuniquefactorsandforcesthatimpingeuponthem;(3)thatmorphogen systemscanuseverydifferentmechanismstomeetsimilarobjectives;(4)thateverymechanism,nomatterhow advantageous,hastrade-offs;and(5)thatunderstandinghowmorphogensystemsworkandbalanceconfl ictingpriorities isimpossiblewithoutanalyzingsystemsasawhole.
Doesitmakesensethatmorphogensystemshaveevolvedsomanydifferentmechanismstoachievesimilargoals? Fromengineeringandevolutionaryperspectives,theanswerisyes.Engineersareveryfamiliarwiththe “nofreelunch” principle i.e.,everymechanismhastrade-offs,andmechanismsthatconferrobustnessinonesettingmightincrease fragilityinanother.Thus,despiteattemptstomakesystemsresistanttomosteverything,highlyengineeredsystemsare inescapablyfragile(CarlsonandDoyle,2002).Furthermore,fragilityisnotallbad infact,fragilityisnecessaryfor adaptability sothe “robust,yetfragile” trade-offisacommonfeatureofcomplexsystems.Giventhatevolutionactsupon preexistingbiologicaltemplatesratherthancleanslates,mechanisticmultiplicityandredundancywouldalsobelogical, necessary,andunavoidable.
1.1.3Howmorphogensaredistributed
Howaremorphogensdistributedtogenerateconcentrationgradients?Asoriginallyenvisioned(Wolpert,1969;Crick, 1970),extracellulardiffusionisapredominantmechanism(Landeretal.,2002;RogersandSchier,2011;Zhouetal., 2012).Theadvantagesofdiffusionaremany.Itissimple,fast,andoccursviarandomwalkratherthanbeingballistic, whichnegatesimpedimentssuchastortuosityoftheextracellularspace(Lander,2007).(Randomwalkisthereasonwhy diffusion fillsamazealmostasfastasitdoesanopenspace.)Thespeedofdiffusioncanalsoexplainhowlong-range gradientscanforminthepresenceofhigh-affinityreceptorsbecauseligand-receptorbindingratesformorphogensare oftenmuchslowerthandiffusion(e.g.,foractivin,ittakes30mintoload0.5%ofavailablereceptors)(Freemanand Gurdon,2002).Accordingly,diffusivityisanimportantpointofregulationandcanoccurbymodifyingthemorphogens themselves(e.g.,vialipidmodification)ortheirbindingtocofactorsandextracellularmatrix(RogersandSchier,2011). Diffusioncanbeusedininterestingwaystogeneratesignalinggradients e.g.,bygivingamorphogenanditsinhibitor differentdiffusionvectorsforfacilitatedtransport(Holleyetal.,1996;Shimmietal.,2005)ordifferentdiffusivities,which generatesdifferentrangesofactivity(Meinhardt,2009).
Clearly,however,mechanismsotherthandiffusionarealsousedtogeneratemorphogengradients.Forretinoicacid, spatiallyregulatedintracellulardegradationleadstoitsgradient(Whiteetal.,2007).Cellularratherthanmolecular mechanismshavealsobeeninvoked,suchastheprogressivedilutionofintracellularmorphogensresultingfromcell division,andtheuseofcellular filopodia-likeextensionsor “cytonemes” (RogersandSchier,2011).Cell-to-cell “transcytosis” hasbeenproposedasanalternativetodiffusion,althoughtranscytosisisdif ficulttocleanlydissociatefrom diffusion,andithasbeenarguedthatthedatasupportingtranscytosiscanbeentirelyexplainedbydiffusion(Landeretal., 2002;KichevaandGonzalez-Gaitan,2008;Zhouetal.,2012).Nevertheless,nondiffusionmechanismscertainlyexistand arelikelytorectifywhateverdeficienciesdiffusionhasinspeci ficsystems(Lander,2007).
Whatdomorphogengradientslooklike?IntheFrench flagmodel,Wolpertillustratedadecliningexponentialfunction (Fig.1.1),andthisturnsouttoaccuratelydescribemanymorphogensystems.Fordiffusivesystems,threeparameters describegradientprofile:morphogenproductionrateor flux,diffusion,andclearance.Whenclearanceoccursviaasink,as postulatedbyCrick(Crick,1970),gradientprofileislinear.Incontrast,uniformclearancewithinatissueleadstoa decliningexponentialgradient.Nonuniformdiffusionorclearancecanleadtootherdistributions,suchaspowerlaw functions(KichevaandGonzalez-Gaitan,2008).
Thesizeofmorphogenetic fields(i.e.,theamountoftissuepatternedbymorphogengradients)canalsobeestimated. Thelengthscale,ordecaylength,foranexponentialgradientisthedistanceoverwhichmorphogenconcentrationfallsby e 1 (w37%).Lengthscaleisdeterminedbydiffusivityandclearancerate,butitisindependentofsynthesisrate,andthe morphogenetic fieldsforbicoidandGurken/EGFRin Drosophila are w3 5timesthelengthscale(Goentoroetal.,2006). Usingreasonableparameters,othershavesuggestedafewhundredmicronsasatheoreticalmaximum,whichmatches invivosituationsreasonablywell(Landeretal.,2009a).
1.1.4Howmorphogensignalingistransducedandinterpreted
Howisextracellularmorphogenconcentrationmeasuredbyacell?Inmostcases,signalingintensityisdeterminedbythe absolutenumberofoccupiedreceptors(DysonandGurdon,1998),althoughinthecaseofhedgehog(Hh),theratioof occupied:unoccupiedreceptorshasbeenimplicatedasthekeydeterminant(RogersandSchier,2011).Intuitively,theuse ofabsoluteratherthanrelativenumbersofoccupiedreceptorswouldbeusefulatlowmorphogenconcentrationsandcould allowforlargermorphogenetic fields.Signalingintensitycanbemodulatedinmanyways,includingviachangestothe extracellularmatrixorreceptornumbers.
Whenabsolutereceptornumbersareused,morphogenreceptorsignalingisdirectlyproportionaltoextracellular morphogenconcentration.Importantly,formostmorphogens,thisproportionalityismaintainedallthewaydowntotheir transcriptionaleffectorsinthenucleus(AsheandBriscoe,2006;RogersandSchier,2011).Alikelyexplanationforthis proportionalityisthelinearratherthanbranchedconstructionofmostmorphogensignalingpathways.Indeed,formany morphogenpathways,thesignaltransducerdoublesasthetranscriptionaleffector(e.g.,SmadforNodalandBMP,Glifor Sonichedgehog/Shh)(AsheandBriscoe,2006).Theabsenceofsigni ficantbranchingorcascadingreducesthepossibilities fornonlinearsignalampli fication.Inthisway,thepositionalvaluesimpartedbyextracellularmorphogenconcentrations aredirectlyandproportionallytransmittedintorespondingcells.
Respondingcellsarealsoquitesensitivetomorphogenconcentration.Forexample,cellscansenselargeconcentration differencesinShhoractivin(25-50X),andrelativelysmallchangesinconcentrationoractivatedreceptornumber(2-3X) aresuf ficientforfatetransformationsormajorshiftsinboundaryposition(FergusonandAnderson,1992;Dysonand Gurdon,1998;AsheandBriscoe,2006).Highsensitivityalsoexpandstherangeoverwhichmorphogenscanact,uptothe pointwheresignalandrobustnessmechanismsareovercomebynoise.Formanygradients,bindingnoiseduetolow receptoroccupancyisalikelylimitingfactor(Landeretal.,2009a).
Timeisanothercrucialfactorforinterpretingmorphogens.Forpracticalreasons,spatialgradientsareoften consideredattheirsteadystates,butthesestatesrepresentoversimplifyingassump tionsinmanycases.Indeed, patterninginvivocanbequitefast(evenlessthanafewhours),andsomegradientsaredecodedduringrising, pre steadystateconditions( BarkaiandShilo,2009 ).Temporalintegrationanddynamicinterpretationsofsignalingcan alsobecritical(SagnerandBriscoe,2017).Insomecases,fatetransformationsca usedbyincreasedconcentrationcanbe mimickedbyincreasedduratio n;thisisparticularlywelles tablishedforShh.Inothersystems,however,cellfatesre fl ect thehighestconcentr ationeverseenbyacellratherthanatemporalintegral( AsheandBriscoe,2006;Rogersand Schier,2011).
1.1.5Howmorphogengradientsareconvertedintosharpboundaries
Theneedtogeneratediscreteandwell-separatedcelltypesisnearlyuniversal,andmorphogensystemshaveevolvedmany differentwaystoconvertgradedextracellular(andintracellular)informationintononlinearfatedecisionsandboundaries. Somemechanismsgeneratenonlinearityinextracellularmorphogendistribution;theseincludefacilitatedtransport, alterationsinmorphogendegradationorclearance,andregulationofreceptoravailability.
However,mostnonlinearconversionmechanismsareintracellular,andmanyaretranscriptional.Thismakessome sensebecausemostmorphogentransductionpathwaysretainlinearityallthewaydowntothenucleus.Nonlinear transcriptionalmechanismsincludecooperativity,differentialbindingsiteaf fi nity,autoregulatorypositivefeedback, feedforwardloops,signswitching(e.g.,fromtranscriptionalactivatortorepressor),andcross-repression(Ashe andBriscoe,2006).Manypositivefeedbackmechanismsalsogeneratebistability,thepropertyofhavingtwopotential stablestatesatsomestimulusconcentr ations.Bistabilityrepresentsaformo fcellmemoryandprovidesrobustness to “on” states,whichenablescellstomaintaintheirfatesaf tertheinducingmorphogenisnolongeravailable.In additiontothesetranscriptionalsharpeningmechanisms, morphogensystemsalsoemploycellular-levelmechanisms tosharpenboundaries,includingsorting,death,andrespeci fi cationofmislocalizedcellsnearboundaries(Ashe andBriscoe,2006).
1.1.6Summary generalprinciplesofmorphogengradients
Morphogengradientsystemsarecomplexbutsharemanycommonfeatures,whichcanbesummarizedasfollows(Rogers andSchier,2011):
1. Morphogensarereleasedfromdynamiclocalizedsources,assemblewithothermolecules,andmoveviadiffusion throughtheextracellularspace.
2. Gradientshapeisdeterminedby fluxfromthesource,diffusivity,andclearancefromtissues.
3. Morphogenconcentrationanddurationaretransmittedlinearlytointracellularmolecules,ultimatelyresultinginthe gradedandproportionalactivityoftranscriptionaleffectors.
4. Transcriptionaleffectorsparticipateincomplexregulatorynetworksthatinvolvepreexistingintrinsicfactors,which ultimatelydeterminetargetgeneresponses.
5. Feedbackmechanismsacttobuffer fluctuationsinmorphogenproduction,regulatesignalinginterpretation,andconfer scalabilityandrobustnesstomorphogen-mediatedpatterning.
1.2Localsignalingcentersandprobablemorphogensinthetelencephalon
Developmentalneurobiologistswererelativelyslowtoadopttheconceptthatmorphogenssecretedfromsignalingcenters canpatterncomplexstructuresoftheembryonicbrain.Thiswasparticularlytrueofthetelencephalon,giventhatthispart oftheembryonicbraingivesrisetothecerebralcortex,longbelievedtobesofunctionallycomplexthatuniquemechanismswouldbeneededtopatternit.Therecognitionofseveralputativemorphogensourcesadjacenttotheearly embryonicforebrain,however,ledtotheproposalthatthesesignalingcentersandmorphogenswerecriticaltothe structuralorganizationofthetelencephalon,justastheyareforotherpartsoftheembryo(Furutaetal.,1997;Groveetal., 1998;Crossleyetal.,2001;RagsdaleandGrove,2001;Ohkuboetal.,2002).Thus,eventhemostfunctionallycomplex partofthebodyispatternedbymechanismsthatarecommonlyusedelsewhereintheembryo.
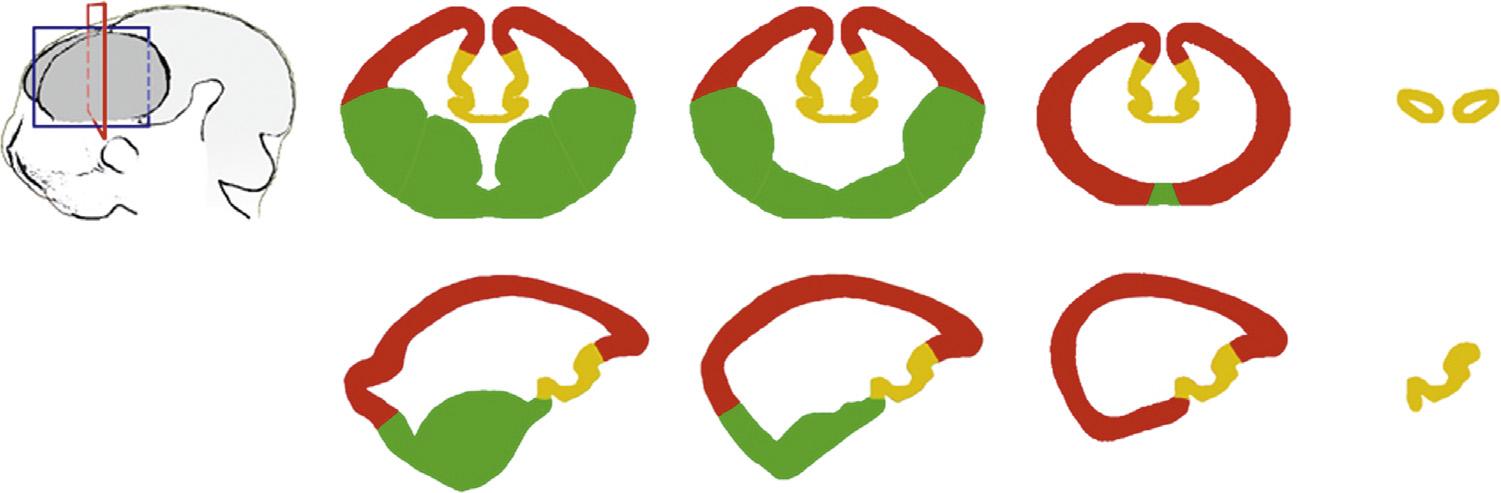
Halfadozenputativesignalingcentershavenowbeenidenti fiedforthetelencephalon,chie flyforthecerebralcortex, andsubstantialevidencesupportsapatterningroleforseveralofthese.Thecandidatesignalingcenterscomprise(1)a sourceofShhfromtheprechordalmesodermthatunderliesthemedialprosencephalicneuralplate(Rubensteinand Beachy,1998);(2)theanteriorneuralridge(ANR),whichappearsbeforeclosureoftheanteriorneuroporeattheedgeof theneuralplateandexpresses Fgf8 (CrossleyandMartin,1995);(3)therostraltelencephalicpatterningcenter(RPC,also knownastheanteriorcerebralpole)formedasthetelencephalicvesiclegeneratestwocerebralhemispheres,also




FIGURE1.2 Threetelencephalicsignalingcenters. (A,B)DorsalviewsoftwoE10.5forebrainsprocessedwithinsituhybridizationtoshowthegenes indicated,anteriortothetop.(C,D)E13.5hemispheresviewedfromthemedial(C)orlateral(D)face,anteriortotheleft. Fgf8 isexpressedattherostral patterningcenter(alsoknownastheanteriorcerebralpoleoracp)(A)and Wnt3a atthecorticalhem(B D). sFrp2 expressionmarkstheantihem,which formsapincershapewiththeWnt3a-expressinghem(D).
expressing Fgf genesofthe Fgf8 subfamily(BachlerandNeubuser,2001);(4)thetelencephalicroofplate,asourceof BMPsignals(Furutaetal.,1997;Monukietal.,2001;Chengetal.,2006);(5)lineallyrelatedsuccessorstotheroofplateat thedorsomedialedgeofeachcerebralcorticalhemisphere thechoroidplexusepithelium(CPE)andcorticalhem, expressingWntandBMPproteins(Furutaetal.,1997;Groveetal.,1998;Hebertetal.,2002;Currleetal.,2005);and (6)the “antihem,” atthejunctionofthedorsalandventraltelencephalon,secretingtheWntinhibitorsFrp2;theEGFfamily membersTgf-a,neuregulins1and3;andFGF7(Assimacopoulosetal.,2003)(Fig.1.2).Wedescribeheretheputative signalingsourcesandtheirconstituentsignalingmoleculesthatcontrolpatterningofbroaddivisionsofthetelencephalon. Anotherchapterinthisvolumediscusses,morespeci fically,theroleoftwoofthesignalingsourcesinpatterningthe neocortexintoamapofdistinctareas.
1.2.1Earlyforebrainpatterning
Theprechordalmesoderm,producingShh,andtheANRinfluencetheearlieststagesofforebrainpatterning.FGF8from theANRupregulatesgeneexpressionofthetranscriptionfactor,Foxg1,whoseexpressionisthe firstmarkeroftelencephalictissue(ShimamuraandRubenstein,1997;RubensteinandBeachy,1998).Shhfrommesodermunderlyingthe medialprosencephalicneuralplatedividesthediencephaliceye fi eldintotwo(Chiangetal.,1996).Furthermore,similarto theactionofShhinthespinalcord,earlyactivityofShhcontributestothespeci ficationofventralcellfatesintheventral telencephalon(Susseletal.,1999;GulacsiandAnderson,2006).Dissimilarfromthecaudalcentralnervoussystem(CNS), ventralizingthetelencephalonappearsalsotorequireFGFsignaling(Gutinetal.,2006;Danjoetal.,2011).
1.2.2TheRPC
ThesignalingcenterwetermtheRPCexpresses Fgf genesofthe Fgf8 subfamily,similarlytotheisthmicorganizer(ISO) atthemidbrain/hindbrainjunction.Atbothsites,FGF8andFGF17havepartlyseparateandpartlycomplementary patterningroles. Fgf8 isexpressedastheRPCforms,andFGF8inducesexpressionof Fgf18 and Fgf17 (Cholfinand Rubenstein,2008),thelatterofwhichisexpressedmorebroadlythan Fgf8 Fgf18 hasamorelimitedexpressiondomain anditstelencephalicrolehasnotyetbeenstudied.AlthoughtheRPCmayarisefromcellsoftheANR,thetwosignaling sourcesaredistinguishabletemporallyandbymorphology.Thatis,theANRisevidentwhentheneuraltubeisopen,but theRPCisidentifiedwhentheanteriorneuroporehasclosedataboutembryonicday(E)9inthemouse.TheANRis criticaltotheinitialpatterningoftheforebrain,theRPCinlaterpatterningofthetelencephalon.
TheRPChasalsobeenreferredtoasthe “commissuralplate,” astructurethatalsoformsanteromedially,butlaterin development,asachannelforthemajorcommissuresofthehemispheres.Indeed,FGF8isneededtopositionthe commissuralplate(Moldrichetal.,2010).TheRPCdoesnotappeartobeaspeci ficprogenitorofthecommissuralplate. FatemappingindicatestheRPCgivesrisetoneuronsthatpopulatetheprefrontalcortexandpartsoftheseptum,aswellas thelikelycommissuralplate(Toyodaetal.,2010;Hochetal.,2015).Evidencedetailedinasubsequentchapterindicates thatFGF8andFGF17,dispersingfromtheRPCasmorphogens,patterntheneocorticalareamap(Fukuchi-Shimogoriand Grove,2001;Gareletal.,2003;CholfinandRubenstein,2007,2008;Toyodaetal.,2010).
1.2.3Thetelencephalicroofplateandcorticalhem
Thetelencephalicroofplatecanbedefi nedasthemidlineofthetelencephalicvesiclebeforeithasdividedintotwo hemispheres.Oncethetwomedialhemisphericwallsaredistinct,bilateralCPEandcorticalhemsbecomeevidentatthe dorsomedialedgeofeachcerebralcortex.TheroofplateproducesseveralmembersoftheBMPfamilyofsignaling molecules,andgeneticablationofeitherthetypeIBMPreceptorBMPRIaoroftheentireroofplatecausesalossof telencephalicCPE(Hebertetal.,2002).Ablationoftheroofplatefurthercausesacorticalphenotypethatresemblesmiddle interhemispheric(MIH)holoprosencephaly(Chengetal.,2006;Monuki,2007).ThecorticalhemsecretesWntsandBMPs fromthedorsomedialedgeofthecorticalprimordium(Furutaetal.,1997;Groveetal.,1998)andisbothnecessaryand suf ficientforspecifyingthehippocampus.Withoutacorticalhem,orifhemWntsignalingissuf ficientlydepleted,the hippocampusfailstodevelop(Galceranetal.,1999;Leeetal.,2000;Yoshidaetal.,2006).SuggestingthatcanonicalWnt signalinginducesthedifferenthippocampal fields,constitutivelyactive b-catenininducescorticalcellstoexpressgenes normallycharacteristicofhippocampus(Machonetal.,2007).Moststriking,anectopicheminducesasmallsecondary, ectopichippocampus(Mangaleetal.,2008).
BMPsignalingalsocontributestotheformationofthehippocampus.InmicedeficientfortwoofthetypeIBMP receptors,BMPsignalingisreducedbutnotabrogated,givenathirdtypeIreceptorisalsopresentinthetelencephalon (Caroniaetal.,2010).Thedouble-mutantmousehasagreatlyreducedhippocampaldentategyrus(DG)comparedwith
controlmiceandaproportionallysmallerpopulationofadultDGneuralstemcells.ModeratereductionsinWntsignaling inthehemalsocauseadiminishedorabsentDG(LiandPleasure,2005).Beyondthehippocampus,thecorticalhemalso regulatesthesizeandpatterningofneocortex(Caronia-Brownetal.,2014).HowWntandBMPsignalingworktogetherin earlyhippocampalandneocorticaldevelopmentstillneedsclari fication.
1.2.4Theantihem
Theantihemliesattheoppositeedgeofth ecerebralcortextothehem,forminganarrowbandsurroundingtheboundary betweenthedorsalandventraltelencephalon.Interestingly,inmicedefi cientinthetranscriptionfactorLhx2,which promotescorticalidentity,boththehemandth eantihemexpandintothevacantterritory( Mangaleetal.,2008 ), suggestinghemandantihemareinsomesenseequivalentstructures.Theantihemexpresses sFrp2,encodingasoluble Wntinhibitor(Kimetal.,2001 ), Fgf7,andtheEGFgenes, Tgf a , Nrg1,and Nrg3 (Kimetal.,2001;Assimacopoulos etal.,2003 ).Thelatterareorthologsof DrosophilaSpitz and Vein,encodingEGFligandsthatcontrolneuronal speci fi cationinthe Drosophila ventralnervecord(Skeath,1998;vonOhlenandDoe,2000).Theantihemisevidentby geneexpressionadayortwoafterthehem,suggestingitispresenttoolateforaroleinearlycorticogenesis. Nonetheless,completelossoftheantiheminthe smalleye (Pax6-defi cient)mutantsuggestspossibleinvolvementinthe corticalpatterningandcellmigrationdefectsthatoccurin smalleye andPax6nullmice.FurtherimplyingthattheEGF familyregulatescorticalregi onalization,EGFinducesamolecularmarkeroflimbiccorticalareas,LAMP,inexplantsof nonlimbiccortex(FerriandLevitt,1995 ).Greaterunderstandingofthespeci fi cfunctionsofthisputativesignaling centerawaitsconditionalg eneticmanipulationsspeci fi ctotheantihem.
1.3BMPsasmorphogensintelencephalicpatterning
Dothegeneralprinciplesofmorphogengradientsapplytothemammaliantelencephalon?Inthenexttwosections,we focusonBMPandFGFsignalingindorsaltelencephalicpatterning,forwhichthereissubstantialevidencethattheanswer is “yes.”
1.3.1PerformanceobjectivesforaBMPgradientinthedorsaltelencephalon
FollowingneuralinductionandneuraltubeclosureatE9inmice,fourdistinctcellfatesdifferentiatealongthedorsoventral (DV)axisofthedorsaltelencephalonbyE12(Fig.1.3).Threeoftheseformdomainsatornearthedorsaltelencephalic midline(DTM) frommedialtolateral,thesearethechoroidplaque,CPE,andcorticalhem.CPEproducesthe cerebrospinal fl uid,andthecorticalhemactsasanorganizerforthehippocampus(Mangaleetal.,2008)andneocortical patterningcenter(Caronia-Brownetal.,2014).(Thechoroidplaqueisnotknowntohaveaspecificfunction.)Lateralto theseisthecortexorcorticalprimordium,whichismuchlargerandconstitutesmostofthedorsaltelencephalon.
Inmice,thecriticalperiodforspecifyingthesedorsaltelencephalicfatesprecedestheonsetofcorticalneurogenesisat E11.ExcessiveCPEandhemformwhenthetranscriptionfactorLhx2isinactivatedbyE8.5,butnotafterE10.5,andthis sameE8.5 E10.5perioddefinesthecriticalperiodforspecifyingcorticalidentity(Mangaleetal.,2008).Forebrain competencyforCPEfatealsocoincideswiththisperiod,basedonculturestudiesofE8.5andE9.5forebraincells(Thomas andDziadek,1993)orE9.5andE10.5dorsalforebrainexplants(Srinivasanetal.,2014).PeakCPEcompetencyin embryonicstem(ES)cell derivedsystemsalsocorrelateswithpreneurogenicneuroepithelialcellsratherthanneurogenic radialglia(Watanabeetal.,2012).Thus,ifaninstructiveBMPgradientexists,itmustexistinthepreneurogenicdorsal telencephalon.
Inadditiontospecifyingcelltypes,whatotherperformanceobjectivesmightaBMPgradientinthedorsaltelencephalonhave?Thespeci fi cationofCPEandcorticalhem fi tswiththeperformanceobjectiveofspecifyingsecondary organizersbecausebothtissueshavespeci fi csignalingfunctions(Mangaleetal.,2008;Lehtinenetal.,2011 ).Another objectivecouldbegradedpatterningofthecortex.Theneurogeniccorticalprimordiumiswellknownforits transcriptionalgradients ratherthanthresholds( Sansometal.,2005 ),andthegradientsimpactcorticalarealizationin matureanimals(Bishopetal.,2000;Mallamacietal.,2000).Onenotableperformanceobjec tivethatthecortexlacksis regeneration.Withtheexceptionofhippocampusandolfactorybulb,signifi cantneuronalregenerationdoesnotoccurin thedorsaltelencephalon,whichmayrefl ectapositiveselectionduringevolu tionforlong-termmemorystorage (Spaldingetal.,2005;Bhardwajetal.,2006).(Theideaisthatneuronalregenerationandreplacementwouldcause lossesofmemory/informationstoredwithinexistingneuronalcircuits.)Thelackofregenerationalsoimpliesthatthe neocortexmusthavegoodnegativefeedbackandmaintenancesystemstogenerateandmaintaintherightnumberof cellsfromthebeginning(Landeretal.,2009b ).








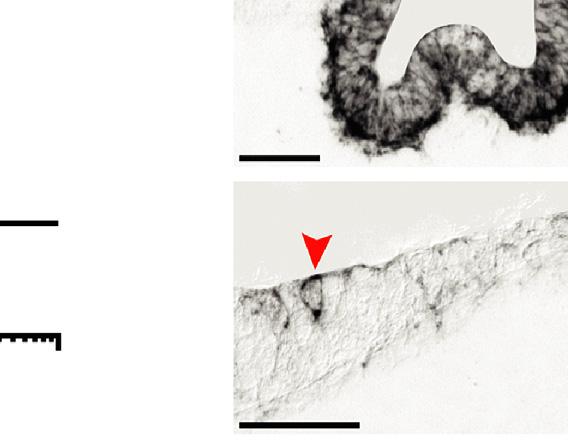
FIGURE1.3 Abonemorphogeneticprotein(BMP)signalinggradientandswitchinthedorsaltelencephalon. (A)Coronalschematicsofthe dorsaltelencephalon.BMPs( green )producedatthedorsalmidlinediffuseoveranaïvedorsaltelencephalicneuroepithelium.Within2 3daysin mice,fourfatesarespeci fi ed choroidplaque( green),choroidplexusepithelium( blue),corticalhem( orange),andcortex(red ).(B)Modi fi edFrench fl agmodel.TheBMPgradientgeneratesthreethresholdsseparatingthefourcelldomains,whichincludecellsthatcontinuetoproduceBMPs,andalso providesgradedpositionalinformationtothecortex.(C)TheBMPsignalinggradientinE10.5dorsaltelencephalon(from Chengetal.,2006).The pSmadgradientisasimpledecliningexponential(decaylength w290um),whichbecomesreducedand fl attenedfollowingroofplateablation (“mutant ”).(D)Schematicsofagradedresponse(gray)andanultrasensitive “switch” (red).(E)UltrasensitivityofE12.5corticalprogenitorstoBMP4 (Msx1 and Msx1-nlacZ RT-qPCRfrom Huetal.,2008 ).(F) Msx1 ultrasensitivityinvivoatE10.5,asevidencedbyitssharpborderinnormalembryos (arrows)andasisolatedhighlyexpressingcellsfollowingroofplateablation(arrowheads)( Msx1 ISHfrom Huetal.,2008 ).
Collectively,theseobservationssuggestthefollowingpotentialperformanceobjectivesforaBMPgradientinthe preneurogenicdorsaltelencephalon(Fig.1.3).TheBMPgradientmightspecifyuptofourdiscretefatesseparatedbythree thresholds,whichincludesthepositioningoftwosecondaryorganizers(CPEandcorticalhem).Nonlinearconversionsof gradedBMPinformationwouldbeneededforthesethresholds.Coincidentally,theBMPorthologuedppisthoughttobe responsibleforthreethresholdsinboththe Drosophila embryoandwingimaginaldisc(Asheetal.,2000;Affolterand Basler,2007).Duringtheneurogenicperiod,BMPgradientsmightalsoregulatetranscriptionalgradientswithinthecortex, whichwouldrequirelinearorsublinearinterpretationsoftheBMPgradient.
1.3.2MidlineexpressionandhomeogeneticexpansionofBMPproduction
IsBMPproductionsuf ficientlylocalizedtogenerateagradientinthedorsaltelencephalon?Beforeandafterneuraltube closureinmice,atleastsixBMPs(BMP2,4,5,6,7,and12,whichisalsoknownasGDF7)aretranscribedbyroofplate orDTMcells(Furutaetal.,1997).TheexpressionepicenterforalloftheseBMPsisthemidline.LocalizedBMPproductionatthemidlinewouldbepredictedtoformapreneurogenicBMPgradientthatishighestatthemidlineandlower morelaterally(Fig.1.3).
TwoadditionalfeaturesofBMPexpressioninthedorsaltelencephalonincreasethespatialandtemporalrangesover whichBMPgradientsmightact.First,someBMPsareexpressedbeyondthemidlineinthecorticalprimordium,andthis
expressionisalsograded(Furutaetal.,1997).Second,earlyBMP-producingcellsoftheroofplateinducelaterBMPproducingCPEandhemcells(Currleetal.,2005),aformof “homeogenetic” induction(i.e.,likeinducinglike)akin tothosedescribedinthemidbrain,spinalcord,and Drosophila (Liemetal.,1995;AlexandreandWassef,2003;Bierand DeRobertis,2015).Ontheotherhand,geneticlineagetracingsuggeststhatlineage-basedmechanismsdonotplayamajor roleinexpandingBMPproduction,atleastwithinprogenitordomains(Currleetal.,2005).Intuitively,thesemechanisms forexpandingBMPproductionwouldbeusefulaccompanimentstotheevolutionaryenlargementofthetelencephalon.
1.3.3BMPsignalinggradientinthedorsaltelencephalon
IsthereaBMPgradientinthedorsaltelencephalon?Asofnow,thereremainsnodirectevidenceforBMPsthemselves havingagradeddistribution.However,thereisevidenceforagradientofBMP signaling basedonthedistributionof phosphorylatedSmad1/5/8(pSmad),thedirectreadoutofBMPsignaling.(Note:Inthedorsaltelencephalon,Smad1and Smad5,butnotSmad8,areprobablytherelevantSmads)(Arnoldetal.,2006).AtE10.5,thepSmadgradientpeaksatthe midline,whereBMPproductionishighest,andexhibitsasimpleexponentialdeclineawayfromthemidline(Fig.1.3C) (Chengetal.,2006)withalengthscaleof290um(Srinivasanetal.,2014).Thehighdorsomedial-to-lowventrolateral (“DV”)orientationofthisgradientisconsistentwiththemidline-centeredBMPproductionmentionedearlier.
Perturbationstothesystemhaveconfirmedtheexistenceofthissignalinggradient.First,geneticroofplateablations, whichreducemidlineBMPproduction,leadtoacorrespondingreductionand flatteningoftheexponentialpSmadgradient (Fig.1.3C)(Chengetal.,2006).Second,BMP4-soakedbeadsindorsaltelencephalicexplantsinduceconcentration-, position-,andorientation-dependentresponsesthatimplyanunderlyingBMPsignalinggradientwithintheexplanted tissue(Huetal.,2008).Lastly,differencesintheEC50 valuesinvitroandborderpositionsinvivoforBMPtargetgenes Msx1 and Msx2 leadtoaBMPlengthscalecalculationof270um,whichagreeswellwiththe290umvaluederivedfrom thepSmadsignalinggradient(Srinivasanetal.,2014).
TheseobservationsontheBMPsignalinggradientinthedorsaltelencephalonhaveseveralimplications.The exponentialratherthanpower-lawshapeofthegradientsuggestsuniformclearanceofBMPsfromdorsaltelencephalic neuroepithelium.Usingtheroughestimatorof3 5timesthecalculatedlengthscale(Srinivasanetal.,2014),theBMP signalinggradientmightthenpattern800 1400um,whichwouldsuf ficefortheentiredorsaltelencephalonat preneurogenicstages.Theshapeofthegradientalsomakesgoodsensebecausemultiplecellfatethresholdsareneeded towardtheDTM,anditiseasiertomakethresholdswheregradientsaresteepest.
TheexponentialpSmadgradientfurthersuggeststhatdorsaltelencephaliccellsinterpretextracellularBMPconcentrationinalinearlyproportionalfashion.Inthe Drosophila embryoandwingimaginaldisc,GFP-DppandpMad distributionsarebothbest fi tbysimpledecliningexponentials,implyingalineargradedrelationship(Kichevaand Gonzalez-Gaitan,2008).AlthoughBMPdistributionsareunknowninthedorsaltelencephalon,nuclearpSmadlevelsin culturedE12.5corticalprogenitorscorrelateinagradedandpositivefashionwithextracellularBMP4concentration(Hu etal.,2008).Theaforementionedexplantstudies(Huetal.,2008)alsoimplythatBMP4-soakedbeadshaveagradedand additiveeffectonBMPsignaling.
1.3.4BMPsasdorsaltelencephalicmorphogens
AreBMPsand/orBMPsignalingnecessaryandsuffi cientfordorsaltelencephalicfates?WhilethinkingaboutBMP signalinginthese “operational ” termsprovideslimitedinsightintohowthesystemactuallyworks,themodelfallsapartin theabsenceofsuchevidence.Fortunately,thereissubstantialoperationalevidencetosupportthemodel.Cellularablations establishedarequirementforBMP-producingroofplatecellsinthespecificationofallthreeDTMfates(i.e.,roofplate, CPE,andcorticalhem)andinnormalcorticalpatterning(Currleetal.,2005;Chengetal.,2006).Importantly, telencephalon-specifi cinactivationsofBMPreceptors(BMPRIaandBMPRIb)demonstratedsimilarrequirementsfor BMPsignaling(Hebertetal.,2002;Fernandesetal.,2007;Caroniaetal.,2010).WhileDTMfatespecificationdoesnot occurfollowingroofplateorBMPreceptorablation,cortexisspeci fied,albeitabnormallypatterned.Thus,DTMfatesare moredependentonBMPsignalingthancortex agradedrequirementforBMPsignalingthatcorrelateswellwithits normalgradientinthedorsaltelencephalon.
InadditiontotherequirementsforBMPsignaling,suf ficiencyisthe sinequanon foraninstructivemorphogen.Initial evidenceforsuf ficiencycamefromexplantstudies,whichdemonstratedtheabilityofBMP4andotherBMPstoimpart DTM-associatedpropertiesuponthecorticalprimordium(apoptosis,proliferation,andgeneexpression)(Furutaetal., 1997).BMP4aloneisalsosuf ficienttorescueCPEfateinroofplate ablatedexplants(Chengetal.,2006),toinduceCPE ectopicallyinwild-typeexplants(Srinivasanetal.,2014),andtoinduceCPEfateinmouseandhumanEScell derived
systems(Watanabeetal.,2012).Morerecently,BMP4hasbeenshowntosuf ficeforconcentration-dependentand temporally-appropriateinductionofCPEandcorticalhemfatesinareducedculturesystem(Watanabeetal.,2016). Additionalevidenceforsuf ficiencycomesfromnestin-drivenexpressionofconstitutivelyactiveBMPtypeIreceptors, whichresultsinexcessivesimpleepitheliumresemblingchoroidplaqueandCPE(althoughperhapsmorechoroidplaquelikebecauselittletonopapillaryhistologywasobserved)(Panchisionetal.,2001).WhenappliedtoE12.5cortical progenitors,BMP4caninduceconcentration-dependentchangesingeneexpressionthatcharacterizeCPEandchoroid plaque(Huetal.,2008)andevendifferentEC50 valuesforDTMgenes(Msx1 and Msx2)thatreflecttheirborderpositions invivo(Srinivasanetal.,2014).These fi ndingssuggestBMP4suf ficiencytospecifymultipleDTMfatesandborder positionsinaconcentration-dependentfashion,asexpectedforamorphogen.
1.3.5LinearconversionofBMPsignalingbycorticalcells
WhiletheDTMisnotrequiredforcorticalfate,itisrequiredfornormalDVpatterningoftheearlycortex.Asdescribed earlier,theneurogeniccorticalprimordiumischaracterizedbymyriadtranscriptionfactor(TF)gradientsratherthan thresholds,andtheorientationsofmanyoftheseTFgradientsalignwiththatoftheBMPsignalinggradient.TFgradients arenotapparentuntilafterE10.5andthereforeafterthespeci ficationofDTMandcorticalfatesandtheonsetofcortical neurogenesis.
AlthoughlinearityofBMP-to-cor ticalTFregulationhasnotbeenrig orouslyestablished,itisdif fi culttoimagine otherwise.ThesimplefactthatcorticalTFexpressionpatternsaregradedimplieslin earinterpretationsofBMP signaling,ifBMPsignalingindeedregulatescorticalTFexpression,andthereisevidenceforthisbeingthecase.The gradientof Emx2 ,anessentialcorticalpatterninggene,matchesthat oftheBMPsignalinggradient(i.e.,italsohasaDV orientation).Correspondingly,the Emx2 genehasaSmad-bindingenhancerthatupregulatesitsexpression( Theiletal., 2002).Thecorticalselectorgene Lhx2 alsohasaDVgradient,andLhx2canbeupregulatedinexplantsatadistance fromBMP4-soakedbeads( Monukietal.,2001).Followingroofplateablation,theEmx2andLhx2gradientsare reducedand fl attenedinafashionthatcorrelatesstronglywiththeabnormalBMPsignalinggradient( Chengetal., 2006).Interestingly,thegradientsofthreeotherTFs(Pax6,Foxg1,andNgn2)withopposingVDpolaritiesarenot affectedbyroofplateablation,andthesameselectivityforDVgradedgeneswasobservedatintermediateBMP4 concentrationsinareducedculturesystem(Watanabeetal.,2016 ).Together,these fi ndingssuggestthatBMPs selectivelyupregulateDVgenesanddosoinalineargradedfashion.
1.3.6NonlinearconversionofBMPsignalingbyDTMcells
Incontrasttocorticalprogenitors,DTMprogenitorsmustsomehowconvertgradedBMPsignalingintothresholdTFand cellfateresponses,andanonlinearconversionmechanism cell-intrinsicultrasensitivitytoBMP4 hasbeendescribed (Fig.1.3D F )(Huetal.,2008 ).Ultrasensitivitydescribesanyprocesstha tdisplaysnonlinearswitch-likebehaviorin responsetoagradedstimulus.Inthedorsaltelencephalon,ultrasensitivitytoBMP4occursat thelevelofthewell-known BMPtargetandDTMgene, Msx1 ,indissociatedcorticalpr ogenitors,inexplantstreatedwithBMP4-soakedbeads, andinnormalandroofplate ablatedcontextsinvivo(Huetal.,2008).Otherfeaturesofthisultrasensitivity phenomenon includingitsmagnitudeandimm ediate-earlylikekinetics suggestitsutilityformakingcrude initialbordersintheDTM,whicharethenre fi nedandsharpened.Themechanismunderlyingthisformofultrasensitivity turnsouttoinvolvemutuallyinhibitoryinterac tionsbetweenBMPsandothersignalingpathways speci fi cally,theFGF andEGFsignalingpathways whichoccursintracellularlyabovethelevelofindividualBMPtargetgenes.In theabsenceofFGF/EGFsignaling, Msx1 (and Msx2)doseresponsestoBMP4becomeentirelygraded(Fig.1.3E) (Srinivasanetal.,2014 ).
Isthereevidenceforlaterre fi nementofbordersinthedorsaltelencephalon?Theanswerhereisalso “ yes. ” A commonmechanismusedtore fi nebordersiscellsortingbetweenneighboringcells,whichresultsfromdifferentialcell af fi nitiesspeci fi edbyselectorgenes.Inthedorsaltelencephalon, Lhx2 actsasaclassicselectorgeneforthe cortex( Mangaleetal.,2008 ).Amongitsselectoractivities,Lhx2speci fi esacorticalaf fi nitystatethatdiffersfromLhx2negativecorticalhemcells.Thesedifferentialaf fi nitiesresultinsharpsegregationsofLhx2-on(cortical)fromLhx2-off (hem)cellsininducedmosaiccontextsinvivoandinvitro,whichsuggestsanLhx2-dependentcellsortingprocessatthe normalcortex-hemboundary.SimilarroleshavebeenascribedtoPax6atalateralcorticalborder(pallial subpallial boundary)( Stoykovaetal.,1997;Gotzetal.,1998 ).
1.3.7Summary theBMPsignalinggradient
Thedescribed findingsleadtothefollowingmodelofhowtheearlydorsaltelencephalonispatternedbyBMPsignaling:
1. SeveralBMPsareproducedbycellsofthemidlineroofplate,withBMPproductionthenextendingtosecondary signalingcentersandintothecorticalprimordiumviahomeogeneticinduction.
2. ThevariousBMPsourcesgiverisetoadecliningexponentialgradientofBMPsignaling,suggestinglinearinterpretationofextracellularBMPconcentrationbydorsaltelencephaliccellsanduniformBMPclearancefromthepreneurogenicneuroepithelium.
3. DTMcellsconvertthegradedBMPsignalinginformationintothreenonlinear,ultrasensitivethresholdsthatseparate choroidplaque,CPE,corticalhem,andhippocampus.
4. DTMcellsuseacell-intrinsicformofultrasensitivitytoBMP4,whichrequiresmutuallyinhibitoryinteractionswith FGF/EGFsignalingasaninitialnonlinearconversionmechanism.
5. Dorsaltelencephalicbordersarethenrefinedbycomplementarymechanismssuchascellsorting,whichisgovernedby selectorgenessuchasLhx2.
6. Aftercellfatesandbordersarespeci fied,BMPsignalingisinterpretedinalineargradedfashiontoregulateDV patterningofthecorticalprimordium.
Manyfeaturesofthesystemremaintobediscoveredandunderstood.Inadditiontoelucidatinghomeogeneticinduction andcell-intrinsicultrasensitivitymechanismsfurther,itwillbeimportanttodeterminewhetherlinksexistbetweenthe BMPsignalinggradientandothergradedphenomenainthecortex,suchascellcyclelength,self-renewalprobability,and neurogenesis.UnderstandingbetterthespecificBMPsinvolved(BMPhomodimersandheterodimers)andtheir interactionswithBMPinhibitors(e.g.,noggin),transporters(chordin)andothermorphogensshouldalsobeilluminating, aswillstudiesonBMPdiffusivity,clearance,anddistribution.Fascinatingquestionsabouttime,scalingofpatterntotissue size,andtelencephalicgrowthregulationalsoremaintobeanswered.Systems-levelapproacheswillberequiredto understandhowthesystemworksandwhythesystemevolvedthewayitdid.Asthebest-studiedmorphogensystemstell us,attainingafullquantitativeunderstandingofdorsaltelencephalicpatterningwillnotbeeasy.Nonetheless,systemslevelunderstandingwillundoubtedlyprovideinsightsintomanyimportantquestions,includinghowmorphogenetic patterningminimizesfragilitieswhilemaximizingrobustnessandtheadaptabilityneededtoincreaseneocorticalsizein humansandotherspecies.
1.4FGF8asamorphogenintelencephalicpatterning
Asnotedpreviously,thereisnowsubstantialevidencethatFGF8andFGF17,fromtheRPC,patterntheneocorticalarea map(Fukuchi-ShimogoriandGrove,2001;Gareletal.,2003;CholfinandRubenstein,2007,2008;Toyodaetal.,2010), andthistopicisthesubjectofasubsequentchapter.MostrelevanthereisevidencethatFGF8actsasaclassicmorphogen topatterntheneocortexandpossiblytheearlier-stagetelencephalon.
IncreasingevidencesupportsanearlyroleforFGF8inthel argerdivisionofthetelencephalicvesicleintoadorsal partthatwillgeneratethecerebralcortexandaventralpa rtthatwillgeneratesubcorti calnuclei.Forexample,in zebra fi sh,knockingdowneither Fgf8 or Fgf8 and Fgf3 expressiontogethercausesreductionorlossofventraltelencephaliccells(Shanmugalingametal.,2000;WalsheandMason,2003).Inmouse, Fgf3 appearslessimportant(Theil etal.,2008),butreduced Fgf8 causesastrikingreductionoftheseptumand lateralandmedialganglioniceminences (LGE,MGE)(Stormetal.,2006).Furthermore,inmicede fi cientin Fgfr1 and Fgfr2 ,theventraltelencephalonisgreatly reduced,re fl ectingashiftfromaventraltoadorsaltelencephaliccellfate(Gutinetal.,2006 ).Finally,characteristically ventralgeneexpressioncanbeinducedbyFGF8indorsaltelencephalicexplants(Kuscheletal.,2003 )andindorsal telencephaloninvivo(Assimacopoulos,Taylor,andGrove,unpublis hedresults).These fi ndingsmaybesurprisingin thecontextofthewell-establishedroleforShhinspecifyingventralcelltypesinthespinalcord(Fuccilloetal.,2006 ). TherelativecontributionofShh,FGF8,andotherfactorshasbeeninvestigatedrecentlybyrecapitulatingventral telencephalicdevelopmentinmouseEScellculture(Danjoetal.,2011).Corticalandsubcorticalcellidentitieswere judgedbypatternsofexpressionofgenescharacteristicofcellsinvivo.ApplicationofmoderatelevelsofShhbeginning earlyintheEScellcultureandcontinuingforseveraldaysinducedLGEprogenitors.StrongerShhsignalingspeci fi ed MGEandcaudalganglioniceminence(CGE)progenitors;however,MGEspeci fi cationalsorequiredFGF8,andthe
CGErequiredFGF15.TheextenttowhichFGF8,FGF15,andShheachcontroltelencephalicventralizationinvivo needsadditionalstudyandislikelytoinvolveanincreasedunderstandingofinteractionsbetweentheShhandFGF signalingpathways,asdiscussedfurtherbelow.
Oncetheneocorticalprimordium(NP)isestablished,FG F8dispersesfromtheRPCthroughtheentireNP,creatinga proteingradientwithanexponential declinefromanteriortoposterior(Fig.1.4 )(Toyodaetal.,2010 ).Ifweassumethat FGF8dispersesbydiffusionfromsourcecells,theexponen tialdeclineimpliesuniformclearanceofFGF8throughout theNP(seeabove).ThishasnotyetbeenshownforthemouseNP,butFGF8dispersionfromasourcefollowedby ubiquitousendocytosisanddegradationoftheproteinhasbeendemonstratedinzebrafi sh( ScholppandBrand,2004; Yuetal.,2009 ).
TheRPCbecomesevidentjustafterneuraltubeclosureataboutE9inthemouse.AtE9.5,whentheFGF8gradient wasquanti fi edfromFGF8immuno fl uorescence,theNPisabout300micronslongfromanteriortoposterior(Fig.1.4D), or70 80cellwidths( Toyodaetal.,2010 ),consistentwiththesizeofaclassicmorphogenetic fi eld(seeabove).The measuredhalf-decayofFGF8intheE9.5NPoccurredoverabout10cellwidths( Toyodaetal.,2010 ).Giventhatas littleasatwofolddifferenceinmorphogenconcentrationcaninducecellsto adoptdifferentfates(GreenandSmith, 1990),thehalf-declineofFGF8comparedwiththetotallengthoftheNPatE9.5suggeststhatseveraldifferentareafates couldbeobtainedfromtheestimatedFGF8gradient.Ectopicsourcesofmyc-taggedFGF8wereintroducedby electroporation,andFGF8-mycimmuno fl uorescenceshowedgradeddispersionfromtheelectroporationsite.FGF8-myc musthavediffusedfromthenewsourceorutilizedanothermechanismfordispersionthatresultsinagradient.Ectopic FGF8-mycsourcesinducedexpressionofdifferentFGF8targetgenesatdifferentdistancesindicatingthatupregulation oftheexpressionofthesetargetgenesrequireddifferentlevelsofFGF8(Toyodaetal.,2010).
Akeyfeatureofamorphogenisthatitactsdirectlyatad istance,ratherthanbyarelayofseveralsignaling molecules.Totestthis,adominantnega tiveFGFreceptorwithhighinvitroaf fi nityforFGF8(Ornitzetal.,1996; Chellaiahetal.,1999;Zhangetal.,2006)waselectroporatedintoth ecentralregionoftheNP(Toyodaetal.,2010).The dominantnegativereceptor,dnFGFR3c,capturedFGF8atadistancefromtheFGF8source,andthereductionofFGF8 inducedcellstoadoptamoreposteriorareaidentity,demonstratingdir ect,long-rangepatterningbyFGF8( Toyoda etal.,2010).TheseobservationssupportFGF8asaclassicdiffusiblemorphogeninneocortex.
1.5Interactionsamongsignalingcentersintelencephalicpatterning
Evidencetodateindicatesthatinteractionsamongsignalingcentersinthetelencephalon(1)regulatetheexpressionof genesencodingspeci ficsignalingmoleculesintheothersignalingcenterand(2)modifytheactivityoftheothersignaling centerbyregulationofadownstreamsignalingcomponent.






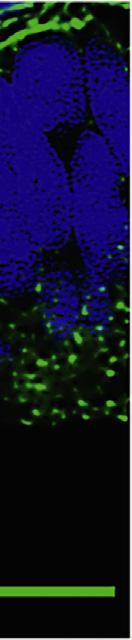
FIGURE1.4 CharacterizationoftheFGF8gradientintheneocorticalprimordiumatE9.5. (A)Sagittalmidlinesectionthroughoneofninemouse brainsusedinquantification.FGF8IFlwasmeasuredinastandardizedsegmentoftheneocorticalprimordium(ncxp)(yellowband;whitelinesmark anteriorandposteriorncxpboundaries).(B,C)FGF8IFl(grayscale)inadigitallystraightenedsegmentshowsanA/Pintensitygradient(B),asdoes FGF8 IFl(false-colored)averagedfromninesegments(C).(D)PlotofmeanFGF8IFlinarbitraryunits(AU)againstdistancefromtheanteriorFGF8source. An IFlintensityplateauinthemostanteriorportion(C)wasnotplottedin(D).Adecliningexponentialcurve(red)was fittedtotheremainingdata(blue).The x and y axesoftheplotinDdonotstartatzerotoalloweasieridentificationofhalf-declinepointsofthegradient.MaximumFGF8IFlintensityis(1), sequentialhalf-declinepointsarelabeled(2),(3),and(4).TheA/Pdistanceofthehalf-declineisabout45microns(seebrokengreenlinesinD),roughly thewidthof10DAPI-stainednuclei(whitearrows,E).Scalebars:0.1mmin(A);0.04mmin(E).
1.5.1FGF8,Shh,andBMPsignaling
Twocompanionpapersdescribedthe firstdirectstudyofinteractionsamongjuxtaposedtelencephalicsignalingcentersin chickandmouse(Crossleyetal.,2001;Ohkuboetal.,2002).ThesestudiesproposedthatFGF8,BMP4,andMGE-derived Shhregulateeachother’sgeneexpression,andtherebythegrowthofthetelencephalicvesicle,inamannersimilarto interactionsofFGF8,BMPs,andShhinthelimbbud(Ohkuboetal.,2002).BMP4wasfoundtorepress Fgf8 and Shh expression,whereasShhrepressed Bmp expressionandmaintainedexpressionof Fgf8.Bothexperimentallyincreasedand decreasedBMPsignalingcorrelatedwithdecreasedgrowthofthetelencephalicvesicles,suggestingthatnormalgrowth requiresabalanceofBMP,FGF8,andShhsignaling(Ohkuboetal.,2002).
1.5.2Cross-regulationofBMP,FGF,andWNTsignaling
OnefunctionofinteractionsbetweentheRPCandthecorticalhemistoregulatetheothersignalingcenter ’sboundaries. IncreasedFGF8signalingdecreases Wnt2b, 3a,and 5a expressioninthehem,withaconsequentreductioninthesizeof thehippocampus.Conversely,BMPsignalingfromthehemregioninbothchickandmousedecreases Fgf8 expression (Ohkuboetal.,2002;Shimogorietal.,2004).Thus, Wnt signalinginthehemisprotectedbyBMPactivitythatholdsback Fgf8 expression.Surprisingly,theBMPantagonist,noggin,cancausewidespreadupregulationof Fgf8 expression, suggestingthatalargepartofthecorticalprimordiumiscompetenttoexpress Fgf8 andwoulddosointheabsenceof activerepression(Shimogorietal.,2004).Giventhatthecorticalselectorgene Lhx2 suppressescorticalhemandantihem tissuefates(Mangaleetal.,2008),theNPmightdefaultintoaconglomerationofhem,antihem,andRPC,wereitnot blockedfromdoingsobyBMPsignalingandLhx2.
Thetranscriptionfactor,EMX2(anorthologof Drosophila emptyspiracles),seemstobeatthehubofinteractions betweenFGFandWntsignalinginthedorsaltelencephalon.FGF8downregulatesexpressionof Emx2,whereasWnt3a upregulatesitsexpression(Fukuchi-ShimogoriandGrove,2001;Caronia-Brownetal.,2014).Emx2,inturn,regulatesthe twosignalingcenters(Fukuchi-ShimogoriandGrove,2003).Micedefi cientin Emx2 haveacombinationofsignaling centerdefects. Fgf8 and Fgf17 expressionincreases,expandingpastnormallimitsoftheRPC,andexpressionof Wnt2b, 3a, 5a,and 8b isreducedinthedorsomedialtelencephalon,includingthehem,asdoestheotherdownstreamcomponents ofcanonicalWntsignalingsuchasLef1(Fukuchi-ShimogoriandGrove,2003).Cellcyclecontrol,likelytobeinfluenced byWntsignaling,isaberrantinthe Emx2 mutanttelencephalon,andthehippocampusisshrunken(Toleetal.,2000b; Muzioetal.,2002,2005;Fukuchi-ShimogoriandGrove,2003;Shimogorietal.,2004;CholfinandRubenstein,2008). ReducingexcessFGF8partiallyrescuesboth Wnt geneexpressionandthehippocampal fields(Shimogorietal.,2004),and reactivatingWntsignalingwithlithiumtreatmentpartiallyrestoresnormalcellcycledynamicsinthe Emx2 mutant(Muzio etal.,2005).WhetherreducedWntsignalinginthe Emx2 mutantmouseiscauseddirectlybylossofEmx2(Muzioetal., 2005),indirectlybyincreasedFGF8signaling(Shimogorietal.,2004),orboth(CholfinandRubenstein,2008)remainsto befullyclari fied.WhatseemsclearisthatthehemandRPChaveoppositeeffectsontheexpressionof Emx2 andthatthis islikelytoshapethecaudomedialhightorostrallowgradientof Emx2 expressionintheNP.Aswillbedescribedina subsequentchapter,theNP Emx2 gradienthasamajorroleinneocorticalareapatterning.
1.5.3InteractionsofShh,FGFs,andGli3
ShhsignalingismediatedbytheGligenes,andinthepresenceofShh,allthreeGlitranscriptionfactorsworkas transactivators.IntheabsenceofShh,however,Gli3remainsinarepressorstate(Gli3R)( Fuccilloetal.,2006 ). Observationsofthe extra-toes mutantmouse,whichlacks Gli3 altogether,indicatethatGli3Rrepressesexpressionof Fgf8 , Fgf17,and Fgf15 ( Aotoetal.,2002;RashandGrove,2007).Thatis,in extra-toes ,theexpressiondomainofeach ofthese Fgf genesgreatlyexpands.Notably,inthe extra-toes mouse,thecerebralcortexhasahighlydisrupted morphology;moreovergeneexpressiontypicaloftheventraltelencephalonextendsintotherostrodorsaltelencephalon (Toleetal.,2000a;Kuscheletal.,2003 ).Thus,undernormalcircumstances,thedorsaltelencephalonisprotectedby Gli3R-mediatedsuppressionfromtheventralizingeffectsofFGFsignaling( Kuscheletal.,2003).
1.6Morphogensinhumanbraindisease
1.6.1HoloprosencephalyandKallmannsyndrome
Amajorhumandisorderassociatedwithdefectivemorphogen-mediatedpatterningisholoprosencephaly(HPE,OMIM 236100),themostcommoncongenitalbirthdefectofthehumanforebrain(1in16,000livebirths,1in250conceptions)
(Monuki,2007).HPEhaslongfascinatedcliniciansandscientistsbecauseofitsstrikingphenotype,whichcanincludea singleforebrain “holosphere” ratherthanhemispheres(Fig.1.5)andmidlinecraniofacialdefectssuchasasinglemidline eye(cyclopia).ThesephenotypesreflecttheprimaryfailuresinmidlineinductionthatdefineHPE(Golden,1998).Human HPEisdividedintotwocategorieswithdistinctphenotypes:(1)classicand(2)middleinterhemispheric(MIH,alsoknown assyntelencephaly).Themore-commonclassicHPEisdividedintoalobar,semilobar,andlobarsubtypesbasedon severity,withalobarbeingthemostsevere.AnewHPEsubtype,referredtoasseptopreopticHPE(Hahnetal.,2010), probablyrepresentsamilder “formfruste” ofclassicHPE.OtherHPEspectrumdisordersincludearrhinencephaly (absenceofolfactorybulbs),septoopticdysplasia(OMIM182230),andKallmannsyndrome(OMIM308700),which combinesanosmiawithhypogonadotropichypogonadismduetodefectsinolfactoryandhypothalamicdevelopment. Humanandanimalgeneticsdirectlyimplicatesignalingbyfourfamiliesofmorphogens NODAL,SHH,FGF,and BMP intheinductionandpathogenesisofHPE(Monuki,2007).Althoughchromosomalabnormalities(mostoften trisomies)aremoreoftenthegeneticculprits,overadozensinglegeneshavenowbeendirectlyassociatedwithhuman HPE(Table1.1)(Solomonetal.,2011;Dubourgetal.,2016).ThesegenesincludeseveralfactorsthataffectNODALand
TABLE1.1 Holoprosencephalygenesimplicatedinmorphogensignaling.
NODALSHHFGF8 Chordin
TDGF1/CRIPTOPTCH1FGFR1 Noggin
FOXH1/FAST1DHCR7 Twsg1
TGIF1GLI2 BmpRIa/Ib
Cyclops DISP1 Squint GAS1 One-eyedpinhead CDON SUFU Smo (ZIC2)(SIX3)(SIX3)(ZIC2) (Megalin) (TGIF1) (Megalin)
Genesorwell-establishedmodifiersfromhumansandexperimentalorganismslinkedtoholoprosencephalyand/orcyclopicphenotypes,groupedby morphogensignalingpathway.Humangenesareshowninbolduppercase;geneswithtentativeplacementsareparenthesizedandlistedtowardsthe bottom.



FIGURE1.5 Morphogeninteractionsinholoprosencephaly. (A,B)Anteriorviewsofthecentralnervoussystemfromanormal13-weekfetus(A)and an18-weekgestationhumanfetuswithalobarholoprosencephaly(B).Thefetuswithholoprosencephalyhasasingleforebrainvesicle(“holosphere”)and acortexthatiscontinuousacrosstheventrobasal,rostral,anddorsalmidlines.(C)Morphogeninteractionnetworkthatcanaccountforclassicholoprosencephaly(HPE)subtypeswithvariableextensiontothedorsalmidline,aswellasMIHHPE,whichselectivelyinvolvesthedorsalmidlineregion. ModifiedfromMonuki,E.S.,Walsh,C.A.,2001.Mechanismsofcerebralcorticalpatterninginmiceandhumans.Nat.Neurosci.4Suppl.,1199 1206; andMonuki,E.S.,2007.Themorphogensignalingnetworkinforebraindevelopmentandholoprosencephaly.J.Neuropathol.Exp.Neurol.66,566 575.
Bmp4
Fgf8
Shh
Nodal
(C)
(B)
(A)
SHHsignalingandmorerecently FGF8 and FGFR1 (Arauzetal.,2010;Rosenfeldetal.,2010;McCabeetal.,2011; Dubourgetal.,2016).Theassociationof FGF8 mutationswithbothmildHPEandKallmannsyndromesupportthe conceptthatthesemalformationslieonadiseasecontinuum.Atthispoint,animalstudiesaloneimplicateBMPsignaling inclassicandMIHforms(Chengetal.,2006;Fernandesetal.,2007).MajorhumanHPEgenesthatdonotencodefor morphogensthemselves(TGIF, ZIC2,and SIX3)regulatemorphogensormorphogensignaling.Forexample, Tgif mutationsdisruptShhandNodalsignaling(Taniguchietal.,2012),while Zic2 mutationsresultindefectivenodeand prechordalplatedevelopment,therebyimpactingShhandNodalsignaling(Warretal.,2008). Six3,ontheotherhand,is requiredforShhtranscriptionalactivation(Gengetal.,2008).
ThefundamentaldefectinHPEisfailedmidlineinduction(Golden,1998),andallfourofthemorphogenfamilies implicatedareexpressedathighestlevelsinmidlinetissues(Monuki,2007).Asdiscussedabove,NODALandSHHare bothmainlyproducedbyventralmidlinetissues(prechordal,preoptic,andhypothalamicregions),FGF8isexpressedin theRPC,andBMPsareproducedbothinventralmidline(BMP7intheprechordalplate)andDTM(roofplate,CPE,and corticalhem).
Importantly,interactionsbetweenthefourmorphogenfamiliescanaccountforhumanHPEphenotypesand,in particular,forthelong-standingconundrumofhowprimarydefectsinventralsignalingcanleadtoDTMfailures (Fig.1.5C)(Monuki,2007).Briefl y,andasdetailedabove,NODALisupstreamofSHH,whichthenengagesinpositive feedbackwithrostralFGF8.Ontheotherhand,dorsalBMPsnegativelyregulatetheSHH-FGF8circuit,whileFGF8can havebothpositiveandnegativeeffectsondorsalBMPs.Thismorphogensignalingnetworkcanthenexplainhow NODAL,SHH,andFGF8signalingdefectscanextendtoincludetheDTMregion,butwhythisextensionisquitevariable (perhapsduetothemorecomplexFGF dorsalBMPinteractions),evenwithinfamiliesharboringidenticalmutations. ThisnetworkcanalsoexplainwhyrostralandventralregionsarenotaffectedinMIHHPE(i.e.,whendorsalBMP signalingisthesuspectedculprit).
1.6.2Gradientsinholoprosencephalyneuropathology
Withregardtothemorphogengradienthypothesis,itisimportanttoconsideraspectsofHPEneuropathologythatare themselvesgraded.Thegradedneuropathologythat firstcomestomindisthegradedinvolvementofventralversusdorsal forebrainregionsinclassicHPE,inwhichventrobasalandrostralregionsaremoreseverelyaffectedthandorsaland posteriorones.ThisspatialgradientissimplesttovisualizeinsemilobarHPE,butitisevidentinmostclassicHPEcases (perhapswiththeexceptionofthemostcompleteformsofalobarHPE).However,thisgradientofneuropathologycannot beexplainedbythespatiotemporalgradientofanysinglemorphogen.Instead,networkinteractionsbetweenmidline morphogensmustbeinvoked(Fig.1.5C)(Monuki,2007).
AnothersetofgradientsobservedinHPE whicharemoredirectlyrelevanttothemorphogengradient hypothesis aretheneuropathologicalgradientsrelativetothemidline.Becauseallofthemorphogensimplicatedin classicHPEareexpressedathighestlevelsinmidlinetissues,itmakessensethatHPEneuropathologyisgreatesttoward themidline(Monuki,2007).Equallyimportant,neuropathologicseveritydiminishesawayfromthemidline (e.g.,neocortexisalwayspresentinsevereclassicHPEcases),andinmilderforms,midlinetissuessuchasolfactorybulbs orseptopreoptictissuesappeartobetheonlystructuresinvolved.ThegradedneuropathologyinMIHHPEisalso consistentwiththemorphogengradienthypothesis.MIHHPEpatientsuniformlyhavetotaltonear-totalabsenceofCPEin thelateralventricles,whiletheirhippocampiareoften,butnotalways,smallanddysplastic(Chengetal.,2006).Thus,as inclassicHPE,MIHHPEneuropathologyismoreseveremedially(CPE)thanlaterally(hippocampus).
1.6.3Gradientsinotherhumanbraindisorders
AlthoughlesswellappreciatedthaninHPE,manydevelopmentaldiseasesexhibitcharacteristicgradientsinneuropathology.Oneexampleisadrenoleukodystrophy(OMIM300100),anX-linkedperoxisomaldisorderthatresultsin widespreaddysmyelination/demyelinationofCNSwhitematter.Withinthecerebrum,adrenoleukodystrophyhasa characteristicneuropathologicalgradient,withposterior(occipital)whitemattermoreseverelyaffectedthananterior (frontal).Anotherexampleislissencephaly(OMIM607423),aseverecorticalmalformation.Thetwogenesmost commonlyassociatedwithlissencephalyare LIS1 and DCX (alsoknownas LISX1),andtheneuropathologiesdueto mutationsinthesetwogenesexhibitopposinggradients LIS1-associatedlissencephalyisposteriorpredominant,whereas DCX mutationscauseanteriorpredominantlissencephaly(GuerriniandMarini,2006).Patterningdisordersofthe brainstemalsodisplayAPgradientsinneuropathology(Barkovichetal.,2009).
Theseandotherexamplesreflectanaspectofdevelopmentalbraindisordersthatremainspoorlyunderstood,but experimentallytractableinanimalmodels theroleofmorphogeneticgradientsindeterminingthegradedpatternsof disease.Whetherprimarymorphogengradientsareresponsibleforthegradedneuropathologiesintheseandother disordersisunknown,andifresponsible,itremainstobeseenwhethertheresponsibilitiesaredirectorindirect (e.g.,morphogengradientsinthedorsaltelencephaloncouldindirectlydeterminethegradedlissencephalyphenotypesby regulatinggradientswithinthecorticalplate).Regardless,carefulexaminationsoftheneuropathologiesinhumansand animalmodelsshouldprovidecluestothepotentialgradientsimpingingupondiseaseprocessesandwhetherlinearor nonlinearconversionsofgradedinformationareinvolved.Inthisregard,understandinghowgradedorswitch-like signalingisachievedcouldalsoimpactotherhumanbraindiseaseslinkedtomorphogens,butnottogradientsofthese morphogensperse(e.g.,theroleofSHHandWNTsignalinginmedulloblastoma).
References
Affolter,M.,Basler,K.,2007.TheDecapentaplegicmorphogengradient:frompatternformationtogrowthregulation.Nat.Rev.8,663 674. Alexandre,P.,Wassef,M.,2003.Theisthmicorganizerlinksanteroposterioranddorsoventralpatterninginthemid/hindbrainbygeneratingroofplate structures.Development130,5331 5338.
Aoto,K.,Nishimura,T.,Eto,K.,Motoyama,J.,2002.MouseGLI3regulatesFgf8expressionandapoptosisinthedevelopingneuraltube,face,andlimb bud.Dev.Biol.251,320 332.
Arauz,R.F.,Solomon,B.D.,Pineda-Alvarez,D.E.,Gropman,A.L.,Parsons,J.A.,Roessler,E.,Muenke,M.,2010.AhypomorphicalleleintheFGF8 genecontributestoholoprosencephalyandisallelictogonadotropin-releasinghormonedeficiencyinhumans.Mol.Syndromol.1,59 66. Arnold,S.J.,Maretto,S.,Islam,A.,Bikoff,E.K.,Robertson,E.J.,2006.Dose-dependentSmad1,Smad5andSmad8signalingintheearlymouseembryo. Dev.Biol.296,104 118.
Ashe,H.L.,Briscoe,J.,2006.Theinterpretationofmorphogengradients.Development133,385 394.
Ashe,H.L.,Mannervik,M.,Levine,M.,2000.DppsignalingthresholdsinthedorsalectodermoftheDrosophilaembryo.Development127,3305 3312. Assimacopoulos,S.,Grove,E.A.,Ragsdale,C.W.,2003.IdentificationofaPax6-dependentepidermalgrowthfactorfamilysignalingsourceatthelateral edgeoftheembryoniccerebralcortex.J.Neurosci.23,6399 6403.
Bachler,M.,Neubuser,A.,2001.ExpressionofmembersoftheFgffamilyandtheirreceptorsduringmidfacialdevelopment.Mech.Dev.100,313 316. Barkai,N.,Shilo,B.Z.,2009.Robustgenerationanddecodingofmorphogengradients.ColdSpringHarb.Perspect.Biol.1,a001990.
Barkovich,A.J.,Millen,K.J.,Dobyns,W.B.,2009.Adevelopmentalandgeneticclassificationformidbrain-hindbrainmalformations.Brain132, 3199 3230.
Bhardwaj,R.D.,Curtis,M.A.,Spalding,K.L.,Buchholz,B.A.,Fink,D.,Bjork-Eriksson,T.,Nordborg,C.,Gage,F.H.,Druid,H.,Eriksson,P.S., Frisen,J.,2006.Neocorticalneurogenesisinhumansisrestrictedtodevelopment.Proc.Natl.Acad.Sci.U.S.A.103,12564 12568. Bier,E.,DeRobertis,E.M.,2015.EMBRYODEVELOPMENT.BMPgradients:aparadigmformorphogen-mediateddevelopmentalpatterning.Science 348. https://doi.org/10.1126/science.aaa5838.
Bishop,K.M.,Goudreau,G.,O’Leary,D.D.,2000.RegulationofareaidentityinthemammalianneocortexbyEmx2andPax6.Science288,344 349. Boveri,T.,1901.Diepolaritätvonovocyte,ei,undlarvedesstrongylocentruslividus.Zool.Jahrb.-Abt.Anat.Ontog.Tiere384.
Briscoe,J.,Small,S.,2015.Morphogenrules:designprinciplesofgradient-mediatedembryopatterning.Development142,3996 4009.
Carlson,J.M.,Doyle,J.,2002.Complexityandrobustness.Proc.Natl.Acad.Sci.U.S.A.99(Suppl.1),2538 2545.
Caronia,G.,Wilcoxon,J.,Feldman,P.,Grove,E.A.,2010.Bonemorphogeneticproteinsignalinginthedevelopingtelencephaloncontrolsformationof thehippocampaldentategyrusandmodifiesfear-relatedbehavior.J.Neurosci.30,6291 6301.
Caronia-Brown,G.,Yoshida,M.,Gulden,F.,Assimacopoulos,S.,Grove,E.A.,2014.Thecorticalhemregulatesthesizeandpatterningofneocortex. Development141,2855 2865.
Chellaiah,A.,Yuan,W.,Chellaiah,M.,Ornitz,D.M.,1999.Mappingligandbindingdomainsinchimeric fibroblastgrowthfactorreceptormolecules. Multipleregionsdetermineligandbindingspecificity.J.Biol.Chem.274,34785 34794.
Cheng,X.,Hsu,C.M.,Currle,D.S.,Hu,J.S.,Barkovich,A.J.,Monuki,E.S.,2006.Centralrolesoftheroofplateintelencephalicdevelopmentand holoprosencephaly.J.Neurosci.26,7640 7649.
Chiang,C.,Litingtung,Y.,Lee,E.,Young,K.E.,Corden,J.L.,Westphal,H.,Beachy,P.A.,1996.Cyclopiaanddefectiveaxialpatterninginmicelacking Sonichedgehoggenefunction.Nature383,407 413.
Cholfin,J.A.,Rubenstein,J.L.,2007.PatterningoffrontalcortexsubdivisionsbyFgf17.Proc.Natl.Acad.Sci.U.S.A.104,7652 7657.
Cholfin,J.A.,Rubenstein,J.L.,2008.FrontalcortexsubdivisionpatterningiscoordinatelyregulatedbyFgf8,Fgf17,andEmx2.J.Comp.Neurol.509, 144 155.
Crick,F.,1970.Diffusioninembryogenesis.Nature225,420 422.
Crossley,P.H.,Martin,G.R.,1995.ThemouseFgf8geneencodesafamilyofpolypeptidesandisexpressedinregionsthatdirectoutgrowthand patterninginthedevelopingembryo.Development121,439 451.
Crossley,P.H.,Martinez,S.,Ohkubo,Y.,Rubenstein,J.L.,2001.CoordinateexpressionofFgf8,Otx2,Bmp4,andShhintherostralprosencephalon duringdevelopmentofthetelencephalicandopticvesicles.Neuroscience108,183 206.
Morphogens,patterningcenters,andtheirmechanismsofaction Chapter|1 19
Currle,D.S.,Cheng,X.,Hsu,C.M.,Monuki,E.S.,2005.DirectandindirectrolesofCNSdorsalmidlinecellsinchoroidplexusepitheliaformation. Development132,3549 3559.
Danjo,T.,Eiraku,M.,Muguruma,K.,Watanabe,K.,Kawada,M.,Yanagawa,Y.,Rubenstein,J.L.,Sasai,Y.,2011.Subregionalspecificationof embryonicstemcell-derivedventraltelencephalictissuesbytimedandcombinatorytreatmentwithextrinsicsignals.J.Neurosci.31,1919 1933. Dekanty,A.,Milan,M.,2011.Theinterplaybetweenmorphogensandtissuegrowth.EMBORep.12,1003 1010.
Driever,W.,Nusslein-Volhard,C.,1988a.ThebicoidproteindeterminespositionintheDrosophilaembryoinaconcentration-dependentmanner.Cell54, 95 104.
Driever,W.,Nusslein-Volhard,C.,1988b.AgradientofbicoidproteininDrosophilaembryos.Cell54,83 93. Dubourg,C.,Carre,W.,Hamdi-Roze,H.,Mouden,C.,Roume,J.,Abdelmajid,B.,Amram,D.,Baumann,C.,Chassaing,N.,Coubes,C.,FaivreOlivier,L.,Ginglinger,E.,Gonzales,M.,Levy-Mozziconacci,A.,Lynch,S.A.,Naudion,S.,Pasquier,L.,Poidvin,A.,Prieur,F.,Sarda,P., Toutain,A.,Dupe,V.,Akloul,L.,Odent,S.,deTayrac,M.,David,V.,2016.MutationalsepctruminholoprosencephalyshowsthatFGFisanew majorsignalingpathway.Hum.Mutat.37,1329 1339.
Dyson,S.,Gurdon,J.B.,1998.Theinterpretationofpositioninamorphogengradientasrevealedbyoccupancyofactivinreceptors.Cell93,557 568. Ferguson,E.L.,Anderson,K.V.,1992.Decapentaplegicactsasamorphogentoorganizedorsal-ventralpatternintheDrosophilaembryo.Cell71, 451 461.
Fernandes,M.,Gutin,G.,Alcorn,H.,McConnell,S.K.,Hebert,J.M.,2007.MutationsintheBMPpathwayinmicesupporttheexistenceoftwo molecularclassesofholoprosencephaly.Development134,3789 3794.
Ferri,R.T.,Levitt,P.,1995.RegulationofregionaldifferencesinthedifferentiationofcerebralcorticalneuronsbyEGFfamily-matrixinteractions. Development121,1151 1160.
Freeman,M.,Gurdon,J.B.,2002.Regulatoryprinciplesofdevelopmentalsignaling.Annu.Rev.CellDev.Biol.18,515 539.
Fuccillo,M.,Joyner,A.L.,Fishell,G.,2006.Morphogentomitogen:themultiplerolesofhedgehogsignallinginvertebrateneuraldevelopment.Nat.Rev. Neurosci.7,772 783.
Fukuchi-Shimogori,T.,Grove,E.A.,2001.NeocortexpatterningbythesecretedsignalingmoleculeFGF8.Science294,1071 1074.
Fukuchi-Shimogori,T.,Grove,E.A.,2003.Emx2patternstheneocortexbyregulatingFGFpositionalsignaling.Nat.Neurosci.6,825 831.
Furuta,Y.,Piston,D.W.,Hogan,B.L.,1997.Bonemorphogeneticproteins(BMPs)asregulatorsofdorsalforebraindevelopment.Development124, 2203 2212.
Galceran,J.,Farinas,I.,Depew,M.J.,Clevers,H.,Grosschedl,R.,1999.Wnt3a-/–likephenotypeandlimbdeficiencyinLef1(-/-)Tcf1(-/-)mice.Genes Dev.13,709 717.
Garcia-Bellido,A.,1975.GeneticcontrolofwingdiscdevelopmentinDrosophila.CibaFound.Symp.0,161 182.
Garel,S.,Huffman,K.J.,Rubenstein,J.L.R.,2003.MolecularregionalizationoftheneocortexisdisruptedinFgf8hypomorphicmutants.Development 130,1903 1914.
Geng,X.,Speirs,C.,Lagutin,O.,Inbal,A.,Liu,W.,Solnica-Krezel,L.,Jeong,Y.,Epstein,D.J.,Oliver,G.,2008.Haploinsuf ficiencyofSix3failsto activateSonichedgehogexpressionintheventralforebrainandcausesholoprosencephaly.Dev.Cell15,236 247.
Goentoro,L.A.,Reeves,G.T.,Kowal,C.P.,Martinelli,L.,Schupbach,T.,Shvartsman,S.Y.,2006.QuantifyingtheGurkenmorphogengradientin Drosophilaoogenesis.Dev.Cell11,263 272.
Golden,J.A.,1998.Holoprosencephaly:adefectinbrainpatterning.J.Neuropathol.Exp.Neurol.57,991 999. Gotz,M.,Stoykova,A.,Gruss,P.,1998.Pax6controlsradialgliadifferentiationinthecerebralcortex.Neuron21,1031 1044.
Grove,E.A.,Tole,S.,Limon,J.,Yip,L.,Ragsdale,C.W.,1998.ThehemoftheembryoniccerebralcortexisdefinedbytheexpressionofmultipleWnt genesandiscompromisedinGli3-deficientmice.Development125,2315 2325.
Green,J.B.,Smith,J.C.,1990.GradedchangesindoseofaXenopusactivinAhomologueelicitstepwisetransitionsinembryoniccellfate.Nature347 (6291),391 394.
Guerrini,R.,Marini,C.,2006.Geneticmalformationsofcorticaldevelopment.Exp.BrainRes.173,322 333.ExperimentelleHirnforschung.
Gulacsi,A.,Anderson,S.A.,2006.ShhmaintainsNkx2.1intheMGEbyagli3-independentmechanism.Cerebr.Cortex16(Suppl.1),i89 95. Gutin,G.,Fernandes,M.,Palazzolo,L.,Paek,H.,Yu,K.,Ornitz,D.M.,McConnell,S.K.,Hebert,J.M.,2006.FGFSignallingGeneratesVentral TelencephalicCellsIndependentlyofSHH.Development.
Hahn,J.S.,Barnes,P.D.,Clegg,N.J.,Stashinko,E.E.,2010.Septopreopticholoprosencephaly:amildsubtypeassociatedwithmidlinecraniofacial anomalies.AJNRAm.J.Neuroradiol.31,1596 1601.
Hebert,J.M.,Mishina,Y.,McConnell,S.K.,2002.BMPsignalingisrequiredlocallytopatternthedorsaltelencephalicmidline.Neuron35,1029 1041. Hoch,R.V.,Clarke,J.A.,Rubenstein,J.L.,2015.FgfsignalingcontrolsthetelencephalicdistributionofFgf-expressingprogenitorsgenerated intherostral patterningcenter.NeuralDev.10,8.
Holley,S.A.,Neul,J.L.,Attisano,L.,Wrana,J.L.,Sasai,Y.,O’Connor,M.B.,DeRobertis,E.M.,Ferguson,E.L.,1996.TheXenopusdorsalizingfactor nogginventralizesDrosophilaembryosbypreventingDPPfromactivatingitsreceptor.Cell86,607 617.
Hu,J.S.,Doan,L.T.,Currle,D.S.,Paff,M.,Rheem,J.Y.,Schreyer,R.,Robert,B.,Monuki,E.S.,2008.BorderformationinaBmpgradientreducedto singledissociatedcells.Proc.Natl.Acad.Sci.U.S.A.105,3398 3403.
Kicheva,A.,Gonzalez-Gaitan,M.,2008.TheDecapentaplegicmorphogengradient:aprecisedefinition.Curr.Opin.CellBiol.20,137 143.
Kim,A.S.,Anderson,S.A.,Rubenstein,J.L.,Lowenstein,D.H.,Pleasure,S.J.,2001.Pax-6regulatesexpressionofSFRP-2andWnt-7binthedeveloping CNS.J.Neurosci.21,RC132.
Kuschel,S.,Ruther,U.,Theil,T.,2003.AdisruptedbalancebetweenBmp/WntandFgfsignalingunderliestheventralizationoftheGli3mutant telencephalon.Dev.Biol.260,484 495.
Lander,A.D.,2007.Morpheusunbound:reimaginingthemorphogengradient.Cell128,245 256. Lander,A.D.,Nie,Q.,Wan,F.Y.,2002.Domorphogengradientsarisebydiffusion?Dev.Cell2,785 796.
Lander,A.D.,Lo,W.C.,Nie,Q.,Wan,F.Y.,2009a.Themeasureofsuccess:constraints,objectives,andtradeoffsinmorphogen-mediatedpatterning ColdSpringHarb.Perspect.Biol.1,a002022.
Lander,A.D.,Gokoffski,K.K.,Wan,F.Y.,Nie,Q.,Calof,A.L.,2009b.Celllineagesandthelogicofproliferativecontrol.PLoSBiol.7,e15. Lee,S.M.,Tole,S.,Grove,E.,McMahon,A.P.,2000.AlocalWnt-3asignalisrequiredfordevelopmentofthemammalianhippocampus.Development 127,457 467.
Lehtinen,M.K.,Zappaterra,M.W.,Chen,X.,Yang,Y.J.,Hill,A.D.,Lun,M.,Maynard,T.,Gonzalez,D.,Kim,S.,Ye,P.,D’Ercole,A.J.,Wong,E.T., LaMantia,A.S.,Walsh,C.A.,2011.Thecerebrospinal fluidprovidesaproliferativenicheforneuralprogenitorcells.Neuron69,893 905. Li,G.,Pleasure,S.J.,2005.Morphogenesisofthedentategyrus:whatwearelearningfrommousemutants.Dev.Neurosci.27,93 99.
LiemJr.,K.F.,Tremml,G.,Roelink,H.,Jessell,T.M.,1995.DorsaldifferentiationofneuralplatecellsinducedbyBMP-mediatedsignalsfromepidermal ectoderm.Cell82,969 979.
Machon,O.,Backman,M.,Machonova,O.,Kozmik,Z.,Vacik,T.,Andersen,L.,Krauss,S.,2007.AdynamicgradientofWntsignalingcontrols initiationofneurogenesisinthemammaliancortexandcellularspecificationinthehippocampus.Dev.Biol.311,223 237.
Mallamaci,A.,Muzio,L.,Chan,C.H.,Parnavelas,J.,Boncinelli,E.,2000.AreaidentityshiftsintheearlycerebralcortexofEmx2-/-mutantmice. Nat. Neurosci.3,679 686.
Mangale,V.S.,Hirokawa,K.E.,Satyaki,P.R.,Gokulchandran,N.,Chikbire,S.,Subramanian,L.,Shetty,A.S.,Martynoga,B.,Paul,J.,Mai,M.V.,Li,Y., Flanagan,L.A.,Tole,S.,Monuki,E.S.,2008.Lhx2selectoractivityspecifiescorticalidentityandsuppresseshippocampalorganizerfate.Science 319,304 309.
McCabe,M.J.,Gaston-Massuet,C.,Tziaferi,V.,Gregory,L.C.,Alatzoglou,K.S.,Signore,M.,Puelles,E.,Gerrelli,D.,Farooqi,I.S.,Raza,J.,Walker,J., Kavanaugh,S.I.,Tsai,P.S.,Pitteloud,N.,Martinez-Barbera,J.P.,Dattani,M.T.,2011.NovelFGF8mutationsassociatedwithrecessiveholoprosencephaly,craniofacialdefects,andhypothalamo-pituitarydysfunction.J.Clin.Endocrinol.Metab.96,E1709 E1718.
Meinhardt,H.,2009.Modelsforthegenerationandinterpretationofgradients.ColdSpringHarb.Perspect.Biol.1,a001362.
Moldrich,R.X.,Gobius,I.,Pollak,T.,Zhang,J.,Ren,T.,Brown,L.,Mori,S.,DeJuanRomero,C.,Britanova,O.,Tarabykin,V.,Richards,L.J.,2010 Molecularregulationofthedevelopingcommissuralplate.J.Comp.Neurol.518,3645 3661.
Monuki,E.S.,2007.Themorphogensignalingnetworkinforebraindevelopmentandholoprosencephaly.J.Neuropathol.Exp.Neurol.66,566 575.
Monuki,E.S.,Porter,F.D.,Walsh,C.A.,2001.Patterningofthedorsaltelencephalonandcerebralcortexbyaroofplate-lhx2pathway.Neuron32, 591 604.
Morgan,T.H.,1901.Regenerationandliabilitytoinjury.Science14,235 248.
Muzio,L.,Soria,J.M.,Pannese,M.,Piccolo,S.,Mallamaci,A.,2005.AmutuallystimulatingloopinvolvingEmx2andcanonicalWntsignalling specificallypromotesexpansionofoccipitalcortexandhippocampus.Cerebr.Cortex15,2021 2028.
Muzio,L.,DiBenedetto,B.,Stoykova,A.,Boncinelli,E.,Gruss,P.,Mallamaci,A.,2002.Emx2andPax6controlregionalizationofthepre-neuronogenic corticalprimordium.Cerebr.Cortex12,129 139.
Ohkubo,Y.,Chiang,C.,Rubenstein,J.L.,2002.CoordinateregulationandsynergisticactionsofBMP4,SHHandFGF8intherostralprosencephalon regulatemorphogenesisofthetelencephalicandopticvesicles.Neuroscience111,1 17.
Ornitz,D.M.,Xu,J.,Colvin,J.S.,McEwen,D.G.,MacArthur,C.A.,Coulier,F.,Gao,G.,Goldfarb,M.,1996.Receptorspecificityofthe fibroblastgrowth factorfamily.J.Biol.Chem.271,15292 15297.
Panchision,D.M.,Pickel,J.M.,Studer,L.,Lee,S.H.,Turner,P.A.,Hazel,T.G.,McKay,R.D.,2001.SequentialactionsofBMPreceptorscontrolneural precursorcellproductionandfate.GenesDev.15,2094 2110.
Ragsdale,C.W.,Grove,E.A.,2001.Patterningthemammaliancerebralcortex.Curr.Opin.Neurobiol.11,50 58. Rash,B.G.,Grove,E.A.,2007.Patterningthedorsaltelencephalon:aroleforsonichedgehog?J.Neurosci.27,11595 11603.
Rogers,K.W.,Schier,A.F.,2011.Morphogengradients:fromgenerationtointerpretation.Annu.Rev.CellDev.Biol.27,377 407.
Rosenfeld,J.A.,Ballif,B.C.,Martin,D.M.,Aylsworth,A.S.,Bejjani,B.A.,Torchia,B.S.,Shaffer,L.G.,2010.Clinicalcharacterizationofindividuals withdeletionsofgenesinholoprosencephalypathwaysbyaCGHrefinesthephenotypicspectrumofHPE.Hum.Genet.127,421 440. Rubenstein,J.L.,Beachy,P.A.,1998.Patterningoftheembryonicforebrain.Curr.Opin.Neurobiol.8,18 26.
Sagner,A.,Briscoe,J.,2017.Morphogeninterpretation:concentration,time,competence,andsignalingdynamics.WileyInterdiscip.Rev.Dev.Biol.6 https://doi.org/10.1002/wdev.271.
Sansom,S.N.,Hebert,J.M.,Thammongkol,U.,Smith,J.,Nisbet,G.,Surani,M.A.,McConnell,S.K.,Livesey,F.J.,2005.Genomiccharacterisationofa Fgf-regulatedgradient-basedneocorticalprotomap.Development132,3947 3961. Scholpp,S.,Brand,M.,2004.EndocytosiscontrolsspreadingandeffectivesignalingrangeofFgf8protein.Curr.Biol.14,1834 1841. Schwank,G.,Basler,K.,2010.Regulationoforgangrowthbymorphogengradients.ColdSpringHarb.Perspect.Biol.2,a001669.
Shanmugalingam,S.,Houart,C.,Picker,A.,Reifers,F.,Macdonald,R.,Barth,A.,Griffin,K.,Brand,M.,Wilson,S.W.,2000.Ace/Fgf8isrequiredfor forebraincommissureformationandpatterningofthetelencephalon.Development127,2549 2561. Shimamura,K.,Rubenstein,J.L.R.,1997.Inductiveinteractionsdirectearlyregionalizationofthemouseforebrain.Development124,2709 2718. Shimmi,O.,Umulis,D.,Othmer,H.,O’Connor,M.B.,2005.FacilitatedtransportofaDpp/ScwheterodimerbySog/Tsgleadstorobustpatterningofthe Drosophilablastodermembryo.Cell120,873 886.
Shimogori,T.,Banuchi,V.,Ng,H.Y.,Strauss,J.B.,Grove,E.A.,2004.EmbryonicsignalingcentersexpressingBMP,WNTandFGFproteinsinteractto patternthecerebralcortex.Development131,5639 5647.
Skeath,J.B.,1998.TheDrosophilaEGFreceptorcontrolstheformationandspecificationofneuroblastsalongthedorsal-ventralaxisoftheDrosophila embryo.Development125,3301 3312.
Solomon,B.D.,Gropman,A.,Muenke,M.,2011.HoloprosencephalyOverview.
Spalding,K.L.,Bhardwaj,R.D.,Buchholz,B.A.,Druid,H.,Frisen,J.,2005.Retrospectivebirthdatingofcellsinhumans.Cell122,133 143. Spemann,H.,Mangold,H.,1924.ÜberInduktionvonEmbryonanlagendurchImplantationartfremderOrganisatoren.Roux’ Arch.F.Entw.Mech.100, 599 638.
Srinivasan,S.,Hu,J.S.,Currle,D.S.,Fung,E.S.,Hayes,W.B.,Lander,A.S.,Monuki,E.S.,2014.ABMP-FGFmorphogentoggleswitchdrivesthe ultrasensitiveexpressionofmultiplegenesinthedevelopingforebrain.PLoSComput.Biol.10,e1003463.
Storm,E.E.,Garel,S.,Borello,U.,Hebert,J.M.,Martinez,S.,McConnell,S.K.,Martin,G.R.,Rubenstein,J.L.,2006.Dose-dependentfunctionsofFgf8 inregulatingtelencephalicpatterningcenters.Development133,1831 1844.
Stoykova,A.,Gotz,M.,Gruss,P.,Price,J.,1997.Pax6-dependentregulationofadhesivepatterning,R-cadherinexpressionandboundaryformationin developingforebrain.Development124,3765 3777.
Sussel,L.,Marin,O.,Kimura,S.,Rubenstein,J.L.,1999.LossofNkx2.1homeoboxgenefunctionresultsinaventraltodorsalmolecularrespecification withinthebasaltelencephalon:evidenceforatransformationofthepallidumintothestriatum.Development126,3359 3370.
Taniguchi,K.,Anderson,A.E.,Sutherland,A.E.,Wotton,D.,2012.LossofTgiffunctioncausesholoprosencephalybydisruptingtheSHHsignaling pathway.PLoSGenet.8,e1002524.
Theil,T.,Dominguez-Frutos,E.,Schimmang,T.,2008.DifferentialrequirementsforFgf3andFgf8duringmouseforebraindevelopment.Dev.Dynam. 237,3417 3423.
Theil,T.,Aydin,S.,Koch,S.,Grotewold,L.,Ruther,U.,2002.WntandBmpsignallingcooperativelyregulategradedEmx2expressioninthedorsal telencephalon.Development129,3045 3054.
Thomas,T.,Dziadek,M.,1993.Capacitytoformchoroidplexus-likecellsinvitroisrestrictedtospecificregionsofthemouseneuralectoderm. Development117,253 262.
Tole,S.,Ragsdale,C.W.,Grove,E.A.,2000a.Dorsoventralpatterningofthetelencephalonisdisruptedinthemousemutantextra-toesJ.Dev.Biol. 217, 254 265.
Tole,S.,Goudreau,G.,Assimacopoulos,S.,Grove,E.A.,2000b.Emx2isrequiredforgrowthofthehippocampusbutnotforhippocampal field specification.J.Neurosci.20,2618 2625.
Toyoda,R.,Assimacopoulos,S.,Wilcoxon,J.,Taylor,A.,Feldman,P.,Suzuki-Hirano,A.,Shimogori,T.,Grove,E.A.,2010.FGF8actsasaclassic diffusiblemorphogentopatterntheneocortex.Development137,3439 3448.
Turing,A.M.,1952.Thechemicalbasisofmorphogenesis.Philos.Trans.R.Soc.Lond.Ser.BBiol.Sci.237,37 72. vonOhlen,T.,Doe,C.Q.,2000.Convergenceofdorsal,dpp,andegfrsignalingpathwayssubdividesthedrosophilaneuroectodermintothreedorsalventralcolumns.Dev.Biol.224,362 372.
Walshe,J.,Mason,I.,2003.UniqueandcombinatorialfunctionsofFgf3andFgf8duringzebrafishforebraindevelopment.Development130, 4337 4349.
Warr,N.,Powles-Glover,N.,Chappell,A.,Robson,J.,Norris,D.,Arkell,R.M.,2008.Zic2-associatedholoprosencephalyiscausedbyatransientdefect intheorganizerregionduringgastrulation.Hum.Mol.Genet.17,2986 2996.
Wartlick,O.,Kicheva,A.,Gonzalez-Gaitan,M.,2009.Morphogengradientformation.ColdSpringHarb.Perspect.Biol.1,a001255.
Watanabe,M.,Kang,Y.-J.,Davies,L.M.,Meghpara,S.,Lau,K.,Chung,C.-Y.,Kathirya,J.,Hadjantonakis,A.-K.,Monuki,E.S.,2012.BMP4sufficiencytoinducechoroidplexusepithelialfatefromembryonicstemcell-derivedneuroepithelialprogenitors.J.Neurosci.32,15934 15945.
Watanabe,M.,Fung,E.S.,Chan,F.B.,Wong,J.S.,Coutts,M.,Monuki,E.S.,2016.BMP4actsasadorsaltelencephalicmorphogeninamouseembryonicculturesystem.Biol.Open5,1834 1843.
White,R.J.,Nie,Q.,Lander,A.D.,Schilling,T.F.,2007.Complexregulationofcyp26a1createsarobustretinoicacidgradientinthezebrafishembryo. PLoSBiol.5,e304.
Wolpert,L.,1969.Positionalinformationandthespatialpatternofcellulardifferentiation.J.Theor.Biol.25,1 47.
Yoshida,M.,Assimacopoulos,S.,Jones,K.R.,Grove,E.A.,2006.MassivelossofCajal-Retziuscellsdoesnotdisruptneocorticallayerorder. Development133,537 545.
Yu,S.R.,Burkhardt,M.,Nowak,M.,Ries,J.,Petrasek,Z.,Scholpp,S.,Schwille,P.,Brand,M.,2009.Fgf8morphogengradientformsbyasource-sink mechanismwithfreelydiffusingmolecules.Nature461,533 536.
Zhang,X.,Ibrahimi,O.A.,Olsen,S.K.,Umemori,H.,Mohammadi,M.,Ornitz,D.M.,2006.Receptorspecificityofthe fibroblastgrowthfactorfamily. ThecompletemammalianFGFfamily.J.Biol.Chem.281,15694 15700.
Zhou,S.,Lo,W.C.,Suhalim,J.L.,Digman,M.A.,Gratton,E.,Nie,Q.,Lander,A.D.,2012.Freeextracellulardiffusioncreatesthedppmorphogen gradientoftheDrosophilawingdisc.Curr.Biol.22,668 675.