John Lacava Visit to download the full and correct content document: https://ebookmass.com/product/the-eukaryotic-rna-exosome-methods-and-protocols1st-ed-2020-edition-john-lacava/
More products digital (pdf, epub, mobi) instant download maybe you interests ...
Chaperones: Methods and Protocols 2nd Edition John M. Walker
https://ebookmass.com/product/chaperones-methods-andprotocols-2nd-edition-john-m-walker/
Flower Development: Methods and Protocols 2nd Edition John M. Walker
https://ebookmass.com/product/flower-development-methods-andprotocols-2nd-edition-john-m-walker/
Exosomes and Microvesicles: Methods and Protocols 1st Edition Andrew F Hill (Eds.)
https://ebookmass.com/product/exosomes-and-microvesicles-methodsand-protocols-1st-edition-andrew-f-hill-eds/
The Renewable Energy Transition: Realities for Canada and the World 1st ed. 2020 Edition John Erik Meyer
https://ebookmass.com/product/the-renewable-energy-transitionrealities-for-canada-and-the-world-1st-ed-2020-edition-john-erikmeyer/
The Precision Farming Revolution: Global Drivers of Local Agricultural Methods 1st ed. 2020 Edition
James E. Addicott
https://ebookmass.com/product/the-precision-farming-revolutionglobal-drivers-of-local-agricultural-methods-1st-ed-2020-editionjames-e-addicott/
Peacebuilding and the Arts 1st ed. 2020 Edition Jolyon Mitchell
https://ebookmass.com/product/peacebuilding-and-the-arts-1sted-2020-edition-jolyon-mitchell/
Doing Business in Chile and Peru: Challenges and Opportunities 1st ed. 2020 Edition John E. Spillan
https://ebookmass.com/product/doing-business-in-chile-and-peruchallenges-and-opportunities-1st-ed-2020-edition-john-e-spillan/
Sustainability in Energy and Buildings: Proceedings of SEB 2019 1st ed. 2020 Edition John Littlewood
https://ebookmass.com/product/sustainability-in-energy-andbuildings-proceedings-of-seb-2019-1st-ed-2020-edition-johnlittlewood/
Maternal Sentencing and the Rights of the Child 1st ed. 2020 Edition Shona Minson
https://ebookmass.com/product/maternal-sentencing-and-the-rightsof-the-child-1st-ed-2020-edition-shona-minson/
Methods in Molecular Biology 2062
John LaCava Štěpánka Vaňáčová
Editors
The Eukaryotic RNA Exosome Methods and Protocols M ETHODSIN M OLECULAR B IOLOGY SeriesEditor
JohnM.Walker
SchoolofLifeandMedicalSciences
UniversityofHertfordshire Hatfield,Hertfordshire,UK
Forfurthervolumes: http://www.springer.com/series/7651
Forover35years,biologicalscientistshavecometorelyontheresearchprotocolsand methodologiesinthecriticallyacclaimed MethodsinMolecularBiology series.Theserieswas thefirsttointroducethestep-by-stepprotocolsapproachthathasbecomethestandardinall biomedicalprotocolpublishing.Eachprotocolisprovidedinreadily-reproduciblestep-bystepfashion,openingwithanintroductoryoverview,alistofthematerialsandreagents neededtocompletetheexperiment,andfollowedbyadetailedprocedurethatissupported withahelpfulnotessectionofferingtipsandtricksofthetradeaswellastroubleshooting advice.ThesehallmarkfeatureswereintroducedbyserieseditorDr.JohnWalkerand constitutethekeyingredientineachandeveryvolumeofthe MethodsinMolecularBiology series.Testedandtrusted,comprehensiveandreliable,allprotocolsfromtheseriesare indexedinPubMed.
TheEukaryoticRNAExosome MethodsandProtocols Editedby JohnLaCava
LaboratoryofCellularandStructuralBiology,TheRockefellerUniversity,NewYork,NY,USA
EuropeanResearchInstitutefortheBiologyofAgeing,UniversityMedicalCenterGroningen,Groningen, TheNetherlands
ŠtěpánkaVaňáčová CentralEuropeanInstituteofTechnology(CEITEC),MasarykUniversity,Brno,CzechRepublic
Editors
JohnLaCava LaboratoryofCellular andStructuralBiology
TheRockefellerUniversity NewYork,NY,USA
EuropeanResearchInstitute fortheBiologyofAgeing UniversityMedicalCenterGroningen Groningen,TheNetherlands S
CentralEuropeanInstitute ofTechnology(CEITEC)
MasarykUniversity Brno,CzechRepublic
ISSN1064-3745ISSN1940-6029(electronic)
MethodsinMolecularBiology
ISBN978-1-4939-9821-0ISBN978-1-4939-9822-7(eBook) https://doi.org/10.1007/978-1-4939-9822-7
© SpringerScience+BusinessMedia,LLC,partofSpringerNature2020
Thechapters5and6arelicensedunderthetermsoftheCreativeCommonsAttribution4.0InternationalLicense (http://creativecommons.org/licenses/by/4.0/).Forfurtherdetailsseelicenseinformationinthechapters. Thisworkissubjecttocopyright.AllrightsarereservedbythePublisher,whetherthewholeorpartofthematerialis concerned,specificallytherightsoftranslation,reprinting,reuseofillustrations,recitation,broadcasting,reproduction onmicrofilmsorinanyotherphysicalway,andtransmissionorinformationstorageandretrieval,electronicadaptation, computersoftware,orbysimilarordissimilarmethodologynowknownorhereafterdeveloped. Theuseofgeneraldescriptivenames,registerednames,trademarks,servicemarks,etc.inthispublicationdoesnotimply, evenintheabsenceofaspecificstatement,thatsuchnamesareexemptfromtherelevantprotectivelawsandregulations andthereforefreeforgeneraluse.
Thepublisher,theauthors,andtheeditorsaresafetoassumethattheadviceandinformationinthisbookarebelievedto betrueandaccurateatthedateofpublication.Neitherthepublishernortheauthorsortheeditorsgiveawarranty, expressorimplied,withrespecttothematerialcontainedhereinorforanyerrorsoromissionsthatmayhavebeenmade. Thepublisherremainsneutralwithregardtojurisdictionalclaimsinpublishedmapsandinstitutionalaffiliations.
ThisHumanaimprintispublishedbytheregisteredcompanySpringerScience+BusinessMedia,LLC,partofSpringer Nature.
Theregisteredcompanyaddressis:233SpringStreet,NewYork,NY10013,U.S.A.
Preface TheRNAexosomecomplexwasfirstdescribedbyPhilipMitchell,togetherwithDavid Tollerveyandcolleagues,in1997(Cell,Vol.91,457–466).Inthisseminalwork,theexosome wasdescribedas“aconservedeukaryoticRNAprocessingcomplexcontainingmultiple3
exoribonucleases.”Sincethen,thefieldofexosomeresearchhasexpandedsteadily.The proliferationofthefieldwasfueledbytheever-growinglistofrolesplayedbytheexosome in(1)thepreciseprocessingofRNAprecursorstomatureformsand(2)theturnoverofRNAs inresponsetoqualitycontrolsurveillanceandhomeostaticRNAdecay.Theexosomewas foundtobeassociatedwithadizzyingarrayofsubstratesandinstance-specificmodesof enzymaticactivity.Themechanismsregulatingitsbiochemistrywerethusswiftlybroughtto thefore,inducingthesearchforadapterproteinsandcomplexesthatcouldimpartfunctional selectivitytotheexosome.This,alongsideabundantepisodesofmysteryandintriguesurroundingthepotentialforexo-andendoribonucleolytic,aswellashydro-andphosphorolytic exosomeactivities.Throughthefunctionsitserves,theexosome’subiquityextendstoallthe domainsoflife.Inbacteriaandarchaea,cognateproteinsandcomplexeshavebeenshownto exhibitexosome-likeactivities,e.g.,polynucleotidephosphorylase/thedegradosomeinbacteriaandinarchaea,amoreorthologouscomplex,alsocalledtheexosome.Soon,itseemed thattheexosomewaseverywhereinRNAbiology—andthesituationremainsmuchthesame today,20yearson,withnosignthatexosomeresearchisyetpeteringout.Indeed,quitethe contrary,theexosomeisunderstoodtobeavaluablebiomedicaltarget.
Thus, TheEukaryoticRNAExosome:MethodsandProtocols isintendedtoprovidea thoroughbasisincontemporaryexosomeresearchforthenewcomer,thisbothintermsof thetechniquesusedandthegeneraldirectionofthefield.Forthosegrizzledveteransof exosomeresearchwhomayhavestuckmostlytotheirfavoritemodelorganism,wehave triedtoprovideacross-sectionofprotocolsrepresentingthediversityofmodelorganisms usedforexosomeresearch,hopefullyempoweringbroaderandmorefrequentcrossorganismcomparisonswithinthesameresearchteam.ThisbookbeginswithanintroductiontoRNAexosomebiomedicalrelevance(Chapter 1)andthenstepsbacktoexaminethe originsofexosomeactivityinbacteria(Chapters 2–3)andarchaea(Chapter 4).Fromthere, methodsofstudyingexosomeRNAsubstratesarecovered(Chapters 5–10),followedbythe studyofexosomeadaptercomplexesandthebroaderinteractionnetworksthatimpart selectivitytopathwaystheexosomeparticipatesin(Chapters 11–16).Finally,structural andmechanisticbiochemicalstudiesarecovered,withemphasisonmethodsthatuse recombinantexpressionandexogenousreconstitution(Chapters 17–24).
S ˇ te ˇ pa ´ nkaandIwouldliketothankallthecontributingauthorsfortheirenthusiasm, professionalism,andscholarship.Asourfirstexperienceeditingabookofthisnature,with itsassociatelearningcurveandtimecommitment,wewereluckytohavethisgroupof contributorstoworkwith.Indeed,therewasalsoaspecial“something”forusinbeingable toeditthisbooktogetherasateam.AdditionalthankstoMs.HuaJiangforproofreading thechaptersandtoDr.JohnWalkerfortheguidanceonhowtoexecuteourtaskseffectively andthepatiencealongthewaytocompletion.
NewYork,NY,USAJohnLaCava Brno,CzechRepublicS
Preface.. ...................................................................v
Contributors .................................................................xi
PART IINTRODUCTIONTOTHE RNAEXOSOMEIN BIOMEDICINE
1TheRNAExosomeandHumanDisease ...................................3
MiloB.Fasken,DerrickJ.Morton,EmilyG.Kuiper,StephanieK.Jones, SaraW.Leung,andAnitaH.Corbett
PART IIPROKARYOTIC RNASESAND EXOSOMES 2TheBacterialCounterpartsoftheEukaryoticExosome: AnEvolutionaryPerspective. .............................................37
SandraC.Viegas,RuteG.Matos,andCecı ´ liaM.Arraiano
3InVitroCharacterizationoftheProkaryoticCounterparts oftheExosomeComplex... .............................................47
RuteG.Matos,SandraC.Viegas,andCecı ´ liaM.Arraiano
4EnzymaticAnalysisofReconstitutedArchaealExosomes .....................63 ElenaEvguenieva-Hackenberg,A.SusannGauernack,LinlinHou, andGabrieleKlug
PART IIIANALYZING EXOSOME SUBSTRATES
5ProtocolsforNorthernAnalysisofExosomeSubstrates andOtherNoncodingRNAs .............................................83 CristinaCruzandJonathanHouseley
6MappingExosome–SubstrateInteractionsInVivo byUVCross-Linking ....................................................105 Cle´mentineDelan-ForinoandDavidTollervey
7GlobalIdentificationofHumanExosomeSubstratesUsing RNAInterferenceandRNASequencing ...................................127 MartaLloret-LlinaresandTorbenHeickJensen
8High-ResolutionMappingof3’ExtremitiesofRNAExosome Substratesby3’RACE-Seq. .............................................147 He´le`neScheer,CarolineDeAlmeida,NataliaSikorska,SandrineKoechler, DominiqueGagliardi,andHe´le`neZuber
9DeterminingmRNAStabilitybyMetabolicRNALabeling andChemicalNucleosideConversion .....................................169 VeronikaA.Herzog,NinaFasching,andStefanL.Ameres
10Thiouridine-to-CytidineConversionSequencing(TUC-Seq) toMeasuremRNATranscriptionandDegradationRates ....................191
AlexandraLusser,CatherinaGasser,LukasTrixl,PaoloPiatti, IsabelDelazer,DietmarRieder,JeffreyBashin,ChristianRiml, ThomasAmort,andRonaldMicura
PART IVEXOSOMESAND THEIR
SUPRAMOLECULAR FUNCTIONAL INTERACTIONS 11RNAExosomesandTheirCofactors. .....................................215
CorneliaKilchert
12PurificationofEndogenousTaggedTRAMP4/5andExosome ComplexesfromYeastandInVitroPolyadenylation-Exosome ActivationAssays........................................................237
DagmarZiga ´ c ˇkova ´ ,VeronikaRa ´ jecka ´ ,andS
13ComparativePoly(A)+RNAInteractomeCaptureofRNA SurveillanceMutants ....................................................255
CorneliaKilchert,SvenjaHester,AlfredoCastello,ShabazMohammed, andLidiaVasiljeva
14PurificationandInVitroAnalysisoftheExosomeCofactors Nrd1-Nab3andTrf4-Air2.. .............................................277 OdilPorrua
15AffinityProteomicAnalysisoftheHumanExosome andItsCofactorComplexes. .............................................291 KingaWinczura,MichalDomanski,andJohnLaCava
16InVitroCharacterizationoftheActivityoftheMammalian RNAExosomeonmRNAsinRibosomalTranslationComplexes .............327 AlexandraZinoviev,ChristopherU.T.Hellen,andTatyanaV.Pestova
PART VRECOMBINANT EXPRESSION STRATEGIESAND STRUCTURAL CHARACTERIZATIONOF EXOSOMES
17NativeMassSpectrometryAnalysisofAffinity-Captured EndogenousYeastRNAExosomeComplexes ..............................357 PaulDominicB.OlinaresandBrianT.Chait
18ChemicalCross-LinkingandMassSpectrometricAnalysis oftheEndogenousYeastExosomeComplexes .............................383 YufeiXiang,ZhuolunShen,andYiShi
19Cryo-ElectronMicroscopyofEndogenousYeastExosomes... ...............401 Jun-JieLiuandHong-WeiWang
20StrategiesforGeneratingRNAExosomeComplexes fromRecombinantExpressionHosts. .....................................417 Eva-MariaWeick,JohnC.Zinder,andChristopherD.Lima
21Reconstitutionof S.cerevisiae RNAExosomeComplexes UsingRecombinantlyExpressedProteins ..................................427 JohnC.ZinderandChristopherD.Lima
22Reconstitutionofthe Schizosaccharomycespombe RNAExosome ..............449 KurtJanuszykandChristopherD.Lima
23ReconstitutionoftheHumanNuclearRNAExosome .......................467
KurtJanuszyk,Eva-MariaWeick,andChristopherD.Lima
24PurificationandReconstitutionofthe S.cerevisiae TRAMP andSkiComplexesforBiochemicalandStructuralStudies ...................491 AchimKeidel,ElenaConti,andSebastianFalk
Index...
Contributors STEFAN L.AMERES IMBA—InstituteofMolecularBiotechnology,ViennaBiocenter(VBC), Vienna,Austria
THOMAS AMORT DivisionofMolecularBiology,Biocenter,MedicalUniversityofInnsbruck, Innsbruck,Austria
CECI ´ LIA M.ARRAIANO InstitutodeTecnologiaQuı ´ micaeBiologicaAntonioXavier,Oeiras, Portugal
JEFFREY BASHIN ZymoResearchCorp.,Irvine,CA,USA
ALFREDO CASTELLO DepartmentofBiochemistry,UniversityofOxford,Oxford,UK
BRIAN T.CHAIT LaboratoryofMassSpectrometryandGaseousIonChemistry,The RockefellerUniversity,NewYork,NY,USA
ELENA CONTI DepartmentofStructuralCellBiology,Max-Planck-Instituteof Biochemistry,Martinsried,Germany
ANITA H.CORBETT DepartmentofBiology,RRC1021,EmoryUniversity,Atlanta,GA, USA
CRISTINA CRUZ EpigeneticsProgramme,BabrahamInstitute,Cambridge,UK
CAROLINE DE ALMEIDA InstitutdeBiologieMole´culairedesPlantes,CentreNationaldela RechercheScientifique(CNRS),Universite´deStrasbourg,Strasbourg,France
CLE ´ MENTINE DELAN-FORINO WellcomeCentreforCellBiology,UniversityofEdinburgh, Edinburgh,UK
ISABEL DELAZER DivisionofMolecularBiology,Biocenter,MedicalUniversityofInnsbruck, Innsbruck,Austria
MICHAL DOMANSKI DepartmentofChemistryandBiochemistry,UniversityofBern,Bern, Switzerland
ELENA EVGUENIEVA-HACKENBERG InstituteforMicrobiologyandMolecularBiology,JustusLiebig-UniversityGiessen,Giessen,Germany
SEBASTIAN FALK DepartmentofStructuralCellBiology,Max-Planck-Instituteof Biochemistry,Martinsried,Germany;MaxPerutzLaboratories,DepartmentofStructural andComputationalBiology,UniversityofVienna,Vienna,Austria
NINA FASCHING IMBA—InstituteofMolecularBiotechnology,ViennaBiocenter(VBC), Vienna,Austria
MILO B.FASKEN DepartmentofBiology,RRC1021,EmoryUniversity,Atlanta,GA,USA
DOMINIQUE GAGLIARDI InstitutdeBiologieMole´culairedesPlantes,CentreNationaldela RechercheScientifique(CNRS),Universite´deStrasbourg,Strasbourg,France
CATHERINA GASSER DepartmentofChemistryandPharmacy,InstituteofOrganic Chemistry,LeopoldFranzensUniversity,Innsbruck,Austria
A.SUSANN GAUERNACK InstituteforMicrobiologyandMolecularBiology,Justus-LiebigUniversityGiessen,Giessen,Germany
CHRISTOPHER U.T.HELLEN DepartmentofCellBiology,SUNYDownstateHealthSciences University,Brooklyn,NY,USA
VERONIKA A.HERZOG IMBA—InstituteofMolecularBiotechnology,ViennaBiocenter (VBC),Vienna,Austria
SVENJA HESTER DepartmentofBiochemistry,UniversityofOxford,Oxford,UK
LINLIN HOU InstituteforMicrobiologyandMolecularBiology,Justus-Liebig-University Giessen,Giessen,Germany
JONATHAN HOUSELEY EpigeneticsProgramme,BabrahamInstitute,Cambridge,UK
KURT JANUSZYK StructuralBiologyProgram,SloanKetteringInstitute,MemorialSloan KetteringCancerCenter,NewYork,NY,USA
TORBEN HEICK JENSEN DepartmentofMolecularBiologyandGenetics,AarhusUniversity, Aarhus,Denmark
STEPHANIE K.JONES DepartmentofBiology,RRC1021,EmoryUniversity,Atlanta,GA, USA;GeneticsandMolecularBiologyGraduateProgram,EmoryUniversity,Atlanta, GA,USA
ACHIM KEIDEL DepartmentofStructuralCellBiology,Max-Planck-Instituteof Biochemistry,Martinsried,Germany
CORNELIA KILCHERT Institutfu¨rBiochemie,Justus-Liebig-Universit € atGießen,Gießen, Germany
GABRIELE KLUG InstituteforMicrobiologyandMolecularBiology,Justus-Liebig-University Giessen,Giessen,Germany
SANDRINE KOECHLER InstitutdeBiologieMole´culairedesPlantes,CentreNationaldela RechercheScientifique(CNRS),Universite´deStrasbourg,Strasbourg,France
EMILY G.KUIPER DepartmentofCancerImmunologyandVirology,Dana-FarberCancer Institute,Boston,MA,USA
JOHN LACAVA LaboratoryofCellularandStructuralBiology,TheRockefellerUniversity, NewYork,NY,USA;EuropeanResearchInstitutefortheBiologyofAgeing,University MedicalCenterGroningen,Groningen,TheNetherlands
SARA W.LEUNG DepartmentofBiology,RRC1021,EmoryUniversity,Atlanta,GA,USA
CHRISTOPHER D.LIMA StructuralBiologyProgram,SloanKetteringInstitute,Memorial SloanKetteringCancerCenter,NewYork,NY,USA;HowardHughesMedicalInstitute, NewYork,NY,USA
JUN-JIE LIU MolecularBiophysicsandIntegratedBioimagingDivision,LawrenceBerkeley NationalLaboratory,Berkeley,CA,USA;DepartmentofMolecularandCellBiology, UniversityofCalifornia,Berkeley,Berkeley,CA,USA
MARTA LLORET-LLINARES DepartmentofMolecularBiologyandGenetics,Aarhus University,Aarhus,Denmark
ALEXANDRA LUSSER DivisionofMolecularBiology,Biocenter,MedicalUniversityof Innsbruck,Innsbruck,Austria
RUTE G.MATOS InstitutodeTecnologiaQuı ´ micaeBiologicaAntonioXavier,Oeiras, Portugal
RONALD MICURA DepartmentofChemistryandPharmacy,InstituteofOrganicChemistry, LeopoldFranzensUniversity,Innsbruck,Austria
SHABAZ MOHAMMED DepartmentofBiochemistry,UniversityofOxford,Oxford,UK; DepartmentofChemistry,UniversityofOxford,ChemistryResearchLaboratory,Oxford, UK
DERRICK J.MORTON DepartmentofBiology,RRC1021,EmoryUniversity,Atlanta,GA, USA
PAUL DOMINIC B.OLINARES LaboratoryofMassSpectrometryandGaseousIonChemistry, TheRockefellerUniversity,NewYork,NY,USA
TATYANA V.PESTOVA DepartmentofCellBiology,SUNYDownstateHealthSciences University,Brooklyn,NY,USA
PAOLO PIATTI ZymoResearchCorp.,Irvine,CA,USA
ODIL PORRUA InstitutJacquesMonod-UMR7592,CNRS,Universite´deParis,Paris, France
VERONIKA RA ´ JECKA ´ CentralEuropeanInstituteofTechnology(CEITEC),Masaryk University,Brno,CzechRepublic
DIETMAR RIEDER DivisionofBioinformatics,Biocenter,MedicalUniversityofInnsbruck, Innsbruck,Austria
CHRISTIAN RIML DepartmentofChemistryandPharmacy,InstituteofOrganicChemistry, LeopoldFranzensUniversity,Innsbruck,Austria
HE ´ LE ` NE SCHEER InstitutdeBiologieMole´culairedesPlantes,CentreNationaldela RechercheScientifique(CNRS),Universite´deStrasbourg,Strasbourg,France
ZHUOLUN SHEN DepartmentofCellBiology,UniversityofPittsburghSchoolofMedicine, Pittsburgh,PA,USA;SchoolofMedicine,TsinghuaUniversity,Beijing,China
YI SHI DepartmentofCellBiology,UniversityofPittsburghSchoolofMedicine,Pittsburgh, PA,USA
NATALIA SIKORSKA InstitutdeBiologieMole´culairedesPlantes,CentreNationaldela RechercheScientifique(CNRS),Universite´deStrasbourg,Strasbourg,France
DAVID TOLLERVEY WellcomeCentreforCellBiology,UniversityofEdinburgh,Edinburgh, UK
LUKAS TRIXL DivisionofMolecularBiology,Biocenter,MedicalUniversityofInnsbruck, Innsbruck,Austria
S
NKA VAN
C
OVA ´ CentralEuropeanInstituteofTechnology(CEITEC),Masaryk University,Brno,CzechRepublic
LIDIA VASILJEVA DepartmentofBiochemistry,UniversityofOxford,Oxford,UK
SANDRA C.VIEGAS InstitutodeTecnologiaQuı ´ micaeBiologicaAntonioXavier,Oeiras, Portugal
HONG-WEI WANG MinistryofEducationKeyLaboratoryofProteinSciences,TsinghuaPekingJointCenterforLifeSciences,CenterforStructuralBiology,SchoolofLifeSciences, TsinghuaUniversity,Beijing,China
EVA-MARIA WEICK StructuralBiologyProgram,SloanKetteringInstitute,MemorialSloan KetteringCancerCenter,NewYork,NY,USA
KINGA WINCZURA SchoolofBiosciences,CollegeofLifeandEnvironmentalSciences, UniversityofBirmingham,Birmingham,UK
YUFEI XIANG DepartmentofCellBiology,UniversityofPittsburghSchoolofMedicine, Pittsburgh,PA,USA
DAGMAR ZIGA ´ C ˇ KOVA ´ CentralEuropeanInstituteofTechnology(CEITEC),Masaryk University,Brno,CzechRepublic
JOHN C.ZINDER StructuralBiologyProgram,SloanKetteringInstitute,MemorialSloan KetteringCancerCenter,NewYork,NY,USA;Tri-InstitutionalTrainingProgramin ChemicalBiology,MemorialSloanKetteringCancerCenter,NewYork,NY,USA
ALEXANDRA ZINOVIEV DepartmentofCellBiology,SUNYDownstateHealthSciences University,Brooklyn,NY,USA
HE ´ LE ` NE ZUBER InstitutdeBiologieMole´culairedesPlantes,CentreNationaldela RechercheScientifique(CNRS),Universite´deStrasbourg,Strasbourg,France
IntroductiontotheRNAExosomeinBiomedicine TheRNAExosomeandHumanDisease MiloB.Fasken,DerrickJ.Morton,EmilyG.Kuiper,StephanieK.Jones, SaraW.Leung,andAnitaH.Corbett
Abstract
TheevolutionarilyconservedRNAexosomeisamultisubunitribonucleasecomplexthatprocessesand/or degradesnumerousRNAs.Recently,mutationsingenesencodingbothstructuralandcatalyticsubunitsof theRNAexosomehavebeenlinkedtohumandisease.Mutationsinthestructuralexosomegene EXOSC2 causeadistinctsyndromethatincludesretinitispigmentosa,hearingloss,andmildintellectualdisability.In contrast,mutationsinthestructuralexosomegenes EXOSC3 and EXOSC8 causepontocerebellarhypoplasiatype1b(PCH1b)andtype1c(PCH1c),respectively,whicharerelatedautosomalrecessive,neurodegenerativediseases.Inaddition,mutationsinthestructuralexosomegene EXOSC9 causeaPCH-like diseasewithcerebellaratrophyandspinalmotorneuronopathy.Finally,mutationsinthecatalyticexosome gene DIS3 havebeenlinkedtomultiplemyeloma,aneoplasmofplasmaBcells.Howmutationsinthese RNAexosomegenesleadtodistinct,tissue-specificdiseasesisnotcurrentlywellunderstood.Inthis chapter,weexaminetheroleoftheRNAexosomecomplexinhumandiseaseanddiscussthemechanisms bywhichmutationsindifferentexosomesubunitgenescouldimpairRNAexosomefunctionandgiverise todiversediseases.
Keywords Pontocerebellarhypoplasia,Retinitispigmentosa,Spinalmotorneuronopathy,Multiple myeloma,RNAexosome,EXOSC2,EXOSC3,EXOSC8,EXOSC9,DIS3,Rrp4,Rrp40,Rrp43, Rrp45,Rrp44
1Introduction
TheRNAexosomeisaconservedribonucleasecomplex,madeof structuralandcatalyticsubunits,whichprocessesand/ordegrades numerousRNAs[1–6].Recently,mutationsinseveralRNAexosomesubunitgeneshavebeenidentifiedinindividualswithtissuespecificdiseases.Inthischapter,weexaminethemutationsingenes encodingRNAexosomestructuralsubunits—EXOSC2, EXOSC3, EXOSC8,and EXOSC9—andthecatalyticsubunit—DIS3—that havebeenlinkedtohumandiseases.
TheRNAexosomeiscomposedoftenconservedcoresubunits thatformaring-likestructure[1, 2, 4, 5](Fig. 1).Theexosome subunitsinhumansaretermed Exosome Component(EXOSC)
JohnLaCavaandStepankaVanacova (eds.), TheEukaryoticRNAExosome:MethodsandProtocols,MethodsinMolecularBiology, vol.2062, https://doi.org/10.1007/978-1-4939-9822-7_1, © SpringerScience+BusinessMedia,LLC,partofSpringerNature2020
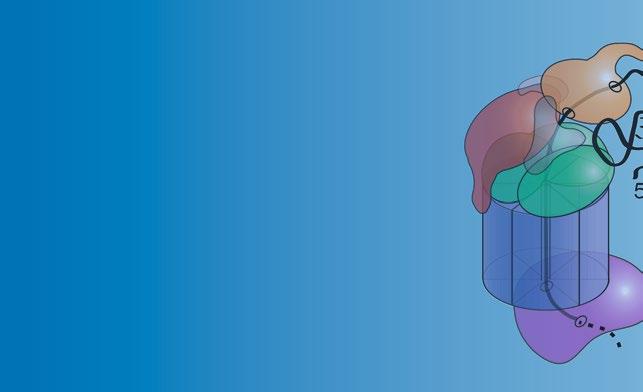
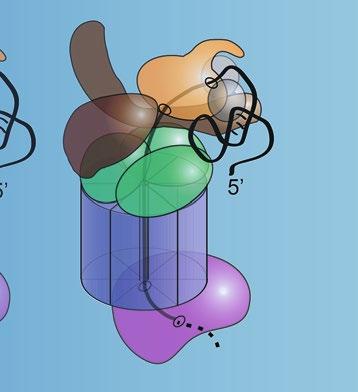
Current Structure of Human RNA Exosome (11 Subunits) with MTREX & MPH6 Exosome Cofactors
Current Structure of Yeast RNA Exosome (11 Subunits) with Mtr4, Mpp6, & Rrp47 Exosome Cofactors
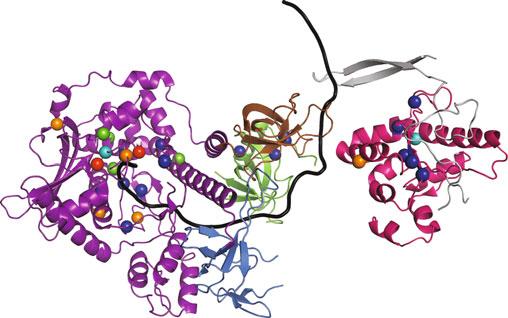
Fig.1 StructureoftheRNAexosome,amultisubunitribonucleasecomplexthatprocesses/degradesmultiple classesofRNA.(a)Totheleft,acartoonrepresentationofoneofthecurrentstructuresofthe11-subunit humanRNAexosomeinassociationwithtwoexosomecofactors(MTREX(MTR4);MPH6(MPP6))isdepicted [7].The10-subunitcoreexosomeiscomposedofathree-subunitcap(EXOSC1/2/3)atthetop,asix-subunit ring(EXOSC4-9)inthemiddle,andribonucleasesubunit,DIS3,atthebottom.Partofeleventhriboexonucleasesubunit,EXOSC10,whichcouldberesolvedinthestructure,associateswiththeEXOSC6ringsubunit. TheMTREX(MTR4)helicasecofactorassociateswiththeEXOSC2capsubunitandtheMPH6(MPP6)cofactor, whereastheMPH6(MPP6)cofactorassociateswiththeEXOSC3andEXOSC1capsubunits.Totherightofthe cartoon,surfacerepresentationsofthis11-subunithumanRNAexosomestructureincomplexwithMTREX (MTR4)andMPH6(MPP6)cofactors(PDB#6D6Q)[7]areshown,depictedintop,side,andreversesideviews. Thestructureofthe10-subunithumancoreexosomerevealsaring-likearchitecturecomposedofthreecap subunits—EXOSC1/Csl4(gray),EXOSC2/Rrp4(teal),andEXOSC3/Rrp40(slateblue),sixPH-likeringsubunits—EXOSC4/Rrp41(orange),EXOSC5/Rrp46(yellow),EXOSC6/Mtr3(marineblue),EXOSC7/Rrp42(salmon red),EXOSC8/Rrp43(magenta),andEXOSC9/Rrp45(firebrickred),andacatalyticbasesubunit,DIS3(brown). Partoftheeleventhcatalyticsubunit,EXOSC10/Rrp6(forestgreen),associateswithEXOSC6/Mtr3.Atthetop ofthecomplex,thecofactor,MTREX(MTR4)(deepolivegreen),associateswithEXOSC2/Rrp4andMPH6 (MPP6),whereasthecofactor,MPH6(MPP6)(hotpink)associateswithEXOSC3/Rrp40.TheDNA/RNAchimera (black)usedtostalltheMTREX(MTR4)helicaseduringunwindingisalsoshowninthestructure.TheEXOSC2/
proteinsandmostexosomesubunitsin S.cerevisiae and Drosophila aretermedRrpproteins[1, 19, 20].The10-subunitcoreexosome comprisesacentral,6-subunitring(EXOSC4-9;Rrp41/42/43/ 45/46/Mtr3),a3-subunitcap(EXOSC1-3;Csl4/Rrp4/Rrp40), andaribonucleasesubunit(DIS3/Dis3/Rrp44),whichassociates withthesix-subunitringatthebase[4, 5, 21]andpossessesboth endoribonucleaseandexoribonucleaseactivity[6, 22–24].Inaddition,theRNAexosomeineukaryotescontainsaneleventhriboexonucleasesubunit,EXOSC10/Rrp6,whichassociateswiththecap [5, 14, 25].Interestingly,EXOSC8/Rrp43,whichisconserved fromyeasttohuman,isapparentlyabsentfrom Drosophila [26].
Structuresofthearchael,yeast,andhumanRNAexosomehave beeninstrumentalindecipheringexosomefunction[4, 5, 7, 13–15, 25, 27–30](see,e.g.,Chapters 4, 21,and 23).InFig. 1a, oneofthecurrent11-subunithumanRNAexosomestructures, whichincludestheDIS3catalyticsubunit,partoftheEXOSC10 catalyticsubunit,andtwoexosomecofactors(MTREX(MTR4); MPH6(MPP6))[7],isdepictedand,inFig. 1b,oneofthe 11-subunit S.cerevisiae RNAexosomestructures,whichincludes Dis3/Rrp44andRrp6catalyticsubunitsandthreeexosomecofactors(Mtr4;Mpp6;Rrp47)[13],isshown.Importantly,these
Fig.1 (continued)Rrp4(teal)andEXOSC3/Rrp40(slateblue)capsubunitsalteredinnovelsyndromeand pontocerebellarhypoplasia1b(PCH1b),respectively[8, 9],EXOSC8/Rrp43(magenta)ringsubunitalteredin PCH1c[10],EXOSC9/Rrp45(firebrickred)ringsubunitalteredinPCH-likedisease[11],andDIS3(brown) catalyticsubunitalteredinmultiplemyeloma[12]arehighlighted(b)Totheleft,acartoonrepresentationof oneofthecurrentstructuresofthe11-subunit S.cerevisiae RNAexosomeinassociationwiththreeexosome cofactors(Mtr4;Mpp6;Rrp47)isdepicted[13].The11-subunitexosomeiscomposedofathreesubunitcap (Csl4/Rrp4/Rrp40)atthetop,asix-subunitring(Rrp41/Rrp42/Rrp43/Rrp45/Rrp46/Mtr3),andtwocatalytic subunits(Dis3/Rrp44andRrp6)[14].TheMtr4helicasecofactorassociateswiththeRrp4capsubunitandthe Mpp6cofactor,theMpp6cofactorassociateswiththeRrp40andCsl4capsubunits,andtheRrp47cofactor associateswithRrp6.Totherightofthecartoon,surfacerepresentationsofthis11-subunityeastRNA exosomestructureincomplexwithMtr4,Mpp6,andRrp47cofactors(PDB#6FSZ)[13]areshown,depictedin top,side,andreversesideviews.The11-subunityeastRNAexosomestructureshowsaring-likeshapewith threecapsubunits—Csl4(gray),Rrp4(teal),andRrp40(slateblue),sixPH-likeringsubunits—Rrp41(orange), Rrp46(yellow),Mtr3(marineblue),Rrp42(salmonred),Rrp43(magenta),andRrp45(firebrickred),andtwo catalyticsubunits,Dis3/Rrp44(brown)andRrp6(forestgreen).Thecofactor,Mtr4(deepolivegreen), associateswithRrp4andMpp6,thecofactor,Mpp6(hotpink),associateswithRrp40,andthecofactor, Rrp47(pink),associateswithRrp6.TheRNA(black)isalsoshowninthestructure.TheDIS3/Dis3/Rrp44 ribonucleasesubunitcontainsbothariboexonuclease(largeoval)andriboendonuclease(smalloval)catalytic site.TheRNAexosomestructuresillustratehowthecatalyticsubunits,DIS3/Dis3/Rrp44andEXOSC10/Rrp6, andtheexosomecofactors,MTREX/Mtr4,MPH6/Mpp6,andRrp47,interfacewiththering-likecoreoftheRNA exosome.Thering-likeRNAexosomestructuresformsacentralchannelthroughwhichRNAisdirectedtothe catalyticsubunit,DIS3/Dis3/Rrp44,forprocessing/degradation.IntheyeastRNAexosome,RNAcanalsogain accesstoDis3/Rrp44inachannel-independentordirectaccessmanner[15–17].Thecolorschemeofthe humanandyeastRNAexosomesubunitsisidentical.ComparisonofthehumanandyeastRNAexosome structuresrevealsthattheRNAexosomestructureishighlyevolutionarilyconservedineukaryotes (FigureadaptedfromMortonetal.[18])
structuresrevealthattheRNAexosomeformsaring-likeshape withacentralchannelthroughwhichRNAsubstratesarethreaded fromthecaptothehexamericringtothecatalyticsubunit,DIS3/ Dis3/Rrp44,atthebaseforprocessing/degradation[4, 5, 7, 21, 25, 31].RNAsubstratescanalsobedirectlytargetedtoRrp6via thecap[14, 15]ordirectlytargetedtoDis3/Rrp44viaachannelindependentordirectaccessmechanism[15–17].Thesestructures alsoprovidedetailsonthespecificmolecularcontactsmadebyeach exosomesubunitwithothersubunitsinthecomplexandexosome cofactors[5, 7, 13–15, 25, 28, 29].TheseelegantRNAexosome structuresthuspermitspeculationaboutthefunctionofspecific aminoacidsintheexosomesubunitsthatarealteredindisease.
TheRNAexosomefunctionsinboththenucleusandthe cytoplasm[32–34],processingnumerousnoncodingRNAs (ncRNAs)inthenucleusanddegradingmanyimproperlyprocessed,“faulty”RNAsinnuclearandcytoplasmicsurveillancepathways[35, 36].Inparticular,theRNAexosomeprocessesrRNAs, snRNAs,andsnoRNAs[1, 2, 19, 37, 38]anddegradesunstable ncRNAs,including cryptic unstable transcripts(CUTs)[39]in buddingyeastand promoterupstream transcripts(PROMPTs) [40]and transcription start site-associatedantisense RNAs (xTSSRNAs)[41]inhumancells.TheRNAexosomealsoturnsover maturetRNA[42, 43].Finally,theRNAexosomedegradesaberrant,improperlyprocessedRNAs,includingpre-mRNAsandhypomodifiedtRNAs[44–49].
Incurrentmodels,specificityoftheRNAexosomefordistinct RNAsubstratesisconferredbydifferentexosomecofactorsthat targettheexosometospecificRNAsforprocessing/degradation [32, 33, 36, 50, 51](see Chapter 11).Keyconserved,nuclear exosomecofactorsincludetheTRAMPcomplex(yeast Tr f4/5Air1/2-Mtr4 Polyadenylationcomplex;humanTENT4B (PAPD5)-ZCCHC7-MTREX(MTR4)complex)[39, 52–55], Mpp6/MPH6(MPP6)[3, 56],Rrp47/C1D[57, 58],andthe NNScomplex(yeastNrd1-Nab3-Sen1complex;humanSETX) [59, 60](Fig. 1).Additionally,inhumans,theNEXTcomplex (Nuclear Exosome Targetingcomplex:MTREX(MTR4)ZCCHC8-RBM7)hasbeenidentifiedasanimportantnuclear exosomecofactor[54].Critical,conservedcytoplasmicexosome cofactorsincludetheSkicomplex(yeastSki2-Ski3-Ski8complex; humanSKIV2L-TTC37-WDR61complex)[61]andSki7/ HBS1L3[32, 62, 63].ThenuclearcofactorMtr4/MTREX (MTR4)andcytoplasmiccofactorSki2/SKIV2LareRNAhelicases thathelptoremodelRNAsubstratesforcompartment-specific exosomecomplexes[34, 64].Thusfar,mutationsingenesencodingcomponentsoftheNEXTcomplex(RBM7)andtheSkicomplex(SKIV2L and TTC37)havebeenlinkedtohumandisease [65–67].
FunctionalstudiesoftheRNAexosomegenesindifferent modelorganismsindicatethatmostRNAexosomegenesareessentialforlife.In S.cerevisiae,allninecorestructuralexosomegenes (RRP4/40/41/42/43/45/46/MTR3/CSL4)andthecorecatalytic exosomegene, DIS3/RRP44,areessential,buttheadditionalcatalyticexosomegene, RRP6,isnotessential[1, 2, 68].Similarly,in S.pombe,studiesindicatethatcoreexosomegenes,including dis3, areessential,but rrp6 isnotessential[69, 70].In Drosophila, severaloftheexosomegenes(dDis3,dMtr3,dRrp6,dRrp41, dRrp42)areessentialandcriticalfornormalflydevelopment [71, 72].Inmice,consistentwiththeothermodelsanalyzed,the Exosc3 geneencodingthemurineRrp40orthologhasbeen reportedtobeessentialforviability,butthedatasupportingthis conclusionremainstobepublished[41].Theseresultssupportthe ideathatalltencoreexosomesubunitsareessentialforviabilityand suggestthattheRNAexosomeisessentialforfunctionincellsfrom yeasttoman.
2MutationsinRNAExosomeGenesLinkedtoDifferentHumanDiseases
Recently,mutationsinfourstructuralexosomesubunitgenes, EXOSC2, EXOSC3, EXOSC8,and EXOSC9,andonecatalytic exosomesubunitgene, DIS3,havebeenlinkedtodifferenttissuespecificdiseases[8–12].Inparticular,mutationsin EXOSC2 have beenlinkedtoanovelsyndromethatcausesretinitispigmentosa andotherphenotypes[9],mutationsin EXOSC3 and EXOSC8 havebeenlinkedtopontocerebellarhypoplasia[8, 10, 73, 74], mutationsin EXOSC9 havebeenlinkedtocerebellaratrophywith spinalmotorneuronopathy[11],andmutationsin DIS3 havebeen linkedtomultiplemyeloma[12, 75–78].
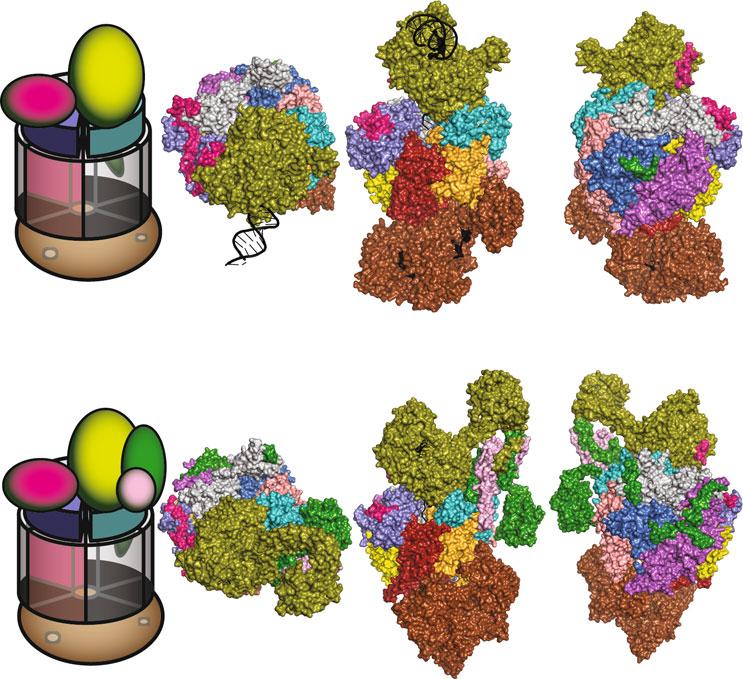
Mostoftheseexosomegenemutationsaremissensemutations thatalterconservedaminoacidswithintheevolutionarilyconservedsubunitsequences(Figs. 2 and 4a).InFigs. 2 and 4a,the domainstructuresofthehumanEXOSC2,EXOSC3,EXOSC8, EXOSC9,andDIS3proteinsareshownthathighlighttheamino acidchangesidentifiedineachproteininindividualswithdisease. Belowthedomainstructures,alignmentsofhuman,mouse, Drosophila and S.cerevisiae EXOSC2/3/8/9/DIS3ortholog sequencesaredepictedthatshowtheconservationoftheresidues alteredindiseaseandthesurroundingsequences.InTable 1,allthe aminoacidsubstitutionsinEXOSC2/3/8/9andthemostprevalentaminoacidchangesinDIS3thathavebeenidentifiedin affectedindividualsaresummarizedalongwiththecorresponding genotypesandphenotypesoftheaffectedindividuals.
As EXOSC2/3/8/9 and DIS3 arepresumedessentialgenesin alleukaryotes,basedpredominantlyondatafromyeastandflies, thefactthattheseexosomegenemutationsleadtodifferenttissue-
Dm
Mm
Dm Rrp40
Hs
Mm EXOSC9 11 RRFLLRA
Dm Rrp45 17 RSFVQLA
Sc Rrp45 12 SKFILEA
Fig.2 AminoacidsubstitutionsidentifiedintheEXOSC2,EXOSC3,EXOSC8,andEXOSC9subunitsoftheRNA exosomeinindividualswithanovelsyndrome,pontocerebellarhypoplasia(PCH),andPCH-likedisease. DomainstructuresofhumanEXOSC2,EXOSC3,EXOSC8,EXOSC9proteinshighlightingtheaminoacid changesidentifiedinaffectedindividuals[8–11, 73, 74, 79, 80].AminoacidchangesintheEXOSC2and EXOSC3capsubunits(showninred),linkedtoanovelsyndromeandpontocerebellarhypoplasiatype1b (PCH1b),respectively,arelocatedintheN-terminaldomain(green),thecentralputativeRNA-bindingS1 domain(blue),ortheC-terminalputativeRNA-bindingKhomology(KH)domain(yellow).Aminoacidchanges intheEXOSC8andEXOSC9ringsubunits(showninred),linkedtopontocerebellarhypoplasiatype1c(PCH1c) andPCH-likedisease(cerebellaratrophywithspinalmotorneuronopathy),respectively,arelocatedinthe PH-likedomain(orange).Belowthedomainstructures,alignmentsofEXOSC2/3/8/9orthologsequencesfrom human(Hs),mouse(Mm), Drosophilamelanogaster (Dm),and S.cerevisiae (Sc)thatsurroundtheevolutionarilyconservedresiduesalteredindisease(highlightedinred,labeledinblackabove)areshown.TheGxNG motif(boxedingreen)presentintheEXOSC2/Rrp4andEXOSC3/Rrp40KHdomainsmayplayastructuralrole, astheGXNGmotifin ScRrp40isburiedattheinterfacebetweentheS1andKHdomains[81].Aminoacid positionsareshownbelowthedomainstructures(FigureadaptedfromMortonetal.[18])
specificdiseasesmaybeconsideredsurprising.Thesedifferentphenotypessuggestthatthedisease-associatedaminoacidchangesin EXOSC2/3/8/9andDIS3supportsufficientexosomeactivityfor viabilitybutcouldhavespecificconsequencesthatvaryindifferent tissuesand/orcelltypes.Priortothediscoveryofthesediseasecausingmutations,itwouldlikelyhavebeenassumedthatany impairmentoftheRNAexosomecomplexwouldhavehadsimilar functionalconsequences.Forthesereasons,consideringhowthe disease-linkedaminoacidsubstitutionsinEXOSC2/3/8/9and DIS3mayaffectRNAexosomefunctioncouldprovideinsights
intowhatexosomeRNAprocessingstepsandprotein-protein interactionsarealtered.InFig. 3a,b,theconservedaminoacids inhumanEXOSC2/3/8/9thatarealteredindiseasearedepicted inthecontextofthe9-subunithumanRNAexosomestructure (PDB#2NN6)[4]andintheisolatedhumanEXOSC2/3/8/9 structures.InFig. 4b,theconservedaminoacidsinDIS3thatare mostcommonlyalteredindiseaseareshownintheisolatedhuman DIS3structurefromtherecent11-subunithumanRNAexosome structure(PDB#6D6Q)[7].Visualizationofthepositionsofthese disease-linkedaminoacidsintheseexosomestructurespermitted usandotherstospeculateonhowtheaminoacidchangesin EXOSC2/3/8/9andDIS3couldalterRNAexosomefunction; detailedbelow.
3EXOSC2Mutations
Mutationsin EXOSC2,whichencodesastructuralcapsubunitof theRNAexosome(Figs. 1a and 2),havebeenlinkedtoanovel syndromecharacterizedbyearlyonsetretinitispigmentosa,progressivesensorineuralhearingloss,hypothyroidism,premature aging,andmildintellectualdisability[9].TheEXOSC2protein containsthreedomains:anN-terminaldomain,aputative RNA-bindingS1domain,andaputativeRNA-bindingKhomology(KH)domain(Figs. 2 and 3b).Exomesequencingoftwo relatedindividualswithdiseaseidentifiedhomozygous EXOSC2 (G30V)mutations—locatedintheN-terminaldomainof EXOSC2(Figs. 2 and 3b;Table 1).Inaddition,exomesequencing ofathirdunrelatedindividualrevealedcompoundheterozygous EXOSC2 (G30V)and EXOSC2 (G198D)mutations—locatedin theN-terminalandKHdomainofEXOSC2,respectively(Figs. 2 and 3b;Table 1).Allthreeaffectedindividualsshowedrelatively milddisease(aliveatages6,44,and28)andborderlineormild cerebellaratrophy.
AnalysisofthehumanRNAexosomestructuresuggeststhat thehighlyconservedG30residueinEXOSC2/Rrp4couldbe importantforintersubunitinteractionswithEXOSC4/Rrp41 (Fig. 3a)[4].StructuralmodelingsuggeststheEXOSC2-G30V substitutioncouldimpairinteractionswithkeyEXOSC4residues (D153,D154,F155)[9].Incontrast,theconservedG198residue inEXOSC2/Rrp4,whichislocatedattheendofa β-strandinthe KHdomain,couldplayastructuralrolewithinEXOSC2/Rrp4 itself(Fig. 3b):theG198Dsubstitutioncouldshortenanddisturb the β-hairpinstructureinEXOSC2[9].Atpresent,littlefunctional analysisoftheEXOSC2/Rrp4variantshasbeenperformed.
Side ViewTop View EXOSC1/
EXOSC3/ Rrp40
EXOSC5 EXOSC5/ Rrp46
EXOSC9/ Rrp45
EXOSC6/ Mtr3
EXOSC2/ Rrp4 EXOSC8/ Rrp43
EXOSC4/ Rrp41
EXOSC7/ Rrp42
EXOSC8/ Rrp43
EXOSC3/ Rrp40
EXOSC9/ Rrp45
EXOSC2/ Rrp4
N-terminal Domain
KH Domain
S1 Domain
PH Domain
EXOSC2/ Rrp4
EXOSC3/ Rrp40
EXOSC8/ Rrp43
EXOSC9/ Rrp45
Fig.3 StructuresoftheEXOSC2,EXOSC3,EXOSC8,andEXOSC9subunitsinthecontextofthestructureofthe humanRNAexosomecomplexandalonethathighlighttheconservedresiduesalteredinanovelsyndrome, pontocerebellarhypoplasia(PCH),andPCH-likedisease.(a)The9-subunithumanexosomestructure(PDB# 2NN6)[4],depictedintopandsideviews,showsribbonrepresentationsoftheEXOSC2/Rrp4(teal),EXOSC3/ Rrp40(slateblue),EXOSC8/Rrp43(magenta),andEXOSC9/Rrp45(firebrickred)subunitsandhighlightsthe conservedresiduesalteredindisease(coloredspheres):EXOSC2—G30andG198(orangespheres)alteredin novelsyndrome,EXOSC3—G31,V80,Y109,D132,G135,A139,G191,andW238(redspheres)alteredin PCH1b,EXOSC8—S272(greensphere)alteredinPCH1c,andEXOSC9—L14(bluesphere)alteredinPCH-like disease,whicharelabeledinblack.TheEXOSC8aminoacidT7(greensphere)islabeledtoshowthe approximatepositionoftheconservedaminoacidA2thatisalteredinPCH1cindividuals;A2couldnotbe labeleddirectlybecauseitwasnotresolvedinthestructure.Transparent,surfacerepresentationsofthe EXOSC1/hCsl4(gray),EXOSC4/Rrp41(orange),EXOSC5/Rrp46(yellow),EXOSC6/Mtr3(marineblue),and EXOSC7/Rrp42(salmonred)subunitsaredepicted.(b)Separate,ribbonrepresentationsoftheEXOSC2/ Rrp4,EXOSC3/Rrp40,EXOSC8/Rrp43,andEXOSC9/Rrp45subunitsthathighlightthedomainsoftheproteins andtheaminoacidsubstitutionsidentifiedindisease.TheEXOSC2-G30Vand-G198DaminoacidsubstitutionsarelocatedintheN-terminaldomain(green)andputativeRNA-bindingKHdomain(yellow),respectively. TheEXOSC3-G31Aand-V80FsubstitutionsarelocatedintheN-terminaldomain(green),theEXOSC3-Y109N,
Mutationsin EXOSC3,whichlike EXOSC2 encodesastructural capsubunitoftheRNAexosome(Figs. 1a and 2),havebeenlinked topontocerebellarhypoplasiatype1b(PCH1b),anautosomal recessive,neurodegenerativediseasecharacterizedbysignificant atrophyoftheponsandcerebellum,Purkinjecellabnormalities, anddegenerationofspinalmotorneurons,startingatbirth (MIM#606489—humangeneslinkedtoMendeliandisordersare cataloguedwithMIMnumbersintheOnlineMendelianInheritanceinMan(OMIM)database(https://www.omim.org))[8, 73, 74].Thecerebellumandponsintegrateinformationfromsensory systems,thespinalcord,andotherpartsofthebraintoregulate motormovements,breathing,andlearningmotorbehavior [90].IndividualswithPCH1balsoshowmuscleatrophy/weakness,microcephaly,developmentaldelay,andbrainsteminvolvement[8, 73, 74].MostindividualswithPCH1bdonotlivepast childhoodandcurrenttreatmentispurelypalliative.LikeEXOSC2, theEXOSC3proteincontainsthreedomains:anN-terminal domain,aputativeRNA-bindingS1domain,andaputative RNA-bindingKHdomain(Figs. 2 and 3b).
ExomesequencingofindividualswithPCH1bfromthirtyeightfamiliesidentifiedseveraldifferent EXOSC3 mutations [8, 73, 74, 91–94](Fig. 2;Table 1).Inparticular,homozygous EXOSC3 (G31A)mutations—locatedintheN-terminaldomainof EXOSC3—wereidentifiedinthirteenindividualswithsevere PCH1b(lifespan 2years)andhomozygous EXOSC3 (D132A) mutations—locatedintheS1domainofEXOSC3—wereidentified ineighteenindividualswithlessseverePCH1b(lifespan 3years) (Figs. 2 and 3b).Inaddition,compoundheterozygous EXOSC3 (D132A)and EXOSC3 (nullallele[frameshift;prematureterminationcodon;indel],Y109N,orA139P)mutationswereidentifiedin fourteenindividualswithseverePCH1b(Figs. 2 and 3b).Finally, compoundheterozygous EXOSC3 (G31A)and EXOSC3 (W238R)mutations—locatedintheN-terminaldomainandKH domainofEXOSC3—wereidentifiedintwoindividualswithsevere PCH1bandhomozygous EXOSC3 (G135E)mutations—located intheS1domainofEXOSC3—wereidentifiedinanindividual withseverePCH1b(Figs. 2 and 3b).
Fig.3 (continued)-D132A,-G135E,-A139P,and-G191Csubstitutionsarelocatedintheputative RNA-bindingS1domain(blue),andtheEXOSC3-W238RsubstitutionislocatedintheKHdomain(yellow). TheEXOSC8-S272TsubstitutionislocatedattheC-terminalendofthePH-likedomain(orange).TheEXOSC8T7residueislabeledtoshowtheapproximatepositionoftheEXOSC8-A2VsubstitutionattheN-terminalend ofthePH-likedomain(orange),astheA2residuecouldnotberesolvedinthestructure.TheEXOSC9-L14P substitutionislocatedattheN-terminalendofthePH-likedomain(FigureadaptedfromMortonetal.[18])
Exosomesubunitvariantslinkedtodisease
EXOSC2G30VG30VHomozygousNovelsyndromea Mild
EXOSC2G198DG198D/ G30V HeterozygousNovelsyndromea Mild
EXOSC3G31AG31AHomozygousPCH1bb Severe
EXOSC3Y80FV80F/ D132A HeterozygousPCH1bb,SPc Mild
EXOSC3Y109NY109N/ D132A HeterozygousPCH1bb Severe
EXOSC3D132AD132AHomozygousPCH1bb Mild D132A/ nulld HeterozygousPCH1bb Severe
EXOSC3G135EG135EHomozygousPCH1bb Severe
EXOSC3A139PA139P/ D132A HeterozygousPCH1bb Severe
EXOSC3G191CG191CHomozygousPCH1bb,SPc Mild
EXOSC3W238RW238R/ G31A HeterozygousPCH1bb Severe
EXOSC8A2VA2VHomozygousPCH1ce Mild
EXOSC8S272TS272THomozygousPCH1ce Severe
EXOSC9L14PL14PHomozygousPCH-likef Mild L14P/ R161X HeterozygousPCH-likef Severe
DIS3R108CR108C/ nullg
DIS3R108SR108S/ nullg
DIS3R467QR467Q/ nullg
DIS3C483WC483W/ nullg HeterozygousMMh
DIS3D487VD487V/ nullg HeterozygousMMh Mild
DIS3D488Ni D488N/ nullg HeterozygousMMh Mild
DIS3D488GD488G/ nullg HeterozygousMMh
Table1 (continued)
DIS3S550FS550F/ nullg
DIS3E665KE665K/ nullg
DIS3H764YH764Y/ nullg
DIS3F775LF775L/ nullg
DIS3R780Ki D780K/ nullg
DIS3R780TR780T/ nullg
HeterozygousMMh
HeterozygousMMh
HeterozygousMMh
HeterozygousMMh
HeterozygousMMh
HeterozygousMMh
DIS3R789WR789W/ nullg HeterozygousMMh
DIS3R820WR820W/ nullg HeterozygousMMh
SummaryofallaminoacidsubstitutionsidentifiedtodateinEXOSC2,EXOSC3,EXOSC8EXOSC9structuralexosome subunitsandthemostcommonaminoacidsubstitutionsidentifiedintheDIS3catalyticexosomesubunitinindividuals withdiseaseandtheassociatedgenotypes,phenotypes,andseverityofdiseaseoftheaffectedindividuals.Atpresent, EXOSC2 mutationshavebeenidentifiedinindividualswithanovelsyndrome[9], EXOSC3 mutationshavebeen identifiedinindividualswithpontocerebellarhypoplasiatype1b(PCH1b)[8, 73, 74, 79, 80], EXOSC8 mutations havebeenidentifiedinindividualswithpontocerebellarhypoplasiatype1c(PCH1c)[10], EXOSC9 mutationshavebeen identifiedinindividualswithcerebellaratrophyandspinalmotorneuronopathy[11],and DIS3 mutationshavebeen identifiedinindividualswithmultiplemyeloma[12, 75–78].Althoughmostaffectedindividualswithexosomesubunit mutationshavereducedlifespanandqualityoflife,milddiseaseherespecificallyindicatesthattheindividualsarestill livingorlivedforseveralyears,whereasseverediseaseindicatesthattheindividualsdiedwithin2years aRetinitispigmentosa,hearingloss,prematureaging,shortstature,mildintellectualdisability bPontocerebellarhypoplasiatype1b cSpasticparaplegia dPrematureterminationcodon,Indel,Frameshift ePontocerebellarhypoplasiatype1c fPontocerebellaratrophy-like:cerebellaratrophywithspinalmotorneuronopathy gDeletionofchromosome13qregioncontaining DIS3 gene hMultiplemyeloma iHotspotmutation
EXOSC3 mutationsthatcauseamild,clinicallydiverseformof PCH1bwithadditionalphenotypeshavealsobeenidentifiedinsix individuals[79, 80].Compoundheterozygous EXOSC3 (V80F) and EXOSC3 (D132A)mutationshavebeenidentifiedintwo affectedindividualsthatexhibitintellectualdisability,spasticparaplegia,andcerebellaratrophy,butnomicrocephalyandanormal brainstem[79].Inaddition,homozygous EXOSC3 (G191C) mutationshavebeenidentifiedinfouraffectedindividualsthat
showspasticparaplegiaandmildcerebellaratrophy,butnomicrocephalyandnormalpons[80].Thus,distinctaminoacidchangesin EXOSC3cancausedifferentfunctionalconsequencesanddisease phenotypes,evenimpactingdifferentregionsofthebrain. AnalysisofthehumanRNAexosomestructuresuggeststhat theevolutionarilyconservedG31,D132,andW238residuesin EXOSC3/Rrp40,locatedintheN-terminal,S1andKHdomain, respectively,maybeimportantforintersubunitinteractionswith EXOSC5/Rrp46andEXOSC9/Rrp45(Fig. 3a)[4].Inparticular, theG31residueinEXOSC3istightlypackedagainstthesurfaceof EXOSC5and,therefore,theEXOSC3-G31Asubstitutioncould impairtheinteractionwithEXOSC5(Fig. 3a)[95].TheD132 residueinEXOSC3islocatedinaloopbetweenstrandsintheS1 domainand,therefore,theEXOSC3-D132Asubstitutioncould impairthefoldingoftheloopandsubsequentlydisturbinteractions withEXOSC5andEXOSC9(Fig. 3a,b)[95].TheW238residue inEXOSC3,whichliesinapocketbetweentheS1andKH domains,couldpositionresiduesinEXOSC3tointeractwith EXOSC9and,therefore,theEXOSC3-W238Rsubstitutioncould weakeninteractionsbetweenEXOSC3andEXOSC9(Fig. 3a,b) [95].TheseanalysessuggestthatPCH1b-associatedsubstitutions inEXOSC3couldimpairinteractionswithotherexosomesubunits,leadingtocompromisedRNAexosomefunctionanddisease.
Somerecentstudieshaveusedmodelorganismstoassessthe functionoftheEXOSC3/Rrp40variantsidentifiedinhumanswith PCH1b.Thestabilityofthebuddingyeastrrp40-W195Rvariant, correspondingtothehumanEXOSC3-W238Rvariant,isreduced comparedtowild-typeRrp40[95].Inaddition,theexpression levelsofthemouseEXOSC3-G31AandEXOSC3-W237Rvariants,correspondingtohumanEXOSC3-G31Aand-W238R,in amouseneuronalcelllinearereducedcomparedtowild-type mouseEXOSC3[95].Thesedatacouldsuggestthatthe EXOSC3substitutionsimpairthefoldingoftheEXOSC3protein. Buddingyeastcellsthatexpresstherrp40-W195Rvariantasthe soleRrp40proteinshowimpairedprocessingofrRNAanddegradationofncRNAssuchasCUTs[95, 96].Inzebrafish,morpholino knockdownof exosc3 inembryosshrinksthehindbrain.Expression ofwild-typezebrafish exosc3 mRNArescuesthishindbraindevelopmentdefect,butmutantzebrafish exosc3 mRNAcorresponding to EXOSC3 (G31A,D132A,orW238R),failtorescuethishindbraindefect[8].Notably,knockdownof exosc3 mostseverelyaffects brachimotorfacialneuronsanddisruptsthestructureofthePurkinjecelllayer/cerebellum[65].Theseresultssupportthenotion thattheaminoacidsubstitutionsinEXOSC3identifiedinindividualswithPCH1bimpairthefunctionoftheRNAexosome.
Mutationsin EXOSC8,encodingahexamericringsubunitofthe RNAexosome(Figs. 1a and 2),havebeenlinkedtopontocerebellarhypoplasiatype1c(PCH1c),anautosomalrecessive,neurodegenerativedisordercharacterizedbypsychomotordeficit, cerebellarandcorpuscallosumhypoplasia,hypomyelination,and spinalmuscularatrophy(SMA)startingatbirth(MIM#606019) [10].IndividualsthatsufferfromPCH1calsoshowseveremuscle weakness,impairedvisionandhearing,andoftendieduetorespiratoryfailure[10].LikePCH1b-affectedindividualswith EXOSC3 mutations,PCH1c-affectedindividualswith EXOSC8 mutations primarilyexhibitdefectsinspinalmotorneuronsandPurkinjecells; however,PCH1c-affectedindividualsalsoshowdefectsinoligodendrogliathatleadtohypomyelination[10].TheEXOSC8subunitcontainsacatalyticallyinert,PH-likeribonucleasedomain (Figs. 2 and 3b).
ExomesequencingoftenindividualswithseverePCH1c(lifespan 2years)revealedhomozygous EXOSC8 (S272T)mutations—locatedattheC-terminalendofEXOSC8—inallten individualsfromtwofamilies(Figs. 2 and 3b;Table 1).Inaddition, homozygous EXOSC8 (A2V)mutationswereidentifiedintwo individualsfromathirdfamilywithlessseverePCH1c(lifespan 2.3years)(Figs. 2 and 3b;Table 1).Examinationofthehuman RNAexosomestructuresuggeststhattheconservedS272residue inEXOSC8/Rrp43,locatedinthePHdomain,couldbeimportant forinteractionswithEXOSC9/Rrp45and/orimpactthecentral channelopeningatthebottomoftheRNAexosomecomplex wheresingle-strandedRNAisfunneled(Fig. 3a,b)[4].ThepositionoftheT7residueattheN-terminusofEXOSC8,whichisthe firstresidueofEXOSC8visibleinthishumanRNAexosomestructure[4],suggeststheconservedA2residueinEXOSC8/Rrp43 couldpotentiallyinteractwithEXOSC1/Csl4orimpactanopeningatthesideoftheRNAexosome(Fig. 3a,b).
AnalysisoftheEXOSC8/Rrp43variantsincellsofPCH1caffectedindividualsshowsthattheexpressionlevelsoftheEXOSC8 proteinvariantsareseverelyreducedcomparedtohealthycontrols. Inparticular,EXOSC8-S272TvariantlevelsarereducedinmyoblastsofaffectedindividualsandEXOSC8-A2Vvariantlevelsare decreasedinfibroblastsofaffectedindividuals[10].Importantly,in myoblastsofaffectedindividualsthatexpressareducedlevelof EXOSC8-S272T,thelevelofEXOSC3isalsoseverelyreduced, suggestingdepletionofEXOSC8leadstoareductionofother exosomesubunitsandpotentiallydepletestheentireRNAexosome complex[10].PCH1c-affectedindividualswithEXOSC8-S272T variantsalsohavemusclewithvariablemyofibersize,moderately decreasedfunctionofmitochondrialrespiratorychaincomplexes,
andbrainandspinalcordwithprofoundlossofmyelin.Notably, fibroblastsofaffectedindividualswithEXOSC8-A2Vvariantsshow increasedlevelsofdevelopmental HOX mRNAsandthe HOTAIR longncRNA,akeyepigeneticregulatorofgeneexpression[65].In addition,myoblastsofaffectedindividualswithEXOSC8-S272T variantsshowelevatedlevelsofsomemyelin-related,AU-richelement(ARE)-containingmRNAs(e.g., MBP, MOBP);AREmRNAs havepreviouslybeenshowntobetargetsoftheRNAexosome [10, 97].ThesedatasuggestthatthereducedEXOSC8variant levelsinPCH1c-affectedindividualscanimpairRNAexosome function,leadingtoaccumulationof HOX andmyelin-related mRNAsandpotentialdisruptionofdevelopmentandmyelinsynthesis.InsupportofaroleforEXOSC8inthebrain,morpholino knockdownof exosc8 inzebrafishembryosdisruptsnormalhindbraindevelopment,alteringthestructuresofthePurkinjecell layer/cerebellumandneuromuscularjunctionandimpairing growthofmotorneuronaxons[10, 65].
6EXOSC9Mutations
Mutationsin EXOSC9,whichlike EXOSC8 encodesahexameric ringsubunitoftheRNAexosome(Figs. 1a and 2),havebeen linkedtoapontocerebellarhypoplasia(PCH)-like,autosomal recessive,neurodegenerativedisorderthatischaracterizedby early-onset,progressivespinalmuscularatrophy(SMA)-like motorneuronopathyandcerebellaratrophy[11].LikePCH1b/ c-affectedindividualswith EXOSC3/8 mutations,affectedindividualswith EXOSC9 mutationsexhibitprogressivemuscleweakness,respiratoryimpairment,andcerebellaratrophy [11].However,unlikePCH1b/c-affectedindividuals,affected individualswith EXOSC9 mutationshavearelativelynormalpons [11].Notably,oneaffectedindividualwithseverediseasealso exhibitedcongenitalfracturesofthelongbones(femurand humerus)[11].LikeEXOSC8,theEXOSC9subunitharborsa singlecatalyticallyinactivePH-likeribonucleasedomain(Figs. 2 and 3b).
Exomesequencingofthreeunrelatedindividualswithaless severeformofthePCH-likedisease(aliveatages1.5,2.3,and 4.5years)revealedhomozygous EXOSC9 (L14P)mutationsinall threeindividuals[11](Figs. 2 and 3b;Table 1).Inaddition, compoundheterozygous EXOSC9 (L14P)and EXOSC9 (R161X)mutationswereidentifiedinafourthunrelatedindividual withasevereformofthePCH-likedisease(lifespan ¼ 1.3years) [11](Fig. 2;Table 1).AnalysisofthehumanRNAexosome structureshowsthattheL14residueislocatedinthefirstalpha helixofEXOSC9/Rrp45andcoulddisturbinteractionswithin EXOSC9/Rrp45(Fig. 3a,b)[4].
ExaminationofEXOSC9proteinexpressionincellsfromindividualswiththePCH-likediseaserevealsthattheEXOSC9-L14P variantlevelisreducedcomparedtohealthycontrols[11].Inparticular,infibroblastsfromanindividualwithhomozygous EXOSC9 (L14P)mutations,theEXOSC9-L14Plevelisreduced to~70%comparedtothewild-typeEXOSC9levelincontrols [11].Moreover,inskeletalmusclefromtheindividualwithheterozygous EXOSC9 (L14P)and EXOSC9 (R161X)mutations,the EXOSC9-L14Plevelisreducedto~55%orlesscomparedtothe wild-typeEXOSC9levelincontrols[11].Thegreaterreductionin EXOSC9variantlevelintheindividualwithheterozygous EXOSC9 mutationscomparedtotheindividualwithhomozygous EXOSC9 mutationscouldexplaintheearlymortalityandadditionalphenotypes(e.g.,bonefractures)inthisindividual.Notably,inpatientderivedfibroblaststhatexpressareducedlevelofEXOSC9-L14P variant,thelevelsofotherexosomesubunits(EXOSC3/8)arenot reducedcomparedtocontrols[11].Incontrast,inPCH1patientderivedfibroblaststhatexpressareducedlevelofEXOSC3-G31A orEXOSC8-A2Vvariant,thelevelsofotherexosomesubunits (EXOSC3/8/9)arereduced[11].Thesedatasuggestthatthe EXOSC9-L14Pvariantmaynotdestabilizetheentireexosome complex,whereastheEXOSC3-G31AandEXOSC8-A2Vvariants maydoso.Diseaseseverityforeachexosomesubunitvariantmay thereforecorrelatewiththedegreetowhichitreducesthestability ofotherexosomesubunits.InsupportforanalternativemechanismbywhichEXOSC9-L14Pimpairstheexosome,inpatientderivedfibroblaststhatexpressEXOSC9-L14P,theamountof EXOSC3subunitdetectedintheexosomecomplexbynativegel analysisisreducedcomparedtocontrols[11].Thisresultsuggests thattheEXOSC9-L14Pvariantmayaltertheassociation/dissociationofspecificexosomesubunits,suchascapsubunits,withthe exosomecomplexandcouldpotentiallyaltertheassembly/disassemblyoftheentireexosomecomplex.
RNA-seqanalysisoffibroblastsfromindividualswithhomozygous EXOSC9 (L14P)mutationsrevealedchangesinthelevelsof ARE-containingtranscripts,increasesinthelevelofdevelopmental HOXC8 mRNA,butnochangeinthelevelofepigeneticregulator HOTAIR ncRNA.ManyofthesignificantlyalteredmRNAsare relatedtodevelopmentalprocessesoftheneuronalsystem [11].RNA-seqofskeletalmusclefromindividualswithheterozygous EXOSC9 mutationsthatexhibitedcerebellaratrophyand bonefracturesshowedchangesinthelevelsofARE-containing transcriptsandmRNAslinkedtomotorneuronopathy(EPHA4, IGHMBP2, VAPB, BICD2,and DYNC1H1)andbonedisease (PLS3, BMPR1B, ACVR2A, DDR2, COL2A1, MC4R,and SEMA4D)[11].
Likereductionof exosc3 and exosc8 inzebrafish,morpholino knockdownorCRISPR/Cas9mutationof exosc9 inzebrafish
reducesheadsizeandhindbraindevelopment,supportingarolefor EXOSC9inthebrain[11].Inparticular,in exosc9 mutants,the brainisoftenmisshapenandthecerebellumandhindbrainare absent[11].Inaddition,in exosc9 mutants,theneuromuscular junctionsdevelopabnormally,withmotoraxonsfailingtomigrate totheneuromuscularjunctions,suggestinganeuronalpathfinding defect[11].The exosc9 mutantsalsoshowdamagedandmisaligned myofibers[11].ThesedatasuggestthatproperRNAexosome functionisrequiredfornormalneuromusculardevelopment.
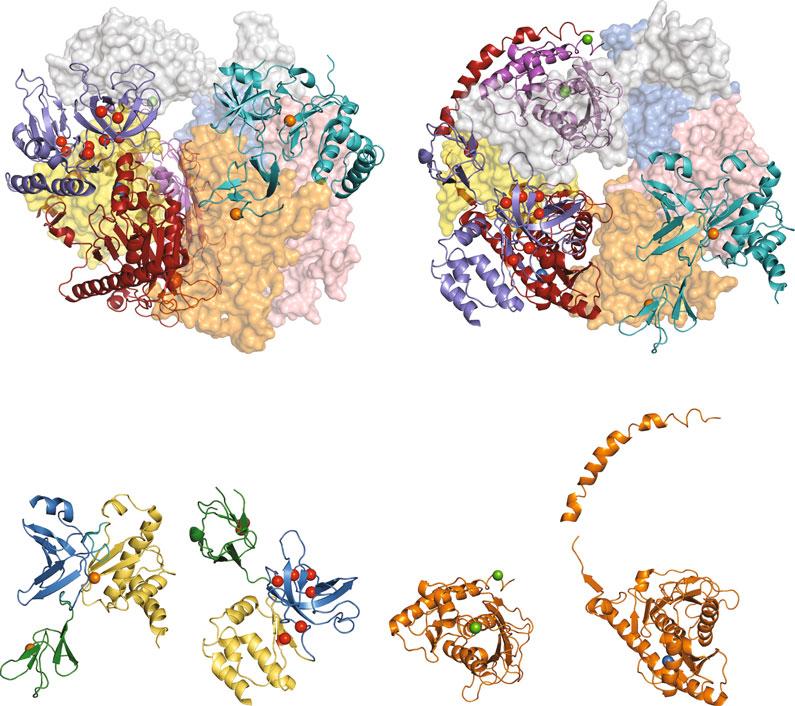
7 DIS3 Mutations Recurrentsomaticmutationsin DIS3,encodingthecatalyticsubunitoftheRNAexosome(Figs. 1a and 4),havebeenlinkedto multiplemyeloma,amalignantneoplasmofthemonoclonal antibody-producingplasmacells(differentiatedBcells)inthe bonemarrowthatcausesanemia,bonelesions,hypercalcemia, renaldysfunction,andcompromisedimmunefunction[12, 75, 76, 82, 98, 99].Multiplemyelomaisanincurable,genetically heterogeneousdiseaseinvolvingchromosometranslocations(e.g., fusionofChr14 IGH enhancerlocustootherchromosomes:t (11;14);t(4;14)),copynumbervariants(e.g.,hyperdiploidy;loss ofchromosomeregions:13q;17p;1p)),andsinglenucleotide variantsof DIS3 andotherimportantgenes(e.g., KRAS; NRAS; BRAF; FAM46C; TP53)[75, 98, 99].Multiplemyelomahasbeen estimatedtoaccountfor1.8%ofnewcancercasesand2.1%of deathsduetocancerin2018intheUnitedStates[100].The catalyticDIS3subunitcontainssixdomains:aCR3motif—importantforcoreexosomeinteraction,anendoribonucleasePIN domain,anexoribonucleaseRNBdomain,andthreeOB-fold RNA-bindingdomains—CSD1,CSD2,andS1[101, 102] (Fig. 4).
Exome,whole-genome,andtargetedsequencingofindividuals withmultiplemyelomainseveralstudiesrevealed141/1246 affectedindividuals(11.3%)withheterozygous DIS3 mutations [12, 75–78, 82–89](Fig. 4a;Table 1).Inmostindividuals,the myelomacellscontainedonealleleof DIS3 withamissensemutationandadeletionofthesecondalleleof DIS3,whichwasdeleted duetolossofthechromosome13qregion[12, 75, 82].Thesecells wereusuallynonhyperdiploidandharboredchromosometranslocations[12, 75, 82].Theaminoacidsubstitutionsidentifiedin DIS3inaffectedindividuals(83intotal)alter70highlyconserved residuesinDIS3thatpredominantlymaptotheexoribonuclease RNBdomainandendoribonucleasePINdomain(Fig. 4a).Notably,hotspotaminoacidsubstitutionsinDIS3occuratresidues D488andR780intheRNBdomain,mostfrequentlyDIS3D488NandDIS3-R780K(Fig. 4a).Importantly,theisolated
Domain
Domain
Domain
Domain
Domain S1 Domain
Fig.4 AminoacidsubstitutionsidentifiedintheDIS3catalyticsubunitoftheRNAexosomeinindividualswith multiplemyeloma.(a)DomainstructureofhumanDIS3proteinhighlightingtheaminoacidchangesidentified inindividualswithmultiplemyeloma[12, 75–78, 82–89].Abovethedomainstructure,theaminoacid substitutionsareshowninthedomainsofDIS3inwhichtheyarelocated.Mostoftheaminoacidsfrequently alteredinindividualswithmultiplemyelomaarelocatedintheexoribonucleaseRNBdomain(purple)and endoribonucleasePINdomain(pink)ofDIS3,butadditionalaminoacidsalteredindiseasearelocatedinother domainsofDIS3:CR3(gray),CSD1(green),andCSD2(brown).Agraphabovetheaminoacidsubstitutions showsthenumberofindividualswithmultiplemyelomathathavebeenidentifiedwitheachDIS3variant.The colorcodeoftheaminoacidsubstitutionsdenotesthenumberofaffectedindividualswitheachDIS3variant: 1(black),2(blue),3(green),4(orange),5+(red).Belowthedomainstructure,alignmentsofDIS3ortholog sequencesfromhuman(Hs),mouse(Mm), Drosophilamelanogaster (Dm),and S.cerevisiae (Sc)thatsurround theevolutionarilyconservedresiduealteredindisease(highlightedinred)areshown.Aminoacidpositionsof thedomainsandthecatalyticresiduesofthePINdomain(∗D146)andtheRNBdomain(∗D487)are highlighted.(b)IsolatedstructureofhumanDIS3fromtherecent11-subunithumanRNAexosomestructure (PDB#6D6Q)[7]thathighlightsthemostfrequentlyalteredresiduesinmultiplemyeloma.Aribbon representationofthehumanDIS3structureshowsthesixdomainsofDIS3—CR3(gray),PIN(pink),CSD1 (green),CSD2(brown),RNB(purple),S1(skyblue)—andhighlightstheconservedresiduesfrequentlyaltered indisease(coloredsphereswithaminoacidsidechains).TheRNA(black)thatenterstheRNBcatalyticsiteis alsoshown.Thecolorcodeoftheresiduesandspheresdenotes,likein(a),thenumberofaffectedindividuals thathavebeenidentifiedwithsubstitutionsattheseresiduepositions:2(blue),3(green),4(orange),5+(red).