
TheEffectof Lactobacillusplantarum ExtracellularVesicles fromKoreanWomeninTheir20sonSkinAging
ChanSongJo 1,†,CheolHwanMyung 1,†,YeoChoYoon 2,BeomHeeAhn 2,JinWooMin 3,WonSangSeo 2,3 , DongHwanLee 4,HeeCheolKang 2,3,YunHoeHeo 5,HyeongChoi 5,InKiHong 5 andJaeSungHwang 1,*,†
Citation: Jo,C.S.;Myung,C.H.; Yoon,Y.C.;Ahn,B.H.;Min,J.W.; Seo,W.S.;Lee,D.H.;Kang,H.C.; Heo,Y.H.;Choi,H.;etal.TheEffect of Lactobacillusplantarum ExtracellularVesiclesfromKorean WomeninTheir20sonSkinAging. Curr.IssuesMol.Biol. 2022, 44, 526–540. https://doi.org/10.3390/ cimb44020036
AcademicEditor:DongchulKang
Received:3December2021
Accepted:17January2022
Published:21January2022
Publisher’sNote: MDPIstaysneutral withregardtojurisdictionalclaimsin publishedmapsandinstitutionalaffiliations.

Copyright: ©2022bytheauthors. LicenseeMDPI,Basel,Switzerland. Thisarticleisanopenaccessarticle distributedunderthetermsand conditionsoftheCreativeCommons Attribution(CCBY)license(https:// creativecommons.org/licenses/by/ 4.0/). Article
1 DepartmentofGeneticEngineering&GraduateSchoolofBiotechnology,CollegeofLifeSciences,KyungHee University,Yongin17104,Gyeonggi-do,Korea;jchansong93@naver.com(C.S.J.); audjoin2@naver.com(C.H.M.)
2 Human&MicrobiomeCommunicatingLaboratory,GFCCo.,Ltd.,Hwasung18471,Gyeonggi-do,Korea; yc.yoon@gfcos.co.kr(Y.C.Y.);bh.ahn@gfcos.co.kr(B.H.A.);seows@gfcos.co.kr(W.S.S.); michael@gfcos.co.kr(H.C.K.)
3 Green&BiomeCustomizingLaboratory,GFCCo.,Ltd.,Hwasung18471,Gyeonggi-do,Korea; jw.min@gfcos.co.kr
4 ClinicalBusinessDivision,KoreaDermatologyResearchInstitute,GFCCo.,Ltd., Sungnam13517,Gyeonggi-do,Korea;dh.lee@gfcos.co.kr
5 R&DComplex,HKKolmarCo.,Ltd.,Seoul30004,Korea;yhheo@kolmar.co.kr(Y.H.H.); jecniclous@kolmar.co.kr(H.C.);inkiaaa@kolmar.co.kr(I.K.H.)
* Correspondence:jshwang@khu.ac.kr
†Theseauthorscontributedequallytothiswork.
Abstract: Extracellularvesicles,whicharehighlyconservedinmostcells,containbiologicallyactive substances.Thevesiclesandsubstancesinteractwithcellsandimpactphysiologicalmechanisms.Theskin isthemostexternalorganandisindirectcontactwiththeexternalenvironment.Photoagingandskin damagearecausedbyextrinsicfactors.Theformationofwrinklesisamajorindicatorofskinagingand iscausedbyadecreaseincollagenandhyaluronicacid.MMP-1expressionisalsoincreased.Dueto accruingdamage,skinagingreducestheabilityoftheskinbarrier,therebyloweringtheskin’sability tocontainwaterandincreasingtheamountofwaterloss. L.plantarum suppressesvariousharmful bacteriabysecretinganantimicrobialsubstance. L.plantarum isalsofoundintheskin,andresearch ontheinteractionsbetweenthebacteriaandtheskinisinprogress.Althoughseveralstudieshave investigated L.plantarum,thereareonlyalimitednumberofstudiesonextracellularvesicles(EV)derived from L.plantarum,especiallyinrelationtoskinaging.Herein,weisolatedEVsthatweresecretedfrom L.plantarum ofwomenintheir20s(LpEVs).Wetheninvestigatedtheeffectof LpEVsonskinagingin CCD986sk.Weshowedthat LpEVsmodulatedthemRNAexpressionofECMrelatedgenes invitro Furthermore, LpEVssuppressedwrinkleformationandpigmentationinclinicaltrials.Theseresults demonstratedthat LpEVshaveagreateffectonskinagingbyregulatingECMrelatedgenes.Inaddition, ourstudyoffersimportantevidenceonthedepigmentationeffectof LpEVs.
Keywords: extracellularvesicles(EVs);exosome;skinaging; Lactobacillusplantarum;pigmentation
1.Introduction
Extracellularvesicles(EVs)arehighlyconservedlipid-membrane-enclosedvesicles foundinmostcells,includingprokaryotes,eukaryotes,andarchaea[1,2].EVscontainavarietyofbiologicallyactivesubstancessuchasproteins,lipids,nucleicacids,andmetabolites. Theyreflectthestateofthecellsfromtheoriginatingmolecules,andcommunicatewith neighboringordistantcells[3].EVsincludeexosomesandmicro-vesicles[4].Exosomes are30–200nmvesiclessecretedfrommulti-vesicularendosomes(MVEs),whichareendosomesthatmakeupnumerousvesiclesandundergofusionwiththeplasmamembrane[5]. Micro-vesiclesare100–1000nminsizeandareproducedthroughbuddingwiththeplasma
membrane[6].EVsderivedfromvariouscellsinteractwithtargetcellsandaffectvarious physiologicalmechanisms,suchastheimmuneresponseandinflammation[7].
Bacterial-derivedEVsareclassifiedintoGram-negativeandGram-positivebacteriaderivedEVs.EVsfromGram-negativebacteriacontainlipopolysaccharides(LPS)[8],and EVsfromGram-positivebacteriacontainlipoteichoicacids(LTA)[9].Bacterial-derivedEVs areinvolvedintransferringantibioticresistanceproteinstootherbacteria[10]andinduce communicationsbetweenbacteria.Theyalsofunctiontoeliminatecompetingbacteriaby deliveringaproteinthatdegradesthepeptidoglycansofcompetingbacteria[11].Additionally, bacterialEVscancausediseaseinthehost[12].Forexample,Staphylococcusaureus-derived EVscauseatopicdermatitisbydelivering α-hemolysintotheskin[13].Consequently,there areanumberofresearchstudiesthatusethesecharacteristicsandoptimizethetransporter mechanismtomediatedrugdeliverytospecificcellsortissues[14–18].
Lactobacillusplantarum isaGram-positivememberofthegenusLactiplantibacillus,is rod-shapedand3–8 µminlength[19],andproduceslacticacid[20]. L.plantarum isfound inmanyfermentedproductsandhasbeenassociatedwithreducingallergicreactionsasa probiotic,andloweringcholesterolandtriglyceridelevels[21–24].Inparticular, L.plantarum suppressesvariousharmfulGram-positiveandGram-negativebacteriabysecretingan antimicrobialsubstancefromthehumangastrointestinaltract[25,26]. L.plantarum,which isanaerotolerantGram-positivebacteria,isalsofoundontheskin,andresearchonthe interactionswiththeskinarecurrentlyinprogress[27–30].However,theeffectof LpEVs onskinwrinkleformationnothavebeenstudied.
Theskinisthelargestorganinthehumanbodyandthemostexternal-facingorgan, indirectcontactwiththeexternalenvironment[31].Photoagingandskindamageoccur becauseofextrinsicfactorssuchasUVandexternalharmfulfactors;theyalsooccurdue tovariousinflammatorycytokinesandintrinsicfactors,suchasROS,thataregenerated duringmetabolism[32].Wrinklesoftheskinareamajorindicatorofskinaging[33].The formationofwrinklesiscausedbyadecreaseintheexpressionofcollagenandhyaluronic acid,whicharecomponentsoftheextracellularmatrix(ECM)[34].Simultaneously,an increaseintheexpressionofMMP-1,ametalloproteinasethatdegradescollagen,stimulates skinwrinkles[35].Inaddition,agingoftheskinreducestheabilityoftheskinbarrier becauseofdamage,therebyloweringtheskin’sabilitytocontainwater,andincreasingthe amountofwaterloss[36].
Variousbacteriaandvirusesresideincoloniesinthestratumcorneumandpores ofthehumanskin.Thiscoexistenceisknownasthemicrobiome[37].Thecomposition ofthemicrobiomedependsontheenvironmentwherethebacteriagrow,aperson’ssex, andage[38,39].Thismicrobiomeresidesontheskinandmaintainsabalancewithother surroundingcommunities,makingitresistantwhenexposedtoexternalpathogens,and italsoservesabeneficialfunctioninpreventinginfectioninthebody[40].Withaging, thecompositionofthemicrobiomechangesduetoexposuretoUVandvariouschemicals. Thischangeinthecompositionofthemicrobiomealsoacceleratesskinaging,becausethe microbiomeandtheskincellsinthebodyinteractwitheachother[41].Substancessuchas proteinsandlipidsaresecreteddifferently,duetovariousenvironmentandstimuli,and aredeliveredthroughEVs[42].
Thisstudyconfirmedtheanti-agingandanti-pigmentationeffectsof LpEVsasdemonstratedin invitro testandclinicaltrials.Wefoundthatthenumberof L.plantarum bacteria intheskinofwomenintheir20swashigherthanforwomenintheir50s,onaverage,and thattheabsenceof L.plantarum wasassociatedwithskinaging.Theresultsconfirmedthat LpEVs,whichwereobtainedfromtheskinofwomenintheir20s,improvesskinagingsuch asskinwrinklingandelasticity.Therefore,theseresultsindicatethatthe LpEVsinyoung skincanbeusedasaneffectiveanti-skinagingagent.
2.MaterialsandMethods
2.1.IsolationofMicroorganisms
Inthisstudy,microorganismswereisolatedfromhumanskinandthefollowing separationmethodwasused:Gauzewasrubbedonthewomen’sskinsuspensions,which wereobtainedbyaddingdistilledwater;then150 µLofeachsamplewerespreadon deMan,Rogosa,andSharpe(MRS)agarplatesunderaerobicconditionat37 ◦C.Single colonieswereobtainedandpurifiedbytransferringthemtonewMRSagarplates.The strainwaskeptinMRSbrothmediumthatcontained30%glycerolat 70 ◦C.
2.2.16SrRNAGeneSequenceandPhylogeneticAnalysis
ThegenomicDNAisolationKit(Geneall,Seoul,Korea)wasusedtoisolatethegenomicDNAofthestrain,accordingtothemanufacturer’sinstructions.The16Sribosomal RNA(rRNA)genewasamplifiedfromchromosomalDNAofastrainusingtheuniversal bacteriaprimersets,andthefullsequencewasassembledwithSeqMansoftwareversion7.1 (DNASTARInc.,Madison,WI,USA).The16SrRNAgenesequencesimilaritiesbetweenthe strainsandotherrelatedLactobacillusspecieswereobtainedfromtheGenBankdatabase. MultiplesequencealignmentswereperformedusingtheCLUSTALXprogramandcalculatedusingthetwo-parameterKimuramethod.Finally,aphylogenetictreewasconstructed withtheneighbor-joiningandmaximum-parsimonymethodsusingtheMEGA7Program.
2.3.MicroorganismPreparation
SinglecoloniesofthemicroorganismsthatbelongedtotheLactobacillusplantarum specieswereinoculatedandpre-incubatedinMRSbrothmedium(KisanBio,Seoul,Korea) at37 ◦Cfor18h.Next,thecultures’ LpEVswererinsedthreetimeswithDPBStoremove residualmedium,andincubatedin10%skimmilk(BD,FranklinLakes,NJ,USA)at37 ◦C for24h.
2.4.ExtracellularVesicleIsolation
L.plantarum,whichwasextractedfromhumanskintissueforuseinthisstudy,was culturedin10%skimmilkandthenExtracellularvesicles(EVs)wereisolated.Indetail,they werecentrifugedat4000× g for10min,andtheEVswerepurifiedwithultra-centrifugation (Hitachi,Chiyoda-ku,Tokyo,Japan)at10,000× g for30min,and150,000× g for2.5h.The EV-richpelletswerere-suspendedatafinalvolumeof100mLwithdistilledwater(DW), andkeptat4 ◦Cinafreezer,afterfilteringwitha0.22 µmbottle-topfilter.
2.5.NanoparticleTrackingAnalysis(NTA)
Nanoparticletrackinganalysis(NTA)wasconductedwithaZetaviewTWIN(Particle Metrix,Meerbusch,DE)toconfirmthediameterandconcentrationoftheextracellular vesicles.Lactobacillusspecies-derivedextracellularvesicles(LpEVs)wereisolatedfrom humanskin,suspendedinfilteredDWat20.15 ◦Candwereirradiatedwithablue-light laserwavelength(λ =488nm).Thesampleconductivitywasperformedat42.19 µS/cmand thefilterwavelengthwasmeasuredwithbackscatterdetection.Samplesweremeasured withdilution(dilutionfactorwas500)ontheequivalentsamplealiquot.Thedatawere analyzedusingZetaViewSoftware(version8.05).
2.6.CellCulture
Thehumandermalfibroblasts(CCD986sk)werepurchasedfromtheAmericanType CultureCollection(Manassas,VA,USA)andincubatedinDMEMhighglucosemedium (WelGeneInc.,Daegu,Korea),supplementedwith10%fetalbovineserum(FBS,WelGene Inc.,Daegu,Korea)and1%penicillin/streptomycin(HycloneLaboratoriesInc.,Logan,UT, USA)at37 ◦C,inanatmospherethatcontained5%CO2
2.7.CellViabilityAssay
Weinvestigatedchangesofviabilityincellsbasedontreatmentwith LpEVs.Thecell viabilitywasdeterminedusingthecellproliferationreagentWST-1(DojindoMolecular TechnologiesInc.,Rockville,MD,USA).Cellswereseededina96-wellplateat1 × 104 cells/wellin200 µLofcompleteconditionedmedium,supplementedwith10%FBSand 1%penicillin/streptomycin.Cellswereincubatedfor18hat37 ◦Cinanatmospherethat contained5%CO2.Cellswerethensimultaneouslytreatedwith0.625%,1.25%,5%,and 10%concentrationsofdose-dependent LpEVs,andincubatedfor24hat37 ◦Cand5%CO2 Afterincubation,200 µL/wellWST-1reagentswereaddedandincubatedfor2hat37 ◦C. Thecellswerethenmeasuredabsorbanceagainstabackgroundcontrolwithamicroplate reader(BioTekInstruments,Inc.,Winooski,VT,USA)at450nm.
2.8.LpEVTreatmentInducesElastaseInhibitoryActivity
ElastaseinhibitoryactivitywasperformedinTris-HCLbuffer(0.2mM,pH8.0). Porcinepancreaticelastase(Sigma-Aldrich,St.Louis,MO,USA)wasdissolvedtomakea 5mg/mLstocksolutionindistilledwater(DW).Assubstrate, N-Succinyl-Ala-Ala-Ala-pnitroanilidewasdissolvedinabufferat1.8mM.TheLpEVsweretreatedandincubated withtheenzymefor20minbeforeaddingasubstratetobeginthereaction.Thefinal reactionmixture(totalvolume200 µL)containedthebuffer.distilledwater(DW)wasused asnegativecontrol.Elastaseinhibitoryactivitywasmeasuredcontinuouslyfor30min immediatelyafteraddingthesubstrateusingaMicroplateReader(BioTekInstruments,Inc., Winooski,VT,USA)in96-wellmicro-plates.
Thepercentageinhibitionforelastaseinhibitoryactivityiscalculatedby:
Elastaseinhibitoryactivity(%)=[(ODcontrol(DW) ODLpEV)/ODcontrol(DW)] × 100
2.9.mRNAExpressionAnalysiswithReverseTranscriptPCR(RT-PCR)
WeperformedaRT-PCRanalysistoinvestigatechangesinmRNAexpression,and todeterminegenesthatwerecorrelatedwithskinelasticityandtreatmentwith LpEVs. ATRIzolreagent(Sigma-AldrichChemicalCo.,St.Louis,MO,USA)wasusedtoextractmRNAfromCCD986skdermalfibroblaststreatedwith LpEVs.Assessmentofthe purityandintegrityofthemRNAwasperformedusingaNanoDrop™ 2000/2000cSpectrophotometer(ThermoFisherscientific,Waltham,MA,USA)andanalyzedat260/280nm. RT-PCRwasconductedwithprimersformatrixmetalloproteinase-1(MMP-1),pro-collagen typeI(COL1A1),andfilaggrin(FLG).Theprimersequencesusedinthisstudyareshown inTable 1 andtheseprimers,whichweredesignedbyourselves,wereused.TheRNA templatewasreverse-transcribedusingamfi-RivertcDNASynthesisPlatinumMasterMix (GenDEPOT,Katy,TX,USA)andamplifiedbyPCRusingaC1000Touch™ thermalcycler (Bio-rad,Hercules,CA,USA).ThePCRprogramincludedaninitialdenaturationat95 ◦C for2min,followedby40cyclesof30sat95 ◦C,90sat62 ◦C,and5minat70 ◦C.Inthis study,allmRNAexpressionexperimentswererepeatedmorethanthreetimes.
Table1. EachprimersequencesandTminformationusedinthisstudy.
PrimerPrimerSequenceTm(◦ C)
Actin Forward5 —CATGAAGTGTGACGTGGACA—3 58 ◦ C Reverse5 —CAGGGCAGTGATCTCCTTCT—3
COL1A1 Forward5 —GACCTCAAGATGTGCCACTC—3 58 ◦ C Reverse5 —CCAGTCTCCATGTTGCAGAA—3 MMP-1 Forward5 —CCCAGCGACTCTAGAAACAC—3 58 ◦ C Reverse5 —GCCTCCCATCATTCTTCAGG—3
Filaggrin Forward5 —GCTGAAGGAACTTCTGGAAAAG—3 62 ◦ C Reverse5 —GCCAACTTGAATACCATCAGAAG—3
2.10.ProteinExpressionAnalysiswithWesternBlot
WeinvestigatedtheeffectsofHyaluronidase2(HAS2),whichisknowntolyase hyaluronicacid invitro,toconfirmtheskinmoisturizingeffectsof LpEVtreatments.Dermal fibroblastswerestimulatedfor24handharvested.Theproteinsincellswereextracted with1 × RIPAbufferandaproteinase/phosphateinhibitorbuffer,andrunon10%SDSPAGEgels.ProteinswerethenblottedonthePVDFmembrane,andimmune-detected withprimaryantibodiesagainstHAS2(ab140671,abcam,Chambridge,UK),andwitha secondaryanti-mouseantibody(ab6728,abcam,Chambridge,UK).AChemiDocimaging systemwasusedfordetection(ChemiDocXRS+,Bio-Rad,Hercules,CA,USA).TheHAS2 proteinvolumewasnormalizedbyactinproteinexpression.
2.11.PreparationofSkinApplicationSolutions
Mannitol5%thatcontainedLactobacillusextracellularvesicleswasusedforanexperimentalgroup,andMannitol5%wasusedasacontrolgroup.Thereafter,phosphate-buffered salinewasaddedandadjustedto100%.Sampleswerestoredat5 ◦Cto25 ◦C.
2.12.VolunteerRecruitmentandSelection
Thetestperiodwasfrom26November2020to24December2020.Ofatotalof20 volunteers,16Koreanwomenweretestedand4droppedout(IRBNumber:KDRI-IRB20936).Thecriteriaforselectingvolunteerswereasfollows:(1)apersonwhovoluntarily wroteandsignedaninformedconsentformaftertheprincipalinvestigator,oraperson delegatedbytheprincipalinvestigator,fullyexplainedandinformedtheresearchsubject; (2)ahealthypersonwithoutacuteorchronicphysicaldiseases,includingskindiseases; (3)asubjectthatcouldcompleteafollow-upduringthetestingperiod.Treatedmethods aredirectlyusedbystudysubjects.Theymixagent1and2,evenlydidonfacetwicea dayinthemorningandevening.Andthen,theymeasuredskinconditionevery2weeks for4weeks.Beforethetest,thevolunteersfirstremovedanywasteordebris;then,after restingat20–24 ◦Cwith40–60%RH,for30min,theytookpicturesusingMARKVuand F-rayequipment.Thereafter,themeasurementsitewaspartitionedandinstrumental measurementwasperformed.Photographyanddeviceevaluationwereperformedinthe samemannerafter2-weekand4-weekperiods.
2.13.SkinContourMeasurement
F-RayandMoiretechniqueswereusedtomeasureskincontours.Theelasticitywas measuredbyshootingatatotalofsevenangles(front,leftandright30◦,45◦,60).An 18-megapixelcamerawasused.
2.14.SkinImageMeasurement
AMARKVu(PSIPLUS,Suwon-si,Korea)wasusedforskinimagemeasurement. AcontinuouslightsourcethatusedfourtypesofLEDs—generallight,polarizedlight, ultravioletlight,andglossylight—wasused.Usingthedevice’sDetailLogicprogram,we wereabletoassess13differentskinconditionssuchaspores,wrinkles,blemishes,and sebum,fromhigh-resolutionphotos.
2.15.SkinWrinkles,Elasticity,andDermalDensityMeasurements
SkinwrinklesweremeasuredusingANTERA3D(Miravex,Dublin,Ireland)whichisa high-resolutionthree-dimensionalimagemeasuringdevicethatobtainedthree-dimensional imagesoftheskinbyusinganopticalmethodandamathematicalalgorithm.Theimage measuredthedepthandwidthoffinewrinkles,theroughnessoftheskin,andthenumber ofpores.ACutometer® MPA580(Courage&Khazaka,Cologne,Germany)wasusedfor skinelasticityanalysis.DermaldensitywasmeasuredusingUltrasound(DermaLabSkin, CORTEXTECHNOLOGY,Hadsund,Denmark)equipment.
2.16.StatisticalAnalysis
SignificancewasconfirmedusingtheMinitab19(Minitab® 19.2,MinitabInc.,State College,PA,USA)program.Thepaired t-testwasusedtocomparethevaluesmeasuredbefore andafterthetest,andthesignificancewasconfirmedatthelevelof p <0.05, p <0.01, p <0.001, throughrepeatedmeasureANOVA,byrepeatingmeasurementsthreeormoretimes.
3.Results
3.1.SrRNAandPhylogeneticAnalysisofLactobacillusplantarum
Wepreviouslyfoundthatthepopulationof L.plantarum wassignificantlyhigherinthe skinofwomenintheir20sthanthoseintheir50s,onaverage(Figure 1).Wehypothesized thatthedecreasein L.plantarum mightberelatedtoskinaging.Toconfirmthishypothesis, wecollectedspecimensfromtheforeheadsofwomenintheir20s,and L.plantarum was isolatedonMRSagarplates.Inordertoidentifywhethertheisolatedstrainwasactually L. plantarum,weextractedgenomicDNAandamplified16SRNAchromosomalDNA.The amplifiedsequencewasassembledusingSeqMansoftwareandaBioEditprogram.The resultswereconsistentwith L.plantarum intheGenBankdatabase(Figure 2),andthisstrain wasusedforsubsequentexperiments.
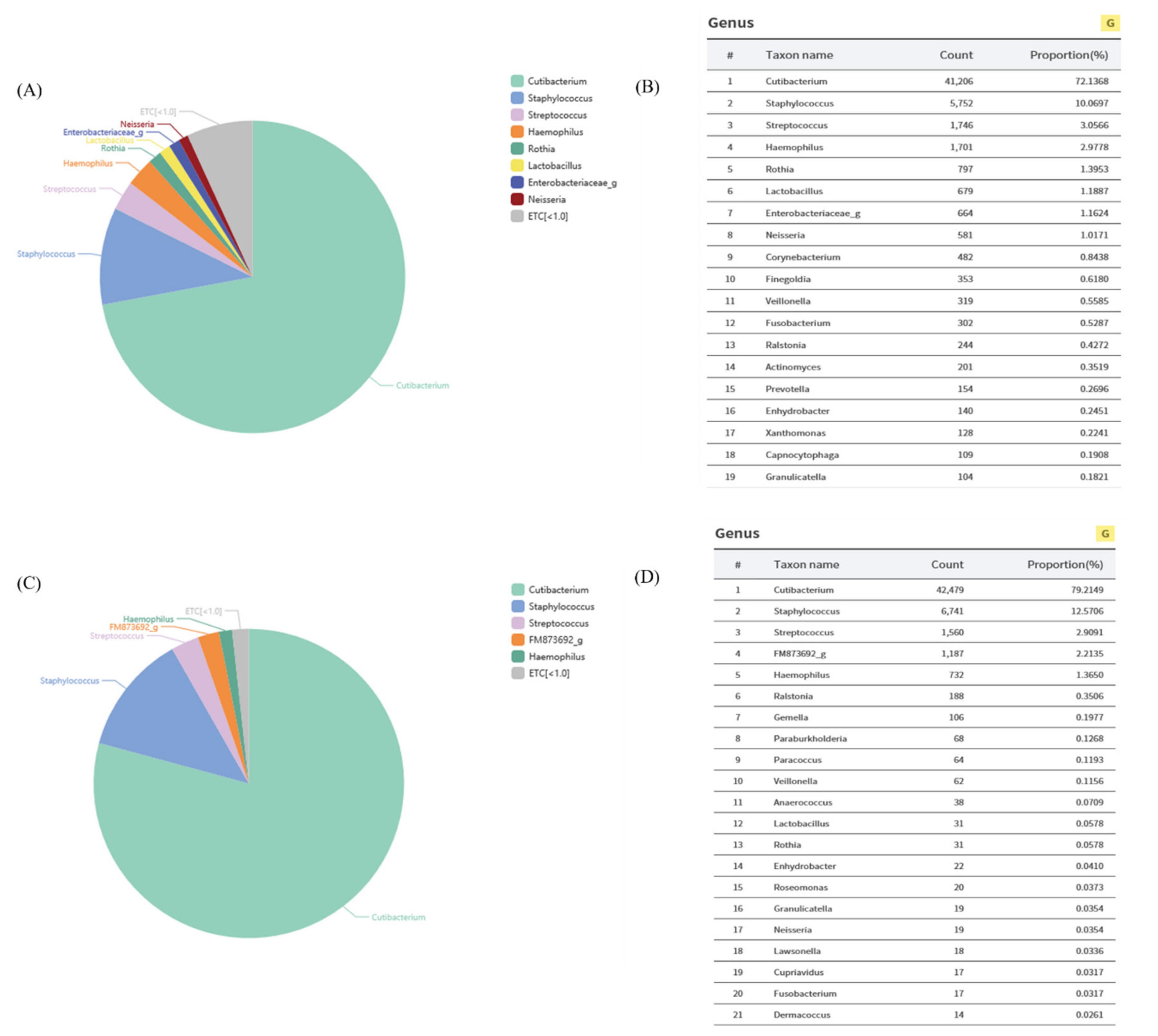

Figure2. Phylogenetictreebasedon16SrRNAgenesequencesof Lactobacillusplantarum isolated fromskinofwomenintheir20 s.
3.2.LactobacillusplantarumActivelySecretesEVs
Weisolatedextracellularvesiclessecretedfrom L.plantarum ofwomenintheir20susing ultracentrifugation,analyzed LpEVswithananoparticletrackinganalysis(NTA)video,andconfirmedtheirsizeanddistribution(Figure 3A).NTAanalysisrevealedthatthe LpEVsexistinexosomeforms(Figure 3B).TheEVparticleshadanaveragediameterof 126.5 ± 56.4nm,anaveragesizeof50–200nm,andtheconcentrationofEVswas 35.86 µg/mL (permL).ThenumberofparticlespermLofEVsandmgofproteinconcentrationwas 9.1 × 109 per1mL,and2.53 × 1011 per1mgofprotein(Figure 3C).Fromtheseresults,we determinedthat L.plantarum isolatedfromtheskinofwomenintheir20sactivelysecretes EVs,withsizesthatrangefrom50to200nm.
3.3.LpEVTreatmentInducesCellProliferationandRegulatesECMDegradation-Associated GeneExpression
Senescentcellsarecharacterizedbytheirinabilitytoproliferate[43].Therefore,we validatedproliferationtoconfirmtheeffectof LpEVsinfibroblasts.Theresultmeansthat the LpEVshaveaneffectonproliferationinfibroblastsatconcentrationsof2.5and5%,but not10%(Figure 4A).WetheninvestigatedECM-relatedgeneexpression.Thecellsthat makeupthedermisaresurroundedbyanextracellularmatrix(ECM)thatconnectsthem andallowsthecellstomaintaintheirshape.First,weirradiatedUVAandthentreated LpEVsinfibroblasts.WeexaminedMMP-1expressionandtheamountofelastasetoconfirm
theeffectof LpEVsonECMdegradation(Figure 4B).ThemRNAlevelofMMP-1,anenzyme thatdegradestheECM,decreasedsignificantlybasedontheassessmentof0.625%of LpEVs. Inaddition,inordertoconfirmtheskinelasticityeffectof LpEVstreatment,fibroblasts weretreatedwith LpEVs,andthentheelastaseactivitywasanalyzed.Asaresult,we foundthat LpEVsincreasedtheinhibitionofelastaseactivity,whichisapeptidase,from approximately20%ataconcentrationof1.25%to40%ata10%concentration(Figure 4C). TheseresultsshowedthattheEVsofwomenintheir20saffectcellproliferation,andcan inhibittheexpressionoractivitiesofECMdegradationenzymesandpeptidase.

Figure3. Purificationandcharacteristicsof Lactobacillusplantarum-derivedextracellularvesicles (LpEVs):(A)arepresentativeframefromoneofthe LpEVs’nanoparticletrackinganalysisvideos. ThepurifiedEVswerediluted1:500indistilledwater;(B)theparticlesizeandnumberof LpEVs determinedbynanoparticletrackinganalysis(NTA);(C)proteinandparticleconcentrationof LpEVs.

Figure4. Evaluationoftheeffectsof LpEVsonMMP-1mRNAexpressionlevelsusingRT-PCRin CCD986sk:(A)cellProliferationof LpEVsinCCD986skdermalfibroblasts.Cellproliferationassays wereperformedoncellstreatedwith LpEVsinadosedependentmanner(** p <0.01);(B)theMMP-1 expressionlevelsinCCD986skafterirradiationofUVAandtreatmentof LpEVs(* p <0.05);(C)the elastaseinhibitoryactivitywasmeasuredinadosedependentmannerinCCD986sk(** p <0.01, *** p <0.001).
3.4.LpEVTreatmentInducesECMProduction-AssociatedGeneExpression WeexaminedmRNAexpressionofType1procollagenfollowing LpEVstreatment inCCD986skdermalfibroblasts,toinvestigateinfluenceof LpEVsonECMproductionas wellasECMdegradation.ThemRNAlevelofType1procollagenincreasedbyafactorof 1.7timescomparedwithTGF-betathatwasusedasapositivecontrol. LpEVsincreased
inadose-dependentmannerandtoabout1.6timesthebaselinemeasurementat10% concentration(Figure 5A).Filaggrinisinvolvedinepidermalhomeostasisandmaintains theskinbarrierfunction.Filaggrinexpressionisgenerallyreducedasagingoccurs[44]. Accordingly,weinvestigatedwhether LpEVsaffecttheexpressionoffilaggrininCCD986sk dermalfibroblasts.Asaresult, LpEVsincreasedthemRNAexpressionoffilaggrinbymore than2timesataconcentrationof2.5–10%(Figure 5B).Inaddition,theproteinexpression ofHAS2,whichinduceshyaluronicacidsynthesisandplaysacrucialroleasacrosslinking ofECM[45],decreasedslightlyatlowconcentrationsofEV(0.625–2.5%),butincreasedby about20–30%at5%and10%concentrations(Figure 5C).Therefore, LpEVsincreasethe expressionofECMcomponentsandtheenzymesrelatedtoECM.

Figure5. Evaluationoftheeffectsof LpEVsoncollagen,filaggrin,andHAS2expressionlevelsin CCD986sk:(A)collagenmRNAexpressionlevels(* p <0.05);(B)filaggrinmRNAlevels(* p <0.05, ** p <0.01);(C)HAS2proteinlevelsinCCD986skafter24hoftreatment(* p <0.05,** p <0.01).
3.5.LpEVTreatmentSuppressesWrinkleFormationinClinicalTrials
Ultravioletradiationordamagecausedbyagingreduceskinelasticityandcause wrinkleformation[46,47].Weconductedclinicalassessmentson16skinwrinklesaround theeyesofwomenintheir50s,onaverage,todeterminewhether LpEVscanrestorethe agingindexinagingskin(Table 2).Weapplied LpEVs(orplaceboEVs)tothewrinkles aroundtheeye,andthewrinklesweremeasuredat0,2,and4weekswiththeAntera3D. Theindentationindexvalue(A.U.),whichdeterminesthedegreeofwrinklesaroundthe eyes,decreasedby8.9%at2weeksand15.89%at4weekscomparedtoweek0,whereas therewasnochangeintheplacebogroup(Figure 6A).Skinelasticityimprovedby14.76% at2weeksandby27.07%at4weeks(Figure 6B).TheAntera3Dimageshowedthat treatmentof LpEVsgraduallydecreasedthedistributionandformationofwrinklesat2and 4weeks.Incontrast,itwasdifficulttoconfirmasignificantwrinklechangeintheplacebo group(Figure 6C).Theseresultssuggestthat LpEVtreatmentimprovesskinelasticityand suppresseswrinkleformation.
Table2. Subjectinformationofclinicaltrials.
SubjectsofClinicalTrials(IRBNumber:KDRI-IRB-20936)*
GenderAgeAverageAge
Female 40’s50’s
Age50 n=6n=10
Totaln=16
*:EachdatapartnerobtainedthenecessaryInstitutionalReviewBoard(IRB)approvalorexemption.

Figure6. Evaluationoftheeffectsof LpEVsonwrinkleformation:(A)eye-wrinkleimprovement assessmentsinclinicaltrials;(B)epidermidiselasticityimprovementresultsinclinicaltrials;and(C) Antera3Dimage(Wrinkle:Small)fromclinicaltrials.
3.6.LpEVTreatmentMoisturizesSkinandEnhancesSkinDensity
Amajorcharacteristicofskinagingisthechangeinmoisturecontentandskindensity. Themoisturecontentoftheskindecreasesastheskinbarrierweakens,andskindensityis alsoreducedduetoadecreaseintheECM[45,48,49].Therefore,wetriedtoassesswhether LpEVscanaffectandimprovethewatercontentanddensityintheskin.Unliketheplacebo group,whichhadnosignificanteffect,the LpEVsincreasedwatercontentby10.79%at 2weeksand21.40%at4weeks(Figure 7A).Weconfirmedthatskindensityincreasedat the2-weekand4-weekassessmentsinbothgroups,usingultrasound(Figure 7B).Image quantificationimagesshowedanincreaseinskindensity,buttheincreasedrateofthe density(39.30%)ofthe LpEVgroupwashigherthanthatoftheplacebo(15.19%)(Figure 7C). Therefore,weconfirmedthat LpEVssuppressthereductioninskinmoisturecontentand increaseskindensity.

Figure7. Evaluationoftheeffectsof LpEVsonmoisturecontentsandskindensity:(A)skinmoisture improvingeffects;(B)ultrasoundimages;and(C)anumericalgraphoftheskindensityimprovementrate.
3.7.LpEVTreatmentSuppressesSkinPigmentationCausedbyAging
Anotherfactorofskinagingisskinpigmentation[50].Weinvestigatedwhetherthe LpEVshadawhiteningeffectthatsuppressedpigmentationcausedbyaging.Asaresult, forpatientstreatedwith LpEVs,unliketheplacebo,thepigmentationofthelesionsites wasdecreasedatthe2-weekand4-weekassessments(Figure 8A,C).Inthe LpEVtreatment group,skindensityimprovedby3.87%at2weeksand8.7%at4weeks(Figure 8B). Therefore,thesedatashowedthat LpEVshaveaneffectonpigmentationcausedbyskin aging.Consequently, LpEVshaveagreatanti-agingeffect.

Figure8. Evaluationoftheeffectsofpigmentationreductionthroughimageanalysis(MarkVu)with LpEVsinclinicaltrials:(A)MarkVuimage;(B)thenumericalgraphoftheskindensityimprovement rateof(A);(C)degreeofreductioninpigmentationinvisualreading.
4.Discussion
SkinagingiscausedbyexternalfactorssuchasUVraysandinternalfactors,which includetelomereshortening[51–53].Recently,studiesontheskinmicrobiomehaveattractedconsiderableinterest,becauseithasbeenidentifiedasafactorthatcanimpact skinaging[54,55].Thedifferenceinthemicrobiomecompositionofyoungandagedskin
suggeststhatthemicrobiomemaybeinvolvedinskinaging[56].Themicrobiomehas directcontactwiththeoutermostskin,andalsointeractswiththeskincellsbysecreting extracellularvesicles(EVs),suchasexosomes,thatcontainbiologicallyactivemolecules[4]. Therefore,wehypothesizedthatdifferencesinthemicrobiomebetweenwomenintheir20s and50s,onaverage,wouldberelatedtoskinaging.
LpEVshaveaneffectonthecellproliferationofCCD986skdermalfibroblasts (Figure 4A). ItisknownthatmanyEVshaveananti-agingeffect,andcanincreaseskindensitybyrestoring orincreasingtheproliferationoffibroblasts[57].Likewise,thedatashowedthattheEVsof L.plantarum increasedcellproliferation.Wetheninvestigatedthe LpEVsinthisexperimentinducedprocessesthatinhibitedECMdegradation(Figure 4B,C)andincreasedtheexpression ofproteinsrelatedtoECMsuchascollagen,filaggrinandHAS2(Figure 5).Basedonour results,wesuggestthat L.plantarum couldbeappliedtohelppreventskinaging.
Weconductedclinicalassessmentsonwomenthatwereintheir50swomen,on average,andconfirmedtheagingindex,whichiscausedbyadecreaseinskinelasticityand wrinkleformation.Wefoundthat LpEVscouldreducewrinkleformation(Figure 6).The lossofmoisturecontentintheskinarisesfromdamagetotheskinbarrier[48].Interestingly, LpEVsincreasedthemoisturecontentoftheskin(Figure 7).However,futurestudiesare requiredtoassesswhetherthe LpEVsrestoretheskinbarrierorthemoisturecontentis increasedbytheECMimprovements,suchascollagenandhyaluronicacid.Inaddition, anothercharacteristiccausedbydamagetotheskinbarrierisanincreaseintheamountof waterlossintheskin.Therefore,itisimportanttoalsoassesstheamountofmoistureloss intheskin.
Wealsodeterminedthat LpEVssuppressedpigmentationcausedbyagingskinfor womenintheir50s,onaverage(Figure 8).Ourresultsshowedthatthe LpEVscaninfluence aging-inducedpigmentation.Furtherstudiesonthedepigmentationeffectcanfurther elucidatetheapplicationsfor LpEVs.
Inthisstudy,wedemonstratedthatwomenintheir20shadahigherpopulationof L.plantarum intheirskinmicrobiomethanwomenintheir50s,onaverage.Additionally, LpEVscouldsuppressagingfactors(Figure 9).Consequently,theresultssuggestthat LpEVs, whicharecomponentsoftheskinmicrobiome,canbeappliedasaneffectiveanti-aging agenttoimproveskinaging,andalsoasaneffectiveanti-pigmentationagent.

Figure9. Theanti-agingeffectsofextracellularvesiclesderivedfrom Lactobacillusplantarum isolated fromtheskinofwomenintheir20s.
AuthorContributions: Conceptualization,J.S.H.,C.S.J.,C.H.M.andY.C.Y.;methodology,C.S.J., H.C.K.,Y.C.Y.andC.H.M.;software,B.H.A.,D.H.L.,Y.H.H.;formalanalysis,J.W.M.,W.S.S.,H.C.K.; investigation,C.S.J.,C.H.M.,I.K.H.andJ.S.H.;Writing—OriginalDraft,C.S.J.andH.C.;Writing— Review&Editing,C.S.J.,C.H.M.andJ.S.H.;supervision,J.S.H.;fundingacquisition,J.S.H.Allauthors havereadandagreedtothepublishedversionofthemanuscript.
Funding: Thisresearchreceivednoexternalfunding.
InstitutionalReviewBoardStatement: Thestudywasconductedaccordingtotheguidelinesof theDeclarationofHelsinki,andapprovedbytheInstitutionalReviewBoardofKoreaDermatology ResearchInstitute(protocolcodeKDRI-IRB-20936anddateofapproval:12January2021).
InformedConsentStatement: Informedconsentwasobtainedfromallsubjectsinvolvedinthestudy.
ConflictsofInterest: Theauthorsdeclarenoconflictofinterest.
Abbreviations
EVextracellularvesicle
ECMextracellularmatrix
LpEVsEVsthatweresecretedfrom L.plantarum ofwomenintheir20s
MMP-1matrixmetalloproteinase-1 COL1A1pro-collagentypeI FLGfilaggrin
References
1. Kim,D.;Kang,B.;Kim,O.Y.;Choi,D.;Lee,J.;Kim,S.R.;Go,G.;Yoon,Y.J.;Kim,J.H.;Jang,S.C.EVpedia:AnIntegratedDatabase ofHigh-ThroughputDataforSystemicAnalysesofExtracellularVesicles. J.Extracell.Vesicles 2013, 2,20384.[CrossRef][PubMed]
2. Woith,E.;Fuhrmann,G.;Melzig,M.F.ExtracellularVesicles—ConnectingKingdoms. Int.J.Mol.Sci. 2019, 20,5695.[CrossRef] [PubMed]
3. Tkach,M.;Théry,C.CommunicationbyExtracellularVesicles:WhereweareandWhereweNeedtoGo. Cell 2016, 164,1226–1232. [CrossRef]
4. Stahl,P.D.;Raposo,G.ExtracellularVesicles:ExosomesandMicrovesicles,IntegratorsofHomeostasis. Physiology 2019, 34, 169–177.[CrossRef]
5. Yu,L.;Zhu,J.;Liu,J.;Jiang,F.;Ni,W.;Qu,L.;Ni,R.;Lu,C.;Xiao,M.AComparisonofTraditionalandNovelMethodsforthe SeparationofExosomesfromHumanSamples. BioMedRes.Int. 2018, 2018,3634563.[CrossRef][PubMed]
6. Bano,R.;Ahmad,F.;Mohsin,M.APerspectiveontheIsolationandCharacterizationofExtracellularVesiclesfromDifferent Biofluids. RSCAdv. 2021, 11,19598–19615.[CrossRef]
7. Lee,Y.;ElAndaloussi,S.;Wood,M.J.ExosomesandMicrovesicles:ExtracellularVesiclesforGeneticInformationTransferand GeneTherapy. Hum.Mol.Genet. 2012, 21,R125–R134.[CrossRef]
8. Tulkens,J.;Vergauwen,G.;VanDeun,J.;Geeurickx,E.;Dhondt,B.;Lippens,L.;DeScheerder,M.;Miinalainen,I.;Rappu, P.;DeGeest,B.G.IncreasedLevelsofSystemicLPS-PositiveBacterialExtracellularVesiclesinPatientswithIntestinalBarrier Dysfunction. Gut 2020, 69,191–193.[CrossRef]
9. Matsuguchi,T.;Takagi,A.;Matsuzaki,T.;Nagaoka,M.;Ishikawa,K.;Yokokura,T.;Yoshikai,Y.LipoteichoicAcidsfrom Lactobacillus StrainsElicitStrongTumorNecrosisFactorAlpha-InducingActivitiesinMacrophagesthroughToll-LikeReceptor2. Clin.VaccineImmunol. 2003, 10,259–266.[CrossRef][PubMed]
10. Schaar,V.;Uddbäck,I.;Nordström,T.;Riesbeck,K.GroupAStreptococciareProtectedfromAmoxicillin-MediatedKillingby VesiclesContainingB-LactamaseDerivedfrom Haemophilusinfluenzae J.Antimicrob.Chemother. 2014, 69,117–120.[CrossRef]
11. Buchon,N.;Broderick,N.A.;Lemaitre,B.GutHomeostasisinaMicrobialWorld:Insightsfrom Drosophilamelanogaster Nat.Rev. Microbiol. 2013, 11,615–626.[CrossRef]
12. Liu,Y.;Defourny,K.A.;Smid,E.J.;Abee,T.Gram-PositiveBacterialExtracellularVesiclesandtheirImpactonHealthandDisease. Front.Microbiol. 2018, 9,1502.[CrossRef]
13. Hong,S.;Choi,E.;Min,T.;Kim,J.;Kim,M.;Jeon,S.G.;Lee,B.;Gho,Y.S.;Jee,Y.;Pyun,B.AnImportantRoleofA-Hemolysinin ExtracellularVesiclesontheDevelopmentofAtopicDermatitisInducedby Staphylococcusaureus PLoSONE 2014, 9,e100499.
14. Walker,S.;Busatto,S.;Pham,A.;Tian,M.;Suh,A.;Carson,K.;Quintero,A.;Lafrence,M.;Malik,H.;Santana,M.X.Extracellular Vesicle-BasedDrugDeliverySystemsforCancerTreatment. Theranostics 2019, 9,8001.[CrossRef]
15. deJong,B.;Barros,E.R.;Hoenderop,J.G.;Rigalli,J.P.RecentAdvancesinExtracellularVesiclesasDrugDeliverySystemsand theirPotentialinPrecisionMedicine. Pharmaceutics 2020, 12,1006.[CrossRef]
16. Surman,M.;Drozdz,A.;St˛epie´n,E.;Przybyło,M.ExtracellularVesiclesasDrugDeliverySystems-MethodsofProductionand PotentialTherapeuticApplications. Curr.Pharm.Des. 2019, 25,132–154.[CrossRef]
17. Saint-Pol,J.;Gosselet,F.;Duban-Deweer,S.;Pottiez,G.;Karamanos,Y.TargetingandCrossingtheBlood-BrainBarrierwith ExtracellularVesicles. Cells 2020, 9,851.[CrossRef]
18. Agarwal,S.;Agarwal,V.;Agarwal,M.;Singh,M.Exosomes:Structure,Biogenesis,TypesandApplicationinDiagnosisandGene andDrugDelivery. Curr.GeneTher. 2020, 20,195–206.[CrossRef][PubMed]
19. Edem,E.E.;Nathaniel,B.U.;Nebo,K.E.;Obisesan,A.O.;Olabiyi,A.A.;Akinluyi,E.T.;Ishola,A.O. Lactobacillusplantarum MitigatesSexual-ReproductiveDeficitsbyModulatingInsulinReceptorExpressionintheHypothalamic-Pituitary-TesticularAxis ofHyperinsulinemicMice. DrugMetab.Pers.Ther. 2021.[CrossRef][PubMed]
20. Passos,F.V.;Fleming,H.P.;Ollis,D.F.;Felder,R.M.;McFeeters,R.F.KineticsandModelingofLacticAcidProductionby Lactobacillusplantarum Appl.Environ.Microbiol. 1994, 60,2627–2636.[CrossRef][PubMed]
21. Nguyen,T.;Kang,J.H.;Lee,M.S.Characterizationof Lactobacillusplantarum PH04,aPotentialProbioticBacteriumwithCholesterolLoweringEffects. Int.J.FoodMicrobiol. 2007, 113,358–361.[CrossRef]
22. Zheng,Z.;Cao,F.;Wang,W.;Yu,J.;Chen,C.;Chen,B.;Liu,J.;Firrman,J.;Renye,J.;Ren,D.ProbioticCharacteristicsof Lactobacillusplantarum E680anditsEffectonHypercholesterolemicMice. BMCMicrobiol. 2020, 20,1–9.[CrossRef]
23. Prakoeswa,C.;Herwanto,N.;Prameswari,R.;Astari,L.;Sawitri,S.;Hidayati,A.N.;Indramaya,D.M.;Kusumowidagdo,E.R.; Surono,I.S. Lactobacillusplantarum IS-10506SupplementationReducedSCORADinChildrenwithAtopicDermatitis. Benef. Microbes 2017, 8,833–840.[CrossRef]
24. Nagata,Y.;Yoshida,M.;Kitazawa,H.;Araki,E.;Gomyo,T.ImprovementsinSeasonalAllergicDiseasewith Lactobacillusplantarum no.14. Biosci.Biotechnol.Biochem. 2010, 74,1869–1877.[CrossRef]
25. Wang,J.;Zeng,Y.;Wang,S.;Liu,H.;Zhang,D.;Zhang,W.;Wang,Y.;Ji,H.Swine-DerivedProbiotic Lactobacillusplantarum Inhibits GrowthandAdhesionofEnterotoxigenic Escherichiacoli andMediatesHostDefense. Front.Microbiol. 2018, 9,1364.[CrossRef]
26. Dinev,T.;Beev,G.;Tzanova,M.;Denev,S.;Dermendzhieva,D.;Stoyanova,A.AntimicrobialActivityof Lactobacillusplantarum againstPathogenicandFoodSpoilageMicroorganisms:AReview. Bulg.J.Vet.Med. 2018, 21,253–268.[CrossRef]
27. Valdez,J.C.;Peral,M.C.;Rachid,M.;Santana,M.;Perdigon,G.Interferenceof Lactobacillusplantarum with Pseudomonasaeruginosa inVitroandinInfectedBurns:ThePotentialuseofProbioticsinWoundTreatment. Clin.Microbiol.Infect. 2005, 11,472–479. [CrossRef]
28. Chen,Y.E.;Fischbach,M.A.;Belkaid,Y.SkinMicrobiota–hostInteractions. Nature 2018, 553,427–436.[CrossRef][PubMed]
29. Tsai,W.;Chou,C.;Chiang,Y.;Lin,C.;Lee,C.RegulatoryEffectsof Lactobacillusplantarum-GMNL6onHumanSkinHealthby ImprovingSkinMicrobiome. Int.J.Med.Sci. 2021, 18,1114.[CrossRef][PubMed]
30. Nam,B.;Kim,S.A.;Park,S.D.;Kim,H.J.;Kim,J.S.;Bae,C.H.;Kim,J.Y.;Nam,W.;Lee,J.L.;Sim,J.H.RegulatoryEffectsof Lactobacillusplantarum HY7714onSkinHealthbyImprovingIntestinalCondition. PLoSONE 2020, 15,e0231268.[CrossRef] [PubMed]
31. Kolarsick,P.A.;Kolarsick,M.A.;Goodwin,C.AnatomyandPhysiologyoftheSkin. J.Dermatol.NursesAssoc. 2011, 3,203–213. [CrossRef]
32. Kammeyer,A.;Luiten,R.M.OxidationEventsandSkinAging. AgeingRes.Rev. 2015, 21,16–29.[CrossRef]
33. Scharffetter–Kochanek,K.;Brenneisen,P.;Wenk,J.;Herrmann,G.;Ma,W.;Kuhr,L.;Meewes,C.;Wlaschek,M.Photoagingofthe SkinfromPhenotypetoMechanisms. Exp.Gerontol. 2000, 35,307–316.[CrossRef]
34. Cole,M.A.;Quan,T.;Voorhees,J.J.;Fisher,G.J.ExtracellularMatrixRegulationofFibroblastFunction:RedefiningourPerspective onSkinAging. J.CellCommun.Signal. 2018, 12,35–43.[CrossRef][PubMed]
35. Pittayapruek,P.;Meephansan,J.;Prapapan,O.;Komine,M.;Ohtsuki,M.RoleofMatrixMetalloproteinasesinPhotoagingand Photocarcinogenesis. Int.J.Mol.Sci. 2016, 17,868.[CrossRef][PubMed]
36. Verdier-Sévrain,S.;Bonté,F.SkinHydration:AReviewonitsMolecularMechanisms. J.Cosmet.Dermatol. 2007, 6,75–82. [CrossRef]
37. Sender,R.;Fuchs,S.;Milo,R.RevisedEstimatesfortheNumberofHumanandBacteriaCellsintheBody. PLoSBiol. 2016, 14, e1002533.[CrossRef]
38. Grice,E.A.;Segre,J.A.TheSkinMicrobiome. Nat.Rev.Microbiol. 2011, 9,244–253.[CrossRef]
39. Orland,C.;Emilson,E.J.;Basiliko,N.;Mykytczuk,N.C.;Gunn,J.M.;Tanentzap,A.J.MicrobiomeFunctioningDependson IndividualandInteractiveEffectsoftheEnvironmentandCommunityStructure. ISMEJ. 2019, 13,1–11.[CrossRef][PubMed]
40. Findley,K.;Grice,E.A.TheSkinMicrobiome:AFocusonPathogensandtheirAssociationwithSkinDisease. PLoSPathog. 2014, 10,e1004436.[CrossRef]
41. Byrd,A.L.;Belkaid,Y.;Segre,J.A.TheHumanSkinMicrobiome. Nat.Rev.Microbiol. 2018, 16,143–155.[CrossRef][PubMed]
42. Macia,L.;Nanan,R.;Hosseini-Beheshti,E.;Grau,G.E.Host-andMicrobiota-DerivedExtracellularVesicles,ImmuneFunction, andDiseaseDevelopment. Int.J.Mol.Sci. 2020, 21,107.[CrossRef]
43. Wang,A.S.;Dreesen,O.BiomarkersofCellularSenescenceandSkinAging. Front.Genet. 2018, 9,247.[CrossRef][PubMed]
44. McGrath,J.A.;Uitto,J.TheFilaggrinStory:NovelInsightsintoSkin-BarrierFunctionandDisease. TrendsMol.Med. 2008, 14, 20–27.[CrossRef]
45. Papakonstantinou,E.;Roth,M.;Karakiulakis,G.HyaluronicAcid:AKeyMoleculeinSkinAging. Derm.-Endocrinol. 2012, 4, 253–258.[CrossRef][PubMed]
46. Baumann,L.SkinAgeinganditsTreatment. J.Pathol.AJ.Pathol.Soc.GreatBr.Irel. 2007, 211,241–251.[CrossRef]
47. Fujimura,T.;Haketa,K.;Hotta,M.;Kitahara,T.LossofSkinElasticityPrecedestoRapidIncreaseofWrinkleLevels. J.Dermatol. Sci. 2007, 47,233–239.[CrossRef]
48. Leyden,J.J.ClinicalFeaturesofAgeingSkin. Br.J.Dermatol. 1990, 122,1–3.[CrossRef]
49. Rawlings,A.V.;Harding,C.R.MoisturizationandSkinBarrierFunction. Dermatol.Ther. 2004, 17,43–48.[CrossRef][PubMed]
50. Helfrich,Y.R.;Sachs,D.L.;Voorhees,J.J.OverviewofSkinAgingandPhotoaging. Dermatol.Nurs. 2008, 20,177–183.
51. Kosmadaki,M.G.;Gilchrest,B.A.TheRoleofTelomeresinSkinAging/Photoaging. Micron 2004, 35,155–159.[CrossRef]
52. Blasco,M.A.TelomereLength,StemCellsandAging. Nat.Chem.Biol. 2007, 3,640–649.[CrossRef]
53. Gilchrest,B.A.;Eller,M.S.;Yaar,M.Telomere-MediatedEffectsonMelanogenesisandSkinAging. J.Investig.Dermatol.Symp. Proc. 2009, 14,25–31.[CrossRef]
54. Li,Z.;Bai,X.;Peng,T.;Yi,X.;Luo,L.;Yang,J.;Liu,J.;Wang,Y.;He,T.;Wang,X.NewInsightsintotheSkinMicrobialCommunities andSkinAging. Front.Microbiol. 2020, 11,2603.[CrossRef][PubMed]
55. Zapata,H.J.;Quagliarello,V.J.TheMicrobiotaandMicrobiomeinAging:PotentialImplicationsinHealthandAge-related Diseases. J.Am.Geriatr.Soc. 2015, 63,776–781.[CrossRef][PubMed]
56. Shibagaki,N.;Suda,W.;Clavaud,C.;Bastien,P.;Takayasu,L.;Iioka,E.;Kurokawa,R.;Yamashita,N.;Hattori,Y.;Shindo,C. Aging-RelatedChangesintheDiversityofWomen’sSkinMicrobiomesAssociatedwithOralBacteria. Sci.Rep. 2017, 7,10567. [CrossRef]
57. Glady,A.;Vandebroek,A.;Yasui,M.HumanKeratinocyte-DerivedExtracellularVesiclesActivatetheMAPKinasePathwayand PromoteCellMigrationandProliferationinVitro. Inflamm.Regen. 2021, 41,4.[CrossRef][PubMed]
