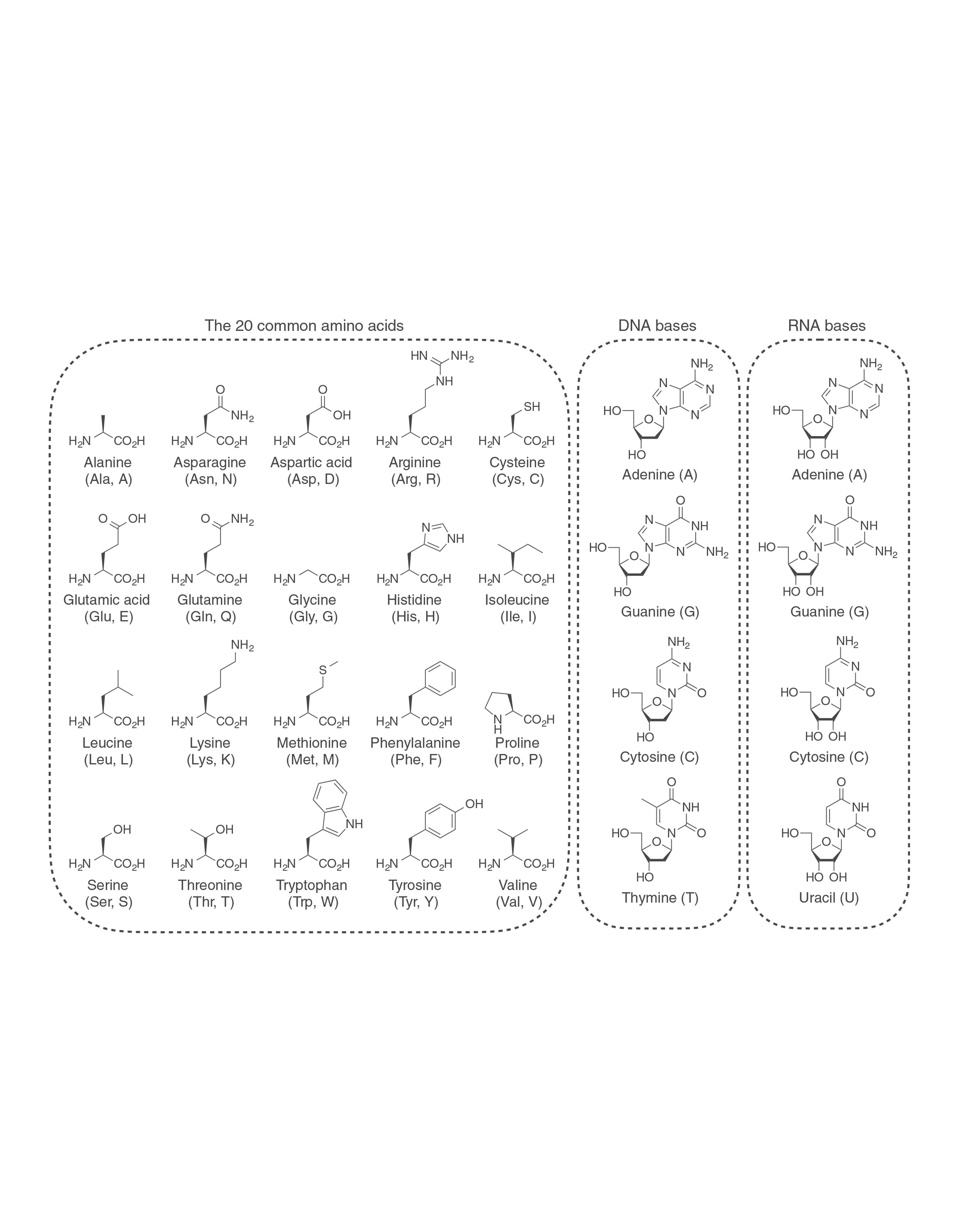
AdvancedChemicalBiology
ChemicalDissectionandReprogrammingofBiologicalSystems
EditedbyHowardC.Hang,MatthewR.Pratt,andJenniferA.Prescher
Editors
HowardC.Hang
ScrippsResearch DepartmentsofImmunology& MicrobiologyandChemistry 10550NorthTorreyPinesRoad LaJolla,CA,USA92037
MatthewR.Pratt
UniversityofSouthernCalifornia DepartmentofChemistry 3430S.VermontAve. CA UnitedStates
JenniferA.Prescher UniversityofCalifornia,Irvine DepartmentofChemistry 1120NaturalSciencesII CA UnitedStates
Coverimage: ©Shutterstock
Allbookspublishedby WILEY-VCH arecarefullyproduced. Nevertheless,authors,editors,andpublisherdonotwarrant theinformationcontainedinthesebooks,includingthisbook, tobefreeoferrors.Readersareadvisedtokeepinmindthat statements,data,illustrations,proceduraldetailsorotheritems mayinadvertentlybeinaccurate.
LibraryofCongressCardNo.: appliedfor BritishLibraryCataloguing-in-PublicationData AcataloguerecordforthisbookisavailablefromtheBritish Library.
Bibliographicinformationpublishedby theDeutscheNationalbibliothek TheDeutscheNationalbibliothekliststhispublicationinthe DeutscheNationalbibliografie;detailedbibliographicdataare availableontheInternetat <http://dnb.d-nb.de>
©2023WILEY-VCHGmbH,Boschstr.12,69469Weinheim, Germany
Allrightsreserved(includingthoseoftranslationintoother languages).Nopartofthisbookmaybereproducedinany form–byphotoprinting,microfilm,oranyothermeans–nor transmittedortranslatedintoamachinelanguagewithout writtenpermissionfromthepublishers.Registerednames, trademarks,etc.usedinthisbook,evenwhennotspecifically markedassuch,arenottobeconsideredunprotectedbylaw.
PrintISBN: 978-3-527-34733-9
ePDFISBN: 978-3-527-82629-2
ePubISBN: 978-3-527-82630-8
CoverDesign AdamDesign,Weinheim,Germany
Typesetting Straive,Chennai,India
Contents
Foreword xv
Preface xvii
AbouttheCompanionWebsite xix
1IntroductiontoAdvancedChemical Biology 1
HowardC.Hang,MatthewR.Pratt,and JenniferA.Prescher
1.1Introduction 1
1.2EnabledbySyntheticandPhysicalOrganic Chemistry 1
1.3GuidedbyBiochemistryandStructural Biology 3
1.4EnhancedbyEngineeringandEvolution 3
1.5ExpandedbyAnalyticalChemistryand “Omics”Technologies 4
1.6ImpactonBiologicalDiscoveryandDrug Development 5
1.7Outlook 5 References 6
2DNAFunction,Synthesis,and Engineering 9 AneeshT.VeetilandYamunaKrishnan
2.1Introduction:AHistoricalPerspective 9
2.1.1TheStructureofDNA 9
2.2NewNucleobasesandUnusualDNA Conformations 11
2.2.1G-QuadruplexDNAStructures 11
2.2.2CircularDNAStructures 11
2.2.3Aptamers 11
2.2.4OtherNucleobases 12
2.3TheModernSynthesisofDNA 13
2.3.1Solid-PhaseDNASynthesis 13
2.3.2Backbone-ModifiedOligonucleotides 15
2.3.2.1PeptideNucleicAcids(PNAs) 16
2.3.2.2MorpholinoNucleicAcids 16
2.4DNASequencing 16
2.4.1ModernMethodstoSequenceDNA 16
2.4.1.1SequencingbySynthesis(SBS) 17
2.4.1.2Third-GenerationDNASequencing 17 2.5DNAEngineering 18
2.5.1DNANanotechnology 18
2.5.2DNA-TemplatedNanoparticleAssembly 19
2.5.3DNANanomachines 20
2.5.4DNANanotechnologyforBiology 20
2.5.5DNA-BasedOrganelleMapping Technology 20
2.5.6DNA-BasedTechnologiesfortheDetectionof EndogenousNucleicAcidsandProteins 22
2.5.6.1Fluorescence InSitu Hybridization (FISH) 22
2.5.6.2DNA-BarcodedAntibodiesforSpatial DetectionofProteins 22
2.5.7DNA-BasedSuperResolutionImaging 22
2.5.8DNA-EncodedLibraries(DEL) 23
2.5.9DigitalDataStorageUsingDNA 23
2.6ToolsforEngineeringDNA 24
2.7SummaryandFutureOutlook 25 Acknowledgments 25 Questions 25 References 26
3ChemicalApproachestoGenome Integrity 31
ElizabethR.Lotsof,SavannahG.Conlon,and SheilaS.David
3.1IntroductionandHistoricalPerspective 31
3.2TypesofDNADamage 32
3.2.1DamagetoNucleobase 32
3.2.1.1Oxidation 32
3.2.1.2Alkylation 33
3.2.1.3Depurination/Depyrimidination 35
3.2.1.4Deamination 35
3.2.1.5DNAMismatches 35
3.2.1.6DNACrosslinks 35
3.2.2DamagetoSugar 36
3.2.3DamagetoPhosphateBackbone 36
3.3TypesofDNARepair 36
3.3.1DirectRepair 36
3.3.2BaseExcisionRepair 38
3.3.3NucleotideExcisionRepair 38
3.3.4MismatchRepair 39
3.3.5Double-StrandBreakandInterstrand CrosslinkRepair 39
3.4IdentificationofSitesofDNADamageand Modification 40
3.4.1TraditionalMethodsforDamage Detection 40
3.4.2SearchingforHotspotsofOxidative Damage–AnOGStory 41
3.4.3SequencingforBulkyAdducts–Cisplatinand PyrimidineDimers 41
3.4.4SequencingforAPSiteandStrandBreaks 44
3.5AssaysthatAllowforMonitoringofthe RepairofDNADamageinCellular Contexts 44
3.5.1LesionReporterAssaystoMonitorBase ExcisionRepair 45
3.5.2LeveragingCell-BasedReporterAssaysto AssessImpactofDNALesionsonReplication andTranscription 47
3.5.3PlasmidReportersMonitoringSeveralDNA RepairPathwaysSimultaneously 47
3.5.4HighlySensitiveFluorescentDNARepair ProbesforClinicalDiagnosticsandImaging inCells 48
3.6SummaryandFutureOutlook 48 Acknowledgments 48 ExamQuestions 48 References 50
4RNAFunction,Synthesis,and Probing 55
AndreasPintado-UrbancandMatthewD.Simon
4.1Introduction 55
4.2ThePrinciplesofRNAChemistry 56
4.2.1TheImpactofa2′ -HydroxylonNucleicAcid Chemistry 56
4.2.2RNABasesandBase-Pairing 56
4.2.3RNASecondaryStructure 58
4.2.4RNATertiaryStructuresandthe Ribosome 58
4.3SynthesisofRNA 58
4.3.1ChemicalSynthesis 58
4.3.2 InVitro Transcription 59
4.4LabelingofRNA 60
4.4.1IntroducingModificationsThroughChemical SynthesisofRNA 60
4.4.2UsingLigationtoIntroduceChemical ModificationsintoRNA 60
4.4.3IncorporationofModifiedBasesintoRNA UsingIVT 61
4.4.4Approachesto3′ -EndLabelRNA 62
4.4.5Approachesto5′ -EndLabelRNA 62
4.5IdentificationandEngineeringofFunctional RNAs 62
4.5.1Aptamers 62
4.5.2Riboswitches 63
4.5.3Ribozymes 63
4.5.4GeneticallyEncodedTagstoLabelRNA 64
4.5.5RNA-BasedTherapeutics 64
4.6TheSequencingofRNA 64
4.6.1ReverseTranscriptionofRNA 65
4.6.2Long-ReadandDirectRNASequencing 65
4.6.3ExtensionsandAlternativeApproachesto RNA-seq 66
4.7TheChemicalProbingofRNAStructure 66
4.7.1In-LineProbingofRNAConformation 67
4.7.2ReagentsforChemicalProbingofRNA ConformationandBase-Pairing 67
4.7.3ReagentsforProbingSolventAccessibility, TertiaryStructure,andHigherOrder Interactions 68
4.8SummaryandFutureOutlook 69 Questions 69 References 69
5ChemicalApproachesto TranscriptionandRNARegulation InVivo 75 TongWuandChuanHe
5.1Introduction/HistoricalPerspective 75
5.2CoreConcepts/LandmarkStudies 75
5.2.1TranscriptionRegulationinEukaryotes 75
5.3TranscriptionRegulationbyChemical TargetingofDNAandtheCoreTranscription Machinery 76
5.3.1Cell-PermeableDNA-TargetingSmall Molecules 76
5.3.2TargetingTranscriptionbyNucleicAcidsand TheirAnalogs 79
5.3.3Small-MoleculeInhibitorsofthe TranscriptionMachinery 80
5.4ChemicalRegulationofTranscriptionvia TargetingofEpigeneticElements 81
5.4.1TranscriptionRegulationThroughTargeting ofHistoneModifications 81
5.4.2DNAMethylationandSmallMolecules TargetingDNAModifications 83
5.5ChemicalApproachestoTarget Post-TranscriptionalRNAMetabolism 86
5.5.1Post-TranscriptionalRNAMetabolism 86
5.5.2RegulatingRNAFunctionbyDirectRNA Binders 89
5.5.3RegulatingRNAFunctionbyTargeting RNA-Binding(Effector)Proteins 91
5.6SummaryandFutureOutlook 91 Questions 92 References 92
6ChemicalBiologyofGenome Engineering 99 CarlosA.VasquezandAlexisC.Komor
6.1IntroductiontoGenomeEditing 99
6.2EarlyGeneticEngineeringExperiments: ChemicalMutagenesis,GeneTransfer,and GeneTargeting 100
6.2.1ChemicalMutagenesisMethods 100
6.2.2GeneTransfer 101
6.2.3GeneTargeting 102
6.3ImprovingPrecisionandProgrammability withDouble-StrandedDNABreaks 103
6.3.1TheDevelopmentofDouble-Stranded Break-ReliantGenomeEditing Technologies 103
6.3.2RepairofDouble-StrandedDNABreaksin MammalianCells 104
6.3.3Meganucleases 105
6.3.4ZincFingerNucleases(ZFNs) 106
6.3.5TranscriptionActivator–LikeEffector Nucleases(TALENs) 108
6.4TheGoldenAgeofGenomeEngineering: CRISPR-BasedGenomeEditing Technologies 108
6.4.1Introduction 108
6.4.2CRISPR-Cas9 109
6.4.3ProgrammabilityImprovements 111
6.4.4EfficiencyImprovements 113
6.4.5SpecificityImprovements 113
6.4.6PrecisionImprovements 115
6.4.7EpigenomeEditing 115
6.5Non-DSB-ReliantGenomeEditing Technologies 116
6.5.1BaseEditing 116
6.5.2PrimeEditing 119
6.6GeneEditingMethodsforSpatialand TemporalControl 119
6.7EthicalImplications,Summary,andFuture Outlook 121
Questions 123 References 123
7PeptideSynthesisand Engineering 135 GordonC.BrownandParamjitS.Arora
7.1Introduction 135
7.2PeptideSynthesis 135
7.2.1SPPSIsOptimizedforStepwise Efficiency 135
7.2.2Nα -protectingGroupsEnsureSingle CouplingoftheIncomingAminoAcid 136
7.2.3PlasticResinsAreUsedDuringSPPS 138
7.2.4TemporaryMaskingofReactiveSideChains IsNecessaryDuringSPPS 138
7.2.5PeptideBondsAreSynthesizedbya CondensationReactionMediatedbya StoichiometricCouplingAgent 140
7.3SecondaryandTertiaryStructuresofAmino Acids 142
7.3.1PeptideBackboneConformations 142
7.3.2BiophysicalDeterminantsofHelixFolding andDesignof α-HelixMimics 143
7.3.3 β-Strandand β-SheetMimics 145
7.3.4ProteinTertiaryStructureMimics 147
7.3.4.1 β-Sheetand β-HairpinMimics 147
7.3.5HelicalTertiaryStructureMimics 149
7.4ConformationallyDefinedPeptidesas ModulatorsofProteinInteractions 150
7.4.1PeptideTherapeutics 151
7.5SummaryandFutureOutlook 157 Questions 157 References 158
8ProteinSynthesisand Engineering 167 MatthewR.PrattandTomW.Muir
8.1Introduction/HistoricalPerspective 167
8.2CoreConcepts/LandmarkStudies 170
8.2.1Cysteine-thioester-BasedLigations:Making thePieces 170
8.2.1.1C-terminalPieces:ChemicalSynthesisof N-terminalCysteinePeptides 170
8.2.1.2C-terminalPieces:RecombinantExpression ofN-terminalCysteinePeptides/ Proteins 170
8.2.1.3N-terminalPieces:ChemicalSynthesisof Thioester-ContainingPeptides 171
8.2.1.4N-terminalPieces:RecombinantExpression ofThioester-ContainingProteins 172
8.2.1.5InternalFragments:PreparationofCysteine andThioester-ContainingPeptides/ Proteins 172
8.2.2AddingMorePieces:MovingBeyond Thioester/CysteineLigations 173
8.2.2.1Desulfurization 174
8.2.2.2Auxiliaries 174
8.2.2.3OtherLigationChemistries 176
8.3PuttingthePiecesTogether:Practical ConsiderationsforLigationReactions 178
8.4ProteinTrans-splicing 178
8.5ExamplesofProteinSynthesis 179
8.5.1Post-TranslationalModifications 179
8.5.1.1CellSignaling 179
8.5.1.2Chromatin 181
8.5.1.3Amyloid-FormingProteins 181
8.5.2ChemicalandBiophysicalProbes 182
8.5.2.1BackboneModifications 182
8.5.2.2SegmentalIsotopicLabeling 182
8.5.3MirrorImageProteins 184
8.5.3.1RacemicCrystallography 184
8.5.3.2MirrorImageDisplay 184
8.5.4ProteinLigationinLivingSystems 184
8.5.5PotentialTherapeuticApplications 184
8.6SummaryandFutureOutlook 185 Questions 185 References 186
9DirectedEvolutionforChemical Biology 193 PuXue,FangGuo,LinzixuanZhang,and HuiminZhao
9.1Introduction 193
9.2Methodologies 195
9.2.1DirectedEvolutionattheProteinLevel 195
9.2.1.1RandomMutagenesis 195
9.2.1.2GeneRecombination 195
9.2.1.3Semi-RationalDesign 197
9.2.2DirectedEvolutionatthePathwayLevel 198
9.2.2.1DirectedEvolutionofaSingleEnzymein aPathway 198
9.2.2.2DirectedEvolutionofanEntire Pathway 199
9.2.3DirectedEvolutionattheGenome Level 199
9.2.3.1AdaptiveLaboratoryEvolution 199
9.2.3.2Genome-ScaleEngineeringStrategies 200
9.2.4ContinuousDirectedEvolution 200
9.2.5ScreeningorSelectionMethods 201
9.2.5.1Selection-BasedTechniques 202
9.2.5.2Screening-BasedTechniques 203
9.3CaseStudies 203
9.3.1DirectedEvolutionofaGlyphosate N -Acetyltransferase 203
9.3.2DirectedEvolutionofaTransaminasefor SitagliptinManufacture 207
9.3.3DirectedEvolutionofaCytokineUsingDNA FamilyShuffling 208
9.3.4EfficientProximityLabelinginLivingCells andOrganismswithTurboID 210
9.3.5BiocatalyticCascadeEvolutionfor ManufacturingIslatravir 211
9.3.6AMulti-FunctionalGenome-WideCRISPR System 212
9.4FuturePerspectivesandConclusion 212 Acknowledgments 213 Questions 213 References 214
10ChemicalBiologyofCellular Metabolism 221 PeterC.GrayandAlanSaghatelian
10.1Introduction/HistoricalPerspective 221
10.2MetaboliteDetectionandQuantitation 223
10.2.1ShotgunMetabolomics 223
10.2.2TargetedMetabolomics 225
10.2.3MetaboliteFluxAnalysis 225
10.2.4UntargetedMetabolomics 227
10.2.5DiscoveringStructurallyNovel Metabolites 228
10.3MetaboliteImagingandSensing 229
10.3.1MassSpectrometryImaging 229
10.3.2ChemicalProbesforMetabolite Imaging 230
10.3.3ProteinandRNAMetaboliteSensors 232 10.4PerturbationofMetaboliteLevels 234
10.4.1Small-MoleculeInhibitorsandDrugsof Metabolism 234
10.4.2EnzymaticPerturbationofMetabolism 235
10.5TheImpactofChemicalBiologyinDisease andDrugDiscovery 236
10.6SummaryandFutureOutlook 237 Questions 238 References 238
11ChemicalBiologyofLipids 243 ScotlandFarley,AlixThomas,AurélienLaguerre, andCarstenSchultz
11.1Introduction 243
11.2IdentificationofBulkLipids 245
11.2.1LipidomicsbyMassSpectrometry 245
11.2.2LipidAnalysisbyThin-Layer Chromatography 247
11.3FixingLipidsinSubcellularSpace 247
11.3.1Protein-BasedTechniquestoLocalize Lipids 248
11.3.2MassSpectrometryImagingofLipids 249
11.3.3LipidDetectionUsingModifiedLipidsas Probes 249
11.4TracingIndividualLipidsviaInCelluloClick Chemistry 250
11.4.1Alkyne/Azide-ModifiedLipidsandClick Chemistry 251
11.4.2BifunctionalLipidDerivatives 251
11.5ToolstoElucidateLipidSignaling 253
11.5.1MetabolicMachineryasaChemicalTool: theAdvantageofChemicalDimerizers 253
11.5.2ReleasingBioactiveLipidswithLight 255
11.6AComprehensiveViewofProtein–Lipid Interactions 256
11.6.1TrifunctionalLipids 256
11.6.2Lipid–ProteinInteractome 258
11.7SummaryandFutureOutlook 258 Questions 259 References 259
12ProteinPosttranslational Modifications 267 SamWhedonandPhilipA.Cole
12.1Introduction 267
12.2FunctionalImpactsofPTMs 268
12.3EvolutionandPTMs 270
12.4MajorClassesofPTMs 270
12.4.1Phosphorylation 270
12.4.2Acetylation 272
12.4.3Ubiquitination 273
12.4.4Methylation 275
12.4.5Glycosylation 275
12.4.6LipidationofProteins 278
12.4.7OxidationofProteins 278
12.4.8MiscellaneousModifications 278
12.5WritersandErasers 280
12.5.1ProteinKinasesandPhosphatases 280
12.5.2AcetyltransferasesandDeacetylases 280
12.5.3UbiquitinLigasesand Deubiquitinases 281
12.5.4MethylationandDemethylases 281
12.5.5Glycosyltransferasesand Glycosidases 282
12.5.6LipidTransferaseandHydrolases 282
12.6StrategiesfortheStudyofPTMs 283
12.6.1Mutagenesis 283
12.6.2GeneticCodonExpansion 283
12.6.3Small-MoleculeProbesandChemical Complementarity 283
12.6.4ChemicalLigation 284
12.6.5ProteinMicroarrays 284
12.7ProteinPTMsinDiseases 284
12.7.1ProteinKinasesandDiseases 285
12.7.2LysAcetylationandCutaneousTCell Lymphoma 285
12.7.3Ubiquitination 285
12.8Summary 286 Questions 286 References 287
13ChemicalGlycobiology 295 AmélieM.Joffrin,AlexanderW.Sorum,and LindaC.Hsieh-Wilson
13.1Introduction 295
13.2TotalChemicalSynthesisof StructurallyDefinedGlycans 297
13.3EnzymaticandChemoenzymaticSynthesisof Glycans 300
13.4ProgrammableandAutomatedGlycan Synthesis 302
13.5SynthesisofGlycopeptidesand Glycoproteins 303
13.6GlycanMicroarrays 305
13.7ChemicalTaggingandRemodelingofCellular Glycans 306
13.8InhibitorsofGlycan-ProcessingEnzymesand GlycanBindingProteins 310
13.9Glycan-TargetedTherapeutics 313
13.10SummaryandFutureOutlook 316 Questions 317 References 317
14TheChemicalandEnzymatic ModificationofProteins 329 NicholasS.Dolan,JohnathanC.Maza, AlexandraV.Ramsey,andMatthewB.Francis 14.1Introduction 329
14.2GeneralConsiderations 329
14.3LysineModification 330
14.4AsparticAcid,GlutamicAcid,and C-TerminalCarboxylateModification 333
14.5TyrosineModification 334
14.6CysteineModification 337
14.7MethionineModification 340
14.8TryptophanModification 341
14.9HistidineModification 343
14.10SerineandThreonineModification 344 14.11N-TerminalModification 344
14.12EnzymaticApproachestoModifying Proteins 347
14.12.1Transpeptidases 347 14.12.2Ligases 348
14.12.3ActivatingEnzymes 349 14.13SummaryandFutureOutlook 350 Questions 350 References 352
15GeneticCodeExpansion 359 PengR.Chen,ShixianLin,andJieP.Li
15.1Introduction 359
15.2GeneticCodeExpansionThroughDirected EvolutionofaaRS/tRNAPairs 359
15.2.1TheDevelopmentoftheMj.TyrRS-tRNA BasedGCESystem 360
15.2.2TheDevelopmentofAdditional aaRS/tRNA-BasedGCESystem 362
15.2.3ThePylRS-tRNAPairasa“one-stop-shop” GCESystem 362
15.2.4GeneticCodeExpansioninMulticellular Organisms 363
15.3GCEwithGenomeRecodingStrainsand/or UnnaturalCodons 364
15.3.1GCEwithGenomeRecodingStrains 364
15.3.2GCEwithFour-BaseCodonsUsing OrthogonalRibosome 366
15.3.3GeneticCodeExpansionwithUnnaturalBase Pairs 366
15.4GCE-basedApplications 366
15.4.1Site-SpecificPosttranslationalModifications (PTMs) 367
15.4.2New“Physical”PropertyEmpoweredby ncAAs 369
15.4.3NewChemicalReactivityDerivedfromncAA andTheirUniqueApplications 369
15.4.4ControlofProteinActivation 372
15.5TherapeuticConjugates 374
15.6Live-AttenuatedVirusandOtherGenetically ModifiedVaccines 375
15.7SummaryandFutureOutlook 376
15.7.1ImprovingtheEfficiency 376
15.7.2ExpandingtheApplications 376
15.7.3ExploringtheTherapeuticPotential 377 Questions 377 References 377
16BioorthogonalChemistry 387 JeremyBaskinandPamelaChang
16.1IntroductionandHistorical Perspective 387
16.2KeyConcepts:Bioorthogonalityand BioorthogonalReactions,ClickChemistry, andtheBioorthogonalMetabolicReporter Strategy 388
16.2.1BioorthogonalityandBioorthogonal Reactions 388
16.2.2ClickChemistry 388
16.2.3TheBioorthogonalMetabolicReporter Strategy 389
16.3TheBeginningsofBioorthogonalChemistry: OximeandHydrazoneFormation 390
16.4TheAzideasaBioorthogonalHandle 391
16.5TheStaudingerLigationofAzidesand Phosphines 392
16.6Cu-CatalyzedAzide–AlkyneCycloaddition (CuAAC)ofAzidesandTerminal Alkynes 393
16.7Strain-Promoted[3+2]Azide–Alkyne Cycloaddition(SPAAC)ofAzidesand Cyclooctynes 395
16.8TheTetrazineLigation:RapidBioorthogonal InverseElectron-DemandDiels–Alder Reactions 396
16.9OtherBioorthogonalLigations 398
16.10Light-ActivatedBioorthogonal Reactions 399
16.11BioorthogonalUncagingandCleavage Reactions 400
16.12MutuallyOrthogonalBioorthogonal Reactions 401
16.13FluorogenicBioorthogonalReagents 402
16.14ApplicationsofBioorthogonal Chemistry 403
16.14.1BioorthogonalNon-canonicalAminoAcid Tagging(BONCAT) 403
16.14.2 InVivo ImagingofGlycans 404
16.14.3TherapeuticApplicationsofBioorthogonal Chemistry 404
16.15SummaryandFutureOutlook 406 Questions 406 References 407
17CellularImaging 415
AmyE.PalmerandLukeD.Lavis
17.1Introduction 415
17.1.1History 415
17.1.2LightandFluorescence 417
17.2Small-MoleculeFluorophores 417
17.2.1Background 417
17.2.2PyrenesandCoumarinFluorophores 417
17.2.3BODIPYDyes 418
17.2.4FluoresceinsandRhodamines 418
17.2.5PhenoxazineandCyanineDyes 419
17.2.6UseasBiomoleculeLabels 419
17.2.7UseasCellularStains 419
17.2.8FluorescentIndicators 420
17.2.9EnzymeSubstrates 421
17.3FluorescentProteins 421
17.3.1Background 421
17.3.2GeneralConsiderationsofFluorescent Proteins 421
17.3.3FluorescentProteinsasBiomoleculeLabels and“Stains” 422
17.3.4FluorescentProteinsasSensors 423
17.3.5FluorescentProteinsasEnzyme Substrates 423
17.4HybridSmall-Molecule–Protein Systems 424
17.4.1Background 424
17.4.2Labels 425
17.4.3Sensors 426
17.5LandmarkStudyI:HarnessingPhotosensitive Fluorophores 426
17.5.1Background 426
17.5.2Super-ResolutionMicroscopy 426
17.6LandmarkStudyII:Ca2+ Imaging 427
17.6.1Background 427
17.6.2Small-MoleculeCa2+ Indicators 427
17.6.3GeneticallyEncodedCa2+ Indicators 428
17.6.4GeneticallyEncodedIndicatorsfor InVivo Imaging 428
17.7SummaryandFutureOutlook 430 Questions 430 References 430
18 InVivo Imaging 435 ZiYaoandJenniferA.Prescher
18.1Introduction 435
18.2BasicConceptsforImaging InVivo436
18.3TheImagingToolbox:ProbesforImaging CellularandMolecularFeatures 438
18.3.1“AlwaysOn”Probes 439
18.3.2“Turn-On”(Activatable)Probes 439
18.3.3GeneticallyEncodedProbes 439
18.4MolecularImagingAcrossthe ElectromagneticSpectrum 440
18.4.1ImagingwithX-rays(CT) 440
18.4.2ImagingwithSound(US) 440
18.4.3ImagingwithRadioWaves(MRI) 441
18.4.4ImagingwithRadionuclides (PET/SPECT) 442
18.4.5ImagingwithOpticalLight (Fluorescence/Bioluminescence) 444
18.4.5.1TargetedFluorophoresandFluorescent Materials 445
18.4.5.2ActivatableProbes 445
18.4.5.3GeneticallyEncodedFluorescentProbes 445
18.4.5.4GeneticallyEncodedBioluminescentProteins (Luciferases) 447
18.4.5.5EngineeredProbesforSensingMetabolites andMolecularFeatures 447
18.5MultimodalityImagingandCombination Probes 449
18.6EmergingAreasinMolecularImaging 450
18.7SummaryandFutureOutlook 450 Questions 451 References 451
19ChemicalBiologyofMetals 459 EvaJ.Ge,PatriciaDeLaTorre,and ChristopherJ.Chang
19.1Introduction 459
19.2MetalsandtheInorganicFoundationsof Life 459
19.2.1MetalComplexesareLewisAcid–Base Complexes 459
19.2.2CrystalFieldTheoryEnablesBonding AnalysisfromMolecularShapeandd Orbitals 460
19.2.3HardSoftAcidBaseTheoryDefines Metal–LigandPreferences 462
19.3Non-RedoxRolesforMetalsinBiology: StructureandLewisAcidCatalysis 462
19.3.1MetalsforStabilizingNucleicAcid Structure 463
19.3.2MetalsasProteinStructuralUnits:Zinc FingerandEFHandMotifs 463
19.3.3MetalsasLewisAcidCatalysts: Metallohydrolases 464
19.4RedoxChemistry:OxygenTransportand ElectronTransferProteins 464
19.4.1OxygenTransportRequiresRedox-Active MetalBinding 465
19.4.2MarcusTheoryandElectronTransfer Proteins 465
19.5RedoxChemistry:MetalloenzymesforRedox CatalysisatOxygen,Nitrogen,and Carbon 467
19.5.1OxygenEvolutioninPhotosynthesis: PhotosystemII 467
19.5.2OxygenReduction:Respirationwith Cytochrome c Oxidase 467
19.5.3OxygenCatalysis:HemeandNon-Heme Iron-DependentOxidations 468
19.5.4NitrogenCycle:Nitrogenasesand Nitrate/NitriteReductases 469
19.5.5BioorganometallicChemistry:Carbon CyclingandVitaminB12 469
19.6MetalsinMedicine:Metallotargets, Metallodrugs,andMetal-BasedImaging Agents 470
19.7EmergingAreasforMetalsinBiology: TransitionMetalSignalingand Metalloallostery 472
19.8ChemicalToolstoStudyMetal Biology 472
19.9SummaryandFutureOutlook 475 Questions 475 References 476
20RedoxChemicalBiology 481
YunlongShiandKateS.Carroll
20.1Introduction 481
20.2Activity-BasedDetectionofCysteine Modifications 484
20.3IndirectProfilingofCysteineOxidation 484
20.4DirectProfilingofCysteineOxiPTMswith ChemoselectiveProbes 486
20.4.1ProfilingProteinSulfenicAcids( SOH) 486
20.4.1.1SulfenicAcidProbes–AHistorical Perspective 486
20.4.1.2ChemicalModelsfortheAssessmentof SulfenicAcidProbes 486
20.4.1.3SelectivityofChemicalProbesforSulfenic Acids 486
20.4.1.4QuantificationofProteinSulfenicAcids 489
20.4.1.5ApplicationofSulfenicAcidProbes 492
20.4.2ProfilingProteinSulfinicAcids ( SO2 H) 492
20.4.3ProfilingProteinPersulfides( SSH) 493
20.5ProbesandBiosensorsforReactiveOxygen SpeciesinCells 494
20.6ConclusionsandOutlook 496 Questions 496 References 497
21Activity-BasedProtein Profiling 503 WilliamH.ParsonsandBenjaminF.Cravatt
21.1Introduction/HistoricalPerspective 503
21.2CoreConcepts/LandmarkStudies 504
21.2.1ProbeDesign 504
21.2.1.1ReactiveGroups 504
21.2.1.2ReporterTags 507
21.2.1.3RecognitionGroup/Linker 508
21.2.2DetectionMethods 509
21.2.2.1Gel-BasedAnalysis 509
21.2.2.2FluorescencePolarization 510
21.2.2.3ImagingofProteinsinCellsand Organisms 511
21.2.2.4QuantitativeProteomicsbyMass Spectrometry 511
21.2.3CommonApplications 512
21.2.3.1ProfilingProteinActivityandAminoAcid ReactivityinBiologicalSystemsof Interest 513
21.2.3.2CompetitiveABPPforLigandDiscoveryand Optimization 513
21.2.3.3TargetIdentificationforLigands 515
21.2.3.4AssignmentofEnzymeFunction 516
21.2.3.5VisualizingEnzymeLocalizationandActivity inLivingCellsandOrganisms 517
21.3SummaryandFutureOutlook 518 Questions 518 References 519
22ChemicalGenetics 527 MichaelS.Cohen
22.1Introduction 527
22.2AS-Protein–OrthogonalMolecular Glues 528
22.3AS-Enzyme–OrthogonalSubstrate Pairs 532
22.3.1ProteinKinases 532
22.3.2ProteinMethyltransferases 535
22.3.3ProteinLysineAcetyltransferases 538
22.3.4PARPs 539
22.3.5Glycosyltransferases 542
22.3.6PTMErasers:LysineDemethylases 544
22.3.7BeyondPTMEnzymes 544
22.4AS-Enzyme–OrthogonalInhibitor Pairs 547
22.4.1ProteinKinases 547
22.4.2OtherEnzymes 549
22.5FinalThoughts 549
22.5.1BeyondBump–Hole 549
Questions 550 References 550
23NaturalProductDiscovery 555 MohammadR.Seyedsayamdost,BrettC. Covington,YifanZhang,andYuchenLi
23.1IntroductionandDefinitions 555
23.2KeyConcept:NaturalProductsare GeneticallyEncoded 556
23.3KeyConcept:StructuralDifferencesBetween NaturalProductsandSyntheticDrugs 558
23.4KeyConcept:TargetSpecificityandLatent Reactivity 559
23.5KeyConcept:NaturalProductDiscoveryand Activity-GuidedFractionation 561
23.6KeyConcept:CrypticBiosyntheticGene Clusters 562
23.7LandmarkStudies:PenicillinandtheGolden AgeofAntibioticDiscovery 563
23.8LandmarkStudies:ActivatingSilent BiosyntheticGeneClusters 565
23.8.1ManipulationofCultureConditions 565
23.8.2ClassicalGenetics 566
23.8.3ChemicalGenetics 567
23.8.4HeterologousExpression 568
23.9SummaryandOutlook 569
Questions 570
References 570
24NaturalProductBiosynthesis 575 EunBinGoandYiTang
24.1Introduction 575
24.2PeptideNaturalProducts 577
24.2.1RibosomallySynthesizedand Post-translationallyModifiedPeptides (RiPPs) 577
24.2.2Non-ribosomalPeptides 579
24.3PolyketideNaturalProducts 582
24.3.1BacterialType-IPolyketides 584
24.3.2BacterialType-IIPolyketides 586
24.4TerpeneNaturalProducts 588
24.5HybridandUnnaturalNaturalProducts 591
24.6SummaryandFutureOutlook 592 Acknowledgment 592 Questions 593 References 594
25ChemicalMicrobiology 597 VictoriaM.Marando,StephanieR.Smelyansky, DariaE.Kim,andLauraL.Kiessling
25.1IntroductionandHistory 597
25.2CellEnvelopeStructureand Biosynthesis 598
25.2.1BacterialCellStructure 598
25.3ChemicalandChemoenzymaticSynthesisfor PathwayElucidation 600
25.3.1Peptidoglycan(PG)Biosynthesis 600
25.3.1.1ReconstructingtheStepsinPGBiosynthesis UsingDefinedSubstrates 600
25.3.1.2AccessingLipidsIandII 602
25.3.2CellEnvelopeComponentsBeyond Peptidoglycan 603
25.3.2.1Gram-NegativeLipopolysaccharides 603
25.3.2.2WallTeichoicAcidBiosynthesis 605
25.3.2.3MycobacterialGalactan 605
25.4TheChemicalBiologyofAntibiotic Action 607
25.4.1PGAssemblyIsTargetedbyDiverse Antibiotics 607
25.4.2PenicillinandOtherAntibioticsInduce Dominant-NegativeEffects 609
25.4.3IdentifyingInhibitorsofEssentialEnzymesIs NotEnough 609
25.4.4IdentifyingAttributesforCompoundUptake inBacteria 610
25.5ChemicalBiologyStrategiesforImagingPG AssemblyandRemodeling 610
25.5.1Antibiotic-BasedPGProbes 611
25.5.1.1AntibioticsthatBindPGIntermediates 611
25.5.1.2ProbesfromAntibioticsthatActonEnzymes thatGeneratePG 612
25.5.2SubstrateAnaloguePGProbes 612
25.6LabelingGlycanCellEnvelope Components 613
25.6.1DiversityandFunctionofBacterial Polysaccharides 613
25.6.2ProbesofBacterialGlycans 614
25.6.3ProbesofLPS:AzKdo 615
25.6.4LabelingMycobacterialGlycans 615
25.6.5TrehaloseAnalogs 616
25.6.6ImagingProbes 616
25.6.7FluorogenicProbes 617
25.7ChemicalProbesAppliedtothe Microbiome 617
25.7.1Microbiome:LookingForward 618
25.8SummaryandFutureOutlook 619 Questions 619 References 620
26ChemicalApproachestoAnalyze BiologicalMechanismsand OvercomeResistanceto Therapeutics 629 RudolfPisa,TommasoCupido,and TarunM.Kapoor
26.1Introduction 629
26.2UsingChemicalInhibitorsasToolstoProbe CellularProcesses 630
26.3UsingResistancetoCharacterizeChemical Inhibitors 632
26.4Crash-TestingDrugs 633
26.5RADD–ResistanceAnalysisDuring Design 635
26.6DesigningInhibitorswithDistinctBinding Modes 636
26.7AddressingDrugResistancewithTargeted ProteinDegradation 639
26.8OvercomingResistancebyUsing CombinationsofDrugs 640
26.9Conclusions 641 Questions 641 References 642
27ChemicalDevelopmental Biology 647 JamesK.Chen
27.1Introduction 647
27.2Small-MoleculeTeratogens 648
27.2.1Cyclopamine 648
27.2.2Thalidomide 650
27.3OptochemicalandOptogeneticProbes 653
27.3.1OptochemicalControlofGene Expression 653
27.3.2OptogeneticControlofCellSignaling 657
27.4LineageTracingTools 660
27.4.1ChemicalControlofGenetic Recombination 661
27.4.2DNABarcodingStrategies 664
27.5Summary 665 Questions 665 References 665
28ChemicalImmunology 669 MatthewE.Griffin,JohnTeijaro,and HowardC.Hang
28.1Introduction 669
28.2ChemicalDissectionofAdaptive Immunity 669
28.3GenerationandChemicalEngineeringof Antibodies 672
28.4AntigenRecognitionbyImmune Cells 673
28.5ChemicalInnovationsforElicitingand DiscoveringAntigen-specificImmune Responses 676
28.6ChemicalModulationofInnate Immunity 678
28.7ChemicalDissectionofImmunity 682
28.8SummaryandFutureOutlook 685 Questions 685 References 685
29ChemicalNeurobiology 695 JohannesMorsteinandDirkTrauner
29.1Introduction 695
29.2Actuation 697
29.2.1NeuropharmacologyHasaStoried History 697
29.2.2MolecularCloningandStructuralBiology HaveRevolutionizedtheField 697
29.2.3CagedLigandsandPhotopharmacology AllowforOpticalControlofNeural Activity 699
29.2.4ChemogeneticsEnablesCell-Specific NeuropharmacologyinBrains 701
29.2.5TetheredPharmacologyOperateson EngineeredReceptorsorNativeReceptorsin GeneticallyModifiedCells 703
29.2.6TetheredPhotopharmacologyCombines GeneticwithOpticalControl 704
29.2.7SyntheticPhotoreceptorsCanBeEngineered ThroughGeneticCodeExpansion 705
29.3Visualization 708
29.3.1ChemicalStainingandImagingMethods HaveLaunchedModernNeuroscience 708
29.3.2CalciumImagingCanBeUsedtoMonitor NeuronalActivity 708
29.3.3VoltageSensingProvidesaDirectPictureof NeuronalActivity 708
29.3.4NeurotransmittersCanBeSensedwith ChemogeneticFRETSensors 708
29.3.5MetalsandGasesintheBrainCanBeSensed withFluorescentProbes 709
29.3.6PositronEmissionTomographyRequiresFast Chemistry 712
29.3.7ProximityLigationEnablesSpatiallyResolved MappingofNeuralNetworks 713
29.4SummaryandOutlook 715 Questions 715 References 715
30Small-MoleculeDrug Discovery
723
LukeL.Lairson
30.1Introduction 723
30.2DiscoveryofChemicalMatter 724
30.2.1Target-BasedDiscovery 724
30.2.2HTS-CompatibleAssayFormats 725
30.2.3Phenotype-BasedDiscovery 727
30.2.4HTS:GeneralConsiderations 728
30.2.5DrugRepurposingandSerendipity 729
30.2.6AlternativeSmall-MoleculeDiscovery Approaches 729
30.3 InVivo Pharmacology:InventionofDrug Candidatesand InVivo Probes 730
30.3.1DrugAbsorption,Distribution,Metabolism, andExcretion 731
30.3.1.1DrugAbsorptionandDistribution 731
30.3.1.2PhysicochemicalPropertiesofDrugs 733
30.3.1.3DrugMetabolismandExcretion 734
30.3.1.4Pharmacokinetics,Pharmacodynamics,and Biomarkers 737
30.3.2MedicinalChemistry 739
30.3.3DrugToxicityandHumanClinical Trials 743
30.4Conclusion 744 Questions 744 References 746
Index 751
Foreword
CarolynR.Bertozzi
Icameofageasascientistduringatimewhentheboundariesbetweenthehistoricallyseparatefieldsofchemistry andbiologywerebeingdismantled.Themolecularbiologyrevolutionofthe1980shadbroughtnewfoundpowerto thelifescientist,allowingbiologicalsystemstobeengineeredandmanipulatedtoanswerquestionsaboutmolecularmechanism,ratherthansimplyobserved.High-resolutionmicroscopyandstructuralbiologytechniquesoffered atomicviewsofbiologicalmolecules,complexes,andmaterials,bringingbiologyeverclosertothescaleatwhich chemistsoperate.Atthesametime,chemistrywaspoweringbiologyatrecordpace:solid-phasepeptideandoligonucleotidesynthesiswererevolutionizingourunderstandingofthesebiomolecules’structuresandfunctions,andalso propellingadvancesingenomesequencingandengineering.Thesyntheticchemist’sabilitytosynthesizecomplex naturalproductsprovidedpharmacologicaltoolsthatrevealedthesecretsofthecell,whileanalyticalchemistrytechnologies,quiteprominentlymassspectrometry,providedunprecedentedclarityonthemolecularcompositionsof biologicalsamples.Thenotionthatchemistscoulddesignmoleculestoprobeorperturbabiologicalprocesswas becomingwidelyrecognizedamongbiologists,andlikewise,historicallyintractablebiologicalproblemshadbecome compellingchallengesforchemists.Inretrospect,mytrainingyears(i.e.thelate1980sandearly1990s)wereafantasticperiodforayoungscientisttopursueresearchattheburgeoninginterfaceofchemistryandbiology!
Sincethoseearlydays,Ihavewatchedthetwofieldscoevolvetocreatethedistinctivedisciplinewenowcallchemicalbiology.Thisevolutionwasnotwithoutfriction.Intheearlydays,veryfewlabspossesseddepthofknowledge andtechnicalknowhowinbothchemistryandbiology.Indeed,itwastherarechemistwhounderstoodtheneedsof biologyandtherarebiologistwhounderstoodthepowerofchemistry;gettingthetwotogetherascollaboratorswas keytoprogressinthefield.Meanwhile,traineeswhosoughttodevelopskillsinbothdisciplineswereoftenmisunderstood,orevenworse,mischaracterizedas“Jacksofalltrades,mastersofnone.”Pioneersatthisexcitinginterface hadtoprovethemselvesseparatelyaschemistsandbiologistswhilealsocreatingtheethosofadistinctivenewfield.
Now,severaldecadesintomyowncareerasachemicalbiologist,Iamdelightedtoseeourfieldplayingacentralrole acrossacademiaandindustry.Wearethegluethatbindschemistsandbiologiststogether,thebilingualinterpreters thatcatalyzecross-pollinationofideasandtechnologies.Andwemakeourownfundamentaldiscoveriesinbiology thatareuniquelyenabledbyourchemicaltools,whilealsodevelopingbiologicaltoolsforbetter,greener,chemical processes.Manybiopharmacompanieswhowereskepticalofourvalueafewdecadesbacknowhostso-namedchemicalbiologygroupsthatcutacrossplatformsandtherapeuticareas.Oursuperpowersasmultidisciplinaryscientists arerecognized,andwearerightfullyinhighdemand.
Whiletheprofessionalpracticeofchemicalbiologyhasbeencodified,themechanismsbywhichwetrainstudents inthisdisciplinecontinuetoevolve.Manyofusacademicsteachcoursesinchemicalbiologythatarerather ad hoc,oftenbasedonprimaryliteraturethathappenstoalignwithourinterests.Asthefieldhasgrowninscopeand participation,sohastheneedformorestructuredandcomprehensiveresourcesonwhichsuchcoursescanbebased. Forthisreason,Iamdelightedtocelebratethisbook, AdvancedChemicalBiology,whichcoversabroadspectrum ofexcitingconceptsandtechnologiesandcapturesboththehistoric,definingmomentsinthefieldaswellasits guidingprinciples.Thetopicscutacrossallthemajorbiomoleculeclassesandhighlighthowchemicalapproaches canpowerfundamentalresearchaswellasclinicaltranslation.Thetextillustratesapplicationsinvariousbranches ofbiology–neuroscience,immunology,cancerbiology,andinfectiousdisease–andshowcasesnewtherapeutic modalitiesarisingfromouruniquebrandofmolecularengineering.Thebook’seditorsandcontributorsareleaders inchemicalbiology,andtheyhavedoneallofusagreatservice.Thisbookwillbeavaluableresourceforboth establishedchemicalbiologistsandmanyfuturegenerationsoftrainees.
Preface
Thefieldofchemicalbiologyisexpandingatarapidpace,withcontinuedadvancesinchemicalmethodologiesand biologicalapplications.Thecommunityofchemicalbiologistsisalsogrowinginnumber,withresearchersnowspanningadiversesetofbackgroundsandinterests.Withthisgrowthcomestheneedtotrainandeducatenewcomersto thefield.Chemicalbiologycourseshavesproutedatinstitutionsaroundtheglobe,andmostdonotuseastandard text.Weweremotivatedtofillthisvoid,providingabookthatiseasilyaccessibletocurrentandfuturegenerationsof chemicalbiologists.Thisisnoeasytask,consideringthebreadthofthedisciplineanditscontinuedevolution.Some unifyingthemeshaveemerged,though,thatwehopedtocaptureinthisbookandprovideahistoricalcontextfor theirdevelopment.Torealizeourvision,wereachedouttoleadersinthefieldfortheirinputongeneratingaresource forthecommunity.Theendproductisthecompilationofthechaptersbetweenthesecovers.
Overall,the AdvancedChemicalBiology textbookshowcaseshowchemicaltoolsandmolecularmethodshavebeen usedtogaininsightintobiologicalsystems.Theinitialchaptershighlightchemicalbiologyinthecontextofthe centraldogma:howmolecular-levelthinkinghasenablednumerousdiscoveriesrelevanttoDNA,RNA,proteins,and metabolites.Subsequentchaptersfeaturetransformativetechnologiesdevelopedwithinthecommunitythatcontinue toenablenewpursuits.Thefinalsectionofthebookillustratestheimpactofchemicalbiologyinthebroaderscientific community,withexamplesfrommicrobiology,immunology,neuroscience,drugdiscovery,andmore.Collectively, thesechaptersunderscorethebreadthofdiscoveryenabledbychemicalapproachesandprovideahistoricalbackdrop forthefield.
AdvancedChemicalBiology isdesignedforentry-levelgraduatestudentsinchemicalbiology,althoughthetext willserveasanexcellentresourceforstudentsinavarietyofchemistry-andbiology-relatedfields,inadditionto advancedundergraduates.Basicknowledgeoforganicchemistryandbiochemistry,uponwhichmuchofchemical biologybuilds,isassumed.Thechaptersarenotintendedtobein-depthreviewsonthesubjectmatter;rather,they serveasbasicprimersfornewcomerstothefield.Eachchapterbeginswithabriefintroductionandhistoricalcontext forthetopic.Thebulkofeachchapteristhendevotedtopresentingkeyconceptsanddevelopmentswithinchemical biology,drawingfromahandfuloflandmarkstudies.Sampleexamquestionsandslidesforinstructionalusearealso included.Sinceeachchaptertopicisnotcoveredin-depth,weexpectthatinstructorswillsupplementthematerials inthisbookwithadditionalexamplesandinformationtobestsuittheirclasses.
Thistextbookwouldnothavebeenpossiblewithoutthehardworkanddedicationofseveralindividuals.Weextend oursincerethankstotheauthorsofeachchapter,whoseworkonthisprojectcoincidedwiththeCOVID-19pandemic. Withouttheireffortsandcommitment,thisbookwouldhavebeenimpossible.Wearealsogratefultotheteamat Wileyforhelpingustonavigatethedevelopmentofateachingtextduringaquiteunprecedentedtime.Last,we wouldliketothankthemanycolleaguesandmentorswhohelpedtosparkourinterestsinthefieldandwhocontinue
xviii Preface
toguideourpaths.Wehopethatthisbooksimilarlycaptivatesthenextgenerationoftraineesandinspiresthemto continuetopushthefrontiersofchemicalbiologyandscientificdiscovery.
11July2022
HowardC.Hang ScrippsResearchInstitute LaJolla,CA92037,USA
MatthewR.Pratt UniversityofSouthernCalifornia LosAngeles,CA90089,USA
JenniferA.Prescher UniversityofCalifornia,Irvine Irvine,CA92697,USA
IntroductiontoAdvancedChemicalBiology
HowardC.Hang 1,2 ,MatthewR.Pratt 3 ,andJenniferA.Prescher 4,5,6
1 ScrippsResearch,DepartmentofImmunology&Microbiology,10550NorthTorreyPinesRoad,LaJolla,CA92037,USA
2 ScrippsResearch,DepartmentofChemistry,10550NorthTorreyPinesRoad,LaJolla,CA92037,USA
3 UniversityofSouthernCalifornia,DepartmentofChemistry,3430S.VermontAve,CA92121,USA
4 UniversityofCaliforniaIrvine,DepartmentofChemistry,1120NaturalSciencesII,CA92697,USA
5 UniversityofCaliforniaIrvine,DepartmentofMolecularBiologyandBiochemistry,3205McGaughHall,CA92697,USA
6 UniversityofCaliforniaIrvine,DepartmentofPharmaceuticalSciences,101TheorySuite100,CA92697,USA
1.1Introduction
Asitsnameimplies,thefieldofchemicalbiology employschemicalprinciplestodissectmechanismsin biologyandpotentiallytranslatethesediscoveriesinto therapeuticapproachesforhealthanddisease.Chemicalbiologyasafieldevolvedfromandmergeddifferent specializedfieldsofinvestigationintoabroadertopic thatencompassesmanyareasofresearch.Onecould arguethattheoriginsofchemicalbiologydateback tothediscovery,characterization,andsynthesisof smallmoleculestodeterminetheirmechanismsof actionandproductionfortherapeuticapplications. Notably,studiesinthelate1800sbyEmilFischerand coworkersledtothesynthesisofindoles,peptides, andmonosaccharidesaswellastheirstereochemical determination[1],whichwashighlightedbytheNobel PrizeinChemistryin1902.Inaddition,PaulEhrlich andcoworkersdevelopedarsphenamine(Salvarsan)as antimicrobialtreatmentforsyphilisintheearly1900s andpioneeredtheconceptofchemotherapyasa“magic bullet”fordiseasetreatment[2].Thesetwolandmark examplesestablishedthefoundationforthesynthesisof smallmolecules,thedeterminationoftheirstructures andmechanismsofactionaswellastheirtherapeutic application.Manyareasofchemistryandbiologyhave evolvedfromthesepioneeringstudiesandhaveculminatedinourcurrentperspectiveonchemicalbiology. Notably,thedesignandsynthesisofspecificchemical probesandhomogeneousbiomoleculesliesattheheart ofchemicalbiology.Itisalsoimportanttonotethat theadvancesinchemicalbiologyhavebeenenabled bymanymajorareasofsciencesuchasphysicaland
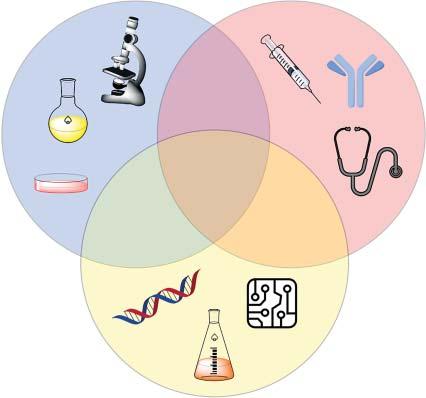
Figure1.1 Chemicalbiologyisatthenexusofbasicscience, medicine,andtechnology.
organicchemistry,biochemistry,structuralbiology,analyticalchemistryaswellasengineeringandevolutionary approaches(Figure1.1),whichwehighlightbelow.
1.2EnabledbySynthetic andPhysicalOrganicChemistry
Theabilityofchemiststounderstandreactivityof moleculesandexploittheseprinciplesforsynthesis hasbeentransformativeforscience[3]andunderlies muchoftheinnovationsinchemicalbiology[4,5] (Figure1.2).Indeed,innovationsinorganicchemistry
AdvancedChemicalBiology:ChemicalDissectionandReprogrammingofBiologicalSystems, FirstEdition. EditedbyHowardC.Hang,MatthewR.Pratt,andJenniferA.Prescher. ©2023WILEY-VCHGmbH.Published2023byWILEY-VCHGmbH. Companionwebsite:www.wiley.com/go/hang
Chemical biology
Basic science
Medicine
Synthetic chemistry
(a)
Physical organic chemistry
(b)
• New synthetic methods
• Access to complex natural products
• Insights into mechanisms of action
• Development of new chemical probes and therapeutic leads
• Development of new bioorthogonal reactions
• Designer chromophores for imaging applications
Figure1.2 Impactofsyntheticandphysicalorganicchemistryonchemicalbiology.(a)Retrosyntheticanalysisofcomplex naturalproductsuchasrapamycin.Source:Nicolaouetal.[6]/AmericanChemicalSociety.(b)Improvedbioorthogonalreactions suchasstrain-promotedazide-alkynecycloaddition[7]aswellasnewchromophoressuchassiliconrhodamine[8].
havegreatlyfacilitatedthesynthesisofcomplexnaturalproducts(Figure1.2a),small-moleculeprobes, andmacromoleculesforfundamentalstudiesand therapeuticapplications[4,5].Forexample,efficient methodsforthechemicalsynthesisofnucleicacids haverevolutionizedmolecularbiology[9],facilitated thedevelopmentofhighlysensitivediagnosticmethods[10],andsupportedthegenerationofprecise vaccines[11](Chapter2).Moreover,thesynthesisof shortoligonucleotideshasenabledstructure–function studies,therapidcloningofgenes(Chapter2)[10], andefficientprogrammablegenomeengineering[12] (Chapter6).Likewise,thechemicalsynthesisofpeptides[13],proteins[14,15],andglycans[16,17]have alsoprovidedimportantaccesstothesebiomolecules forstructure–activitystudiesaswellasthegenerationofdiagnosticsandtherapeutics(Chapters7,8, 13,15,17,24,26,and30).Ofnote,thesite-specific installationofbiophysicalprobesandposttranslationalmodificationsontopeptidesandproteinshas revealedfundamentalprinciplesofproteinfolding, structure,andfunction(Chapters7,8,and15).Alternatively,thesynthesisofglycanshasyieldedhomogeneous materialstoexploretheirfunctionaswellasimportant imaginganddiagnosticagentssuchasfluorine-18-2fluoro-2-deoxy-D-glucose(F18-FDG)(Chapter13).
Beyondthesynthesisofbiomolecules,advances inphysicalorganicchemistrysuchasthehard–soft acid–baseandmolecularorbitaltheories(Figure1.2b)
[18]haveledtothedevelopmentofnewchemical reactionsandprobestoexplorebiology.Forexample, understandingtherelativereactivityofaminoacidside chainswithdifferentchemotypeshasyieldedefficient bioconjugationmethodsformodifyingnativeproteins (Chapter14).Alternatively,thedevelopmentofchemicalreactionsthatareorthogonaltotheendogenous reactivityincellsandyetcompatiblewithbiological conditionshasaffordedavarietyof“bioorthogonal” reactionsforthemodificationofdiversebiomolecules andsmallmoleculeswithuniquefunctionality (Figure1.2b)(Chapter16).Moreover,understanding thestereo-electroniceffectsofchemicalmodifications onchromophoreshasyieldedawiderangeofimaging reagentsforvisualizingmanybiologicalprocessesin cellsandanimals(Figure1.2b)(Chapters17and18). Thesechromophorescanalsobetunedtobinddifferent metalstoexploretheirabundanceanddynamicsinbiologicalsystems(Chapter19).Furthermore,theunique reactivityofdifferentchemotypescanbeharnessedfor selectiveprofilingofvariousredoxstates(Chapter20) andbiochemicalactivitiesofproteins(Chapter21).
Inadditiontoreactionandprobedevelopment,the totalsynthesisofcomplexnaturalproductsandtheir analogshasaffordedimportantreagentstodetermine theirmoleculartargetsandmechanismsofaction [19],whichhasledtomoreprecisetherapeuticsfor humandiseases.Alandmarkexampleofthesestudies isthediscovery,synthesis(Figure1.2a),andtarget
identificationofrapamycin,whichrevealedmammaliantargetofrapamycin(mTOR)[20,21],asakey kinasethatregulatescellulargrowthandmetabolism (Chapter25).Althoughrapamycinfrom Streptomyces hygroscopicus wasoriginallyexploredasananti-fungal agent,itexhibitedpotentimmunosuppressiveactivity onTcellsandwasultimatelyapprovedbytheFederal DrugAdministration(FDA)tomitigatethesideeffects oforgantransplantation(Chapter4).Thesubsequent characterizationofmTORasthemechanistictarget ofrapamycin[20,21]andthediscoveryofitsphosphatidylinositol3-kinase-relatedkinaseactivityledto thedevelopmentofmorespecificandpotentmTOR kinaseinhibitorstotreatcancerandothermetabolic diseasesinhumans(Chapter30).
1.3GuidedbyBiochemistry andStructuralBiology
Thedesignanddevelopmentofspecificchemical probestoperturbandvisualizebiologicalsystemshas beenguidedbyinnovationsinbiochemistry[22]and structuralbiology(Figure1.3)[25,26].Forexample,the studyofenzymereactionmechanisms[22]allowed thedevelopmentofspecificchemicalprobesfor activity-basedproteinprofiling(ABPP)(Figure1.3a) (Chapter21).Alternatively,theadvancesinX-ray
crystallographyhaveallowedstructure-baseddesign ofimportantsmall-moleculeprobesandtherapeutics(Figure1.3b).Moreover,thedesignoforthogonal “bump-and-hole”enzyme–substratepairs(Chapter22) wasfacilitatedbyX-raystructuresofdifferentenzymes andproteinfamilies.Inaddition,structuralstudiesof largemulti-domainproteincomplexessuchaspolyketidesynthases(PKSs)havehelpedtodeconvolutethe biosynthesisofnaturalproductsandprovidednew opportunitiestoengineerthesepathways(Chapter24). Morerecently,advancesincryo-electronmicroscopy haveshedlightonthestructuresofmembraneproteinsandlargercomplexes[27],whichhasenabledthe designanddevelopmentofadditionalchemicalprobes andtherapeutics.Furthermore,theestablishmentof robustproteinstructurepredictionmethodshasprovidedimportantcomputationaltoolsforexploringsmall molecule–proteininteractionsaswellas denovo design ofnovelproteinswithdiversefunctions[28].
1.4EnhancedbyEngineering andEvolution
Aschemistsandbiologistsbegantounderstand thestructureandfunctionofbiomolecules,this collaborationallowedthedesignofnovelsystemswith improvedornewfunctions(Figure1.4).Forexample,
Biochemistry Fluorophosphonate (FP)-biotin Mechanism-/activity-based chemical probe(s)
(a)Enzyme mechanism(s)
Structural biology
• Biomolecule mechanisms of action
• Development of selective antagonists or agonists
• Design of mechanism-/activitybased chemical probes
• Biomolecule atomic structures
• Development of selective antagonists or agonists
• Design of mechanism-/activitybased chemical probes
Figure1.3 Impactofbiochemistryandstructuralbiologyonchemicalbiology.(a)Understandingenzymereactionmechanisms hasaffordedactivity-basedprobessuchasFP-biotin.Source:Liuetal.[23]/TheNationalAcademyofSciences.(b)Structural biologyandcomputationalmethodshaveenabledstructure-baseddesignofselectivechemicalprobesandtherapeuticssuch asHIV-1proteaseinhibitor.Source:Swainetal.[24]/TheNationalAcademyofSciences.
(b)
Synthetic HIV-1 protease (PDB: 7HVP)
Engineering
Evolution
• Engineering proteins with novel functions (b)
• Rational protein design and mutagenesis
• Evolving proteins with novel function
• Random mutagenesis and selection or screening
Figure1.4 Examplesofengineeringandevolutionaryapproachesinchemicalbiology.(a)Advancesinproteinengineeringhave enabledthedesignanddevelopmentofproteinswithnovelactivitysuchascatalyticantibodiesforstereoselectiveDiels–Alder reaction.Source:AdaptedfromGouverneuretal.[29].(b)Directedevolutionhasalsoaffordedproteinswithnovelfunctions suchasP450enzymeswithcyclopropanationactivity.Source:AdaptedfromCoelhoetal.[30].
protein-engineeringmethodswereemployedtogeneratecatalyticantibodiesthatcouldexecutechemical reactionslikenaturalenzymesorentirelynewreactions (Figure1.4a)(Chapter28).Alternatively,directedevolutionapproachescombiningrandommutagenesisin combinationwithhigh-throughputselectionorscreeningmethodsweredevelopedtoidentifyunpredicted andnovelproteinvariantswithuniqueorimproved properties(Figure1.4b)(Chapter9).Ofnote,protein engineeringanddirectedevolutionapproacheshave beenemployedtoestablishgeneticcodonexpansionfor thesite-specificincorporationofnon-canonicalamino acidswithuniquereactivityintospecificproteinsand wholeorganisms(Chapter15).Beyondthesesynthetic biologyexamples,proteinengineeringanddirected evolutionapproacheshavealsobeeninstrumental ingeneratingfluorescentproteins(Chapter17)and reporterenzymes(Chapter18)withimprovedcellular and invivo imagingproperties.
1.5ExpandedbyAnalytical Chemistryand“Omics”Technologies
Chemicalbiologyhasalsobeensignificantlyenabled andexpandeduponwithimprovedanalyticalmethods andinstrumentation(Figure1.5).Thedevelopmentof rapidandinexpensivenucleicacidsequencingmethods hasbeentransformativeforilluminatingthegenome
ofmanyorganismsandhasallowedcomparative genomicsofhealthyanddiseasestates(Figure1.5) (Chapters2–6).Theextensionofthesemethodsto singlecellanalyseshasrevealedspatialandtemporalphenotypesofdiversebiologicalprocessesandis revolutionizingbiologyandmedicine[31].Inparallel, theadvancesinmassspectrometry[32]andnuclear magneticresonancespectroscopy[33]havegreatly improvedthedetectionandstructuralcharacterization ofmacromoleculesandmetabolites(Figure1.5).For example,thehigh-throughputfragmentationanddetectionpeptidesbymassspectrometryalongwithaccurate computationalassemblymethodshavefacilitatedthe large-scalecomparativeanalysisofproteins[32]and theirposttranslationalmodifications(Chapter12).In addition,theunionofmassspectrometrywithchemical affinityprobesandABPP(Chapter21)hasfacilitated theidentificationofsmallmolecule–proteintargets forimprovedpharmacologyanddrugdevelopment (Chapters26and30).Furthermore,thesesignificant advancesinanalyticalchemistryhaveallowedthe large-scalecomparativeanalysisofcellularmetabolites (Chapter10)andlipids(Chapter11)incells,tissues,and wholeorganismsaswellascomplexnaturalproducts (Chapter23).Collectively,theselarge-scalemethods foranalyzingthegenome,transcriptome,proteome, andmetabolomeofcellsandorganismsareproviding importantmethodsfordissectingcomplexbiology systemsanddiseases.
Cytochrome p450 CYP120A1 (PDB: 3QI8)
Evolved
(PDB: 3QI8)
Catalytic
Analytical chemistry
(a)
Genomics
• Improved detection of metabolites, peptides, and proteins for metabolomics and proteomics (b)
• Large-scale profiling of biological systems (cells, tissues, and organisms)
• Discovery of novel biomolecules
• Development of selective chemical probes
• Improves detection of nucleic acids for genomics
• Large-scale profiling of biological systems (cells, tissues, and organisms)
• Discovery of novel genetic variants
• Development of biomarkers
Figure1.5 Impactofanalyticalchemistryandlarge-scalemethodsonchemicalbiology.(a)Betteranalyticalmethodshave allowedimproveddetectionofbiomoleculesformetabolomicsandproteomics.(b)Enhancednucleicaciddetectionand sequencingmethodshavesignificantlyexpandedthescopeandimpactofgenomics.
1.6ImpactonBiologicalDiscovery andDrugDevelopment
Innovationsinchemicalbiologyhaveilluminatedspecificareasofbiologyandarefuelingthedevelopmentof newtherapeutics.Sincetheoriginaldiscoveryofpenicillin[34],chemicalapproachesandnewprobeshave helpedtoelucidatefundamentalbiosyntheticpathways inbacteriaandhavefacilitatedthedevelopmentofnew antibiotics(Chapter25).Likewise,chemicalbiology approacheshaveaidedinthedissectionofcomplex signalingpathwaysineukaryoticcellsandthedeterminationofmechanismsofactionandresistancefor newsmall-moleculedrugcandidates(Chapter26). Chemicalbiologyhasalsohelpedtouncoverimportantdevelopmentalpathwaysinwholeorganismsand characterizedetrimentalsideeffectsofdrugmolecules (Chapter27).Sincethebirthofimmunologyasfield, chemistryhasplayedakeyroleinestablishingthe principlesoftheadaptiveimmuneresponseandhas alsoaffordednewtoolsforlarge-scaleimmuneprofiling aswellasthenextgenerationofadjuvantmolecules (Chapter28).Neurosciencehasalsobenefittedfrom theadvancesinchemicalbiology,astheengineering ofnovelprotein–ligandpairshasaffordedmethodsfor cell-specificperturbationsandimaging invivo,which
hasbeeninstrumentalindeconstructingneuronal circuitsandmodulatinganimalbehavior(Chapter29). Finally,themultitudeofchemicalbiologyapproachesto discovernovelbioactivesmallmoleculesandelucidate theirmechanismsofactionhasgreatlyimprovedthe overallpipelinefordrugdiscovery(Chapter30).
1.7Outlook
Wehavebeenfortunatetowitnessandparticipateinthe evolutionofchemicalbiologyasamulti-disciplinary fieldthatintegratesdifferentfieldsofbasicscienceto understandbiologyanddisease.Wegreatlyappreciate theremarkablecontributionsofthechapterauthors andaregratefulfortheirinsightfulperspectivesoneach areaofchemicalbiology,whichwehopewillbehelpful andinspirethenextgenerationofscientists.Aswelook forwardtothefuture,remarkableadvancesinsynthetic chemistrycontinuetoprovideaccesstomorecomplex moleculesforinvestigation,whilenewandimproved instrumentationfromanalyticalchemistrywillallowfor moresensitiveandhigh-throughputanalysesofdiverse biomolecules.Excitingly,machine-learningandartificialintelligencemethodshavealreadybeguntoprovide newapproachestodesignandsynthesizebiomolecules
moreefficientlyandwithnovelproperties[35].The unionoftheseadvanceswith“omics”technologies shouldprovidenewopportunitiestorealizethepromise ofpersonalizedmedicinefordifferentdiseases.Aswe achievenewmilestonesinchemistryandbiologyfor
References
1 Kunz,H.(2002).EmilFischer–unequalledclassicist,masteroforganicchemistryresearch,and inspiredtrailblazerofbiologicalchemistry. Angew. Chem.Int.Ed. 41(23):4439–4451.
2 Stern,F.(2004).PaulEhrlich:thefounderof chemotherapy. Angew.Chem.Int.Ed. 43(33): 4254–4261.
3 Corey,E.J.andChengX-m(1989). TheLogicof ChemicalSynthesis.NewYork:Wiley,436pages.
4 Schreiber,S.L.,Kotz,J.D.,Li,M.etal.(2015). Advancingbiologicalunderstandingandtherapeutics discoverywithsmall-moleculeprobes. Cell 161(6): 1252–1265.
5 Schreiber,S.,Kapoor,T.M.,Gn,W.,andWiley,I. (2007). ChemicalBiology:FromSmallMolecules toSystemsBiologyandDrugDesign.Weinheim: Wiley-VCH.
6 Nicolaou,K.C.,Chakraborty,T.K.,Piscopio,A.D. etal.(1993).Totalsynthesisofrapamycin. J.Am. Chem.Soc. 115(10):4419–4420.
7 Agard,N.J.,Prescher,J.A.,andBertozzi,C.R.(2004). Astrain-promoted[3+2]azide-alkynecycloaddition forcovalentmodificationofbiomoleculesinliving systems. J.Am.Chem.Soc. 126(46):15046–15047.
8 Koide,Y.,Urano,Y.,Hanaoka,K.etal.(2011). Evolutionofgroup14rhodaminesasplatforms fornear-infraredfluorescenceprobesutilizing Photoinducedelectrontransfer. ACSChem.Biol. 6(6):600–608.
9 Caruthers,M.H.(2013).Thechemicalsynthesisof DNA/RNA:ourgifttoscience. J.Biol.Chem. 288(2): 1420–1427.
10 Mullis,K.,Faloona,F.,Scharf,S.etal.(1986). SpecificenzymaticamplificationofDNAinvitro: thepolymerasechainreaction. ColdSpringHarbor Symp.Quant.Biol. 51(Pt1):263–273.
11 Sahin,U.,Kariko,K.,andTureci,O.(2014). mRNA-basedtherapeutics–developinganew classofdrugs. Nat.Rev.DrugDiscovery 13(10): 759–780.
12 Doudna,J.A.(2020).Thepromiseandchallenge oftherapeuticgenomeediting. Nature 578(7794): 229–236.
globalhealth,wehopethatnewinnovationsinchemical biologywillcontinuetoexpandbeyondhumanhealth andprovidekeysolutionsforothermajorchallenges facingourplanet,includingfoodsecurity,energy production,andclimatechange.
13 Merrifield,B.(1986).Solidphasesynthesis. Science 232(4748):341–347.
14 Thompson,R.E.andMuir,T.W.(2020).Chemoenzymaticsemisynthesisofproteins. Chem.Rev. 120(6): 3051–3126.
15 Dawson,P.E.andKent,S.B.(2000).Synthesisof nativeproteinsbychemicalligation. Annu.Rev. Biochem. 69:923–960.
16 Pardo-Vargas,A.,Delbianco,M.,andSeeberger,P.H. (2018).Automatedglycanassemblyasanenabling technology. Curr.Opin.Chem.Biol. 46:48–55.
17 Cheng,C.W.,Wu,C.Y.,Hsu,W.L.,andWong,C.H. (2020).Programmableone-potsynthesisofoligosaccharides. Biochemistry 59(34):3078–3088.
18 Anslyn,E.V.andDougherty,D.A.(2006). Modern PhysicalOrganicChemistry.Sausalito,CA:University Science.
19 Nicolaou,K.C.,Snyder,S.A.,andCorey,E.J.(2003). ClassicsinTotalSynthesis.Weinheim:Wiley-VCH, xix,639pages:illp.
20 Schreiber,S.L.(1991).Chemistryandbiologyof theimmunophilinsandtheirimmunosuppressive ligands. Science 251(4991):283–287.
21 Sabatini,D.M.(2017).Twenty-fiveyearsofmTOR: uncoveringthelinkfromnutrientstogrowth. Proc. Natl.Acad.Sci.U.S.A. 114(45):11818–11825.
22 Fersht,A.(1999). StructureandMechanisminProteinScience:AGuidetoEnzymeCatalysisandProtein Folding.NewYork:W.H.Freeman,xxi,631pages: illustrationsp.
23 Liu,Y.,Patricelli,M.P.,andCravatt,B.F.(1999). Activity-basedproteinprofiling:theserine hydrolases. Proc.Natl.Acad.Sci.U.S.A. 96(26): 14694–14699.
24 Swain,A.L.,Miller,M.M.,Green,J.etal.(1990). X-raycrystallographicstructureofacomplex betweenasyntheticproteaseofhumanimmunodeficiencyvirus1andasubstrate-basedhydroxyethylamineinhibitor. Proc.Natl.Acad.Sci.U.S.A. 87(22):8805–8809.
25 Hendrickson,W.A.(1991).Determinationofmacromolecularstructuresfromanomalousdiffractionof synchrotronradiation. Science 254(5028):51–58.