nutrients
Article DysregulationofNeuronalGenesbyFetal-Neonatal IronDeficiencyAnemiaIsAssociatedwithAltered DNAMethylationintheRatHippocampus
Yu-ChinLien 1 ,DavidECondon 1 ,MichaelKGeorgieff 2 ,RebeccaASimmons 1,3, *and PhuVTran 2, *
1 CenterforResearchonReproductionandWomen’sHealth,PerelmanSchoolofMedicine,TheUniversityof Pennsylvania,Philadelphia,PA19104,USA;ylien@pennmedicine.upenn.edu(Y.-C.L.); dec986@gmail.com(D.E.C.)
2 DepartmentofPediatrics,UniversityofMinnesotaSchoolofMedicine,Minneapolis,MN55455,USA; georg001@umn.edu
3 Children’sHospitalofPhiladelphia,Philadelphia,PA19104,USA
* Correspondence:rsimmons@pennmedicine.upenn.edu(R.A.S.);tranx271@umn.edu(P.V.T.); Tel.: +1-215-662-3269(R.A.S.);Tel.: +1-612-626-7964(P.V.T.)
Received:17April2019;Accepted:22May2019;Published:27May2019
Abstract: Early-lifeirondeficiencyresultsinlong-termabnormalitiesincognitivefunctionand affectivebehaviorinadulthood.Inpreclinicalmodels,theseeffectshavebeenassociatedwith long-termdysregulationofkeyneuronalgenes.Whilelimitedevidencesuggestshistonemethylation asanepigeneticmechanismunderlyinggenedysregulation,theroleofDNAmethylationremains unknown.TodeterminewhetherDNAmethylationisapotentialmechanismbywhichearly-lifeiron deficiencyinducesgenedysregulation,weperformedwholegenomebisulfitesequencingtoidentify lociwithalteredDNAmethylationinthepostnatalday(P)15iron-deficient(ID)rathippocampus,a timepointatwhichthehighestlevelofhippocampalirondeficiencyisconcurrentwithpeakiron demandforaxonalanddendriticgrowth.Weidentified229differentiallymethylatedlociandthey weremappedwithin108genes.Amongthem,63and45genesshowedsignificantlyincreasedand decreasedDNAmethylationintheP15IDhippocampus,respectively.Toestablishacorrelation betweendifferentiallymethylatedlociandgenedysregulation,themethylomedatawerecomparedto ourpublishedP15hippocampaltranscriptome.Bothdatasetsshowedalterationofsimilarfunctional networksregulatingnervoussystemdevelopmentandcell-to-cellsignalingthatarecriticalfor learningandbehavior.Collectively,thepresentfindingssupportaroleforDNAmethylationin neuralgenedysregulationfollowingearly-lifeirondeficiency.
Keywords: hippocampus;DNAmethylation;DNAsequencing;iron;neurobiology;transcriptome; micronutrientdeficiency;neuroplasticity
1.Introduction
Fetalandneonatal(early-life)irondeficiencywithorwithoutanemiaaffectsmorethan30% ofpregnantwomenandpreschoolagechildrenworldwide,andresultsinlong-termcognitiveand behavioralabnormalities[1–8].Wehavepreviouslyinvestigatedtheeffectsofearly-lifeirondeficiency usingaratmodel,wherebypupsweremadeiron-deficient(ID)fromgestationalday2through postnatalday(P)7byprovidingpregnantandnursingdamswithanIDdiet,afterwhichthey wererescuedwithaniron-sufficient(IS)diet.Thismodelofmaternal-fetalirondeficiencyresults ina50%reductioninbrainironconcentrationbyP7[9],theageatwhichratbraindevelopment approximatesthatofafull-termhumannewborn[10,11].Thedeficitinbrainironcontentissimilarto
Nutrients 2019, 11,1191;doi:10.3390/nu11051191www.mdpi.com/journal/nutrients 1
2.3.WholeGenomeBisulfiteSequencingandLibraryPreparation
GenomicDNAfromISandIDhippocampiwasisolatedusinganAllPrepDNA/RNAMiniKit (Qiagen).WGBSwasperformedusingapreviouslypublishedprotocol[33].Briefly,1 μgofgenomic DNAwasfragmentedinto~300bpfragmentsusingaM220CovarisUltrasonicator(Covaris,Woburn, MA,USA).SequencinglibrariesweregeneratedusingaNEBNextgenomicsequencingkit(New EnglandBiolabs,Ipswich,MA,USA)andligatedwithIlluminamethylatedpairedendadaptors. Librarieswerebisulfite-convertedusinganImprintDNAmodificationkit(MilliporeSigma,St.Louis, MO,USA),andthesizeof300–600bpwasselectedusingthePippinPrepDNAsizeselectionsystem (SageScience,Beverly,MA,USA).LibrarieswerethenamplifiedusingPfu-TurboCxHotstartDNA polymerase(AgilentTechnologies,SantaClara,CA,USA).Paired-endlibrariesweresequencedto100 bponanIlluminahiSeq2000.ThreebiologicalreplicatesforeachgroupwereperformedinWGBS. WGBSdataareavailableontheGeneExpressionOmnibusunderGSE98064.
2.4.IdentificationofDMRsUsingtheDefiantProgram
DMRswereidentifiedbyourin-housedevelopedDefiant(DMRs:Easy,Fast,Identificationand ANnoTation)programbasedonfivecriteria,asdescribedpreviously[30].Briefly,adapterswere trimmedfromthereadsusingacustomClanguageprogram.Trimmedreadswerealignedagainst theratgenome(rn4).Whenreadsoverlappedatabase,themethylationstatusfromread1wasused. MethylationdataattheCandGinaCpGpairweremergedtoproducetheestimateforthatlocus. DMRsweredefinedwithaminimumcoverageof10inallsixsamples, p-value < 0.05,andaminimum methylationpercentagechangeof10%.SincetheDefiantprogramdidnotuseapre-definedborder toidentifyDMRs,the p-value < 0.05cutoff onlyinfluencedthewidthsandquantityofDMRs.The Benjamini–Hochbergapproachwasappliedformultipletestingtoobtainfalsediscoveryrate(FDR, q-values).GeneswereassignedtotheDMRsbasedonapromotercutoff of15kbtothetranscription startsite,withthedirectionoftranscriptiontakenintoaccount.
2.5.Bioinformatics
Theknowledge-basedIngenuityPathwayAnalysis® (IPA,Qiagen,Germantown,MD,USA)was employedtoidentifynetworks,canonicalpathways,molecularandcellularfunctions,andbehavioral andneurologicaldysfunctionsusingaP15DNAmethylationdatasetfromWGBS.Themicroarray datasetfromapriorstudy[34]wasalsoanalyzedbyIPA.IPAmapsgenenetworksusinganalgorithm basedonmolecularfunction,cellularfunction,andfunctionalgroup.Fisher’sexacttestwasusedto calculatethesignificanceoftheassociationbetweengenesinthedatasetsandtheanalyzedpathways orfunctions.
3.Results
3.1.Early-LifeIronDeficiencyInducedDifferentialDNAMethylationintheRatHippocampus
WeperformedwholegenomecytosinemethylationbisulfitesequencingonP15ID(n = 3)and IS(n = 3)rathippocampi.Todeterminewhetherirondeficiencyaltersthegenome-widepatternof DNAmethylationinthedevelopinghippocampus,DNAmethylationat1000randomlyselectedloci werecomparedbetweenIDandISsamplestogeneratearepresentativeheatmap.Thisunsupervised clusteringapproachshowedconsistentpatternsofmethylationacrossallsamples,withoutanoverall shifttowardhypo-orhypermethylationintheIDgroup(Figure 1a).Todeterminewhetheriron deficiencyinduceschangesinDNAmethylationatalocus-specificlevel,a ≥ 10%methylationchange with p-value < 0.05wasusedasaninclusioncriterion[30].Weidentified229DMRs(Figure 1bandTable S1),including58%intergenic,26%intronic,and11%exonicregions(Figure 1c).Approximately4%of DMRswerelocatedinpromoterregions.TheseDMRsmappedtowithin15kbofthetranscription startsiteof108geneswith63hypermethylatedand45hypomethylatedlociinIDcomparedtoIS hippocampi(Table 1).
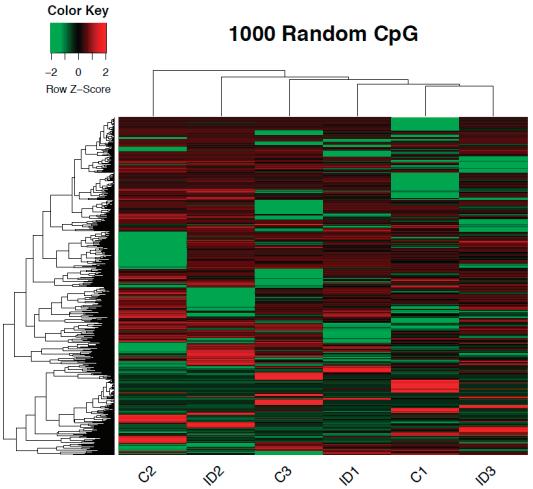
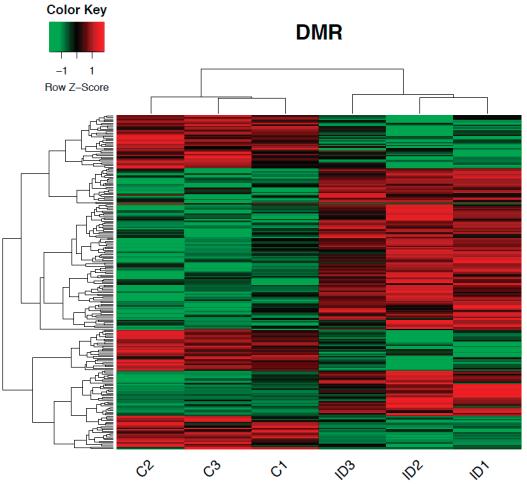
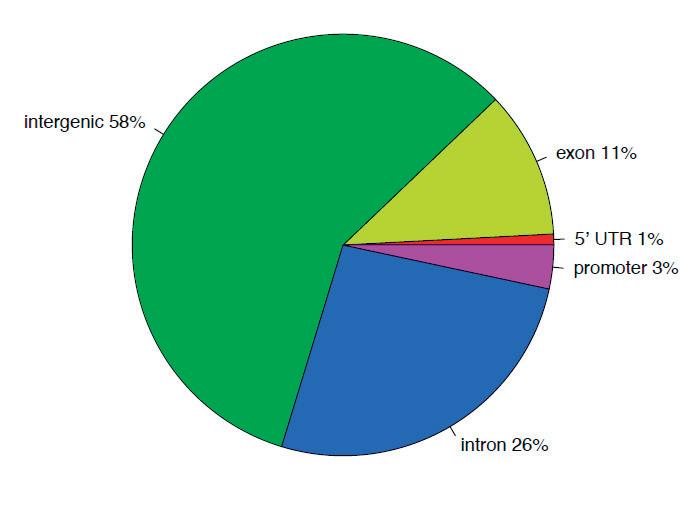
Figure1. DNAmethylomeofthepostnatalday(P)15rathippocampus.(a)Anunsupervisedclustering heatmapof1000randomlyselectedlocishowinganabsenceofbiasinglobalmethylationbetween iron-sufficient(IS)andiron-deficient(ID)hippocampi.Eachrowintheheatmapcorrespondstodata fromasinglelocus.Thebranchingdendrogramatthetopcorrespondstotherelationshipsamong samples.Hyper-andhypomethylationareshownonacontinuumfromredtogreen,respectively. (b)Heatmapofdifferentiallymethylatedregions(DMRs)showingsignificantdifferencesincytosine methylationbetweenIS(labeledC1-3)andID(labeledID1-3)hippocampi.Eachrowintheheatmap correspondstodatapointfromasinglelocus,whereascolumnscorrespondtoindividualsamples.The branchingdendrogramcorrespondstotherelationshipsamongsamples,asdeterminedbyclustering usingthe229identifiedDMRs.Hyper-andhypomethylationareshownonacontinuumfromredto green,respectively.(c)PiechartrepresentingthelocationandproportionofDMRs.Thegenebody includedexonsandintrons.Thepromoterwaslimitedto15kbupstreamfromthetranscriptionalstart site.The5 -untranslatedregionbeganatthetranscriptionstartsiteandendedbeforetheinitiation sequence.Theintergenicregioniscomprisedoftheregionsnotincludedintheabovedefinedregions.
Table1. CpGmethylationwithinthe15kbpromoterregionofgenesintheP15iron-deficient rathippocampus.
Nutrients 2019, 11,1191
Table3. OverlappingcanonicalpathwaysoftheP15DNAmethylomeandP15microarraydatasets.
MethylomeAnalysisMicroarrayAnalysis
IngenuityCanonical Pathways p-value Differentially MethylatedGenes p-valueDifferentiallyExpressedGenes
NitricOxideSignalingin theCardiovascular System
CellularEffectsof Sildenafil(Viagra) 0.004
Cardiac β-Adrenergic Signaling 0.005
cAMP-Mediated Signaling 0.005
AxonalGuidance Signaling 0.006
RelaxinSignaling0.007
ReelinSignalingin Neurons 0.010
G-ProteinCoupled
ProteinKinaseA Signaling 0.013
BreastCancerRegulation byStathmin1 0.017
SynapticLong-Term Potentiation 0.021
GustationPathway0.023
SpermMotility0.023
GNRHSignaling0.032
SignalingbyRhoFamily GTPases 0.034
MolecularMechanisms ofCancer 0.042
MelatoninSignaling0.048
EphrinBSignaling0.049
CACNA1C,PRKAR1B, PDE2A,GUCY2C
CACNA1C,PRKAR1B, PDE2A,GUCY2C
CACNA1C,PRKAR1B, PDE2A,PDE6C 0.036
CAMK2B,MC3R, PRKAR1B,PDE2A, PDE6C 0.000
ARHGEF15,ITSN1, SLIT3,MKNK1, PRKAR1B,EPHB1, SRGAP2 0.003
PRKAR1B,PDE2A, GUCY2C,PDE6C 0.008
ARHGEF15,ARHGEF3, MAP3K11 0.004
CAMK2B,MC3R, PRKAR1B,PDE2A, PDE6C
CAMK2B,Ptpn14, TNNI1,PRKAR1B, PDE2A,PDE6C 0.000
CAMK2B,ARHGEF15, ARHGEF3,PRKAR1B 0.000
ITPR2,PIK3R3,KDR,PTPN11,PRKAA1, GUCY2D,ITPR1,CAMK4,PRKAG1, PDE2A,PDGFC
MYH3,CACNG8,ITPR2,ADCY3,GPR37, GUCY2D,ITPR1,ADCY2,PLCE1, CAMK4,PRKAG1,PDE2A
ADCY3,PKIG,ADCY2,PRKAG1,PDE2A, PPP2R2A,PPP1R11
GABBR1,CHRM3,CAMK4,VIPR1, PDE2A,Htr5b,CHRM2,CNGA2, CAMK2A,GNAI3,ADCY3,HRH3,PKIG, ADCY2,LHCGR,OPRM1,GRM6
CXCL12,PIK3R3,TUBB,EPHA3,ROBO1, PLCE1,DPYSL5,RTN4R,RTN4,GNAI3, FZD4,PDGFC,BAIAP2,SEMA4F,CXCR4, NRAS,CFL1,PTPN11,NTRK2,PRKAG1
PIK3R3,ADCY3,PTPN11,GUCY2D, ADCY2,PRKAG1,PDE2A, NFKBIA,GNAI3
PAFAH1B1,PIK3R3,PTPN11,APP,MAPT, ARHGEF9,APBB1
PIK3R3,GABBR1,CHRM3,CAMK4, VIPR1,PDE2A,Htr5b,NFKBIA,CHRM2, CAMK2A,GNAI3,NRAS,PDPK1, ADCY3,HRH3,PTPN11,ADCY2, PRKAG1,GRM5,LHCGR,OPRM1,GRM6
ITPR2,PLCE1,NFKBIA,CNGA2,GNAI3, PYGB,ADCY3,PTPN11,ITPR1,ADCY2, PTPRF,TGFBR1,PPP1R1B,YWHAB, PPP1R11,DUSP12,PTPRN,CDC25A, PTPN2,PTPRO,H3F3A/H3F3B,CAMK4, PDE2A,PTPN23,CAMK2A,BAD,DUSP5, PTPN12,PRKAG1
ITPR2,PIK3R3,TUBB,CAMK4,PPP2R2A, CAMK2A,GNAI3,STMN1,NRAS, ADCY3,PTPN11,ITPR1,ADCY2, ARHGEF9,PRKAG1,PPP1R11
CAMK2B,CACNA1C, PRKAR1B 0.000 NRAS,ITPR2,GRINA,ITPR1,PLCE1, CAMK4,PRKAG1,GRM5,GRIN1, CAMK2A,GRM6,PPP1R11
PRKAR1B,PDE2A, PDE6C 0.000 CACNG8,ITPR2,ADCY3,CACNB4, CACNA2D1,P2RX5,ITPR1,ADCY2, PRKAG1,PDE2A,P2RY1,CACNA1H
MAP3K11,PRKAR1B, PDE2A 0.002 ITPR2,PAFAH1B1,ITPR1,PLCE1, CAMK4,PRKAG1,PDE2A, CNGA2,CACNA1H
CAMK2B,MAP3K11, PRKAR1B 0.000
CACNG8,ITPR2,CACNB4,CAMK4, CAMK2A,GNAI3,CACNA1H,NRAS, ADCY3,CACNA2D1,GNRHR,ITPR1, ADCY2,PRKAG1
ARHGEF15,ARHGEF3, MAP3K11,EZR 0.010 BAIAP2,CFL1,RHOT2,PIK3R3,PTPN11, RHOB,CDH1,ARHGEF9,PLD1,RHOV, GNAI3,STMN1
CAMK2B,ARHGEF15, ARHGEF3,JAK3, PRKAR1B 0.000
CAMK2B,PRKAR1B 0.021
ITSN1,EPHB1 0.022
RASGRF1,RHOT2,PIK3R3,CDC25A, CASP9,NFKBIA,CAMK2A,BAD,GNAI3, FZD4,NCSTN,NRAS,RALBP1,ADCY3, PTPN11,RHOB,HIF1A,ADCY2,CASP3, TGFBR1,CDH1,ARHGEF9, PRKAG1,RHOV
PLCE1,CAMK4,PRKAG1, CAMK2A,GNAI3
CXCL12,CXCR4,CFL1,CAP1,GNAI3
2019, 11,1191
diagnosisandtreatment,indicatingtheneedforadequateironduringcriticalperiodsofbrain development.Inpreclinicalmodels,theseeffectshavebeenascribedinparttothepersistence ofabnormalitiesinmonoaminesignaling,myelination,neuralmetabolism,andtheexpressionof neuroplasticity-associatedproteinsintoadulthood[20,56–58].Themolecularmechanismsunderlying thesepersistentchangeshavenotbeenfullyelucidated.Thepresentstudygoeswellbeyond previousstudiesbysystematicallyanalyzingthealterationofDNAmethylationinducedbyearly-life irondeficiencyusingawholegenomebisulfitesequencingapproach.Consistentwithprevious transcriptomicanalysis,thechangesinDNAmethylationintheIDhippocampusmappedtofunctional networksthatareimportantforneuronalplasticity.
DNAmethylationisanimportantepigeneticmechanismregulatinggeneexpression,oftenacross thelifespan.Methylationatgenomicregionshasdifferentinfluencesongenetranscriptionalactivity dependingonthelocationofDNAmethylation.Inthepresentstudy,differentialmethylationwashighly enrichedatintergenicregions(58%)intheIDhippocampus.Thisoutcomeissimilartoourprevious findingsinpancreaticisletsofanintrauterinegrowthrestrictionratmodel[59],whereapproximately 65%ofDMRswerelocatedinintergenicregions,aswellastoothermodelsofearly-lifeadverse environments[33,60–62].Theseconservedintergenicregionsmayrepresentimportantenhancersor cis-regulatorysitesinregulatinggeneexpression[43,63,64].Thus,theseDMRsmightaccountfora substantiallyfewernumberoflociwithDMRscomparedtoanumberofdifferentiallyexpressedgenes inthemicroarraydatasetandasmalloverlapbetweenthesetwodatasets.Ourdataalsoshowed thatapproximately37%ofDMRsintheIDhippocampuswerelocatedingenebodies(26%and11% inintronsandexons,respectively).DNAmethylationingenebodiesisgenerallyassociatedwith highergeneexpressionindividingcells[65],incontrasttotheregulatoryeffectofDNAmethylation inpromoterregions.However,thisassociationisnotseeninnon-dividingcells[27].Althoughnot manycellsinthehippocampusareactivelydividingatP15,theseDNAmodificationsmighthave occurredduringtheperiodofactiveproliferationintheprenatalperiodwhenthepregnantdamand fetuswereiron-deficient.AdditionalDMRanalysisatatimepointwhenthedevelopinghippocampus undergoesactiveproliferationwillprovidefurtherinsightintothisnotion.DNAmethylationin genebodiesmaydefinetheexonboundaries,regulatealternativepromotersingenebodies,and regulatemRNAsplicingandalternativesplicing[65–68].Wanetal.(2013)showedthattissue-specific DMRsarepreferentiallylocatedinexonsandintronsofprotein-codinggenes[69].Thesebiologically relevantDMRsareenrichedinalternativelysplicedgenesandasubsetofdevelopmentalgenes.Itis possiblethattherealeffectofDNAmethylationintheP15IDhippocampusiswithinthesedomains. Ourmicroarrayanalysis[34]wasinsufficienttoprobesucheffects.Finally,irondeficiency-induced intragenicDMRscouldmodifypotentialgeneenhancers[43,44,70,71].TheintragenicDMRsintheID hippocampusmaydirectlycontributetoneuralgenedysregulationbymodifyingtheaccessibilityof alternativesplicesitesorpromoters.Theseanalysesconstituteapotentialdirectionforfuturestudy.
OurWGBSanalysisoftheIDhippocampusidentifiedpertinentsignalingpathwaysthatcould underlietheneurobehavioralabnormalitiesassociatedwithearly-lifeirondeficiency.TheDNA methylomeshowedthatDNAmethylationatgenesregulatingcAMP-mediatedsignalingandprotein kinaseAsignalingwassignificantlyalteredintheP15IDrathippocampus(Table 2).Bothpathways playcriticalrolesinregulatingLTP,aswellastheplasticityofaxonalguidanceresponses[72,73].In addition,thepredictedchangestothe β-adrenergicsignalingandnitricoxidesignalingpathwaysin theIDhippocampuswouldlikelyresultinloweractivitiesofcAMP,cGMP,proteinkinaseC(PKC), mitogen-activatedproteinkinase(MAPK),andN-methyl-D-aspartate(NMDA)receptors[74,75]. Likewise,alteredRhoGTPasesignalingcouldchangetheaxonalresponsestoguidancecuesand affectneuronalconnectionsandLTPformation.TheRhofamilyofGTPasesplaysakeyrolein theformationofLTPbyregulatingcellularprocesses,includingaxonoutgrowthandgrowthcone dynamics[76,77].Thiseffectisconsistentwithandprovidesamolecularbasisforourprevious findingofabnormaldendritogenesisandsynaptogenesisintheseIDrats[17].Ourstudyalsorevealed thealterationofthereelinsignalingpathwayintheIDhippocampus.Reelinregulatesneuronal
Nutrients 2019, 11,1191
23. Tran,P.V.;Carlson,E.S.;Fretham,S.J.;Georgieff,M.K.Early-lifeirondeficiencyanemiaaltersneurotrophic factorexpressionandhippocampalneurondifferentiationinmalerats. J.Nutr. 2008, 138,2495–2501. [CrossRef]
24. Tran,P.V.;Kennedy,B.C.;Lien,Y.C.;Simmons,R.A.;Georgieff,M.K.Fetalirondeficiencyinduceschromatin remodelingattheBdnflocusinadultrathippocampus. Am.J.Physiol.Regul.Integr.Comp.Physiol. 2015, 308,R276–R282.[CrossRef]
25. Tahiliani,M.;Koh,K.P.;Shen,Y.;Pastor,W.A.;Bandukwala,H.;Brudno,Y.;Agarwal,S.;Iyer,L.M.;Liu,D.R.; Aravind,L.;etal.Conversionof5-methylcytosineto5-hydroxymethylcytosineinmammalianDNAbyMLL partnerTET1. Science 2009, 324,930–935.[CrossRef]
26. Hou,H.;Yu,H.Structuralinsightsintohistonelysinedemethylation. Curr.Opin.Struct.Biol. 2010, 20, 739–748.[CrossRef]
27. Moore,L.D.;Le,T.;Fan,G.DNAmethylationanditsbasicfunction. Neuropsychopharmacology 2013, 38,23–38. [CrossRef]
28. Weber,M.;Hellmann,I.;Stadler,M.B.;Ramos,L.;Pääbo,S.;Rebhan,M.;Schübeler,D.Distribution,silencing potentialandevolutionaryimpactofpromoterDNAmethylationinthehumangenome. Nat.Genet. 2007, 39,457–466.[CrossRef]
29. Stevens,M.;Cheng,J.B.;Li,D.;Xie,M.;Hong,C.;Maire,C.L.;Ligon,K.L.;Hirst,M.;Marra,M.A.;Costello,J.F.; etal.Estimatingabsolutemethylationlevelsatsingle-CpGresolutionfrommethylationenrichmentand restrictionenzymesequencingmethods. GenomeRes. 2013, 23,1541–1553.[CrossRef]
30. Condon,D.E.;Tran,P.V.;Lien,Y.C.;Schug,J.;Georgieff,M.K.;Simmons,R.A.;Won,K.J.Defiant:(DMRs: Easy,fast,identificationandANnoTation)identifiesdifferentiallyMethylatedregionsfromiron-deficientrat hippocampus. BMCBioinf. 2018, 19,31.[CrossRef]
31. Fretham,S.J.;Carlson,E.S.;Georgieff,M.K.Theroleofironinlearningandmemory. Adv.Nutr. 2011, 2, 112–121.[CrossRef]
32. Tran,P.V.;Fretham,S.J.;Wobken,J.;Miller,B.S.;Georgieff,M.K.Gestational-neonatalirondeficiency suppressesandirontreatmentreactivatesIGFsignalingindevelopingrathippocampus. Am.J.Physiol. Endocrinol.Metab. 2012, 302,E316–E324.[CrossRef]
33. Sheaffer,K.L.;Kim,R.;Aoki,R.;Elliott,E.N.;Schug,J.;Burger,L.;Schübeler,D.;Kaestner,K.H.DNA methylationisrequiredforthecontrolofstemcelldifferentiationinthesmallintestine. GenesDev. 2014, 28, 652–664.[CrossRef]
34. Carlson,E.S.;Stead,J.D.;Neal,C.R.;Petryk,A.;Georgieff,M.K.Perinatalirondeficiencyresultsin altereddevelopmentalexpressionofgenesmediatingenergymetabolismandneuronalmorphogenesisin hippocampus. Hippocampus 2007, 17,679–691.[CrossRef]
35.Chilton,J.K.Molecularmechanismsofaxonguidance. Dev.Biol. 2006, 292,13–24.[CrossRef]
36.Dickson,B.J.Molecularmechanismsofaxonguidance. Science 2002, 298,1959–1964.[CrossRef]
37. Egea,J.;Klein,R.BidirectionalEph-ephrinsignalingduringaxonguidance. TrendsCell.Biol. 2007, 17, 230–238.[CrossRef]
38. Rohani,N.;Canty,L.;Luu,O.;Fagotto,F.;Winklbauer,R.EphrinB/EphBsignalingcontrolsembryonicgerm layerseparationbycontact-inducedcelldetachment. PLoSBiol. 2011, 9,e1000597.[CrossRef]
39. Shamah,S.M.;Lin,M.Z.;Goldberg,J.L.;Estrach,S.;Sahin,M.;Hu,L.;Bazalakova,M.;Neve,R.L.;Corfas,G.; Debant,A.;etal.EphAreceptorsregulategrowthconedynamicsthroughthenovelguaninenucleotide exchangefactorephexin. Cell 2001, 105,233–244.[CrossRef]
40. Rex,C.S.;Chen,L.Y.;Sharma,A.;Liu,J.;Babayan,A.H.;Gall,C.M.;Lynch,G.DifferentRhoGTPase-dependent signalingpathwaysinitiatesequentialstepsintheconsolidationoflong-termpotentiation. J.Cell.Biol. 2009, 186,85–97.[CrossRef]
41. Malenka,R.C.;Nicoll,R.A.Long-termpotentiation—Adecadeofprogress? Science 1999, 285,1870–1874. [CrossRef]
42.Nicoll,R.A.ABriefHistoryofLong-TermPotentiation. Neuron 2017, 93,281–290.[CrossRef]
43. Blattler,A.;Yao,L.;Witt,H.;Guo,Y.;Nicolet,C.M.;Berman,B.P.;Farnham,P.J.GloballossofDNA methylationuncoversintronicenhancersingenesshowingexpressionchanges. GenomeBiol. 2014, 15,469. [CrossRef]
44. Singer,M.;Kosti,I.;Pachter,L.;Mandel-Gutfreund,Y.Adiverseepigeneticlandscapeathumanexonswith implicationforexpression. NucleicAcidsRes. 2015, 43,3498–3508.[CrossRef]
Nutrients 2019, 11,1191
88. Branco,M.R.;Ficz,G.;Reik,W.Uncoveringtheroleof5-hydroxymethylcytosineintheepigenome. Nat.Rev.Genet. 2011, 13,7–13.[CrossRef]
89. Szulwach,K.E.;Li,X.;Li,Y.;Song,C.X.;Wu,H.;Dai,Q.;Irier,H.;Upadhyay,A.K.;Gearing,M.;Levey,A.I.; etal.5-hmC-mediatedepigeneticdynamicsduringpostnatalneurodevelopmentandaging. Nat.Neurosci. 2011, 14,1607–1616.[CrossRef]
90. Hahn,M.A.;Qiu,R.;Wu,X.;Li,A.X.;Zhang,H.;Wang,J.;Jui,J.;Jin,S.G.;Jiang,Y.;Pfeifer,G.P.;etal. Dynamicsof5-hydroxymethylcytosineandchromatinmarksinMammalianneurogenesis. Cell.Rep. 2013, 3, 291–300.[CrossRef]
91. Al-Mahdawi,S.;Virmouni,S.A.;Pook,M.A.Theemergingroleof5-hydroxymethylcytosinein neurodegenerativediseases. Front.Neurosci. 2014, 8,397.[CrossRef]
© 2019bytheauthors.LicenseeMDPI,Basel,Switzerland.Thisarticleisanopenaccess articledistributedunderthetermsandconditionsoftheCreativeCommonsAttribution (CCBY)license(http://creativecommons.org/licenses/by/4.0/).
Article ASlow-DigestingCarbohydrateDietduringRat PregnancyProtectsOffspringfromNon-Alcoholic FattyLiverDiseaseRiskthroughtheModulationof theCarbohydrate-ResponseElementandSterol RegulatoryElementBindingProteins
RafaelSalto 1 ,ManuelManzano 2 ,MaríaDoloresGirón 1, *,AinaraCano 3 ,AzucenaCastro 3 , José DámasoVílchez 1 ,ElenaCabrera 1 andJosé MaríaLópez-Pedrosa 2
1 DepartmentofBiochemistryandMolecularBiologyII,SchoolofPharmacy,UniversityofGranada,Campus deCartuja,18071Granada,Spain;rsalto@ugr.es(R.S.);e.damaso@go.ugr.es(J.D.V.); elenacc_20@hotmail.com(E.C.)
2 AbbottNutritionR&D,AbbottLaboratories,18004Granada,Spain;manuel.manzano@abbott.com(M.M.); jose.m.lopez@abbott.com(J.M.L.-P.)
3 OWLMetabolomics,ParqueTecnológicodeBizkaia,48160Deiro,Spain; acano@owlmetabolomics.com(A.C.);acastro@owlmetabolomics.com(A.C.)
* Correspondence:mgiron@ugr.es;Tel.: +34-958-246363
Received:12February2019;Accepted:11April2019;Published:14April2019
Abstract: High-fat(HF)andrapiddigestive(RD)carbohydratedietsduringpregnancypromote excessiveadipogenesisinoffspring.Thiseffectcanbecorrectedbydietswithsimilarglycemic loads,butlowratesofcarbohydratedigestion.However,theeffectsofthesedietsonmetabolic programmingintheliversofoffspring,andthelivermetabolismcontributionstoadipogenesis,remain tobeaddressed.Inthisstudy,pregnantinsulin-resistantratswerefedhigh-fatdietswithsimilar glycemicloadsbutdifferentratesofcarbohydratedigestion,HighFat-RapidDigestive(HF–RD)diet orHighFat-SlowDigestive(HF–SD)diet.Offspringwerefedastandarddietfor10weeks,andthe impactofthesedietsonthemetabolicandsignalingpathwaysinvolvedinliverfatsynthesisand storageofoffspringwereanalyzed,includingliverlipidomics,glycogenandcarbohydrateandlipid metabolismkeyenzymesandsignalingpathways.Liversfromanimalswhosemotherswerefedan HF–RDdietshowedhighersaturatedtriacylglyceroldepositswithlowercarbonnumbersanddouble bondcontentscomparedwiththeHF–SDgroup.Moreover,theHF–RDgroupexhibitedenhanced glucosetransporter2,pyruvatekinase(PK),acetylcoenzymeAcarboxylase(ACC)andfattyacid(FA) synthaseexpression,andadecreaseinpyruvatecarboxylase(PyC)expressionleadingtoanaltered liverlipidprofile.TheseparameterswerenormalizedintheHF–SDgroup.Thechangesinlipogenic enzymeexpressionwereparalleltochangesinAktPKBphosphorylationstatusandnuclearexpression incarbohydrate-responseelementandsterolregulatoryelementbindingproteins.Inconclusion,an HF–RDdietduringpregnancytranslatestochangesinliversignalingandmetabolicpathwaysin offspring,enhancingliverlipidstorageandsynthesis,andthereforenon-alcoholicfattyliverdisease (NAFLD)risk.ThesechangescanbecorrectedbyfeedinganHF–SDdietduringpregnancy.
Keywords: earlyprogramming;hepaticlipogenesis;insulin-resistantpregnancy;metabolicflexibility; non-alcoholicfattyliverdisease;slowdigestingcarbohydrates
1.Introduction
Humanshavedevelopedmetabolicadaptationstopromoteenergystorageinperiodsoffasting. Theyareespeciallysuitedtostimulatingfatstoragefromothernutrientsandcarbohydratesasan
Nutrients 2019, 11,844
larddiet(HF;20.5%fat,24.2%protein,41.5%carbohydratesand7.9%fiberperweight)priortomating forsixweeks.ControlanimalswerefedastandardreferencedietAIN93M.Ratswerematedforthree daysandthenrandomlyassignedtooneoftheexperimentaldietsduringgestation:ahigh-fatdiet containingslow-digestingcarbohydrates(HF–SDgroup),ahigh-fatdietcontainingrapid-digesting carbohydrates(HF–RDgroup)oranAIN93Gdietforthereferencegroup.Compositionofthethree dietsusedduringgestationisdescribedinMartinetal.[6](SupplementaryTableS1).Alldietswere preparedatAbbottNutritionR&Dfacilities.Atthedelivery,allanimalswerefedAIN93Gdiets regardlessofthedietconsumedduringpregnancy.Onday21afterdelivery,allpupswereweaned ontotheAIN93Mdietandhousedforsevenadditionalweeks(Figure 1).Attheoutcome,insulinand glucagonserumconcentrationswereassayed(SupplementaryTableS2).
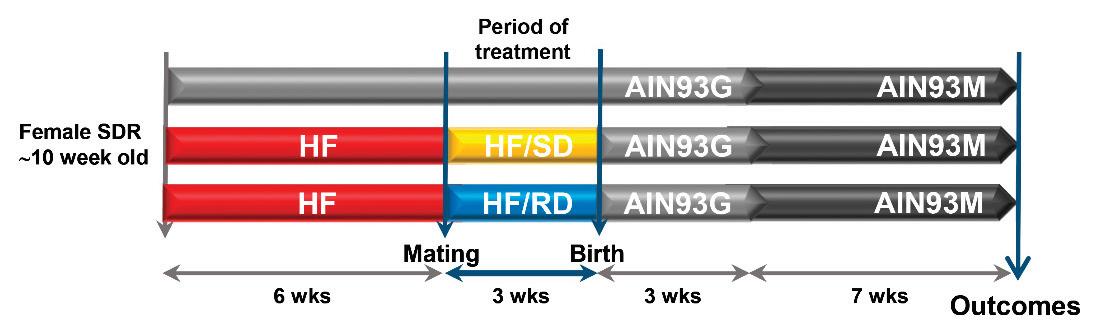
Figure1. Experimentalmodel.Virginratswereassignedtooneofthreeexperimentalgroups:reference damsfedastandardrodentdietbeforematingandthroughoutpregnancy;damsfedahigh-fatdiet (HF)sixweeksbeforematingandthenanHFdietcontainingeithercarbohydrateswithahigh(HF–RD) orlow(HF–SD)digestionratethroughoutpregnancy.Atdelivery,alltheanimalswerefedthestandard rodentdietfortheremainderofthestudy(10weeks).SDR,Sprague-Dawleyrats.
2.3.LiverLipidomicsAnalysis
AbsoluteconcentrationofTAGsintheliverwasmeasuredusingatriglycerides-LQkit(Spinreact, Barcelona,Spain).Metaboliteprofileswereanalyzedaspreviouslydescribed[7,8].Briefly,two separateUHPLC-time-of-flight(TOF)–MS-basedplatforms(AgilentTechnologies,SantaClara,CA, USA)analyzingmethanolandchloroform–methanolliverextractswerecombinedtosemi-quantify lipidspecies.Non-esterifiedfattyacyls,bileacidsandlysoglycerophospholipidswereanalyzedin themethanolextractplatform.Forthis,methanolwasaddedtothefrozenlivertissue(30:1, v/w), andthismixturewashomogenizedwithaPrecellys24grinder(Precellys,Montigny-le-Bretonneux, France),followedbyproteinprecipitation.Themethanolusedforextractionwasspikedwithinternal standardsnotfoundinlivertissueusingthesamemethod.Afterbriefvortexmixing,sampleswere incubatedovernightat 20 ◦ C.Supernatantswerecollectedanddriedaftercentrifugationat 16,000× g for15min.Thedriedextractswereresuspendedinmethanol,centrifugedat16,000× g for5minand supernatantswerecollectedandtransferredtovialsforUHPLC–MS(AgilentTechnologies,Santa Clara,CA,USA)analysis.
Thechloroform–methanolextractplatformprovidedcoverageoverglycerolipids, glycerophospholipids,sterollipidsandsphingolipids.LivertissueswerehomogenizedinthePrecellys 24grinderbymixingwithchloroform–methanol(2:1, v/v)andsodiumchloride(50mmol/L)(overall ratio1:30:3, w/v/v),followedbyproteinprecipitation.Theextractionsolventwasspikedwithinternal standardsnotdetectedusingthesamemethod.Afterbriefvortexmixing,sampleswereincubatedat 20 ◦ Cfor1h.Aftercentrifugationat16,000× g for15min,thelowerorganicphasewascollected andthesolventremoved.Thedriedextractswerethenresuspendedinacetronitrile–isopropanol(1:1), centrifugedat16,000× g for5minand,finally,supernatantsweretransferredtovialsforUHPLC–MS analysis.LipidnomenclaturefollowstheLIPIDMAPSconvention, www.lipidmaps.org
ThiscombinedanalysiswasestablishedforrodentlivertissuebyOWLMetabolomics(Derio, Spain).DataobtainedwiththeUHPLC–MSwereprocessedwiththeTargetLynxapplicationmanager forMassLynx4.1(WatersCorp.,Milford,MA,USA).Intra-andinter-batchnormalizationfollowedthe






















