9 minute read
Alessandro Rosa
STUDY OF THE ROLE OF RNA-BINDING PROTEINS IN THE NEURODEGENERATIVE DISEASE AMYOTROPHIC LATERAL SCLEROSIS
Alessandro Rosa
Advertisement
RESEARCH AREA: Genetics, biology and pathophysiology of eukaryotes
Department of Biology and Biotechnology “Charles Darwin” alessandro.rosa@uniroma1.it
Our laboratory studies the mechanisms underlying the motor neuron disease Amyotrophic Lateral Sclerosis (ALS). Mutations or altered activity in some RNA-binding proteins (RBPs) have been causally linked to this incurable disease. These include the RBP FUS, which is mutated in patients affected by an aggressive form of familial ALS, and TDP-43, which is mutated in a subset of patients with familial ALS and has been found in pathological inclusions of the majority of sporadic ALS patients. The general goal of our project is to gain insights into the toxic functions acquired by mutant/altered ALS-linked RBPs, with a specific focus on the alteration of the cross-regulatory network of RBPs that is in place in motoneurons (MNs). We take advantage of human induced Pluripotent Stem Cells (iPSCs) carrying a pathogenic mutation in the FUS gene (FUSP525L) and their isogenic control (FUSWT), previously raised in our lab (Lenzi et al., Disease Models and Mechanisms 2015; De Santis et al., Stem Cell Reports 2017). These cells can be effectively differentiated into MNs using a novel protocol developed in our lab (De Santis et al., Stem Cell Research 2018; Garone et al., JoVE 2019), providing an in vitro platform for ALS disease modelling. Main findings were then validated in a mutant Fus mouse model (FusΔ14 knock-in) thanks to a collaboration with Prof. Pietro Fratta (UCL, London, UK). We also use simpler cellular models, consisting in HeLa cells ectopically expressing fluorescently labeled RBPs, for the characterization of biomechanical properties of granules and aggregates by a novel microscopy tool.
This annual report is relative to months 19-24 of the project. During this period, we have completed the characterization of the molecular mechanism by which mutant FUS expression in human motor neurons causes an aberrant upregulation of the levels of another neural RNA-binding protein, HuD (also known as ELAVL4), resulting in a publication (Garone et al., Communication Biology 2021). We have also raised novel constructs for studying the biomechanical properties of ALS RBP-containing granules/aggregates by background-deflection Brillouin microscopy.
The Aim 1 of the project was to define the molecular mechanisms and functional consequences of mutant FUS and HuD interplay.
We had previously shown that mutant FUS binds the 3’UTR of HuD mRNA (De Santis et al., Cell Reports 2019), but the consequences of this gain of function remained unexplored.
During this funding period we have completed the characterization of a novel aberrant mechanism that results from the competition between mutant FUS and the RBP FMRP (encoded by the Fragile X mental retardation gene, FMR1) for HuD 3’UTR binding. FMRP binding on the 3’UTR of HuD was initially predicted using a bioinformatics tool. Several RBPs showed indeed interaction propensity with this sequence, including some RBPs involved in the regulation of translation. Among these candidates, we focused on FMRP, which is a known negative regulator of translation. Native RNA immunoprecipitation (RIP) was then used to validate the physical association between FMRP and the HuD transcript. FMRP immunoprecipitation from FUSWT hiPSC-derived motor neuron extracts enriched the HuD mRNA. Notably, HuD mRNA levels were reduced in FMRP immunoprecipitated samples from FUSP525L MNs. Moreover, by taking advantage of a luciferase construct carrying the HuD 3’UTR, we found that reporter activity was strongly reduced when FMRP was overexpressed, while co-expression of FUSP525L, but not FUSWT , partially reverted such negative regulation. Finally, we have set up an in vitro competition assay in which fragments corresponding to the 3’UTR of HuD, in vitro transcribed as biotinylated RNAs and purified with streptavidin beads, showed binding of FMRP, thus validating the RIP results, and competition by mutant FUS. Collectively, these results suggested that FMRP could act as negative regulator of HuD translation in MNs by direct 3’UTR binding. Mutant FUS may intrude in this function, possibly by competition for 3’UTR binding, resulting in increased HuD protein levels. An important functional consequence of increased HuD protein levels is upregulation of its target transcripts, including the known targets NRN1 (Neuritin1, also known as CPG15) and GAP43. We accordingly found increased levels of both NRN1 and GAP43 in human iPSC-derived FUS mutant motor neurons and in the Fus mutant mouse model. Notably, NRN1 is known to be highly expressed in developing MNs, where its overexpression increased axonal outgrowth and neurite branching (Javaherian et al., Neuron 2005). Moreover, previous work showed that GAP43 is upregulated downstream of increased HuD during axon regeneration upon sciatic nerve injury, and its overexpression induces prolonged nerve sprouting and causes death of adult motor neurons in a transgenic mouse model (Yoo et al., Journal of neurochemistry 2013; Aigner et al., Cell 1995; Harding et al, Eur J Neurosci 1999). Interestingly, by taking advantage of microfluidics devices, we found increased axon branching and arborization, and faster growth upon injury, in FUS mutant motor neurons generated by human iPSC differentiation and obtained from the Fus mutant mouse model. These axonal phenotypes could be rescued by dampening NRN1 levels in FUSP525L motor neurons. In siRNA-NRN1-treated cells cultured in microfluidics devices we indeed observed reduced number of axon branches and junctions and reduced axon growth during regeneration, compared to non-targeting control siRNAs. We have thus demonstrated that this phenotype is a consequence of increased NRN1.
These results have been recently published (Garone et al., Communications Biology 2021).
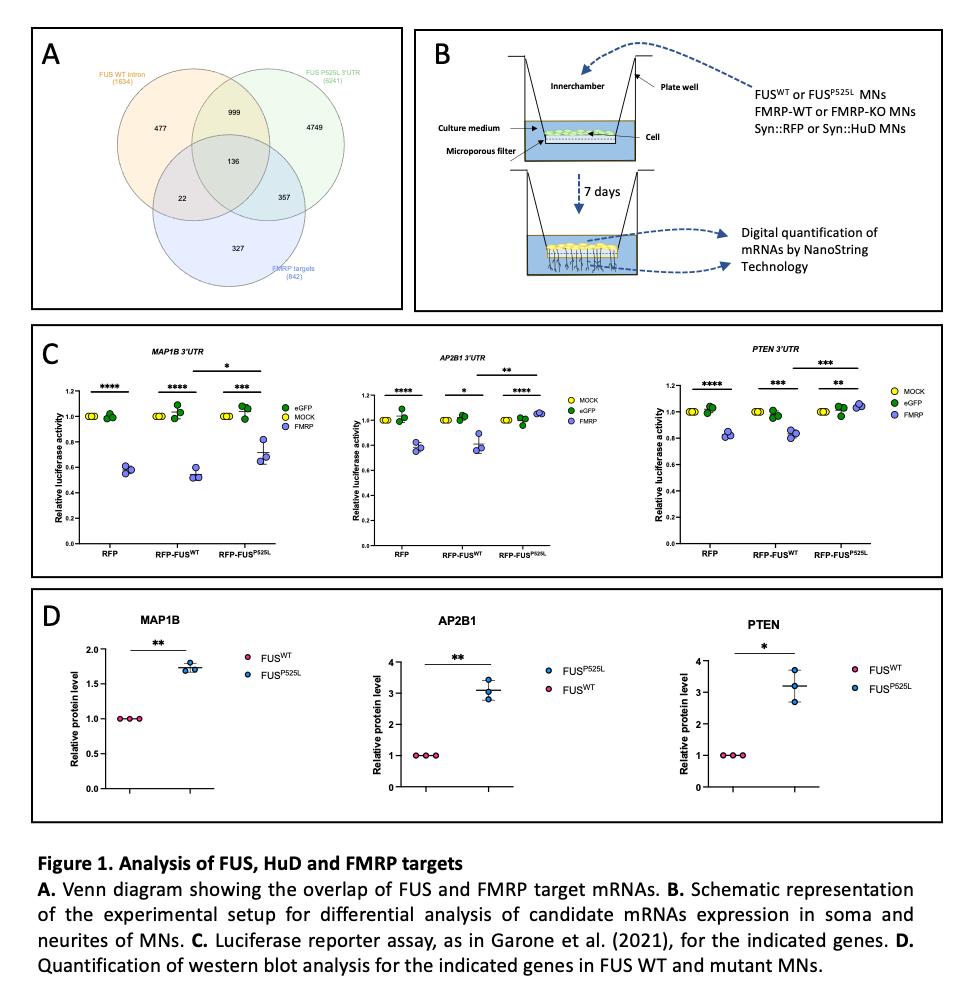
The Aim 2 of the project was to globally assess the impact of mutant FUS on its interactors. An interesting result of the work published during the previous funding period (Garone et al., Scientific Reports 2020) was that cytoskeleton proteins are altered in FUS mutant motoneurons. Interestingly, changes in the proteome were correlated with altered binding of mutant FUS on its target genes, with FUSWT showing preferential binding to introns, while FUSP525L to 3’UTRs. Notably, both FUS, HuD and FMRP are RBPs with a role in RNA transport and localization along neurites. Taken together with the results of Aim 1, these findings prompted us to include FMRP among the RBPs studied in this project. Thus, despite the original plan was to perform a ribosome profiling in whole cells, we have instead decided to understand which mutant FUS targets are differentially localized in MNs due to impaired FMRP activity. FUS interactome data (De Santis et al., Cell Reports 2019) were interrogated to select targets bound in the intronic regions by FUSWT and in the 3'UTR by FUSP525L. This list (1135 targets) was cross-referenced with HITS-CLIP FMRP data (Darnell et al., Cell 2011), resulting in an overlap of 136 targets (Figure 1A). Gene ontology (GO) term enrichment analysis on such set of common FMRP and FUS targets revealed categories related to cytoskeletal protein binding (molecular function), neuron projection development and morphogenesis (biological properties), neuron projection, dendrite, growth cone, synapse and postsynapse (cellular component). Thus, according with our previous work (Garone et al., Scientific Reports 2020; Garone et al., Communication Biology 2021), FMRP and FUS interactomics analysis support the hypothesis that altered RBP network in ALS MNs contribute to impaired mRNAs transport to a subcellular site, a function often compromised in brain diseases. The DISEASE bioinformatics tool (Pletscher-Frankild et al., Methods 2015) was then used to assess the association of the 136 candidates to neurodevelopmental and/or neurodegenerative disorders, resulting in a subset of 71 genes with high z-score. These
targets were profiled in soma and neurite compartments of WT and mutant FUS MNs by taking advantage of cell culture inserts and the NanoString technology. N-counter automated assay by NanoString allows to count the native RNA molecules without amplification, employing fluorescent barcodes that allow for the direct, digital detection of distinct mRNA targets in a single run. It only requires nanoscale amounts of RNA and has a detection sensitivity of 1 copy per cell (Salerno and Rosa, Stem Cells International 2020). Genes downregulated in the neurites of mutant FUS were enriched for categories related to synapse assembly, synaptic growth at the neuromuscular junction, and positive regulation of synaptic transmission, while we found categories related to neuron development, differentiation, and morphogenesis among upregulated transcripts. We then repeated this analysis on HuD overexpressing (Syn::HuD; Garone et al., Communications Biology 2021) and FMRP-KO (Brighi et al., Cell Death and Disease 2021) MNs. Collectively, this work resulted in 12 common targets that: i) show increased levels in FUS mutant, Syn::HuD, and FMRP-KO neurites; ii) are direct interactors of FUSP525L in the 3’UTR, FUSWT in intronic regions, and FMRP; iii) had been previously involved in diseases of the nervous system. Among them, we focused for further validation on 3 candidates: PTEN, MAP1B and AP2B1. As shown in Figure 1C, a luciferase reporter fused to the 3’ UTR of PTEN, MAP1B or AP2B1 showed decreased activity in presence of FMRP and this effect was partially (MAP1B) or completely (PTEN and AP2B1) rescued by mutant (but not WT) FUS co-expression. Finally, il all cases western blot analysis showed upregulated protein levels (Figure 1D).
Collectively, these findings have clarified that altered crosstalk between multiple RBPs, namely FUS, HuD and FMRP, has a significant impact on common targets involved in nervous system diseases in terms of their localization and translation. These results are included in a manuscript in preparation.
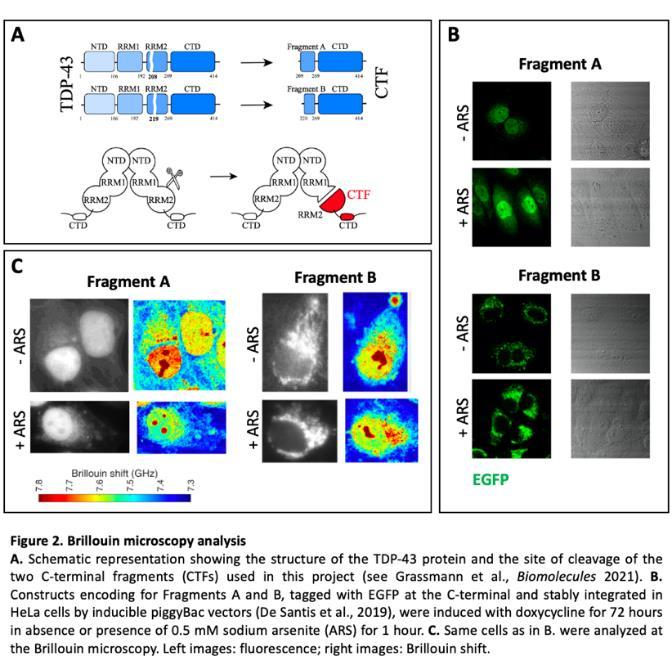
The Aim 3 of the project was to take advantage of the Brillouin microscope to characterize the biomechanical properties of granules/aggregates containing ALSrelevant RBPs. This activity takes advantage of a non-contact and label-free imaging method, named backgrounddeflection Brillouin (BDB) microscopy, to investigate the three-dimensional intracellular biomechanical properties of whole cells at a sub-micron resolution (Antonacci et al., Communications Biology 2018; De Santis et al., Cell Reports 2019). During this funding period, we have used the BDB to study TDP-43 C-terminal fragments (CTFs) that are commonly found in post-mortem ALS patients’ brain samples. We have focused on two CTFs that we have recently characterized by molecular docking and additional molecular dynamics simulations, indicated as Fragment A and Fragment B in Grassmann et al., (Biomolecules 2021) (Figure 2A). As shown in Figure 2B (- ARS panels), we observed a striking difference in the localization of the two fragments, expressed as fluorescently tagged proteins in HeLa cells. In particular, in most cells, Fragment A shows a diffuse distribution in both nucleus and cytoplasm, with few small puncta in some cells, while Fragment B is excluded from the nucleus and localized in speckles in the cytoplasm. Upon induction of oxidative stress by sodium arsenite treatment for one hour, while Fragment A remained diffuse, Fragment B formed bigger condensates (Figure 2B, + ARS panels). Characterization of the biomechanical properties of Fragment A and B at the BDB (Figure 3C) suggest that subcellular compartments containing Fragment B are stiffer than those containing Fragment A. Thus, individual TDP-43 CTFs, even if diverging only for few aminoacids, might differently contribute to pathological aggregation.
Publications
Garone, M.G., Birsa, N., Rosito, M., Salaris, F., Mochi, M., de Turris, V., Nair, R.R., Cunningham, T.J., Fisher, E.M.C., Morlando, M., Fratta, P., Rosa, A. ALS-related FUS
mutations alter axon growth in motoneurons and affect HuD/ELAVL4 and FMRP
activity. Commun Biol 2021 4, 1025. doi:10.1038/s42003-021-02538-8. IF: 6.268
Research Group Collaborations
Maria Giovanna Garone, PhD student Beatrice Silvestri, PhD student Chiara D’Antoni, Graduate fellow Pietro Fratta, Nicol Birsa; UCL Institute of Neurology, London (UK)
Giancarlo Ruocco, Andrea Armirotti, Maria Rosito, Valeria de Turris, Beatrice Salvatori, Debora Salerno,
Claudia Testi; Fondazione Istituto Italiano di Tecnologia






