Bacterial Diversity in Invertebrates: A 16S rRNA Nanopore Sequencing Project

LaShawn Bowe, Felicia Silva, Dr. Barbara Murdoch
Eastern Connecticut State University, Willimantic CT USA
Introduction
• Invertebrates are animals that lack a backbone. They are the largest group of animal species.
• The invertebrate microbiome refers to the collection of all microorganisms living in and on the host.
• Microbiomes are important for immune function, mental health, digestion, and more.
• 16S rRNAgene sequencing identifies bacteria by analyzing the sequences of the 16S ribosomal gene, which is a reliable marker for bacterial identification and phylogeny.
• Nanopore sequencing is a DNA sequencing method that utilizes tiny protein pores (nanopores) to identify the sequence of DNA or RNA
Hypothesis
Giant Mesquite Bug (Thasus gigas) will show higher bacterial diversity than the Whip Spider (Amblypygid).
1. Giant Mesquite Bug & Whip spider


Methodology
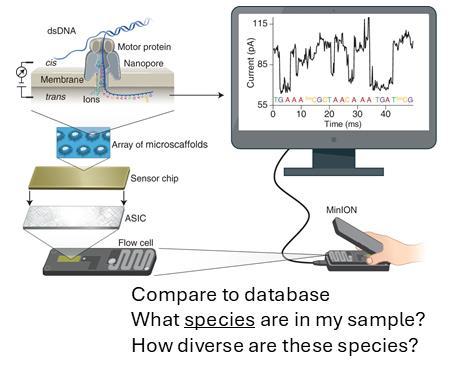
Nanopore sequencing:
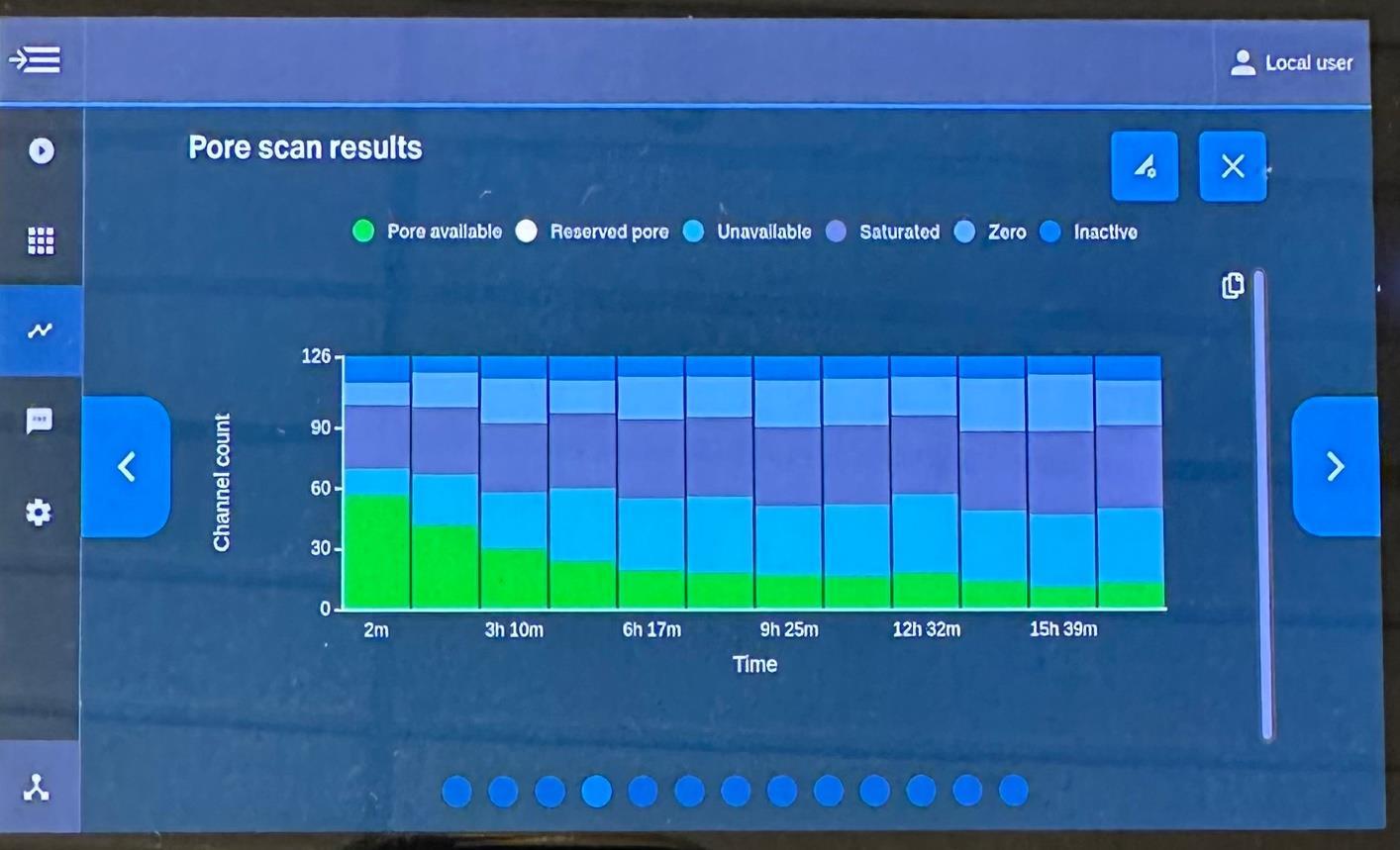
• Utilizing Oxford Nanopore sequencing on Flongle device. DNA was analyzed for 16S rRNA gene
• Taxonomic analysis was conducted through EPI2ME bioinformatics
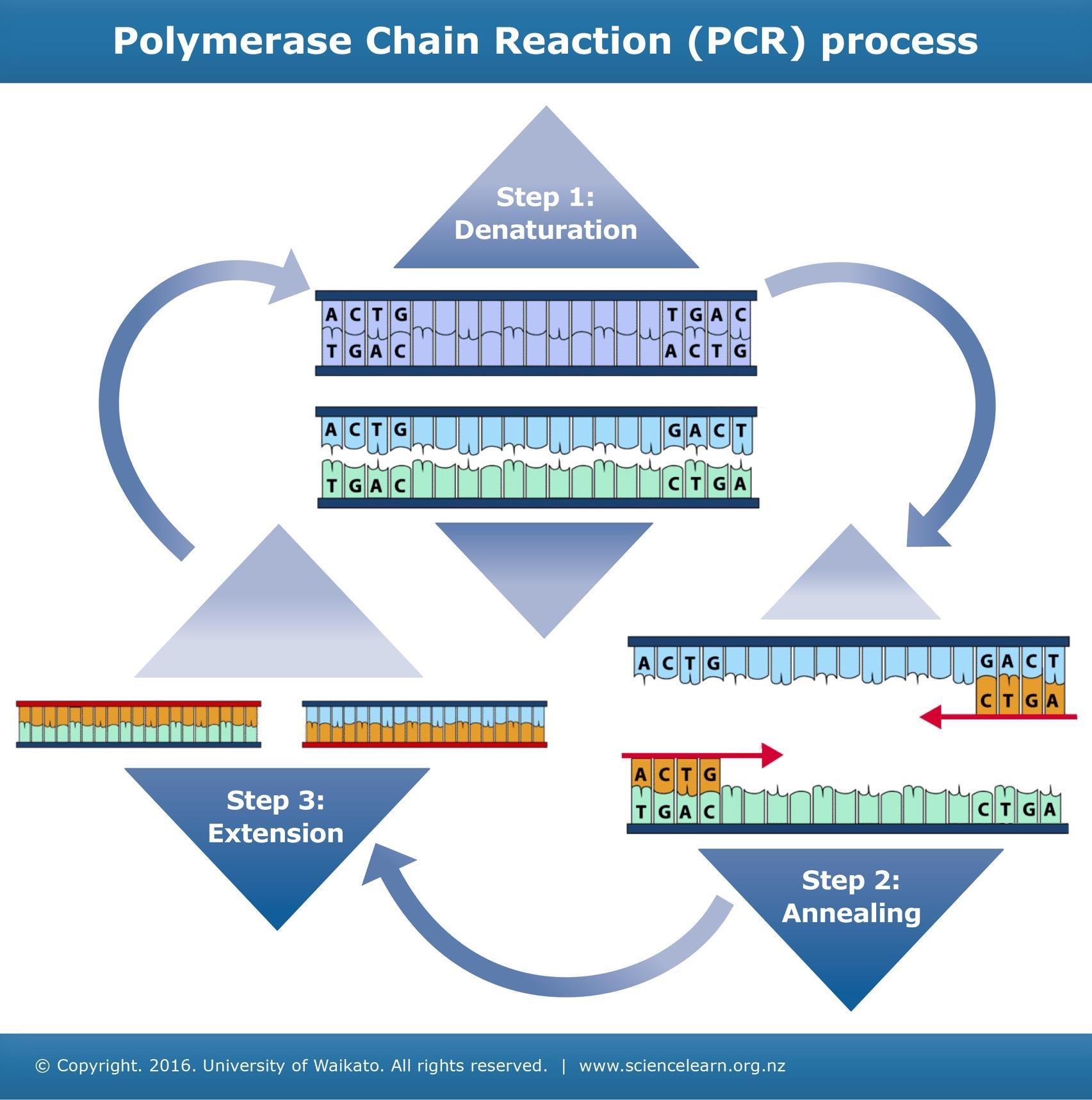
Amplification of bacterial DNA:
• DNA was isolated from T. gigas and Amblypygid
• 16S rRNA genes were amplified
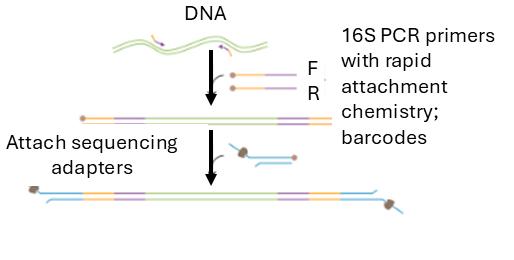
Results
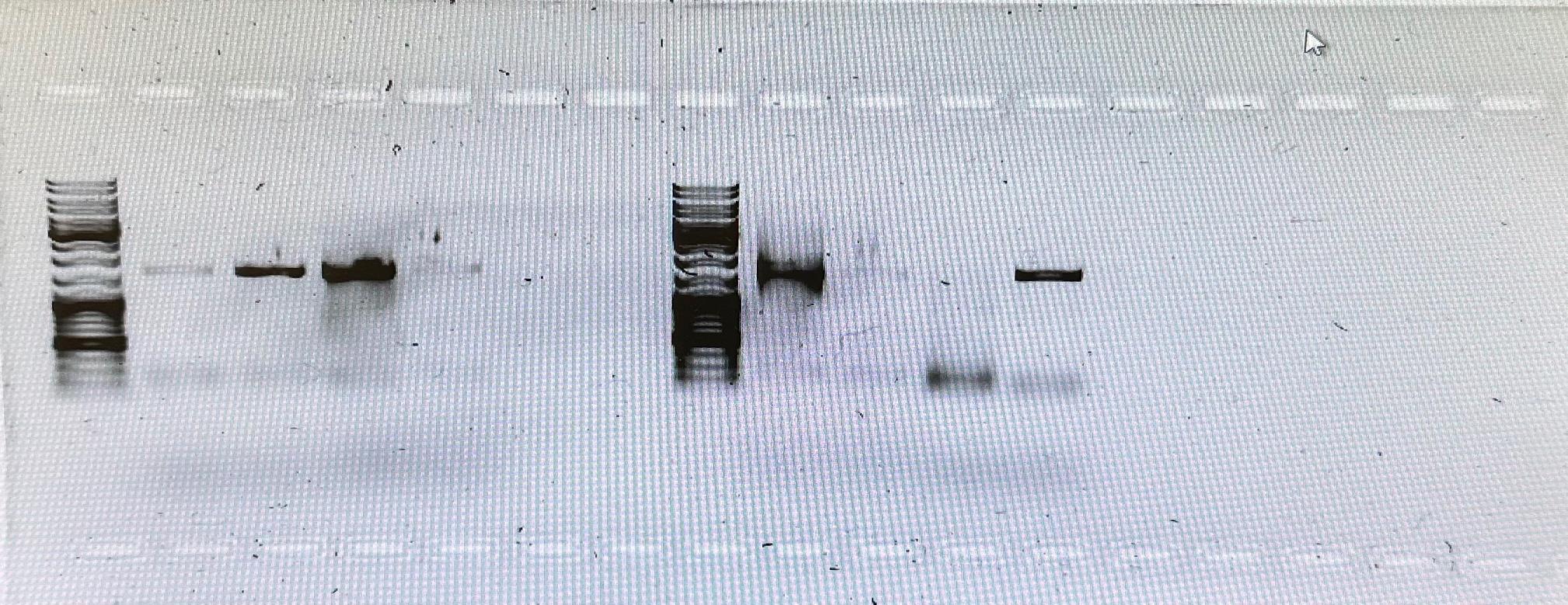
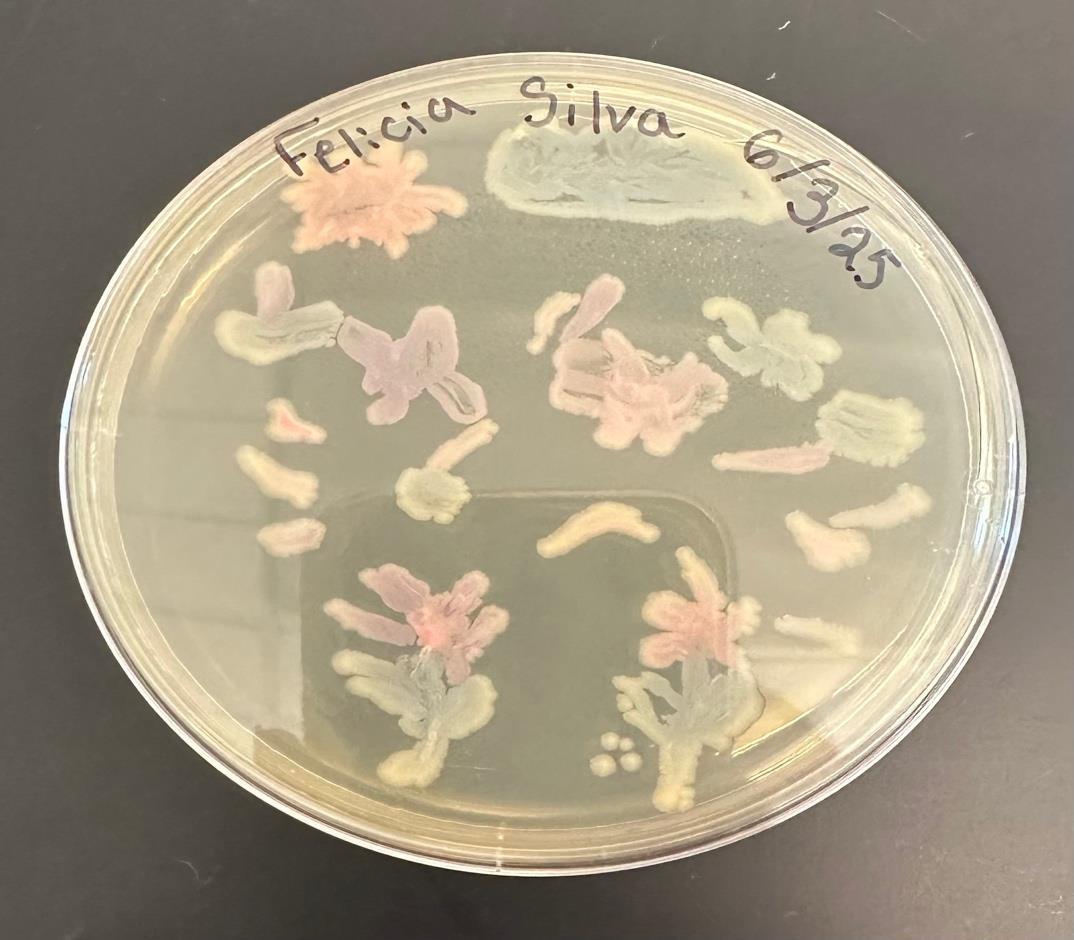
Figure 5. Amplified invertebrate 16S rRNA gene from PCR

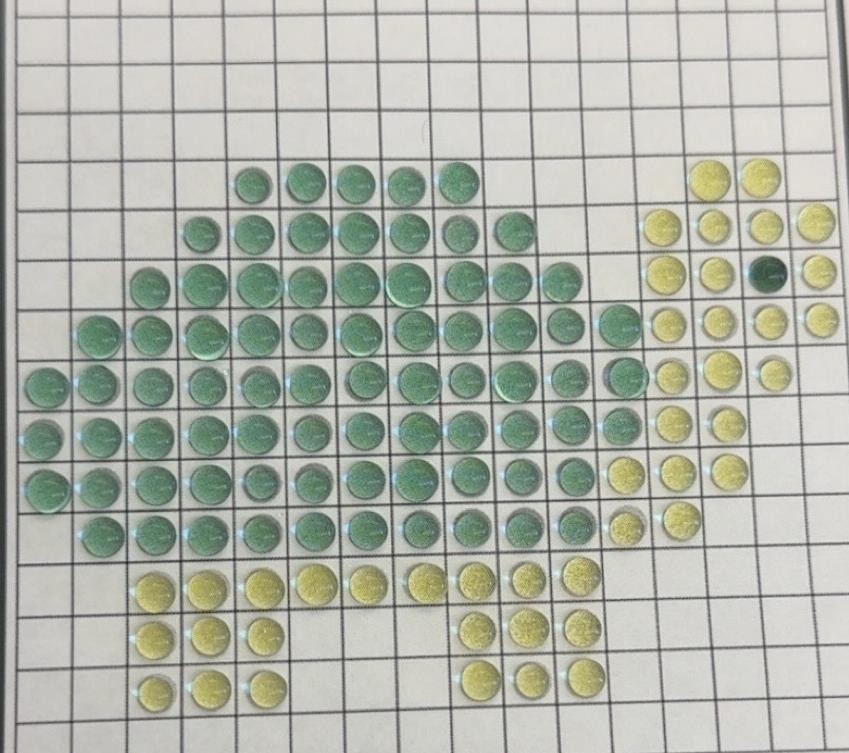
3. Sequenced Bacteria from Invertebrates at Genus Level
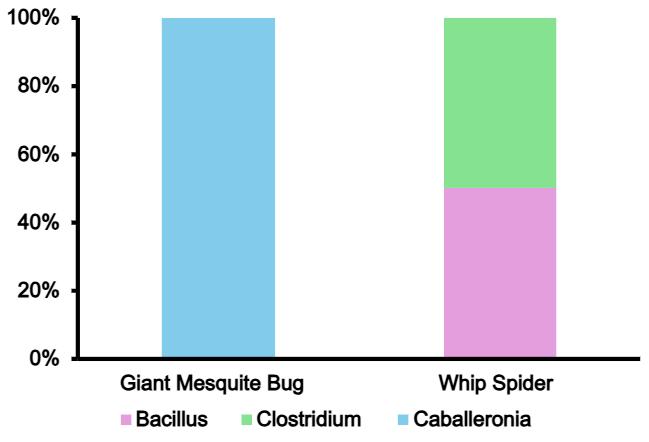
2. Sequenced Bacteria from Invertebrates at Phylum Level
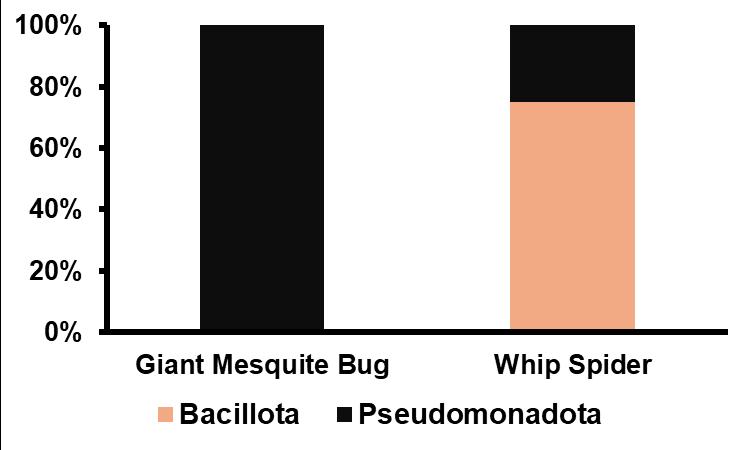
of
in microbiome of invertebrates. Analysis done through EPI2ME bioinformatics.
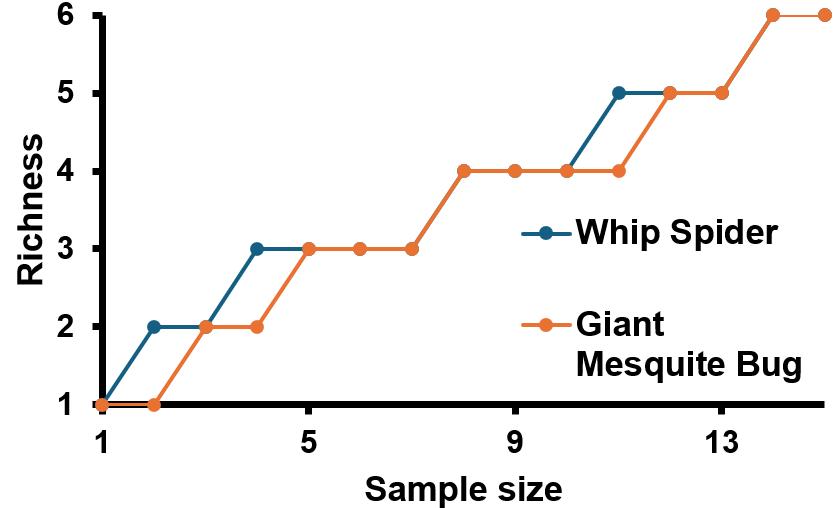
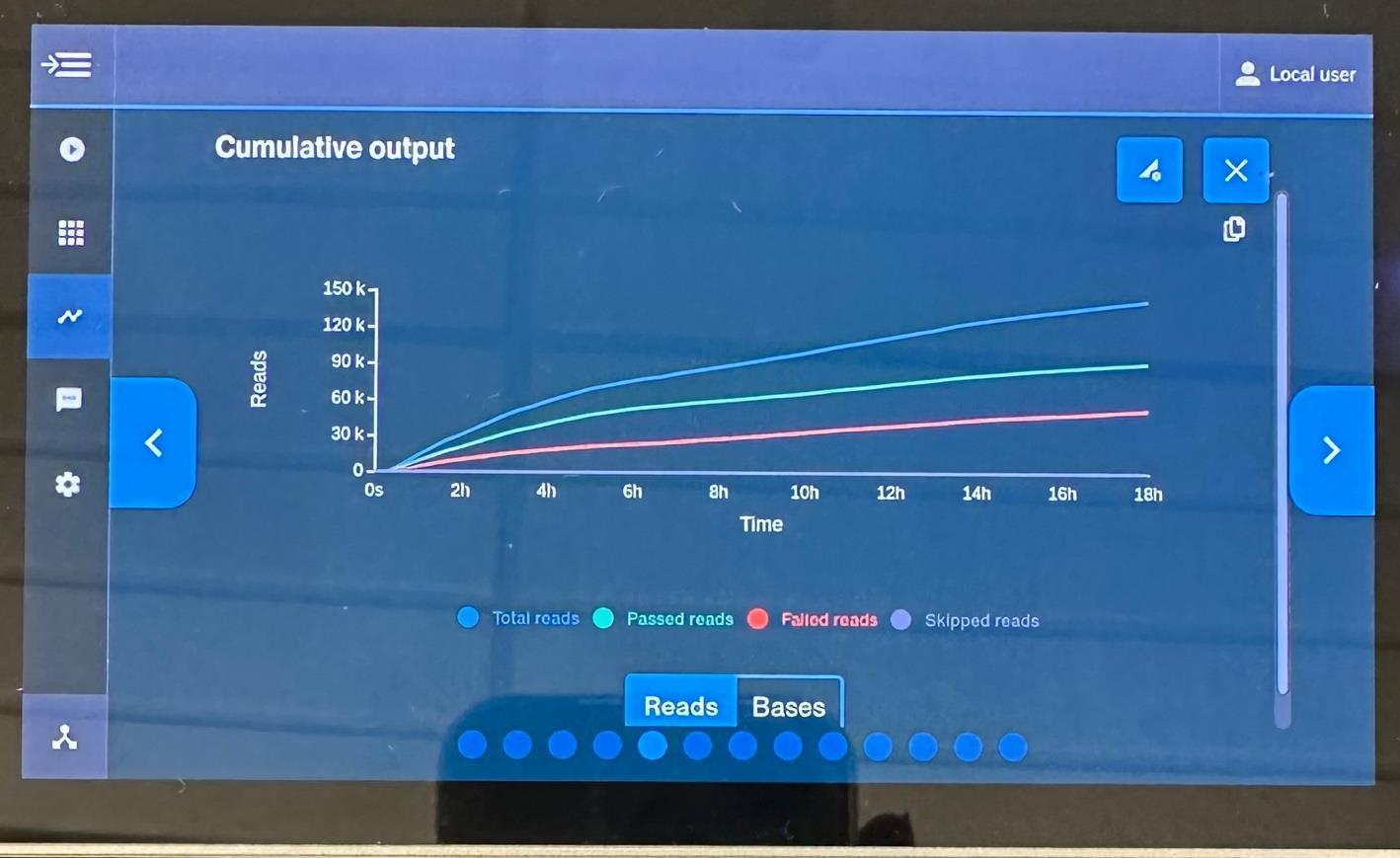
9. Rarefaction Graph. Amount of different bacteria found over the course of the sequencing run shown using EPI2ME bioinformatics.
Conclusions
Not enough data to support hypothesis.
Acknowledgements
We thank Dr. Murdoch for lecture notes and the experience; Dr. Graham for field work to collect samples; Zachary Kelly for his help throughout this process; Gemini, a large language model trained by Google
