
3 minute read
AI & Pathogen Spillover Prediction
CERI Data Science on diverse international panel discussions in Oxford and Berlin providing insights from pathogen spillover dynamics to AI for infectious disease modelling
In the last month, our work within the CERI Data Science Unit was showcased across two international discussion panels contributing to insightful public discourse on pathogen spillover dynamics and AI for infectious disease modelling.
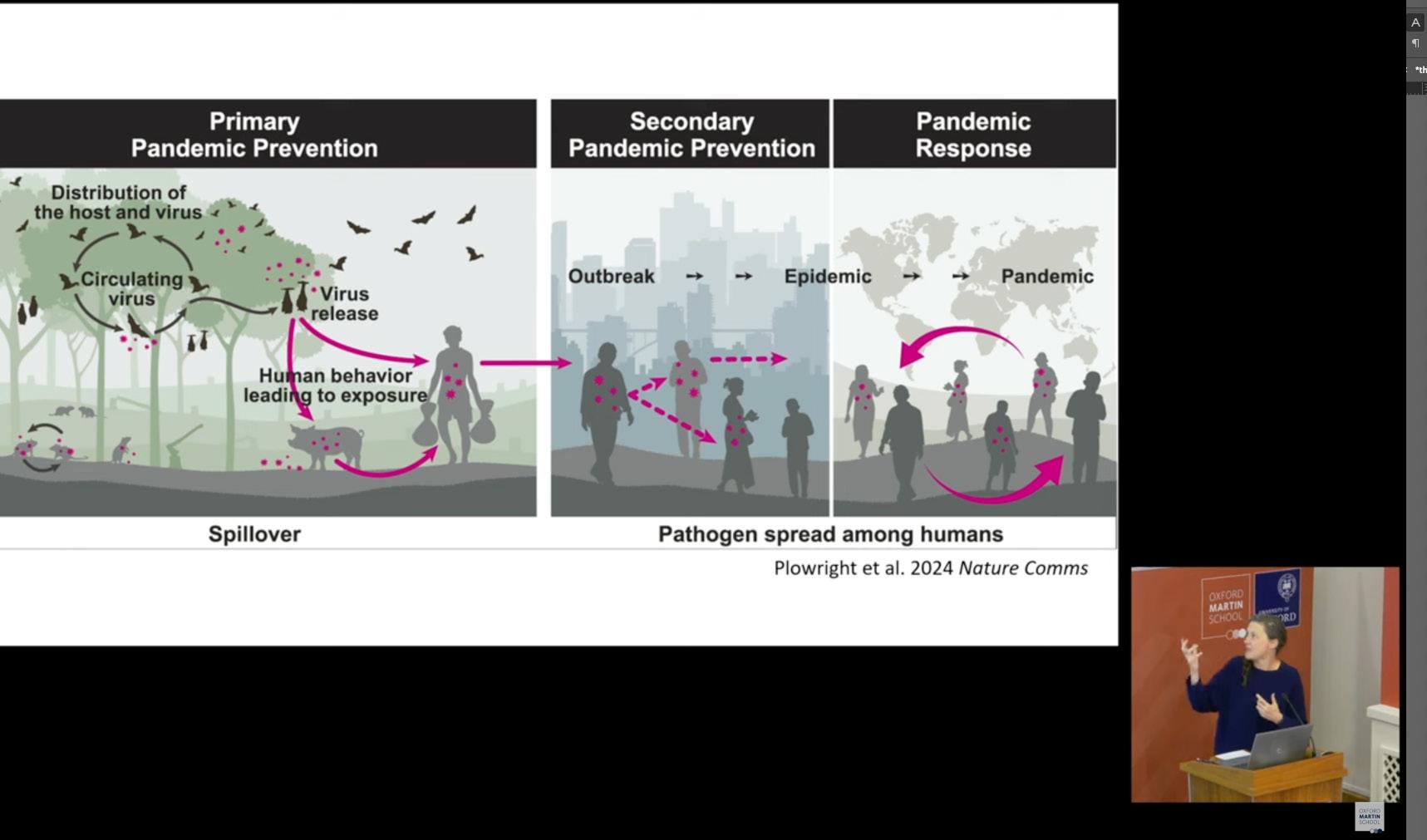
First, the head of CERI’s Data Science Unit, Dr. Houriiyah Tegally, was present at the Oxford Martin School of Oxford University, presenting on an expert panel entitled “What turns spillovers into epidemics?”. Chaired by Professor Jessica Metcalf from Princeton University, this hybrid discussion involved global experts to explore how AI, genomics, climate and ecological data are transforming our ability to predict emerging infectious diseases, featuring leading voices in pathogen genomics, ecology, virology, and data science from Oxford, Glasgow, Edinburgh, Pirbright, and Stellenbosch University.
The panel delved into various critical aspects of understanding why zoonotic disease spillover events sometimes lead to large sustained human-to-human transmission and epidemics, and sometimes not. For instance, the panel discussed the traits of successful pandemic pathogens, emphasizing real-world examples such as the Oropouche virus, which recently escalated from localized spillovers to a widespread epidemic due to a combination of genetic adaptation, increased ecological suitability in urban-sylvatic interfaces, and high human mobility. Experts highlighted that while spillovers are common, the transition to pandemics depends on key factors like viral evolution, host availability, and environmental conditions. Surveillance systems, though imperfect, must be more dynamic and strategically resourced, including global data-sharing and prioritizing localized outbreak monitoring.
The panel stressed the need for better understanding of pathogen adaptation, especially transmission modes, as these changes can undermine existing interventions. Promising future strategies that were discussed include proactive surveillance using multi-model risk estimates and AI-driven genomic analysis to predict and respond to emerging threats.
Our Data Science work was then presented at the most recent WHO Hub Pandemic and Epidemic Intelligence Innovation Forum. This online panel session, attended by over 300 participants, was centred around leveraging AI for infectious disease modelling and public health decision making.
The panel of experts, including Dr. Houriiyah Tegally from CERI, highlighted that the convergence of artificial intelligence (AI) and public health is opening new frontiers in our ability to understand, predict, and respond to infectious disease threats. Speakers explored how AI is transforming infectious disease modelling, enabling more accurate predictions, real-time risk assessments, and adaptive policy responses.
Houriiyah presented an integrated AI-driven modelling framework for genomic epidemiology and pandemic risk mapping. This work combines diverse data sources, genomes, epidemiological, environmental, and demographic data, with advanced phylodynamic, ecological, and epidemiological models. AI methods, including anomaly detection, deep learning, and large language models (LLMs), are enhancing both insight generation and model efficiency. Applications range from rapid variant detection and disease distribution prediction to estimating human migration via satellite imagery. These innovations are directly supporting outbreak response by enabling faster, more localized disease risk mapping and streamlining genomic surveillance outputs for public health decision-makers.
These initiatives are part of CERI’s commitment towards broad scientific communication, ensuring that our research work not only reaches far and wide, but is also directly put into context of immediate public health applications.
This panel discussion followed the recent AI in Infectious Diseases paper at Nature 2025.
Access full paper in Nature: Kraemer, M.U.G., Tsui, J.LH., Chang, S.Y. et al. Artificial intelligence for modelling infectious disease epidemics. Nature 638, 623–635 (2025). https://doi.org/10.1038/s41586-024-08564-w

Watch the Panel Discussion: ‘What turns spillovers into epidemics?’ - https:// www.youtube.com/watch?v=V5EX3DRzyvs










