Molecular characterization of the HMTp210 gene of Avibacterium paragallinarum and the proposition of a new genotyping method as alternative for classical serotyping
Rianne Buter1, Anneke Feberwee1, Sjaak de Wit1,7, Annet Heuvelink1, Ana da Silva2, Rodrigo Gallardo2, Edgardo Soriano Vargas3, Stefan Swanepoel4, Arne Jung5, Mathias Tödte6 & Remco Dijkman1

1Royal GD, P.O. Box 9, 7400 AA Deventer, The Netherlands, 2Department of Population Health and Reproduction, School of Veterinary Medicine, University of California, 4008 VM3B, Davis, CA 95616, 3Centro de Investigación y Estudios Avanzados en Salud Animal, Facultad de Medicina Veterinaria y Zootecnia, Universidad Autónoma del Estado de México, Toluca, México, 4Deltamune, Unit 34, Oxford Business Park, 3 Bauhinia Street Highveld Eco-Park,Centurion,South Africa, 5Klinik für Geflügel, University of Veterinary Medicine, Hannover, Germany, 6Tierarztpraxis MMT, Kothen, Germany, 7Department of Population Health, Faculty of Veterinary Medicine, Utrecht University, the Netherlands.
Introduction
Avibacterium paragallinarum (AVP) is the etiological agent of infectious coryza (IC) in chickens and characterized by acute respiratory distress and severe drop in egg production. Vaccination is important in the control of IC outbreaks and the efficacy of vaccination is dependent on AVP serovars included in the vaccine. Classical serotyping of AVP is laborious and hampered by the poor availability of antigens and antisera. The hemagglutinin, important in classical serotyping is encoded by the HMTp210 gene. HMTp210 gene analysis has shown to have potential as alternative for classical serotyping. The aim of the present study was to further investigate the potential of sequence analyses of nucleotides 1-~1200 of region 1 of the HMTp210 gene, the HMTp210 hypervariable region and the concatenated sequences of both fragments as alternative for classical serotyping.

Methods
• In total 123 sequences were used including:


- 82 cultures from vaccine strains and Page and Kume reference strains and AVP field isolates originating from Asia, Africa, Europe and Latin America (58 of these 82 were with known Page and/or Kume serovar). 41 full HMTp210 coding sequences obtained from GenBank.
• Phylogenetic analysis (Maximum Parsimony tree, MP) using BioNumerics of sequences of HMTp210; concatenated sequences from region 1 and the hypervariable region (HVR) (Figure 1).
• Within and between cluster variation was calculated and presented as percentage identity.
• Genotypes (GT) were defined from clusters of isolates with a nucleotide sequence identity of ≥98%.
• Evaluation of discriminating power and correlation between GTs and Page and/or Kume servars.
Results
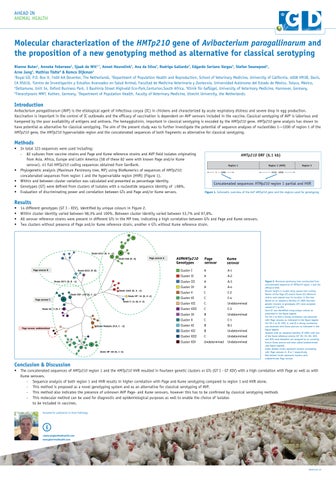
• 14 different genotypes (GT I - XIV), identified by unique colours in Figure 2.
HMTp210
ORF (6,1 kb)

Region 1 Region 2 (HVR) Region 3 1 - ~ 1200
3316 - 4950
Concatenated sequences HTMp210 region 1 partial and HVR
Figure 1. Schematic overview of the AvP HMTp210 gene and the regions used for genotyping
• Within cluster identity varied between 98,3% and 100%. Between cluster identity varied between 53,7% and 97,8%.
• All serovar reference strains were present in different GTs in the MP tree, indicating a high correlation between GTs and Page and Kume serovars.
• Two clusters without presence of Page and/or Kume reference strain, another 4 GTs without Kume reference strain.
Conclusion & Discussion
Figure 2. Maximum parsimony tree constructed from concatenated sequences of HMTp210 region 1 and the HMTp210 HVR. Branch length is scaled using square root scaling. Names of the Page (P) and/or Kume (K) reference strains were placed near its location in the tree. Based on an sequence identity of ≥98% fourteen genetic clusters or genotypes (GT) were assigned, named GT I to XIV. Each GT was identified using unique colours as presented in the figure legend.
For GTs I to XIII a strong correlation was observed with Page serovars as indicated in the figure legend. For GTs I to VI, VIII, X, and XI a strong correlation was observed with Kume serovars as indicated in the figure legend.
Isolates with an sequence identity of <98% with one of the Kume reference strains (GT VII, IX, XII, XIII and XIV) were therefore not assigned to an currently known Kume serovar and were called Undetermined (see figure legend). Green dotted circles represent clusters correlating with Page serovars A, B or C respectively. Red dotted circles represent clusters with undetermined Page serovar.
• The concatenated sequences of HMTp210 region 1 and the HMTp210 HVR resulted in fourteen genetic clusters or GTs (GT I - GT XIV) with a high correlation with Page as well as with Kume serovars.
- Sequence analysis of both region 1 and HVR results in higher correlation with Page and Kume serotyping compared to region 1 and HVR alone.
- This method is proposed as a novel genotyping system and as an alternative for classical serotyping of AVP.