7 minute read
Clincal Feature
Clinical Feature
The Australian Centre for Oral Oncology Research and Education has been awarded $2.23 million in funding to continue their work into the genomic profile of head and neck cancers and precancers.
Professor Camile Farah, executive director of the Australian Centre for Oral Oncology Research and Education (ACORE), says his team was absolutely delighted to receive the funding, which is through the Australian Government’s $20 billion Medical Research Future Fund, and part of the Genomics Health Futures Mission. “We are very grateful for the funding. It’s very difficult obtaining large, competitive research grants these days, with the running average for success in the National Health and Medical Research Council general round at about 10%. The competitive nature of the funding cycles has increased significantly in recent years, and the quality required to pass the bar for success is extremely high.”
The project
The project “Evaluating clinically relevant biomarkers to improve early detection and treatment of head and neck cancer” will be undertaken in collaboration with colleagues at the Peter MacCallum Cancer Centre, Genomics for Life, Icon Cancer Centre, and CSIRO. Camile established ACORE as a private research centre to leverage his academic, clinical and industry partnerships for the advancement of oral oncology translational research, personalised patient care, patient and clinician education and professional development, and overall advocacy for patients with head and neck cancer. “The premise of this grant builds on my previous work and that of my colleagues in genomic medicine,” Camile explains. “Mostly taking what we understand about the molecular aspects of head and neck cancers, specifically oral cancers and pre cancers, and trying to translate that to clinical practice by mapping the genomic profile of these lesions and then matching that to a potential treatment for a patient; a concept called precision genomic medicine.” Camile says currently what happens is cancers and pre cancers are treated the same way for different patients, generally in a “one size fits all” approach. “Currently we don’t really take any regard for the genetic profile of the patient or the molecular profile of the tumour,” he explains. “What we’re proposing is that instead of just looking at the clinical features of a lesion or the patient risk factors, moving forward we would additionally consider the genetic profile of the patient and the molecular profile of the lesion and personalise a treatment for that particular patient. Of course, that implies there are differences in molecular profiles, which we’ve shown previously in our research. That also implies that if you can find these molecular differences, you can act on them. In essence, that’s what we’ll be exploring with this round of funding.” Why the research is so important
Camile says a “one size fits all” approach doesn’t provide the best outcomes for patients. “(We hope) to increase survival rates, making sure patients don’t get recurrences of their tumours and that they respond to the correct treatment, whether that be a drug or surgery or other therapy. The ultimate goal is to improve outcomes for patients and that is why our research is so important. The survival rates of head and neck cancer with traditional treatment these days is just over 50% overall. We believe that is still very poor and would like to improve that, so by understanding the molecular profile of a particular tumour and tailoring a treatment solution to that particular patient and lesion, we believe we can improve survival outcomes for patients.”
Message for dental colleagues
“When we started this research, people called it ‘pie in the sky’ work, because they didn’t really understand what we were doing,” Camile says. “I think we’ve got to the stage where people understand a little bit more about genomic medicine. This has occurred more recently with COVID, as more people have been hearing about genomic sequencing of COVID, so suddenly people have taken more of an interest in genomic sequencing, what it means, how it works and why it’s so powerful. “What I would like my dental colleagues to understand is that this is getting closer and closer to clinical practice,” he adds. “As we move forward, there’s a need for the dental profession to understand the wider implications of personalised and precision medicine on healthcare generally as patients really take up these approaches in earnest. Our focus can’t continue to be just on the patient’s dentition, as there are broader implications for the profession in understanding genomic medicine because it will impact on our patients. I would really like to see the dental profession champion the precision medicine approach for the benefit of their patients.” “Medical research is like running a marathon, but we’re getting to the stage where we can implement some of the findings that we’ve been working on together over the last 10-15 years and translating them into clinical practice. This new funding will accelerate that significantly,” he adds.
Clinical Case
A case in point directly relevant to the dental profession relates to a 57-year-old male who was referred for a recurring non-homogeneous oral leukoplakia on the right antero-ventral tongue (Fig 1). He reported being a non-smoker but consumed alcohol irregularly. His leukoplakia had first appeared some four years earlier and had been treated with various modalities on several occasions with frequent recurrences, and had previously demonstrated mild to moderate dysplasia on histopathology (Fig 2).
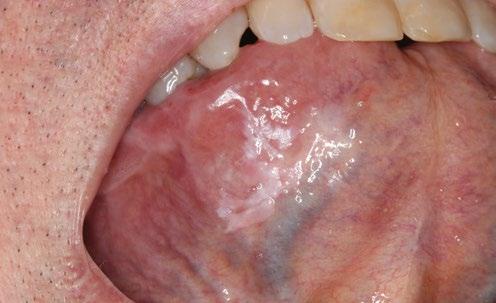
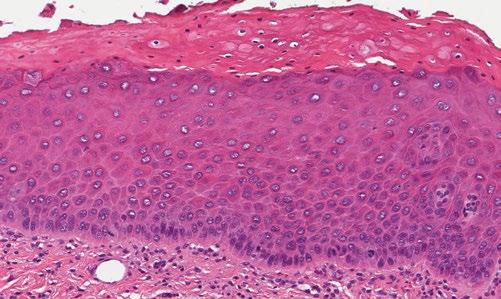
Figure 1 - Top left, Figure 2 - Bottom Left
Assessment of the genomic profile of the leukoplakia was used to understand its biology, to appreciate the reason for recurrences, to understand the malignant potential of the lesion, and finally, to decide if surgical excision or other therapy was the most appropriate approach for future management. Next generation DNA and RNA sequencing was undertaken on the lesion. This demonstrated somatic mutations in tumour suppressor genes CDKN2A and TP53 and oncogene TERT. The lesion demonstrated low Tumour Mutational Load, no Homologous Recombination Deficiency, and no somatic Copy Number Variations (Fig 3). This information categorized the leukoplakia within the “classical” subtype of oral cancers which accounts for only 22% of these tumours. CDKN2A encodes tumour suppressor protein p16 which is involved in cell growth and cell cycle control, and specifically blocks progression from G1 to S phase of the cell cycle by inhibiting Cyclin D1, resulting in cell senescence and subsequently dysplasia. Inactivation of protein p53, encoded by gene TP53, results in alterations to cell growth arrest, apoptosis and disturbed cell cycle due to cellular stress such as DNA damage. The “classical” profile of this leukoplakia clarified that the lesion was not of the more common “basal” or “mesenchymal” subtypes accounting for 42% and 35% of oral cancers respectively. “Basal” subtypes demonstrate NOTCH1 inactivation and co-mutation of HRAS-CASP8, while the “mesenchymal” subtype demonstrates modifications to immune-related genes. “M” class lesions are DNA mismatch repair proficient and thought to occur in a p53-independent tumourigenesis pathway in patients without a significant history of smoking or alcohol consumption. Finally, the “atypical” subtype is commonly seen in HPV-associated lesions. The leukoplakia was surgically excised and the patient is under long term surveillance. In the future, patients could be treated with pharmacotherapeutics either systemically or delivered to the lesion locally. This could result in silencing of an oncogene such as TERT. Alternatively, the patient could receive recombinant gene therapy against TP53 to replace the mutant gene with a wildtype equivalent in order to restore normal p53 activity. These therapies are currently being explored in clinical trials. It is clear that understanding the genomic profile of a lesion as articulated in this example will become vitally important, so the most appropriate therapy can be selected for an individual patient and lesion.
Molecular Analysis
• Clinical actionable (druggable/predictive/prognostoc and/or with diagnostic/classification implications in the disease primary site and histology in which it has been identified • Variants of this gene in this primary site/histology are established as clinically actionable (druggable/ predictive/prognostoc and/or with diagnostic/classification implications), however the specific sequence variants is not one of the currently reported variants in this gene in the histology
SOMATIC DNS MUTATION DETECTED (POSITIVE)
CDKN2A c.238C>T p.R80*
Tumour Suppressor Predicted Driver Mutation
Reference Sequence: NM_001195132.1 CDKN2A Exon 2. Mean Coverage: 10525X. Variant Aliele Fraction: 5%
TP53 c.742C>T p.R248W
Tumour Suppressor Predicted Driver Mutation
Reference Sequence: NM_000546.5 TP53 Exon 7. Mean Coverage: 8481X. Variant Aliele Fraction: 6%.
TERT c.1-124C>T Promoter
Oncogene Known Driver Mutation
Reference Sequence: NM_198253.2 TERT (Promoter) Mean Coverage: 349X. Variant Aliele Fraction: 12%
DNA Variants
Somatic RNA Fusion
146 Genes Negative Mean Coverage: 8609X.
51 Genes Negative Total Reads: 6771436X.
Tumor Mutational Load
Homologous Recombination Deficiency
TML Low Mutation Load per MB: <1.0
HRD Not Detected
Figure 3










