4 minute read
Maternal SMCHD1 Regulation of Hox Gene Expression and Patterning in the Mouse Embryo
Natalia Benetti
This year Natalia wrote a paper on her work on maternal SMCHD1 and Hox genes which is currently under review at Nature Communications. A preprint of this manuscript is available online: https://dx.doi.org/10.1101/2021.09.08.459528.
In addition to being a Residential Tutor for biology at St Hilda’s College, I am a third year PhD student in Professor Marnie Blewitt’s Laboratory in the Epigenetics and Development Division at the Walter and Eliza Hall Institute of Medical Research and the Department of Medical Biology at The University of Melbourne. The Blewitt lab investigates epigenetic mechanisms of gene silencing during embryonic development.
Genetic information is inherited through generations in the form of DNA, which is contained in the nucleus of every cell. DNA carries all the information required for an organism to develop from a single cell and survive as a fully developed organism. As such, all cells in an adult organism contain the same genes, as encoded by their DNA sequence. A single-celled zygote is able to differentiate into different cell and tissue types as it grows due to different genes being switched on and off at the right times during development. This differential gene expression is controlled by epigenetic mechanisms, whereby chemical modifications are made to DNA or proteins it is wrapped around to alter gene expression without changing the DNA sequence itself (reviewed in Li, 2002).
Everybody in our lab works on understanding the molecular function of a protein called SMCHD1, which my supervisor Marnie discovered during her PhD (Blewitt et al., 2005, Blewitt et al., 2008). SMCHD1 is a biologically important epigenetic gene silencer, such that female mouse embryos which lack SMCHD1 protein die mid-gestation and mutations in SMCHD1 are associated with human disease (reviewed in Jansz et al., 2017).
My PhD project is specifically focused on investigating the role of SMCHD1 in silencing a set of developmentally important genes during embryonic development called Hox genes. Hox genes are highly evolutionarily conserved such that they are present in all animals and are required for the correct patterning of the anterior-to-posterior axis of the developing embryo (Lewis, 1978). These genes need to be switched on and off at precisely the right times during mammalian embryonic development for correct skeletal patterning to form, for example ensuring that thoracic elements have ribs and that cervical elements don’t (reviewed in Mallo et al., 2010). Mice which lack SMCHD1 protein exhibit abnormal skeletal patterning caused by aberrant Hox gene expression (Jansz et al., 2018).
One of the questions I aimed to answer in my PhD was whether SMCHD1 has a maternal effect on Hox genes. Upon conception, all of the cytoplasm (the entire cell apart from the nucleus, containing all proteins) comes from the mother’s egg,
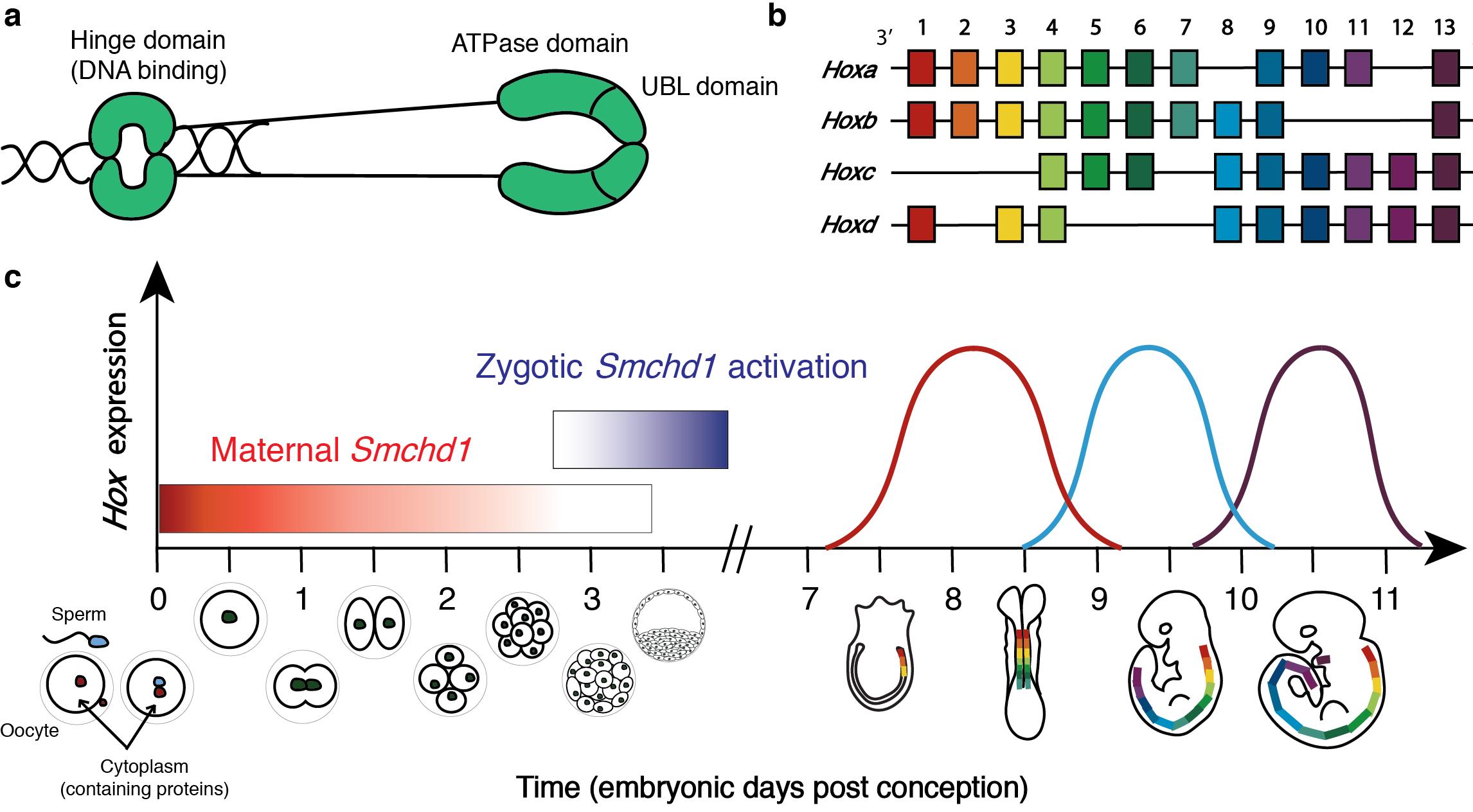
- while the sperm contains only half the DNA of a normal somatic cell and no cytoplasm (the egg also supplies half the DNA of a somatic cell) (reviewed in Kim and Lee, 2014). Hence, until the zygote’s own genes are activated (which is around the 2-cell stage for most genes in mice (Schulz and Harrison, 2019), but at the 32-cell stage for SMCHD1 (Wanigasuriya and Gouil et al., 2020)), the growing zygote is reliant upon the proteins supplied by the mother’s egg to grow. Proteins which are present in the oocyte and are required for normal development are called maternal effect proteins. To test whether maternal SMCHD1 regulates Hox genes we generated a mouse model in which the Smchd1 gene was deleted in the oocyte of the mother, hence there was no SMCHD1 protein present in the cytoplasm of the oocyte. We crossed these female mice to males which had normal Smchd1 genes, so embryos yielded from this cross had no SMCHD1 protein until their own Smchd1 gene (inherited from the father) was activated. Comparing to appropriate controls which had maternal SMCHD1, we saw that Smchd1 maternally deleted embryos consistently exhibited precocious Hox gene activation, leading to abnormal skeletal patterning later in development. This is a very unexpected finding because maternal SMCHD1 is only present until embryonic day (E) 2.75 in the mouse, yet we observed altered Hox gene expression much later in development at E8.5 and altered skeletal patterning at E17.5.
SMCHD1 is an epigenetic silencer protein which silences Hox genes during mouse embryonic development. (a) Cartoon depiction of the SMCHD1 protein as a homodimer, showing the C-terminal DNA binding hinge domain and the N-terminal ATPase and ubiquitin-like (UBL) domains. (b) Diagram showing the genomic organisation of Hox genes in mammals. Mammals have four paralogous clusters of Hox genes 1-13, which are temporally expressed in a co-linear fashion according to their chromosomal position. (c) Summary of the timeline of murine embryonic development, with annotations showing temporal Smchd1 and Hox gene expression. The location of Hox gene expression along the developing anterior-to-posterior axis is the same as their chromosomal order such that Hox1 paralogues are expressed most anteriorly and Hox13 paralogues are expressed most posteriorly. 48






