
2 minute read
ABSTRACTS
SESSION9.2:STRENGTHENINGOFANTIMICROBIALRESISTANCE(AMR)SURVEILLANCE
Throughlaboratoryantibiograms
Advertisement
Toatu Tebuka , Saketa Salanieta , Uluiviti Vasiti , Naivalu Taina , Babiau Raikabakaba Asaeli , Baleivanualala e a a f a Sakiusa ,BuadromoEka ,LeongMargaret ,ColeRussell ,VareaLalomilo aPacific Community (SPC), Public Health Division, Narere, Suva, Fiji Islands. bPacific Islands Health Officers Association (PIHOA), Hawaii, Honolulu, USA cDepartment of Pathology and Medical Laboratory Science, Fiji National University, Suva, Fiji Islands. dWorld Health Organization (WHO), South Pacific, Suva, Fiji Islands. eOtago University, New Zealand. fPacific Pathology Training Centre, Wellington, New Zealand.
Introduction: AMR surveillance and control is a challenge inmostPICTspartlyduetothefollowingreasonsweakAMR surveillance, lack of collaboration between microbiology and pharmacy in developing drug formulary on specific organisms, lack of antibiogram database, ineffective national Drug and Therapeutics Committee and weak Infection Prevention Control (IPC) both in hospital and communitysettings.
SPCanditspartners(FNU,PIHOA,PPTC&WHO)developed a training curriculum to assist Labs in the region in implementing quality Antimicrobial Susceptibility Testing (AST) that will contribute to the development of a quality antibiogramtoguideinformeddecisions.
Objective: The objective of this approach is to strengthen quality implementation of quality microbiology processes to produce an accurate and reliable antibiogram that will guideinformeddecisionsandactionstotackleAMRissues intheregion.
Method: From 2019 to 2022 a standardise training curriculum focusing on strengthening quality microbiology diagnostic procedures to produce quality ASTwasrolledouttofewselectedPacificcountries.In2023 a developed antibiogram database was rolled out to countries which participated in the training to produce an accurate and reliable antibiogram on the susceptibility of antibiotics used to treat infection The generated information on the antibiogram would be used to guide informeddecisionstoaddressAMR.
Results:
Conclusion:
Compilationandreportingofanantibiogramdatatoguide informed decision is a challenge in most laboratories in PICTs.Majorityofcountriesstillrelyheavilyonpaper-based system. The developed excel user-friendly antibiogram database has been rolled out to three (3) piloted site countries and already beneficial outcomes are noted in these countries. The trend in sensitivity pattern over time cannowbemadequicklyusingthesimplefunctionalityof thetool.
SPEAKERPROFILE: MrTebukaToata
Mr Tebuka Toatu is from Kiribati and has been working with The Pacific Community (SPC) for the past twelve years. Prior to joining SPC, he had been a Director of Laboratory Services for over ten years at the Ministry of Health and Medical Services in Kiribati.

Mr Toatu is a Medical Laboratory Scientist by profession and holds a Bachelor of Applied Science in Medical Laboratory Science from RMIT University in Melbourne with majors in Advance Cytopathology and Haematology. In 2008, he completed his post graduate studies doing a Master of Science in Medicine from the University of Sydney
Currently, Mr Toatu is working with the Pacific Community at SPC Public Health Division as a Pacific Public Health Surveillance LabNet Coordinator providing laboratory strengthening activities in PICTs but focusing more on strengthening AMR surveillance and control through microbiology capacity training in the region.
SESSION 9.3: GENOMIC DIVERSITY OF MULTI-DRUG RESISTANT GRAM-NEGATIVE AND GRAM-POSITIVE BACTERIA IN SUVA, FIJI
HawkeyJ,PrasadA,PrasadV,VakatawaT,Young-SharmaTEMW,JenneyAWJ,NaiduR, StewardsonAJ,LoftusMJ,PelegAY
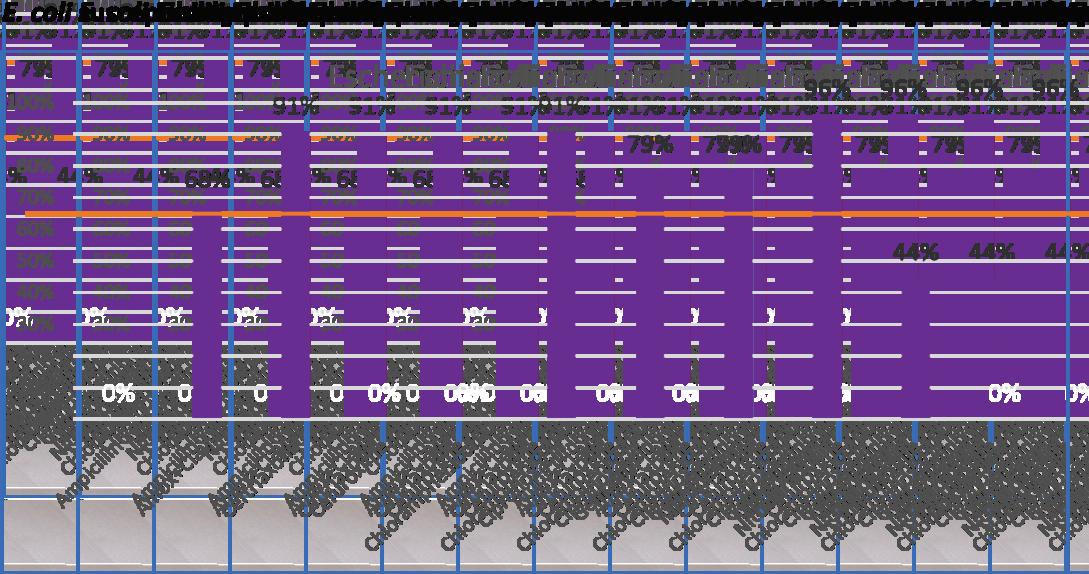
Introduction: Antimicrobial resistance (AMR) is a global health threat, with third-generation cephalosporinresistant(3GCR)andcarbapenem-resistantinfectionsofparticularconcern.Thereis currentlyalackofgenomicdataonAMRorganismsinthePacificregion.Thisstudyaimstoaddress thisgapbyexaminingthegeneticdiversityofacollectionof450clinicalisolatescollectedbetween July2020-Feb2021fromSuva,Fiji.
Methods: Genomes were sequenced using Illumina and assembled with Unicycler Species and sequence types (ST) were determined using PathogenWatch; AMR genes were assigned using AMRFinderPlus. Nine isolates underwent additional long-read sequencing to determine the locationofcarbapenemasegenes.
Results: Of 450 sequenced isolates, 362 were Gram-negative and 88 were Gram-positive bacteria. For Gram-negative bacteria, there were 126 3GCR isolates and 107 carbapenem-resistant Acinetobacter and Pseudomonas. Klebsiella pneumoniae was highly diverse, with most STs represented by a single genome; there was less diversity in Escherichia coli, 42% of genomes belongedtoST131orST69 ThemediannumberofAMRgeneswaslowerinsensitivestrains(1-6vs917,genusdependent).FourteenESBLgenesweredetected;CTX-M-15wasthemostcommon(22%of genomes, all Enterobacterales). Eighteen carbapenemase genes were detected, primarily in Acinetobacter and Pseudomonas. OXA-23, OXA-66 and NDM-1 were the most prevalent, found in 10%, 18% and 17% of genomes respectively Phylogenetic analysis of two dominant clones (A baumannii ST2 and P aeruginosa ST773) revealed Fiji-specific clades comprising highly related strains.ForGram-positivebacteria,StaphylococcusaureusST1wasdominant;phylogeneticanalysis revealedasingleFijianclade,howeverstrainswererelativelydistinctfromoneanother
Conclusions: This study provides crucial genomic data on AMR organisms in Fiji, highlighting the diversity of 3GCR and carbapenem resistant species in the region. Local transmission of two carbapenem-resistantcloneswithinFijiwasobserved,underscoringtheimportanceoflocalspread oftheseresistantstrains.
SPEAKERPROFILE: DrJaneHawkey
Dr Jane Hawkey is a Research Fellow in the Department of Infectious DiseasesatMonashUniversity SheundertookherPhDattheUniversity of Melbourne, where she developed a novel genomic method for detecting bacterial transposases in whole genome sequencing data, to study how transposases have impacted the on-going evolution of Shigella species. Jane’s current research uses her genomics skills to study the evolution and spread of antimicrobial resistance in Gram negative bacterial pathogens, with a specific focus on developing new methodsfortrackingthetransmissionofplasmids.


