
4 minute read
Central Research
When CRISPR meets Dark Genome: Epigenetic Editing
Innovation happens fast in the Biotech industry, and technologies are being superseded for new technology even faster than before. When we are getting used to gene therapy, a new technology is getting attention and sizable investing: epigenetics.
What exactly is epigenetics?
Conrad Waddington coined the term “epigenetics” 81 years ago to describe phenotypic variation that does not result from changes in genotype. Since then, significant progress has been made in understanding epigenetic mechanisms for gene control, including chromatin remodeling, DNA methylation at CpG islands, and post-translational modifications (such as methylation, acetylation, citrullination, and phosphorylation) of the N-terminal tails protruding from the core histones that package genomic DNA.
By orchestrating internal and environmental signaling cues into transcription independent of the underlying genetic code, epigenetic regulatory mechanisms play a central role in nearly all cellular phenomena.
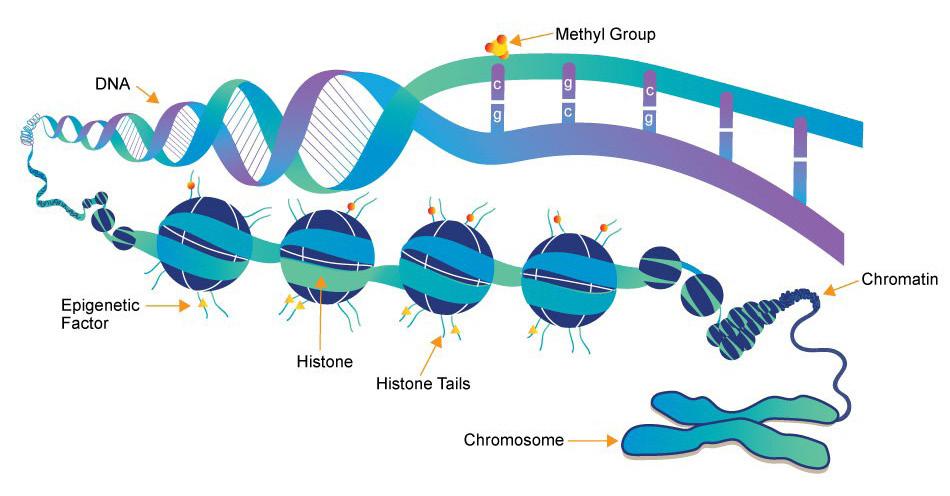
In general, the epigenome is a collection of heritable and nonheritable sequence independent biological molecules that converge to modulate chromatin structure, genome function, and gene expression patterns. Epigenomic regulation occurs through a complex interplay of proteins that bind to genomic DNA, biochemical modifications to DNA and histones, and structural changes that allow regulatory proteins to access DNA. Even though projects like ENCODE and the Roadmap Epigenomics Mapping Consortium have accelerated our understanding of epigenetic states in health and disease, the extent to which epigenetic alterations are drivers or consequences of disease is frequently unknown.
What is Epigenetic Editing?
The use of epigenetic enzymes to rebuild the localized epigenetic environment of an internal genomic region, usually to regulate transcription, is known as epigenetic editing. The use of nuclease-null deactivated (or dead) CRISPR/Cas systems (dCas) to perturb non-coding nucleic acid elements, such as promoters, enhancers, and transcription factor binding motifs, as well as non-coding RNAs, to gain insights into their functional roles and the resulting epigenetic changes, has significantly accelerated epigenetic editing progress.
Many important factors still limit the development of epigenome editing, preventing its use in humans. Like conventional genome editing, the clinical utility of epigenome editing is limited by targeted delivery options, potential off-targeting, and efficacy. Many factors influence these issues, including the identity of the targeted tissue, the chromatin context of the therapeutic gene of interest, and the epigenome editor’s copy number.
So, who´s who in the epigenome editing game?
Chroma Medicine, Tune Therapeutics, Epic Bio, and Navega Therapeutics have raised a total of $ + 200 million in funding, joining Sangamo Therapeutics, Inc. and Encoded Therapeutics Inc. in the quest to make epigenome editing a clinical reality. These startups are primarily developing platforms based on catalytically inactive CRISPR systems linked to effector domains that regulate gene expression. Unlike currently marketed epigenetic cancer drugs, which act on a genome-wide scale and have dose-limiting toxicities, the specificity of epigenome editors promises to open up a wide range of new indications beyond oncology.
In a landscape crowded with small-molecule inhibitors, monoclonal antibodies, gene therapies, small interfering RNAs, antisense oligonucleotides, and traditional gene and base editing, epigenome editors’ ability to restore genes silenced in disease in a tunable and durable manner may prove to be a key therapeutic niche.

How far are we from getting this into the clinic?
Although the road ahead will be long and difficult, epigenome editing therapies will provide several novel therapeutic options. They show promise for monogenic diseases where the target genes outnumber the AAV gene therapy payload capacity, and a healthy endogenous gene can be upregulated or downregulated (depending on the condition). Perhaps the most compelling therapeutic applications are restoring gene expression in congenital diseases of genome imprinting (for example, DiGeorge syndrome), autosomal dominant diseases of haploinsufficiency, or downregulating cancer cell activity, conditions that traditional gene therapy may not be able to treat.
Adrian Rubstein
Founder of Ax3.Bio, leading life sciences investment strategy and advisory company.

References
1. Quratulain Babar, Ayesha Saeed, Tanveer A. Tabish, Sabrina Pricl, Helen Townley, Nanasaheb Thorat, Novel epigenetic therapeutic strategies and targets in cancer, Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease,Volume1868, Issue 12, 2022. Retrieved from: https://doi.org/10.1016/j.bbadis.2022.166552.
2. Jennifer Khirallah, Maximilan Eimbinder, Yamin Li, Qiaobing Xu, Clinical progress in genome-editing technology and in vivo delivery techniques, Trends in Genetics, Volume 39, Issue 3, 2023, Pages 208-216. Retrieved from: https://doi.org/10.1016/j.tig.2022.12.001.
3. Fine-tuning epigenome editors. Nat Biotechnol 40, 281 (2022). Retrieved from: https://doi.org/10.1038/s41587-022-01270-w
4. Rittiner Joseph, Cumaran Mohanapriya, Malhotra Sahil, Kantor Boris, Therapeutic modulation of gene expression in the disease state: Treatment strategies and approaches for the development of next-generation of the epigenetic drugs. Frontiers in Bioengineering and Biotechnology. Retrieved from: https://www.frontiersin.org/ articles/10.3389/fbioe.2022.1035543










