SCI ENCE BIOTODAY

LIFE SCIENCE • BIO-INFORMATICS • MICROBial CHEMISTRY • CLINICAL TRIALS • COMMERCIAL DEVELOPMENt
ISSUE30



 Karen Southern Editor in chief
Karen Southern Editor in chief
Editor
Karen
Southern
karen.southern@distinctivegroup.co.uk
Design
Distinctive Media Group Ltd, 3rd Floor, Tru Knit House, 9-11 Carliol Square, Newcastle, NE1 6UF Tel: 0191 580 5990 distinctivegroup.co.uk
Advertising
Distinctive Media Group Ltd, 3rd Floor, Tru Knit House, 9-11 Carliol Square, Newcastle, NE1 6UF Tel: 0191 580 7163
e: helen.flintoff@distinctivegroup.co.uk distinctivegroup.co.uk

foreword
UK in strong position for investment and growth
It’s all about ‘location, location, location’ as far as life sciences investors are concerned.
Despite a slowdown from recent record highs, last year’s investment volumes were still 13% higher than 2020.
The findings, in JLL’s 2023 Europe Life Sciences Cluster Outlook, show that location definitely matters.
The report shows Cambridge and Oxford as leaders in ‘physical’ capital’ such as funding and real estate while Paris and London lead on ‘human capital’ such as access to talent.
Medium to long-term investment prospects for the sector also remain strong, with growth at pharma and biotech firms, new company launches, and increased use of research contractors driving demand and rental growth. Fingers crossed that the impetus continues, as the UK is also forecast to become the world’s second biggest innovator in medical technology outside the US.
Findings from Snedden Campbell say a rise in post-pandemic investment has put the UK on a path to unprecedented growth, with collaboration between industry and universities in the ‘Golden Triangle’ of
London, Oxford and Cambridge creating a ‘dynamic focus’ for MedTech development to rival Biopharma’s success
CEO Ivor Campbell, whose firm recruits globally for Medtech, points out that shortlists for senior executive positions are now dominated by British-based applicants, with his company placing more senior science professionals in Cambridge alone than it previously did in the whole of the UK.
The ‘Oxford cluster’, meanwhile, includes the Jenner Institute – which developed the Oxford-AstraZeneca vaccine – the Harwell science park, and gene-sequencing equipment manufacturer Oxford Nanopore, one of the largest firms to emerge from the UK’s life-sciences ecosystem.
Ivor says, “Cambridge has come from almost nowhere in diagnostics, six or seven years ago, to being dominant in what we do. The university, the local authority and national government have been able to put an infrastructure together. It’s easy to get to, it’s not a bad place to be and London is nearby.
“With London doing the money and Oxford and Cambridge doing the science, we have the infrastructure for a globally dominant sector.”
| BIOSCIENCE TODAY | 3 www.biosciencetoday.co.uk | foreword |
Distinctive Media Group Ltd or BioScience Today cannot be held responsible for any inaccuracies that may occur, individual products or services advertised or late entries. No part of this publication may be reproduced or scanned without prior written permission of the publishers and BioScience Today.
All the world’s a microbe
features
‘Avatar’ motion tech aids advances in disease research

| BIOSCIENCE TODAY | 4 | contents | 12
Addressing the language tax in life sciences 20
3 Foreword
4-5 Contents
7 LAB TECHNOLOGY
Scientists have made the first bone marrow ‘organoids’ capturing the key features of human bone marrow.
16 LIFE SCIENCE & CAREERS

Staff in the pharmaceutical and life sciences sector who are working from home may be damaging their pay and career prospects.
8 - 9 MACHINE LEARNING
AI-powered simulations pair drugs with cancer patients. Finding solutions to complex diseases is top of the agenda for a collaboration between a leading UK innovation engine and a Budapest-based biotech firm.
10 - 11 COMMERCIAL DEVELOPMENT
A joint venture between Canary Wharf Group (CWG) and Kadans Science Partner (Kadans) has submitted a detailed planning application for Europe’s largest and most technologically advanced commercial health and life sciences building.
26 - 27 BIONANOSCIENCE
Bionanoscience ‘will precipitate a Fifth Industrial Revolution’. Professor Jonathan Heddle is set to embark on a new era in Bionanoscience at Durham University, thanks to a £4.8million Leverhulme International Professorship award.
32 - 33 Drug development & discovery
With the help of artificial intelligence, Swedish researchers have succeeded in designing synthetic DNA that controls the cells’ protein production.
38 - 39 CLINICAL TRIALS
Global community will assess environmental impact of clinical trials.
40 - 41
COGNITIVE DECLINE
Games and stimulation mitigate cognitive decline in older adults. Older people may be able to boost their working memory with a new approach that couples online therapeutic games with a non-invasive brain stimulation technique.
43 NEUROLOGY
The brains of three different species of stranded dolphins show classic markers of human Alzheimer’s disease, according to the most extensive study into dementia in odontocetes (toothed whales).
| BIOSCIENCE TODAY | 5 | contents |
28 34
contents
biosciencetoday.co.uk / issue 30 /
There’s more to collagen than cosmetics...
/
Nishchay Shah Chief technology officer and Business Head, Emerging Products, at CACTUS Labs

As well as his executive leadership role, Nishchay is responsible for global technology strategy and staffing, overseeing technology and innovation across products and brands. He manages a large department focused on product management, software development, UX, DevOps, Digital innovation, and Machine Learning. Having successfully led both B2C and B2B product teams in the past, he has a thorough understanding of the end-to-end product and technology life cycles. Nishchay has a master’s degree from the University of Bridgeport, Connecticut, specialising in Computer Networks and Database Systems.
Gen Li
President and founder of Phesi
Dr. Li was previously Head of Productivity for Pfizer Worldwide Clinical Development, a position he assumed following Pfizer’s acquisition of Pharmacia, where Dr. Li delivered the first implementation of productivity measurement for clinical development. While at Pharmacia and Pfizer, Dr. Li significantly contributed to the Centre for Medicines Research (CMR) International database for pharmaceutical R&D performance, assuring the collection of key clinical trial parameters as representative of the critical path for delivery. He was also instrumental in creating the KMR productivity mode. Previously, Dr. Li led the creation of the first computer-automated resource management system at BristolMyers Squibb.
 Robin May
Robin May
FSA Chief Scientific Adviser and Professor of Infectious Disease at the University of Birmingham

A Wolfson Royal Society Research Merit Fellow and Fellow of the American Academy of Microbiology, Professor May specialises in research into human infectious diseases, with a particular focus on how pathogens survive and replicate within host organisms. He was appointed as Gresham Professor of Physic in May 2022, where he delivers free lectures to the public on medicine, health and related sciences.
to
advertise or contribute to the next edition

advertising: liz.hughes@ distinctivegroup.co.uk
editorial: karen.southern@ distinctivegroup.co.uk
| BIOSCIENCE TODAY | | industry contributors | | BIOSCIENCE TODAY 6 SCI ENCE BIOTODAY
Miniature
‘bone marrows in a dish’ improve anti-cancer treatments
Scientists have made the first bone marrow ‘organoids’ capturing the key features of human bone marrow. The technology, devised by teams from Oxford University and the University of Birmingham, is the subject of a patent application filed by University of Birmingham Enterprise.
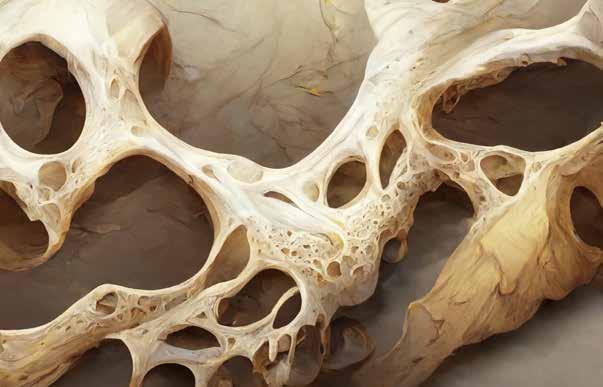
The method will enable the screening of multiple anti-cancer drugs at the same time, as well as testing personalised treatments for individual cancer patients.
A study, published in the journal Cancer Discovery, describes the method, which results in an organoid that faithfully models the cellular, molecular and architectural features of myelopoietic (blood cell producing) bone marrow.
The research also showed that the organoids provide a micro-environment that can accept and support the survival of cells from patients with blood malignancies, including multiple myeloma cells, which are notoriously difficult to maintain outside the human body.
Dr Abdullah Khan, a Sir Henry Wellcome Fellow at the University of Birmingham’s Institute of Cardiovascular Sciences and first author of the study, said: “Remarkably, we found that the cells in their bone marrow organoids resemble real bone marrow cells not just in terms of their activity and function, but also in their architectural relationships - the cell types ‘self-organize’ and arrange themselves within the organoids just like they do in human bone marrow in the body.”
This life-like architecture enabled the team to study how the cells in the bone marrow interact to support normal blood cell production, and how this is disturbed in bone marrow fibrosis (myelofibrosis), where scar tissue builds up in the bone marrow, causing bone marrow failure. Bone marrow fibrosis can develop in patients with certain types of blood cancers and remains incurable.
Senior study author Professor Bethan Psaila is a haematology medical doctor and research Group Leader at the Radcliffe Department of Medicine, University of Oxford. She said: “To properly understand how and why blood cancers develop, we need to use experimental systems that closely resemble how real human bone marrow works, which we haven’t really had before. It’s really exciting to now have this terrific system, as finally, we are able to study cancer directly using cells from our patients, rather than relying on animal models or other simpler systems that do not properly show us how the cancer is developing in the bone marrow in actual patients.”
Dr Khan also added, “This is a huge step forward, enabling insights into the growth patterns of cancer cells and potentially a more personalised approach to treatment. We now have a platform that we can use to test drugs on a ‘personalised medicine’ basis.
“Having developed and validated the model is the first crucial step, and in our ongoing collaborative work we will be working with others to better understand how the bone marrow works in healthy people, and what goes wrong when they have blood diseases.”
| BIOSCIENCE TODAY | 7 | lab technology |
“We now have a platform that we can use to test drugs on a ‘personalised medicine’ basis.”
AI-powered simulations pair drugs with cancer patients
Finding solutions to complex diseases is top of the agenda for a collaboration between a leading UK innovation engine and a Budapest-based biotech firm.
Cancer Research Horizons is part of Cancer Research UK, the world’s largest private funder of cancer research. It has partnered with Turbine to use its Simulated Cells™ platform in identifying target patient populations who could benefit from CDC7* inhibitor therapy with Cancer Research Horizons’ lead compound CRT’2199.
CRT’2199 originates from Cancer Research Horizons’ Therapeutic Innovation.** CDC7 is a protein which plays a vital role in the regulation of cell division in normal cells. However, dysregulation of CDC7 can lead to the formation of cancer cells, and overexpression of this protein is correlated with poor clinical prognosis in diverse cancers of significant unmet patient need.
Despite its role in the progression and outcome in many cancers, no CDC7 inhibitors have progressed to Phase III trials, and a clear picture is lacking on what types of cancer could be effectively and safely treated by inhibiting CDC7.
The partnership aims to change this. Using Turbine’s AI-powered simulation approach, Turbine and Cancer
Research Horizons will inhibit CDC7 in digital cancer cells that represent different patient populations, to determine which cancer types and patient populations are most likely to respond to treatment with CRT’2199.

Turbine, which leverages computational simulations to solve complex questions in oncology, will receive a revenue share of Cancer Research Horizon’s future revenues from the CDC7 inhibitor program upon successful commercialisation in exchange for identifying and validating a disease positioning strategy.
Turbine’s Simulated Cell technology uses machine learning to train digital versions of cancer cells to behave in the same way that real cancer cells would, enabling simulations to show how cancer cells react to different triggers, such as transcriptomic changes and anticancer drugs.*** Predictions based on this biological understanding provide invaluable insights at any point of the drug discovery and development process and can guide subsequent real-life experiments that increase the likelihood of success for a project.

8 | BIOSCIENCE TODAY | | machine learning |
Diagram showing SimCell Loop
Multiple companies have relied on Simulated Cells to inform their pipeline decision-making, including Bayer and two top-20 pharma companies that have leveraged the technology to generate multiple predictions that are currently in clinical validation.
“Turbine’s technology provides a unique opportunity to gather insights into cancer cell behaviour at a scale and speed which isn’t possible to achieve in a traditional drug

discovery setting,” said Dr. Daniel Veres, Chief Science Officer and co-founder of Turbine. “We’re looking forward to working with Cancer Research Horizons to identify suitable patient populations for its CDC7 inhibitors and accelerate their clinical development, in the name of hopefully bringing important new therapeutic options to patients that need them as quickly as possible.”
Tony Hickson, on behalf of Cancer Research UK and Cancer Research Horizons, said: “We know that CDC7 inhibitors hold enormous potential as a class of anti-cancer therapeutics, but the problem so far has been finding the right patients who could benefit from them.
“This is why we are excited to be partnering with Turbine to develop novel patient selection strategies for our CDC7 inhibitor compounds. Bolstered by Turbine’s unique capabilities, we hope to gain novel insights into the activity of these drug candidates, bringing closer the day when we could see them reach the clinic to benefit patients who need them.”
For more information, visit cancerresearchhorizons.com.
NOTES
*Cell Division Cycle 7-related Kinase. CDC7 is an enzyme in humans that is encoded by the CDC7 gene. It is critical for the regulation of the cell cycle at the point of chromosomal DNA replication. In cancer, overexpression of the CDC7 gene is often associated with poor clinical outcomes.
**https://www.cancerresearchhorizons.com/licensing-opportunities/small-moleculecdc7-inhibitors
***These hypotheses are then validated using conventional methods.
9 | BIOSCIENCE TODAY | | machine learning |
Turbine AI founders, from left, Kristof Szalay, Ph.D., Founder & CTO, Szabolcs Nagy, Co-founder & CEO, and Daniel Veres, MD, Ph.D., Co-founder & CSO.
“Turbine’s technology provides a unique opportunity to gather insights into cancer cell behaviour at a scale and speed which isn’t possible to achieve in a traditional drug discovery setting.”
‘World-class’ life sciences ecosystem plan for Canary Wharf

It is claimed that the 23- storey 823,000 sq ft tower, in London, will set a new benchmark in the development of life sciences laboratories and workspace, creating a unique vertical campus.

The building represents the first phase, with outline planning consent already secured for the wider currently undeveloped 3.5 hectare site located next to the Elizabeth Line, which has capacity to deliver 3.5 million sq ft of laboratory and affiliated space.
The building’s innovative stacking infrastructure has the potential to provide laboratory space on every floor. The ground and first floors will also offer community spaces, a café and break out areas for conferences and meet ups. It’s The ground floor will also be open to the public to help make life sciences more approachable and less closed off. At the top of the building there will be a sky lounge and terrace for the tenants.
The building will also incorporate dedicated clinical and chemical waste facilities, multiple goods-lifts with oxygen monitors, freezer farms and specialised receiving and storage areas for hazardous, clinical and chemical goods. Its central service provision will include Nitrogen (gaseous & liquid), CO2 and compressed air.
A low-carbon strategy will be implemented across the development, including a high degree of insulation, optimised façade, energy efficient MEP systems and the provision of renewable sources and highly efficient photo voltaic panels.
Heat pumps and heat recovery systems will also be utilised, making the building one of the largest all-electric lab spaces in the world, in line with net zero ambitions.
Shobi Khan, CEO of Canary Wharf Group, explained: “We have been developing our vision for a world-class life sciences hub at Canary Wharf since 2019, and this is a significant milestone
10 | BIOSCIENCE TODAY | | commercial development |
A joint venture between Canary Wharf Group (CWG) and Kadans Science Partner (Kadans) has submitted a detailed planning application for Europe’s largest and most technologically advanced commercial health and life sciences building.
Life science lab concept plans.
in our journey. This project will continue the transformation of Canary Wharf, providing a sustainable, mixed-use environment for the next generation of life science offices.”

Michel Leemhuis, CEO of Kadans Science Partner, added: “Kadans prides itself on being a leading contributor to the evolution of the life sciences sector and this impressive new building at North Quay is a great example of how we are driving the development of innovative state-of-the-art lab buildings. By capitalising on our previous experience and using lessons learned, we are able to make a significant step forward in the development of life science buildings. It marks a step change in the way ecosystems for knowledge-intensive businesses are designed and developed.”
Elie Gamburg, Design Principal, Kohn Pedersen Fox Associates (KPF), said: “Connection breeds innovation – great
discoveries often result from the confluence of ideas among seemingly disparate disciplines. For North Quay, we sought to design a spatial ecosystem to promote interdisciplinary collaboration in a dense urban environment.
“Traditional lab buildings have been low-rise campuses, but North Quay generates an innovative new paradigm –the vertical campus. Technical areas and social spaces are intermingled in a sustainable development that provides a dynamic place for discovery and invites the public in. An active waterfront, reinterpreting the Quayside docks, is created for an evolving Canary Wharf.”
Three international airports, Heathrow, Gatwick and City, are all accessible within 10-40 minutes and it is also less than 30 minutes to King’s Cross, White City and three significant funders of life science research in the UK – UKRI, Wellcome Trust and Cancer Research UK.
Several science-led businesses are already located in Canary Wharf, with Genomics England, one of the world’s most cutting-edge life science organisations, moving into One Canada Square. Proximity to the Medicines and Healthcare products Regulatory Agency (MHRA) as well as organisations like UCL Management School and the General Pharmaceutical Council make Canary Wharf an attractive location. Level39, a world-leading tech hub, has a vibrant community of 180 start-up and scaleup companies at Canary Wharf including a number of med tech start-ups.
11 | BIOSCIENCE TODAY | | commercial development |
23- storey 823,000 sq ft tower plans.
“We have been developing our vision for a world-class life sciences hub at Canary Wharf since 2019, and this is a significant milestone in our journey.”

12 | BIOSCIENCE TODAY | | artificial intelligence |
Addressing the language tax in life sciences

As a researcher, there are few things more vital to your reputation than your publishing record. But what if your native language is not English? Is this a significant problem? How can it be mitigated? Nishchay Shah, CTO of CACTUS, investigates.
According to data compiled by Scott Montgomery in Does Science Need a Global Language, more than three-quarters of scientific papers are published in English, and in some fields, this figure exceeds 90%.
Whether your objective is tenure, a research grant, a book deal, or just an improved reputation in your department, a string of high-profile journal articles are the currency you require to trade your way to the top. Because the global publishing language is English, this success depends on being able to articulate yourself well in what may not be your first language.
Considering the need to publish in English, not being a native English speaker could be holding 95% of the world’s population back. It’s
like a language tax levied by the native Englishspeaking world on the non-native population, one that is paid with increased effort and a higher rate of rejection.
IMPACT OF THE LANGUAGE TAX
In a 2018 study published in Written Communication, Hanauer et al. investigated the added burden faced by Mexican and Taiwanese researchers when writing research articles in English. They found that non-native English speakers had an average 24% increase in difficulty, 10% rise in dissatisfaction and 22% more anxiety when it came to manuscript preparation. The researchers maintain that the additional burden of English as a second language constitutes a linguistic injustice — a barrier to science that needs to be addressed.
13 | BIOSCIENCE TODAY | | artificial intelligence |
The primary problem for non-native English speakers is that writing a research paper in English takes them longer. On the other hand, the surge in research output means they need to do more to keep up with developments in their field. An article published in Nature points to bibliometrics showing an annual 8-9% increase in published scientific papers in recent decades. In fact, over a million biomedical research papers are added to PubMed every year, which is approximately two papers per minute. Keeping up with such an overwhelming volume of research is challenging for all researchers, but even more so for those who must read and understand this in English, their non-native language. Once non-native English speakers begin writing, grammatical woes can slow down the process even further ─ English grammar is notoriously fickle. While even native English-speaking researchers may struggle with parallelism or infinitives, navigating these complex grammar rules in a second language is much more difficult. In competitive research fields, taking longer to publish may mean another researcher beating you to the punch.
DESK REJECTION AND THE LANGUAGE TAX
The language tax increases the risk of desk rejection for three key reasons. The first, and most obvious, is that if the author’s writing and grammar skills are not as strong as their academic expertise, the manuscript could be difficult to read or understand. The second reason for rejection is that a non-native English speaker might find it more difficult to express their ideas in English, resulting in a manuscript that does not showcase or convey their research effectively. The third reason for desk rejection is not complying with the rules and regulations required to submit successfully, such as the journal’s style guide and referencing format; these guidelines are usually produced in English, making it more difficult to understand and follow.
However, in a 2013 study published in BioScience called Predicting Publication Success for Biologists, William F. Laurance et al. suggested that native English speakers, those who published earlier in their careers, and males have only ‘minor advantages’ in the long term. The biggest benefit identified by the paper is a good pre-PhD publication record, and its findings are sound in that regard. It’s important to note here that this paper looked at long-term career success in relation to a researcher’s publishing record and not their likelihood of getting into print in the first instance.
Seeing success on your first try is still far more common if you are a native English speaker. So, while getting published early is seen as the most important indicator of success, achieving this requires a high degree of familiarity and quick mastery of the English language. To my knowledge, research is still to be done on whether an author is more likely to be published at pre-PhD stage if their native language is English.
OVERCOMING THE LANGUAGE TAX
Editing tools and services offer a way to bridge the language gap for non-native English speaking researchers by getting their writing up to the standard a journal requires. However, while there are many tools available to check spelling and grammar online, they are usually not designed to support

14 | BIOSCIENCE TODAY |
| artificial intelligence |
“Considering the need to publish in English, not being a native English speaker could be holding 95% of the world’s population back. It’s like a language tax levied by the native English-speaking world on the non-native population, one that is paid with increased effort and a higher rate of rejection.”
scientific content and, therefore, are not the best choice to improve academic writing. Moreover, there are few tools that help the author as they work on the all-important first draft and even fewer that conduct the kind of technical checks required to ensure a manuscript complies with journal-specific submission guidelines. Getting their manuscript written, polished and through to the final stage of the process is something many researchers struggle with.
Specialised assistive writing tools, based on artificial intelligence (AI) and machine learning, emerge as the natural next step. For example, Paperpal’s AI-powered assistive writing technology can offer language suggestions while a researcher writes, offering feedback and tips to improve grammar, punctuation, style, and readability. With real-time suggestions to polish academic writing, the tool can help speed up the overall writing process and reduce the need for extensive revision, which instils a sense of confidence in the researcher.
It also allows journals to support researchers with English as a second language by giving them the opportunity to check their manuscripts against journal-specific requirements, such as key declarations, style and structural issues, or inconsistencies in references, figures and tables. By hosting this AI tool on their website for researchers to use during the submission process, journals can also ensure they don’t miss out on significant research that may be rejected due to often avoidable oversights.
While the increasing popularity and success of such software has raised the question of AI potentially replacing writers completely, most academics feel that the creativity, experience and technical expertise humans can work into their writing cannot be replicated by AI.

Artificial intelligence is a transformative technology for academic writing — as important for academics attempting
to achieve publication as the moment when the typewriter was replaced with the word processor or snail mail with email. I hope it will help the academic community put an end to the language tax for good, democratising scientific publishing and making English language journals significantly more accessible to non-native speakers.
Nishchay Shah is CTO and Head of Emerging Products at CACTUS, the company behind research writing and publishing tool Paperpal

15 | BIOSCIENCE TODAY |
NISHCHAY SHAH
| artificial intelligence |
75% of employers in pharmaceuticals and life sciences say WFH could limit career prospects
Staff in the pharmaceutical and life sciences sector who are working from home may be damaging their pay and career prospects.

That’s according to a spotlight on the industry A Reluctant Workforce: What impact are ‘Reluctant Returners’ having on the Pharmaceutical & Life Sciences sector?
In its report – which included the results of an in-depth survey across nine European countries of 3000 employees and 2750 employers in leadership roles – Unispace found that a staggering 75% of employers believed that working from home would limit the career prospects of staff in some way. Forty-two per cent indicate that promotion opportunities will be negatively impacted and 35% say bonuses will be hit. This data suggests that while employees may feel they can work effectively remotely, on a longer-term basis, staff are needed in the workplace for their own development as much as the company’s prospects.
DESIRE FOR GREATER CAREER PROGRESSION
When asked what they thought would encourage employees back to ‘in-office working’, 33% of employers in the sector said flexing start times, 29% stated paying for employees’ travel and 24% said having separate spaces for collaborative and quieter work would entice people back.
When employees were asked the same question, however, there was a greater appetite for incentives to return to the office, with 71% interested in flexi starting times, 77% saying free drinks and snacks would entice them to return, and
78% stating they were interested in access to training and development programmes. Based on these figures, it seems that employers may have misjudged the wants and needs of their workforces, particularly when it comes to the training and development aspirations.
Claire Shepherd, of Unispace, says: “As a sector that often requires more complex workspaces than any other, including laboratories and research and development centres, it’s perhaps little surprise that employers would need many of their employees to be present in the workplace for a significant amount of their working week.
“There would also be a need to create a sense of equity and collaboration, between those who have to be in the office working in laboratories and those who do not. This difference could drive a sense of presenteeism, limiting the career prospects of those who are reluctant to return.
“While the world of work as we know it has changed for good, our research does strongly indicate that the office itself is by no means redundant, particularly for a sector that cannot make a complete shift to remote working.
“But employees need to be shown that there are clear opportunities for career progression and in-office training programmes if employers hope to encourage staff back to the office to enable cross-discipline collaboration and drive impactful engagement.”
16 | careers | | BIOSCIENCE TODAY |
Support for STEM professionals returning to work
A new scheme has been launched for STEM professionals on a career break who want to return to the industry.
Consultancy Indicatura is working with STEM Returners to source candidates and support with mentoring and careers coaching.
The scheme is open to all levels from graduates to senior consultants with an engineering background, who have an interest in understanding how physical assets are configured, maintained and used.

Ian Kennedy, of Indicatura, said: “We are excited to be working with STEM Returners to help in the effort to eliminate the difficult barriers that candidates face returning to work after a career break, and are proud to say that we wholeheartedly value the experience and skills that returners have which is so often overlooked.”
STEM Returners was launched in 2017 by Natalie Desty after she saw how hard it was for people to return to STEM after a career break. They work with companies to facilitate returnships, which allow candidates to be re-integrated into an inclusive environment.
Natalie added: “We are delighted to be launching this new returner scheme with Indicatura to support highly skilled people back into the industry they love. There is a known shortage of skills across STEM industries, and we know that people who have had a career break are faced with an uphill task of getting back into the profession.”
In STEM Returners’ annual survey - The STEM Returners Index – 66% of STEM professionals on a career break said
they are finding the process of attempting to return to work either difficult or very difficult and that nearly half (46%) of participants said they felt bias because of a lack of recent experience.
STEM Returners offers candidates work experience and mentoring during their placement, as well as helping them adjust to life back in work.
The scheme has also helped boost diversity in host organisations. STEM Returners are 46% female and 44% from ethnic minority groups, compared to 14% female and 9% from ethnic minority groups working in industry. For details, see stemreturners.com/placements
Zoological biobank boost for research
A Cambridge-based biotech company has donated 2D-barcode scanners, software, and barcoded sample tubes to the Zoological Society of London (ZSL) to help create a unique biobank of zoological specimens for use by researchers around the world.
ZSL biobank manager Louise Gibson explained: “We needed a simple and robust method for critical DNA and tissue samples from our wide range of specimens, that would be easy to store, easy to retrieve and stand the test of time.
“Following consultation with the sample management specialists at Ziath, we decided to use 2D-barcoded sample tubes that can be stored in racks in a deep freeze. In addition, we will use Ziath’s high performance 2D-barcode scanners and Samples™ software to record and track the locations of the stored samples in a more efficient manner.
Assembled over the last 40 years, the ZSL collection includes samples donated from many diverse sources, including tissue biopsies from animals with unusual diseases, zoological objects seized by regulatory authorities, and endangered species specimens from various parts of the world.
The donation, by Instrumentation control and information management specialist Ziath, recognises the research value of the impressive collection.
Staff at ZSL’s Institute of Zoology aim to catalogue, process and store the growing collection to safeguard the integrity of the samples and open the invaluable resource to researchers.
“Some of our samples will require freezing and so we chose a system in which we are confident that the specimen identification labels will not be compromised. We cannot afford the risks of having to destroy samples because labels are unreadable.”

Louise added: “The aim of this project is to sift through our vast accumulation of specimens, identify them correctly, catalogue them and make biologically relevant samples available to researchers in other academic institutions worldwide. We feel that these samples hold immense value to researchers of zoonotic diseases which may one day become medically relevant to human health.”
For further information ZSL’s scientific research, see zsl.org/ science. More information about Ziath is at ziath.com/blog/ item/43-biobanking-choosing-sample-management-system
17 | news | | BIOSCIENCE TODAY |
THE EVOLUTION AND POTENTIAL OF ELISA
ELISA (Enzyme-Linked Immunosorbent Assay) is a platebased assay technique that allows for the detection and quantification of an analyte in a sample. ELISA utilises highly specific antibodies produced during an immune response, along with a colorimetric reaction and uses this mechanism to measure the concentration of the target antigen. Even when present in small concentrations, ELISAs can detect a wide range of targets, including hormones, peptides, proteins, and antibodies. The four major types of ELISA are indirect, direct, sandwich and competitive [1]. Despite its type, ELISA has been adapted across a wide range of fields due to its reliability, specificity, and stability. Commercial ELISA kits rapidly developed to meet a growing demand, becoming a widely applied tool in biotechnology, the food industry, vaccine development, clinical diagnosis, toxicology, and the pharmaceutical industry.
Compared to other immunoassays, ELISA exhibits greater sensitivity due to its unique enzyme amplification feature. The antibody-antigen coupling method, on the other hand, enhances specificity by linking the antibody’s paratope at the binding location with the antigen’s corresponding epitope in a highly specific manner. Despite the advantages this method offers, ELISA presents some challenges that future research should aim to overcome. Firstly, ELISA follows repetitive procedures, increasing the chances of user error. This method also requires centralised laboratory equipment, such as a spectrophotometer or microplate reader. Similarly, a high sample volume is required. In fact, the nanomolar detection limit of ELISA hinders the identification of many protein biomarkers [2,3].
Recent advances in ELISA-derived technology, however, have highlighted its potential, especially for in vitro
diagnosis and point-of-care settings. ELISA typically consists of four basic components: a solid substrate, a recognition component, a signal amplification component, and a readout method. The characteristics of each of these components significantly influences the performance of the assay. Consequently, advances in ELISA technology strongly rely on enhancing one or more of these components [2].
SOLID SUBSTRATES INNOVATIONS
Generally, ELISA takes place on a solid substrate surface comprising immobilise capture antibodies that isolate reactants from non-reactants. Most ELISAs are performed on a microwell plate, which in fact, require plate coating, blocking, and repeated washing and incubation steps. Thus, making the process tedious and time-consuming. Most importantly, the risk of errors is higher as multiple steps are involved in the process. Several studies have now optimised the blocking process to yield better results, a crucial step to reduce non-specific binding. Among these innovations, blocking using molecules such as PDDA or PAA resulted in impaired binding of non-specific molecules and increased absorption capacity of the capture antibody [3].
Nevertheless, the widespread use of ELISA for epidemiological research have highlighted the urgent need for on-site rapid detection. Paper-based ELISA is now a common practise. In fact, we have seen this technology for monitoring COVID-19, Monkeypox, HIB, and has been adapted for pregnancy testing. Yet, the use of paper substrates goes a long way and offer the possibility to develop more complex biosensors. In the past 10 years, different 3D microfluidic paper devices have been developed to overcome the limitations of lateral flow tests. For instance, a vertical flow-based paper immunosensor with a sample pad with different pore sizes was proposed for the detection of Influenza virus H1N1 [4]. To reduce the time required to complete a conventional ELISA, magnetics beads (MBs) have also been tested as a solid substrate owing to their ability to accelerate the diffusion process, and the possibility to isolate components of interests using a magnetic force. In fact, MS-based ELISA has been shown to detect different biomarkers in complex samples within 40 minutes. MB-based ELISAs have been proposed as an effective, rapid tool for the monitoring of human anti-SARSCoV-2 antibodies [2].
RECOGNITION ELEMENT INNOVATIONS
While antibodies have demonstrated sufficient recognition capacity, molecular imprinted polymers (MIPs) and nucleic acids have been incorporated into ELISA-derived technologies due to their unique cost, stability, specificity, and affinity. Despite the number of antibodies available, their use is limited. Recently, aptamer-based ELISA has been proposed as a solution due to the ability of aptamers to recognise and bind a wider range of targets. Competitive aptamer-based ELISA, for example, rely on a target
18 | BIOSCIENCE TODAY | | abbexa |
‘Abbexa is a dedicated manufacturer and worldwide supplier of biological tools such as ELISA kits, antibodies, proteins, enzymes and other reagents and products designed for use in research’.
molecule competing with the labelled molecule to bind to the aptamer, and has been shown particularly useful for the detection of small molecules. This type of assay has been employed for the detection of Aflatoxin B1 (AFB1) using AFB1 aptamers. Compared to conventional ELISA, this method exhibited higher sensitivity and reduced costs. Similarly, sandwich aptamer-based ELISA has been developed for the detection of infectious particles such as HCV. While aptamer-based ELISA is currently used for research purposes, its broad applicability has enough potential to further develop this technology [2,5].
Nucleic acids have also been used for the detection of polymerase chain reaction products (PCR) using ELISA. A typical PCR-ELISA protocol would include labelled digoxin to denature the PCR product, hybridising the PCR products to the capture probe, adding an enzyme-conjugated antibody specific for digoxin, adding the colorimetric substrate, and measuring the absorbance on a plate reader. While this process is expensive and time-consuming; it shares the high sensitivity and specificity of PCR and the batch detection ability of ELISA. In fact, the reliability of PCR-ELISA is about 100 times higher than that of agarose electrophoresis. Thus, the PCR-ELISA is frequently used for quantification and qualification of known bacteria, fungi, parasites, and viruses [2].
SIGNAL AMPLIFICATION INNOVATION
Just as the other components of ELISA, signal amplification in relation to ELISA sensitivity cannot be overstated. Commonly, ELISA rely on enzymes such as HRP that catalyse substrates and result in colorimetric changes that can be measured. One alternative to natural enzymes is the use of nanozymes: nanomaterials with enzyme-like catalytic activity, in many cases more efficient than natural enzymes. An adaptation of the ELISA format using nanozymes is the metal-organic framework-linked immunosorbent assay (MOFLISA). This assay utilises the increased efficiency of nanozymes for more accurate and sensitive assays. In the Plasmonic ELISA, the enzyme controls the aggregation of gold nanoparticles, generating coloured solutions with a distinct tonality. This format increases accessibility to the high-sensitive ELISA format, by not requiring specialised lab equipment. Qualitative results using this format can be detected by eye, and quantitative results require only a
luminometer. For example, a smartphone-based plasmonic Immunoassay Reader, which uses a smartphone camera as the luminometer, has been shown to produce results comparable with conventional ELISA [7]. Another adaptation of the ELISA format, CLIA, uses a chemiluminescent detection method with higher sensitivity than ELISA, is rapid, and detectable with a luminometer [2,6].
The Author: Pilar Ruiz, Scientific Marketing Communications Assistant at Abbexa
REFERENCES
[1] M. Alhajj and A. Farhana, “Enzyme Linked Immunosorbent Assay (ELISA),” PubMed, 2020. https://www.ncbi.nlm.nih.gov/books/NBK555922/
[2] P. Peng et al., “Emerging ELISA derived technologies for in vitro diagnostics,” TrAC Trends in Analytical Chemistry, vol. 152, p. 116605, Jul. 2022, doi: 10.1016/j.trac.2022.116605.
[3] S. Hosseini, P. Vázquez-Villegas, M. Rito-Palomares, and S. O. MartinezChapa, “Advantages, Disadvantages and Modifications of Conventional ELISA,” SpringerBriefs in Applied Sciences and Technology, pp. 67–115, Dec. 2017, doi: 10.1007/978-981-10-6766-2_5.
[4] N. Colozza, V. Caratelli, D. Moscone, and F. Arduini, “Origami Paper-Based Electrochemical (Bio)Sensors: State of the Art and Perspective,” Biosensors, vol. 11, no. 9, p. 328, Sep. 2021, doi: 10.3390/bios11090328.
[5] J. Bala, S. Chinnapaiyan, R. K. Dutta, and H. Unwalla, “Aptamers in HIV research diagnosis and therapy,” RNA Biology, vol. 15, no. 3, pp. 327–337, Feb. 2018, doi: 10.1080/15476286.2017.1414131.
[6] S. R. Herbin, D. G. Klepser, and M. E. Klepser, “Pharmacy-Based Infectious Diseases Management Programs Incorporating CLIA-Waived Point-of-Care Tests,” Journal of Clinical Microbiology, Feb. 2020, doi: 10.1128/jcm.00726-19.
ELISA KITS FOR ACCURATE DETECTION AND EFFECTIVE RESEARCH
Abbexa offers a broad range of ELISA Kits. Ready to use, built for precision and reproducibility. Have confidence in your results. Fully validated.
www.abbexa.com

19 | BIOSCIENCE TODAY | | abbexa |
20 | bio-informatics | | BIOSCIENCE TODAY |
‘Avatar’ motion tech aids advances in disease research
AI and motion capture technology are helping track the progression of rare genetic diseases affecting movement - and could ultimately be used to monitor common medical conditions like strokes.

In two ground-breaking studies, published in Nature Medicine, AI and clinical researchers have combined human movement data from wearable tech with a powerful new medical AI technology to identify clear movement patterns, predict future disease progression and significantly increase the efficiency of clinical trials in two very different rare disorders, Duchenne muscular dystrophy (DMD) and Friedreich’s ataxia (FA).
DMD and FA are degenerative, genetic diseases that affect movement and eventually lead to paralysis. There are currently no cures for either disease, but researchers hope these results will significantly speed up the search for new treatments.
Scientists hope that as well as using the technology to monitor patients in clinical trials, it could also one day be used to monitor or diagnose a range of common diseases affecting movement behaviour such as dementia, stroke and orthopaedic conditions.
Tracking the progression of FA and DMD is normally done through intensive testing in a clinical setting. These papers offer a significantly more precise assessment that also increases the accuracy and objectivity of the data collected.
The researchers estimate that using these disease markers mean that significantly fewer patients are required to develop a new drug when compared to current methods. This is particularly important for rare diseases where it can be hard to identify suitable patients.
Senior and corresponding author of both papers, Professor Aldo Faisal, from Imperial College London’s Departments of Bioengineering and Computing, who is also Director of the UKRI Centre for Doctoral Training in AI for Healthcare, and the Chair for Digital Health at the University of Bayreuth (Germany), and a UKRI Turing AI Fellowship holder, said: “Our approach gathers huge amounts of data from a person’s full-body movement – more than any neurologist will have the precision or time to observe in a patient.
21 | bio-informatics | | BIOSCIENCE TODAY |
“Our AI technology builds a digital twin of the patient and allows us to make unprecedented, precise predictions of how an individual patient’s disease will progress. We believe that the same AI technology working in two very different diseases, shows how promising it is to be applied to many diseases and help us to develop treatments for many more diseases even faster, cheaper and more precisely.”
MOVEMENT FINGERPRINTS
Co-author of both studies Professor Richard Festenstein, from the MRC London Institute of Medical Sciences and Department of Brain Sciences at Imperial said: “Patients and families often want to know how their disease is progressing, and motion capture technology combined with AI could help to provide this information. We’re hoping that this research has the potential to transform clinical trials in rare movement disorders, as well as improve diagnosis and monitoring for patients above human performance levels.”
In the DMD-focused study, researchers and clinicians at Imperial, Great Ormond Street Hospital and University College London trialled the body worn sensor suit in 21 children with DMD and 17 healthy age-matched controls.

The children wore the sensors while carrying out standard clinical assessments (like the 6-minute walk test) as well as going about their everyday activities like having lunch or playing.
In the FA study, teams at Imperial, the Ataxia Centre, UCL Queen Square Institute of Neurology and the MRC London Institute of Medical Sciences worked with patients to identify key movement patterns and predict genetic markers of disease. FA is the most common inherited ataxia and is caused by an unusually large triplet repeat of DNA, which switches off the FA gene. Using this new AI technology, the team were able to use movement data to accurately predict the ‘switching off’ of the FA gene, measuring how active it was without the need to take any biological samples from patients.
In both studies, all the data from the sensors was collected and fed into the AI technology to create individual avatars and analyse movements. This vast data set and powerful computing tool allowed researchers to define key movement fingerprints seen in children with DMD as well as adults with FA, that were different in the control group. Many of these AI-based movement patterns had not been described clinically before in either DMD or FA. Scientists also discovered that the new AI technique could also significantly improve predictions of how individual patients’ disease would progress over six months compared to current gold-standard assessments. Such a precise prediction allows to run clinical trials more efficiently so that patients can access novel therapies quicker and help dose drugs more precisely.
INVALUABLE FOR CLINICAL TRIALS
This new way of analysing full-body movement measurements provide clinical teams with clear
22 | BIOSCIENCE TODAY |
“Our AI technology builds a digital twin of the patient and allows us to make unprecedented, precise predictions of how an individual patient’s disease will progress .”
- Professor Aldo Faisal, Imperial College London
| bio-informatics |
Professor Aldo Faisal is pictured with Luchen Li wearing the sensor suit. (Thomas Angus/Imperial College London).
disease markers and progression predictions. These are invaluable tools during clinical trials to measure the benefits of new treatments.
The new technology could help researchers carry out clinical trials of conditions that affect movement more quickly and accurately. In the DMD study, researchers showed that this new technology could reduce the numbers of children required to detect if a novel treatment would be working to a quarter of those required with current methods.
Similarly, in the FA study, the researchers showed that they could achieve the same precision with 10 of patients instead of over 160. This AI technology is especially powerful when studying rare diseases, when patient populations are smaller. In addition, the technology allows to study patients across life-changing disease events such as loss of ambulation whereas current clinical trials target either ambulant or non-ambulant patient cohorts.
Co-author on both studies Professor Thomas Voit, Director of the NIHR Great Ormond Street Biomedical Research Centre (NIHR GOSH BRC) and Professor of Developmental Neurosciences at UCL GOS ICH, said: “These studies show how innovative technology can significantly improve the way we study diseases day-to-day.
“The impact of this, alongside specialised clinical knowledge, will not only improve the efficiency of clinical trials but has the potential to translate across a huge variety of conditions that impact movement. It is thanks to collaborations across research institutes, hospitals, clinical specialities and with dedicated patients and families that we can start solving the challenging problems facing rare disease research.”
Joint first author on both studies, Dr Balasundaram Kadirvelu, post-doctoral researcher at Imperial’s Departments of Computing and Bioengineering, said “We were surprised to see how our AI algorithm was able to spot some novel ways of analysing human movements. We call them ‘behaviour fingerprints’ because just like your hand’s fingerprints allow us to identify a person, these digital fingerprints characterise the disease precisely, no matter whether the patient is in a wheelchair or walking, in the clinic doing an assessment or having lunch in a café.”
The research was funded by a UKRI Turing AI Fellowship to Professor Faisal, NIHR Imperial College Biomedical Research Centre (BRC), the MRC London Institute of Medical Sciences, the Duchenne Research Fund, the NIHR Great Ormond Street Hospital (GOSH) BRC, the UCL/UCLH BRC, and the UKRI Medical Research Council.
With thanks to Caroline Brogan, Imperial College London. View original article here
REFERENCES
“Wearable full-body motion tracking of activities of daily living predicts disease trajectory in Duchenne muscular dystrophy” by Ricotti et al., published 19 January 2023 in Nature Medicine.
“A wearable motion capture suit and machine learning predict disease progression in Friedreich’s ataxia” by Kadirvelu et al., published 19 January 2023 in Nature Medicine. The two papers highlight the work of a large collaboration of researchers and expertise, across AI technology, engineering, genetics and clinical specialties. These include researchers at Imperial, the UKRI Centre in AI for Healthcare, the MRC London Institute of Medical Sciences (MRC LMS), UCL Great Ormond Street Institute for Child Health (UCL GOS ICH), the NIHR Great Ormond Street Hospital Biomedical Research Centre (NIHR GOSH BRC), Ataxia Centre at UCL Queen Square Institute of Neurology, Great Ormond Street Hospital, the National Hospital for Neurology and Neurosurgery (UCLH and UCL/UCL BRC), the University of Bayreuth, the Gemelli Hospital in Rome, Italy, and NIHR Imperial College Research Facility.

23 | BIOSCIENCE TODAY |
| bio-informatics |
Patients’ sensors feed into the motion capture technology (Thomas Angus/Imperial College London)
Synthetic routes to pharmaceuticals greatly expanded’
Crystallographers provide medicinal chemists with 1,800 additional pharmaceutical building blocks, leading to new and more effective treatments.
A search of the Cambridge Structural Database (CSD) has found nearly 1800 conglomerate crystal structures — molecules that have spontaneously enriched chirality upon crystallization representing 38% of the predicted chiral conglomerate compounds contained within the CSD. Research published by the American Chemical Society (JACS Au) identified the hidden conglomerate pool, which augments the limited biological chiral pool of synthetic building blocks used by medicinal chemists in drug synthesis.
These finding open new synthetic routes to existing drugs and could lead to new drugs for more effective treatment of disease.
The research, from a team led by Mark Walsh and Matthew Kitching of Durham University, published in JACS Au, introduces a new and potentially unlimited pool of chiral molecules outside of the chiral molecules derived from the natural world - the biological chiral pool. This pool is used by medicinal chemists to introduce chirality to their molecules. Most biological substances are chiral, including protein binding sites, so drugs that bind to these receptors must also be chiral. Famously the drug Thalidomide was withdrawn from the market when one enantiomer was found to cause birth defects, only 20 years later did scientists find that the other chiral form is safe.
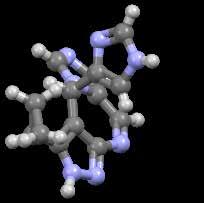
One way used by medicinal chemists to obtain enriched chiral molecules during synthesis is by conglomerate
Juergen Harter, Chief Executive Officer, CCDC

crystallization (where molecules spontaneously crystallise into single enriched crystals) or by using chiral compounds from the limited chiral pool. This research adds a significant alternative source of chirality.
The researchers first mined the ~1.2 million crystal structures in the CSD for those in Sohncke space groups with the potential to be chiral. This produced a subset of over 21,000 crystal candidates that then had their synthesis examined (by reference to the primary literature) to identify those that were produced by conglomerate crystallisation. 1800 compounds were identified that were synthesised by racemic methods but spontaneously crystallised in an enriched chiral form.
“We hope that the curation of this list of conglomerate crystals aids the development of preferential crystallisation and spontaneous deracemization protocols, while also furthering the understanding of the formation of conglomerate crystal behaviour,” Mark says.
“I’m delighted to see that the value of the 1.2 million crystal structures in the CSD is once again being utilised in another scientific field beyond crystallography. Medicinal chemists can now use the CSD to greatly widen their options to introduce chirality to their molecules than previously” adds Dr Jürgen Harter, CEO of CCDC.


relevant crystal structure in the CSD that was identified as a conglomerate. An antimicrobial (CSD Refcode: VEFFAP). Courtesy of the The Cambridge Crystallographic Data Centre (CCDC)).
The collaborative research between the University of Durham and the CCDC has recently been published (Identifying a Hidden Conglomerate Chiral Pool in the CSD - Mark P. Walsh, James A. Barclay, Callum S. Begg, Jinyi Xuan, Natalie T. Johnson, Jason C. Cole, and Matthew O. Kitching, JACS Au Article ASAP DOI: 10.1021/jacsau.2c00394).
24 | BIOSCIENCE TODAY | | pharmaceuticals |
Scientists turn up heat on physics phenomenon
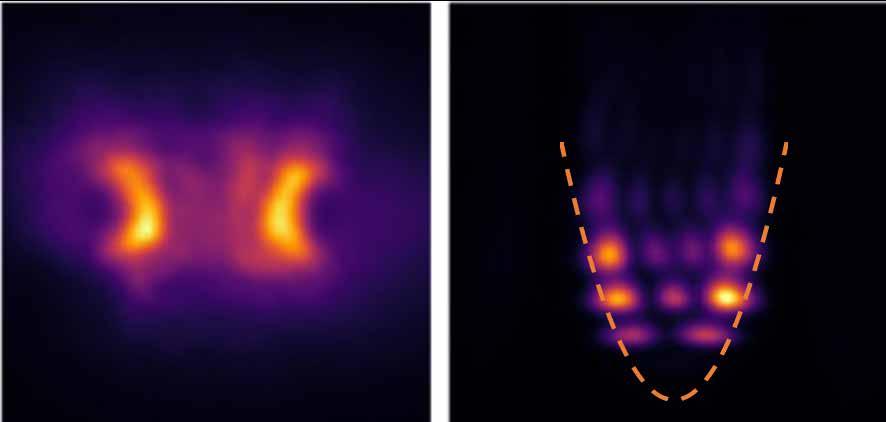
A ‘quantum harmonic oscillator’ has been made at room temperature by a team led by the University of St Andrews.
The structure can control the location and energy of quantum particles which could, in future, be used to develop new technologies including OLEDs and miniature lasers. The research, conducted in collaboration with scientists at Nanyang Technological University in Singapore, was published in Nature Communications. It used an organic semiconductor to produce polaritons which show quantum states, even at room temperature.
Polaritons are quantum mixtures of light and matter that are made by combining excitations in a semiconductor material with photons, the fundamental particles that form light. To create polaritons, the researchers trapped light in a thin layer of an organic semiconductor (the kind of light-emitting material used in OLED smartphone displays) 100 times thinner than a single human hair, sandwiched between two highly reflective mirrors.
Polaritons, like moisture in the air, can condense and form a type of liquid. The researchers corralled this quantum liquid
within a pattern of laser beams to control its properties. This made the fluid oscillate with a series of harmonic frequencies that resemble the vibrations of a violin string. The shape of these quantised states of vibration matched those of a ‘quantum harmonic oscillator’.
One of the project leaders, Dr Hamid Ohadi of the School of Physics and Astronomy at St Andrews, said: “This is a textbook problem that we look at with our students in our quantum physics courses. We used to think that one needs sophisticated cooling methods to see these oscillators. We found that this fundamental physics phenomenon can be seen at room temperature too.”
His colleague Professor Graham Turnbull added: “By studying this quantum oscillator we are learning how to control the location and movement of polaritons. In the future, we hope to exploit this knowledge to develop new quantum technologies for environmental sensing, or new types of OLEDs and miniature lasers.”
Professor Ifor Samuel, also part of the project team in St Andrews, said: “One of the most remarkable aspects of this study is that we excite the sample in one place, but see (polariton) lasing in another, showing that a quantum mixture or light and matter can travel macroscopic distances. This could be useful not only for lasers, but also for solar cells.”
The research was funded by UK Engineering and Physical Sciences Research Council (EPSRC) and the Scottish Funding Council (SFC).
The paper ‘Optically trapped room temperature polariton condensate in an organic semiconductor’ is published in Nature Communications.
25
Left, the trapped quantum fluid as seen under a microscope and (right) the shapes of the individual harmonic oscillation states of the quantum fluid when the fluid is trapped in a dip in the intensity of the laser beams (dashed line).
| BIOSCIENCE TODAY | | quantum physics |
“We used to think that one needs sophisticated cooling methods to see these oscillators. We found that this fundamental physics phenomenon can be seen at room temperature too.”
Bionanoscience ‘will precipitate a Fifth Industrial Revolution’
Currently based at Malopolska Centre of Biotechnology (MCB), Jagiellonian University in Poland, Professor Heddle’s research into bionanoscience aims to understand, design and build artificial and natural biological nanomachines. Living systems are built and maintained through the action of countless biological machines such as enzymes which are made from protein. They exist at the nanoscale (a nanometre being one thousand times smaller than the width of a human hair) and many act like tiny robots.
Working together, they are responsible for defining features of cells such as self-repair and autonomous motion. Taking
inspiration from nature’s nanomachines Professor Heddle hopes to build artificial cell-like ‘nanorobots’ that are biocompatible and biodegradable and capable of carrying out useful tasks not seen or even not possible in nature.

For example, such nanomachines could one day act as ‘gatekeepers’ for our bodies; identifying and destroying cancer, acting as new drug delivery systems and even slow the aging process.
The highly competitive Leverhulme International Professorships enable universities to attract globally leading scholars to take up permanent professorial posts in the UK. The funding will help establish the Centre for Programmable Biological Matter at Durham University, supporting a team of early career researchers and PhD students and building a solid foundation for further development of this exciting new area of research at the university.
Professor Martin Cann, Head of Biosciences Department at Durham University, said: “I am delighted that Jonathan is joining the University. We see this award as just the beginning, his arrival is part of an exciting plan to build on our world class bio-capabilities and provide the basis for further expansion of our research goals.”
26 | BIOSCIENCE TODAY | | bionanoscience |
Professor Jonathan Heddle is set to embark on a new era in Bionanoscience at Durham University, thanks to a £4.8million Leverhulme International Professorship award.
Professor Jonathan Heddle
“Professor Heddle hopes to build artificial cell-like ‘nanorobots’… capable of carrying out useful tasks not seen or even not possible in nature.”
Professor Heddle completed his PhD in biochemistry at the University of Leicester. After obtaining the prestigious Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowship for Research, he conducted structural biology research at Yokohama City University in Japan.
Professor Heddle then created his own laboratory at the Tokyo Institute of Technology, and again at the RIKEN research centre in Wak , Japan, before moving to Jagiellonian University in Poland.
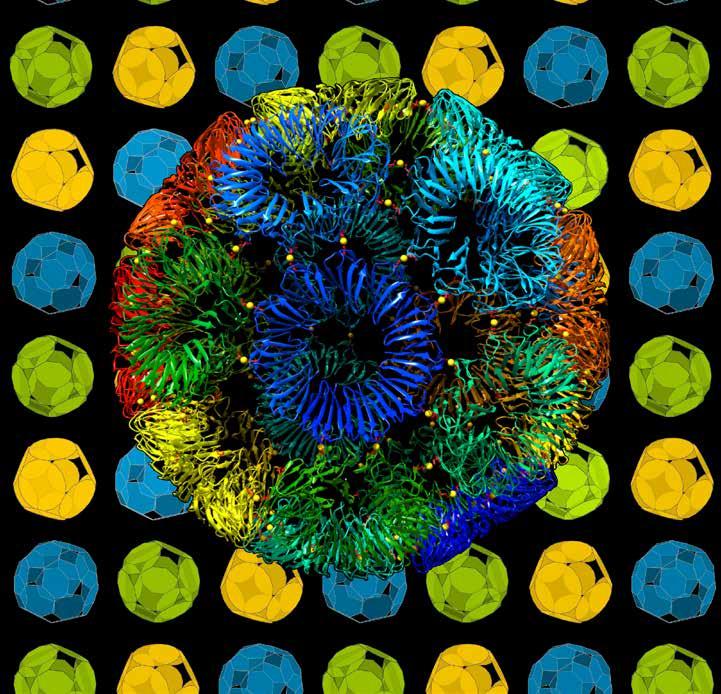
Originally from the Bishop Auckland area, Professor Heddle has a long history of successful collaboration with Durham University, pushing research frontiers in bionanoscience. This has included work aimed at understanding viral protein machinery, to build programmable protein nanocages, which can be used in biomedicine.
Professor Heddle said: “Bionanoscience is a fast growing and ground-breaking area of research which will precipitate a Fifth Industrial Revolution resulting in far-reaching new capabilities in materials, energy production and therapeutics. I look forward to building on the excellent facilities and highly interdisciplinary work being carried out at Durham University’s Biophysical Sciences Institute to develop an internationally renowned centre in this area.”
Professor Anna Vignoles, Director of the Leverhulme Trust, added: “The Trust is thrilled to award Professor Heddle a Leverhulme International Professorship. He is undertaking exciting and internationally important work in bionanoscience. This class of innovative, cutting-edge research will enable the UK to be world-leading in this area.”
More information at heddlelab.org.
27 | BIOSCIENCE TODAY | | bionanoscience |
Image of the RAP-cage with each constituent ring a different colour. Image credit: Heddle Lab, Craig S. Kaplan and Bernard Piette.
28 | BIOSCIENCE TODAY | | microbial chemistry |
All the world’s a microbe

How have humans ‘domesticated’ microbial chemistry? How did ‘chemical warfare’ among microbes lead to the evolution of antibiotics? How can we harness the amazing power of microbes for our future good? At a recent Gresham College lecture, acclaimed microbiologist and Professor of Physic, Robin May, examined some of the surprising ways these tiny organisms affect our daily lives.
Microbial chemistry makes bread rise and cheese mature. It turns grapes into wine. Microbes help make engine fuel, life-saving antibiotics and nano-particle sunscreens. Without fungi and bacteria, the world would sink under its own waste within days, since only these microbes have the ability to degrade complex polymers such as the lignin in plants. How can we harness this amazing power.
Might we be able to harness this amazing power of microbial degradation to help remove the human-made plastic mountain, or clean up toxic waste sites?
Hidden out of sight, beyond the resolution of the human eye, one could be forgiven for thinking that the microbial world is a tranquil place. However, nothing could be further from the truth. Depending on where it is taken from, a gram of soil may contain anything from 1 million to 1 billion bacteria, together with a similar number of fungal cells and perhaps 10100 times as many viruses.
Within this extraordinarily crowded environment, competition is intense and so to maintain their ecological niche, microbes have evolved chemistry skills that exceed anything yet achieved in even the most advanced pharmaceutical laboratory. In this lecture we take a scenic tour of some spectacular microbial molecular wizardry and witness how this unseen chemistry impacts the human world for both good and ill.
FOOD, FERMENTATION AND THE MAGIC OF LIFE WITHOUT OXYGEN
We start our journey with the most fundamental part of biological chemistry –how to gain useful energy from food. All living cells share a common ‘energy currency’ – the molecule Adenosine Triphosphate (ATP). This biological equivalent of the dollar is the high-
energy ‘fuel’ that cells use to drive movement, build complex molecules, or power elaborate behaviours like cell division.
Regardless of whether you are a carnivorous lion or a wood-decomposing fungus, ultimately the food you eat is converted into ATP in order to power all of the energyrequiring functions of that organism. The most efficient way to do this is so-called ‘aerobic respiration’; a process that uses oxygen to break down complex molecules in food, creating lots of ATP and releasing carbon dioxide as a by-product.
Most higher organisms, including humans, are strictly dependent on aerobic respiration – if humans are deprived of oxygen, the consequences are swift and irreversible. However, many microbes have evolved alternative pathways that allow them to continue to create ATP from nutrients even in the absence of oxygen.
Although less efficient than aerobic respiration, some of these alternative fermentation pathways turn out to have major benefits for us. Every time you eat bread or consume an alcoholic drink, you are benefiting from the product of one form of microbial fermentation; ethanol. Equally, an alternative fermentative process leads to the production of lactic acid, rather than ethanol, and in so doing underpins our production of yoghurt, kefir and many cheeses.
Over thousands of years, humans have ‘domesticated’ particular lineages of yeast or bacteria to maximise productivity of these different biochemical pathways, thus resulting in organisms that are adapted in ways that are useful for human food production; producing higher alcohol levels in beer or adding flavour-enhancing complex molecules to cheeses, for instance.
29 | microbial chemistry | | BIOSCIENCE TODAY |
THE CHEMISTRY OF KILLER MICROBES
Sometimes the clever chemistry of microbes in food is altogether less desirable. Each year, in the UK alone there are around 2.4 million cases of food poisoning. In some cases, such as norovirus infection, the disease is a result of the live organism setting up home in your body. More often, however, foodborne disease arises not from the presence of the microbe itself, but from the potent toxins that the organism releases.
One of the most remarkable and deadly of these is botulinum toxin, produced by the bacterium Clostridium botulinum. This widespread soil microbe can contaminate food during production. As an obligate anaerobe, C. botulinum is unable to grow in the presence of oxygen, but when oxygen is removed – for instance when food is vacuum sealed or canned – the bacterial spores can ‘awaken’ and start growing. As they do so, they produce botulinum toxin - the most toxic molecule known, with a lethal dose of less than two nanograms (two millionths of a gram) per kg. Although deadly when consumed in food, as with many poisons, botulinum toxin has valuable medical uses when used with extreme care. As a potent muscle relaxant it is used in the treatment of uncontrollable muscle spasms, post-stroke disability and, famously, to reduce wrinkles caused by aging. In fact, microbial chemistry is capable of creating a bewildering and terrifying array of toxins, often with a precision that is as breathtaking as it is deadly. The bacterium Vibrio parahaemolyticus, for instance, is infamous as the cause of acute shellfish-associated gastrointestinal disease. The hallmark nausea, vomiting and bloody diarrhoea are a result of the bacterium injecting a set of potent cellular toxins directly into intestinal cells, using an elegant ‘bacterial needle’ called the Type III Secretion System. Remarkably, this weapon is normally inactive, but becomes activated only when the bacterium detects human bile salts, present in the human intestine; a precision ‘failsafe’ mechanism that means the bacterium only spends energy on this hugely demanding piece of chemistry when it
is useful to do so.
Chemical toxin warfare is, of course, not the sole preserve of bacteria. In fact the prize for elegant toxin chemistry probably belongs to the fungi, which between them produce a dazzling array of precision poisons. These range from the amatoxins, produced by the aptly named ‘Death Cap’ mushroom, which specifically block an early step in protein synthesis, to the neurotoxin ergotamine, produced by Claviceps fungi that grow on cereal crops. Perhaps the most ‘subtle’ fungal toxin, though, is coprine, produced by Coprinus atramentaria. This molecule is not in itself toxic, but inhibits an enzyme called acetaldehyde dehydrogenase. However, since this enzyme is critical to detoxify alcohol, consuming these mushrooms at the same time as alcohol can prove deadly – explaining the fungus’ common name of ‘Tippler’s Bane’.
MICROBIAL WEAPONS, HUMAN MEDICINES

From our human-centric perspective, we often think of microbes as being ‘out to get us’. But of course on an evolutionary scale, humans are insignificant to most microbes and in fact their major competitors are other microbes. Within that crowded soil particle, or indeed myriad other microbiologically-rich habitats, resources are scarce and competition intense. So it is perhaps no surprise that many microbes resort to chemical warfare in order to maintain their ecological foothold. This microbe-on-microbe battleground has, however, yielded one of the most transformative discoveries in the history of medicine: antibiotics.
The idea of harnessing microbial warfare for human benefit has a long history. Indeed Louis Pasteur commented, “if we could intervene in the antagonism observed between some bacteria, it would offer perhaps the greatest hopes for therapeutics”. He was right, but it took until 1928, and Alexander Fleming’s observation of the fungus Penicillium rubrum inhibiting the growth of neighbouring bacterial colonies, that the first natural antibiotic, penicillin, was discovered (although it was not successfully purified until 1942).
We now know that many bacterial species produce antibiotics, although it is clear that we have barely scratched the surface in terms of their discovery. One bacterial family, the Streptomycetes dominate this landscape. Typically found in soils rich in organic matter, such as forest leaf litter, these bacteria have become masters of secondary metabolism, the process by which complex, but nonessential, molecules are created.
Many of these secondary metabolites are targeted at other microbes and consequently Streptomycetes are the source of over two thirds of antibiotics currently in use by humans. These range from relatively simple molecules such as chloramphenicol, typically used in topical treatments such as eye drops, to eye-wateringly complex chemicals like daptomycin. Most antibiotics are now produced at least partially synthetically in order to ensure consistency of production. However it is somewhat humbling to realise that a process that requires distillation, condensation, purification and numerous other energy-intensive chemistry steps within large factories with highly skilled staff can be carried out faster and more efficiently within a single bacterial cell.
MATERIAL MANUFACTURING ON A MICROBIAL SCALE
Professor Robin May is Gresham Professor of Physic and Professor of Infectious Disease at the University of Birmingham. As the FSA’s Chief Scientific Adviser, he provides expert scientific advice to the UK government and plays a critical role in helping to understand how scientific developments will shape the work of the FSA, as well as the strategic implications of any possible changes.
In our final stop on this microscopic voyage of chemical exploration, we turn our attention to materials science. Human innovation in materials has transformed our world, particularly over the last 100 years. Steel, concrete, glass and - that ubiquitous and now insidious material – plastic are everywhere. Our buildings, clothes, vehicles and homes depend on these materials, and there is an insatiable demand for materials that are stronger, more intricate, lighter, more durable, biodegradable…the list is endless. Might microbes be able to help?
One major advantage of microbial chemists over human ones is their ability to venture to destinations that no
30 | microbial chemistry | | BIOSCIENCE TODAY |
PROF ROBIN MAY
human can go, such as radioactive or heavily polluted areas. This holds particular promise because often the contaminating chemicals are themselves of value – if only they could be ‘harvested’ in a safe way. The bacterium Geobacter sulfurreducens is one organism being investigated for this purpose. Geobacter is covered in long protein filaments that can ‘capture’ heavy metals such as uranium. Remarkably, these filaments are conductive and act as tiny electricity wires, providing electrical energy to the bacterium and, in doing so, precipitating the uranium out of groundwater, making it less likely to spread.

This microbial clean-up act is not restricted only to radioactive metals. Over recent years, detailed investigations of soils, rivers, oceans and caves have turned up a diverse range of microbes that show potential for dealing with many human problems. The bacterium Pseudomonas putida can digest polyurethane foam, a material used widely in buildings and mattresses that was
thought to be non-biodegradable. Several fungi, such as Pleurotus pulmonarius, are able to degrade petroleum oil spills. And perhaps the greatest prize of all – the recently discovered bacterium Ideonella sakaiensis has a remarkable ability to degrade polyethylene tetrapthalate (PET); the component of drink bottles and numerous other disposable plasticware that is responsible for the growing ‘microplastic mountain’ that threatens biodiversity worldwide.
Perhaps the most exciting aspect of microbial material chemistry, though, lies in our ability to adapt the natural biosynthetic pathways of these organisms via genetic modification in order to create high-value chemical products. These range from delicate fibres, such as spider silk (recently produced by genetic engineering of the marine bacterium Rhodovulum sulfidophilum) to the carbonbased ‘wonder material’ graphene, which researchers in the Netherlands have coaxed the bacterium Shewanella to produce from graphite. Given our rapidly advancing capabilities for genetic engineering in fungi and bacteria, together with the ever-expanding catalogue of microbial species with useful features, it seems a fair bet that the future of chemistry will involve a lot of microbiology.
Watch the previous lectures in this series, All The World’s A Microbe, at here gresham.ac.uk/watch-now/series/microbes
You can also read more about Gresham College’s free public lectures since 1597 here gresham.ac.uk/about-us
31 | microbial chemistry | | BIOSCIENCE TODAY |
“Microbes have evolved chemistry skills that exceed anything yet achieved in even the most advanced pharmaceutical laboratory.”
AI tailors artificial DNA for future drug development
With the help of artificial intelligence, Swedish researchers have succeeded in designing synthetic DNA that controls the cells’ protein production.
The technology, developed at Chalmers University of Technology, can contribute to the development and production of vaccines, drugs for severe diseases, as well as alternative food proteins much faster and at significantly lower costs than today.
How our genes are expressed is a process that is fundamental to the functionality of cells in all living organisms. Simply put, the genetic code in DNA is transcribed to the molecule messenger RNA (mRNA), which tells the cell’s factory which protein to produce and in which quantities.
Researchers have put a lot of effort into trying to control gene expression because it can, among other things, contribute to the development of protein-based drugs. A recent example is the mRNA vaccine against Covid-19, which instructed the body’s cells to produce the same protein found on the surface of the coronavirus. The body’s immune system could then learn to form antibodies against the virus. Likewise, it is possible to teach the body’s immune system to defeat cancer cells or other complex diseases if one understands the genetic code behind the production of specific proteins
Most of today’s new drugs are protein-based, but the techniques for producing them are both expensive and slow, because it is difficult to control how the DNA is expressed. In 2021, a research group at Chalmers,
led by Aleksej Zelezniak, Associate Professor of Systems Biology, took an important step in understanding and controlling how much of a protein is made from a certain DNA sequence.


“First it was about being able to fully ‘read’ the DNA molecule’s instructions. Now we have succeeded in designing our own DNA that contains the exact instructions to control the quantity of a specific protein”, Aleksej says about the research group’s latest important breakthrough.
DNA MOLECULES MADE-TO-ORDER
The principle behind the new method is similar to when an AI generates faces that look like real people. By learning what a large selection of faces looks like, the AI can then create completely new but natural-looking faces.
It is then easy to modify a face by, for example, saying that it should look older, or have a different hairstyle. On the other hand, programming a believable face from scratch, without the use of AI, would have been much more difficult and time-consuming.
Similarly, the researchers’ AI has been taught the structure and regulatory code of DNA. The AI then designs synthetic DNA, where it is easy to modify its regulatory information in the desired direction of gene expression. Simply put, the AI is told how much of a gene is desired and then ‘prints’ the appropriate DNA sequence.
“DNA is an incredibly long and complex molecule. It is thus experimentally extremely challenging to make changes to it by iteratively reading and changing it, then reading and changing it again. This way it takes years of research to find something that works. Instead, it is much more effective to let an AI learn the principles of navigating DNA.
“What otherwise takes years is now shortened to weeks or days”, says first author Jan Zrimec, a research associate at the National Institute of Biology in Slovenia and past postdoc in Aleksej’s group.
The researchers have developed their method in the yeast Saccharomyces cerevisiae, whose cells resemble mammalian cells. The next step is to use human cells. The researchers have hopes that their progress will have an impact on the development of new as well as existing drugs.

32 | BIOSCIENCE TODAY | | drug development & discovery |
Aleksej Zelezniak, Associate Professor, Department of Biology and Biological Engineering, Chalmers University of Technology Research associate Jan Zrimec
“Protein-based drugs for complex diseases or alternative sustainable food proteins can take many years and can be extremely expensive to develop. Some are so expensive that it is impossible to obtain a return on investment, making
them economically nonviable. With our technology, it is possible to develop and manufacture proteins much more efficiently so that they can be marketed”, Aleksej concludes.
The authors of the study are Jan Zrimec, Xiaozhi Fu, Azam Sheikh Muhammad, Christos Skrekas, Vykintas Jauniskis, Nora K. Speicher, Christoph S. Börlin, Vilhelm Verendel, Morteza Haghir Chehreghani, Devdatt Dubhashi, Verena Siewers, Florian David, Jens Nielsen and Aleksej Zelezniak.
The researchers are active at Chalmers University of Technology, Sverige; National Institute of Biology, Slovenia; Biomatter Designs, Lithuania; Institute of Biotechnology, Lithuania; BioInnovation Institute, Denmark; King’s College London, UK.
Read the full study at Controlling gene expression with deep generative design of regulatory DNA

33 | BIOSCIENCE TODAY | | drug development & discovery |
“Most of today’s new drugs are protein-based, but the techniques for producing them are both expensive and slow, because it is difficult to control how the DNA is expressed. ”
Illustration credit: Yen Strandqvist, Chalmers

34 | BIOSCIENCE TODAY | | clinical trials |
There’s more to collagen than cosmetics...
Big strides are being made in collagen clinical development, particularly in treatment of age-related conditions. Dr Gen Li, president and founder of Phesi, discusses the increase in collagen research, and its clinical and biological applications.

According to the World Health Organisation, one in six people worldwide will be 60 years old or over by 2030, and the number of persons aged 80 or older is expected to triple between 2020 and 2050, reaching 426 million.
Advances in medical science and technology have greatly improved public health and increased life expectancy, but the natural process of ageing has consequences spanning from a loss of elasticity in the skin to stiff
joints and brittle bones, which affect patients’ quality of life.
As the proportion of older people worldwide is growing, interest in - and awareness of - agerelated conditions has grown.
As a result, the clinical development community has turned its attention to medicines to treat age-related conditions, address quality of life issues, and prevent complications for ageing people.
35 | BIOSCIENCE TODAY | | clinical trials |
The study of collagen is one such area. Collagens are a group of structural proteins that make up a crucial component of connective tissue and exist throughout the human body to improve its strength. Collagens are the most abundant protein in the human body, accounting for nearly a third of all proteins. Our bodies gradually produce less collagen over time and collagen fibres become weaker, which has a subsequent impact on joint and skin health.
The cosmetic industry has picked up on the potential for collagen in skincare and beauty products – but use cases for collagen don’t end here. The wider potential for collagen to improve the health of older persons has captured attention.
As a result, the global market for collagen supplements alone is expected to surpass $2 billion in revenue by 2030 The burgeoning popularity of collagen-based products prompted Phesi to review the evolution of clinical trials over the past decade that have concentrated on collagen and its potential therapeutic applications.
Our analysis of 2,358 clinical trials involving collagen showed a steady and linear growth in the number of trials from 2010 onwards, increasing by 142% between 2010 and 2019. The COVID-19 pandemic caused an inevitable decline in 2020, but the number of collagen trials quickly recovered and returned to pre-pandemic levels in 2021.
Interestingly, our research found that less than 5% of collagen clinical trials in the past 12 years relate to cosmetic applications, demonstrating that interest in applications to mitigate serious age-related conditions is high.
However, commercial support for clinical research is limited, with only 27% of collagen-related trials receiving industry funding. If clinical research into collagen can maintain its current momentum and benefit from greater funding, we have the potential to uncover a wealth of new and innovative therapies. These could include treating skin and bone diseases, as well as improving treatments for wound care and arthritic joints.
The discovery of new treatments for age-related conditions would mark a significant milestone and benefit the lives of millions. For instance, as the most common joint disease, osteoarthritis affects more than more than 500 million people worldwide, with women disproportionately impacted.

Osteoarthritis is more frequent in countries with established market economies where there is an aging population and a higher prevalence of obesity. There is currently no cure aside from joint replacement surgery, but clinical trials are underway to examine how collagen could treat symptoms such as joint inflammation and stiffness, which occur when cartilage wears down.
To help better the industry’s understanding of the use cases of collagen and bring more effective treatments to market faster, clinical trial design must be informed by patientcentric data and predictive analytics.
Data from historical and ongoing clinical trials will reduce the cost of development, minimise patient burden, and accelerate the speed that new therapies become available to patients in need – transforming patients’ quality of life.
To optimise clinical trials and eliminate avoidable protocol amendments – which are both costly and time-consuming –researchers’ understanding of the target patient population must be data-driven rather than perception-led.

Equipped with the right approach to data, the R&D industry has the opportunity to conduct cheaper, more effective clinical trials, and develop innovative treatments for agerelated conditions.
Details at phesi.com
36 | clinical trials | | BIOSCIENCE TODAY |
“If clinical research into collagen can maintain its current momentum and benefit from greater funding, we have the potential to uncover a wealth of new and innovative therapies.”
Dr Gen Li, President & Founder, Phesi
Flatworm-inspired medical adhesives stop blood loss
Every year around two million people die worldwide from haemorrhaging or blood loss.
Uncontrolled haemorrhaging accounts for more than 30% of trauma deaths. To stop the bleeding, doctors often apply pressure to the wound and seal the site with medical glue. But what happens when applying pressure is difficult or could make things worse? Or the surface of the wound is too bloody for glue?
Drawing inspiration from nature, researchers from McGill University have developed a medical adhesive that could save lives, modelled after structures found in marine animals like mussels and flatworms.
“When applied to the bleeding site, the new adhesive uses suction to absorb blood, clear the surface for adhesion, and bond to the tissue providing a physical seal. The entire application process is quick and pressure-free, which is suitable for non-compressible haemorrhaged situations, which are often life-threatening,” says lead author Guangyu Bao, a recently graduated PhD
student under the supervision of Professor Jianyu Li of Department of Mechanical Engineering.
In putting the new technology to the test, the researchers found that the adhesive promotes blood coagulation. The adhesive can also be removed without causing re-bleeding or even left inside the body to be absorbed. “Our material showed much better-improved safety and bleeding control efficiency than other commercial products. Beyond bleeding control, our material could one day replace wound sutures or deliver drugs to provide therapeutic effects,” says senior author Professor Jianyu Li.
REFERENCE
“Liquid-infused microstructured bioadhesives halt non-compressible hemorrhage” by Guangyu Bao et al. was published in Nature Communications.
World-first test for new heart attack drug
Professor Jon Townend, Consultant Cardiologist at University Hospitals Birmingham, Honorary Professor of Cardiology in the Institute of Cardiovascular Sciences at the University of Birmingham, and Chief Investigator of the trial said: “We look forward to starting this exciting trial of a new drug for heart attacks which are still only too common.
A potential new drug to improve the long-term outcomes for heart attack patients will be trialled in the UK.
With the signing of a partnership between the University of Birmingham and Acticor Biotech, a new clinical trial called LIBERATE will take place in two acute care hospitals in the UK: the Queen Elizabeth Hospital, Birmingham and the Northern General Hospital, Sheffield.
The randomised, double-blind Phase 2b LIBERATE study will recruit more than 200 patients to test the tolerance and the efficacy of glenzocimab 1000 mg, versus placebo, to reduce heart damage following a myocardial infarction (MI), commonly known as a heart attack.
The trial will be run at the University of Birmingham with expert clinicians from the Institute of Cardiovascular Sciences and University Hospitals Birmingham NHS Foundation Trust. Bringing together experience in running multi-site clinical trials from the CTU and expertise in heart diseases, the team will see whether glenzocimab will reduce the amount of dead heart tissue in patients following an ST segment elevation myocardial infarction (STEMI), the most serious type of heart attack.
“Although immediate opening of the blocked coronary artery by angioplasty in cases of heart attack is now routine, significant heart damage still occurs. Glenzocimab reduces clot formation and laboratory findings have been impressive. There are strong reasons to believe that this new drug may improve outcomes and this randomized blinded trial is the right way to test this theory.”
Dr Mark Thomas, Associate Professor of Cardiology at the University of Birmingham and Honorary Consultant Cardiologist, who designed the trial and led its development, said: “This trial will help us establish whether glenzocimab is a safe and effective drug for preventing the kind of clotting that can lead to serious damage to the heart following a heart attack. We’re delighted to work with Acticor to see whether this new class of drug has the potential to improve the outcomes of our patients with heart attacks. While the immediate care provided for a heart attack is effective for improving patient survival, there is more we can do to prevent long-term damage to the heart.”
Glenzocimab, a humanized monoclonal antibody (mAb) fragment directed against the platelet Glycoprotein VI (GPVI), was developed by Acticor Biotech. This new drug, currently being trialled for strokes, stops the functioning of platelets that cause abnormal clotting of blood. While platelet function is normally important to stop bleeding, this drug specifically targets only dangerous clotting inside damaged blood vessels (thrombosis).
37 | news | | BIOSCIENCE TODAY |
The partnership between the University of Birmingham and Acticor Biotech will see patients with heart attacks treated with glenzocimab, a promising new class of drug, for the first time.
Global community will assess environmental impact of clinical trials
The collaborative initiative will publish a publicly available methodology for calculating and comparing the carbon footprint of centralised and decentralised clinical trials.
A global, not-for-profit alliance that advocates for greater collaboration in life sciences R&D, has announced the launch of the Clinical Trials Environmental Impact Community of Interest (CoI) to measure and compare the carbon footprint of centralised (traditional sitebased) and decentralised clinical trials.

The initiative, by The Pistoia Alliance, was started by member Syneos Health, which submitted it to the Alliance for further development. The new CoI also work closely with member organisation, the Sustainable Health Coalition (SHC) in support of its work with the Sustainable Markets Initiative (SMI). The Pistoia Alliance and the SHC will further align with the SMI as its efforts in this space develop.
In phase one, the Community of Interest is working to collectively agree the parameters and variables that will be measured. The second phase will see it develop a methodology for calculating the carbon footprint of both trial models. This methodology will be published as a publicly available tool to deliver a cross-industry standard for quantifying the carbon impact of a trial. The CoI currently has 25 members spanning Japan, US, and Europe, including top 10 pharma companies, CROs, academic representatives.
Noolie Gregory, of Syneos Health, explained: “We are committed to delivering on our environmental goals and finding ways to make clinical development activities more sustainable.
“Through this initiative, we can drive toward a tangible outcome that will make a material improvement in how we assess the carbon impact of trials and make recommendations for the reduction of this footprint. We are looking forward to working with our peers and colleagues to share our knowledge.”
There is growing pressure from patients, governments, healthcare payers and providers to reduce the environmental impact of the life sciences. In the future, it can be expected there will be legislation that will necessitate pharma and research companies adjust their behaviors and develop new ways to measure and report on the environmental impact of their operations.
To prepare for this change, the Alliance believes a key starting point is company collaboration on assessing
the impact of trials. Clinical trials are complex and have many moving parts – typically spanning multiple geographies, jurisdictions, and hundreds of different sites. To create a robust and accurate methodology that benefits the industry will require a broad mix of experts and opinions from both the life sciences and sustainability fields.
“Shared frameworks have to be created to allow individuals and companies to be efficient and effective in quantifying the impact of their own clinical trials against an industry-wide standard,” commented Dr Bert Hartog, Senior Director of Clinical Innovation at Janssen. “At Janssen, we are always looking for ways to reduce our impact on the environment whilst continuing to innovate and produce life-changing therapies. We are very keen to get involved in collaborative projects such as the new Pistoia Alliance COI because we recognise that they are the best way to make lasting change happen in our industry.”
“Decentralised trials reduce the burden on the patient and ensure trials are far more patient centric. Whilst quantifying money and time saved is relatively straightforward, accurately measuring the carbon saving requires a more detailed approach to produce a standard that can be used by all stakeholders – from sponsors to CROs to technology vendors,” commented Thierry Escudier, Strategic Leader for Empowering the Patient at Pistoia Alliance. “There’s lots of great work being done in the industry, and the Alliance hopes to work alongside these other groups. The scale and complexity of the issue makes it perfectly placed for the Alliance to tackle – it’s multidisciplinary, global, and industry-wide. We are now calling for all those interested – including research organizations, investigators, pharma companies, and patient groups –to get involved.”
The Pistoia Alliance is seeking experts in sustainability, clinical development, and other relevant fields to join the Community of Interest. Individuals and organizations interested in the initiative should email Thierry Escudier at ProjectInquiries@PistoiaAlliance.org
For more information on the Pistoia Alliance, visit pistoiaalliance.org
38 | BIOSCIENCE TODAY |
| clinical trials |

39 | BIOSCIENCE TODAY | | clinical trials |
Games and stimulation mitigate cognitive decline in older adults
Older people may be able to boost their working memory with a new approach that couples online therapeutic games with a non-invasive brain stimulation technique.
Working memory is critical for people to function well in everyday life. This volatile form of memory holds and manipulates a finite amount of information over a short time interval, enabling people to interact with their environment in an effective and efficient manner. Working memory typically declines with age, with the decline in its capacity causing daily difficulties in people with Parkinson’s disease, dementia, and stroke.
Scientists and clinicians from the University of Birmingham, UK, Dalhousie University in Nova Scotia, Canada, and the University of Trento, Italy, have devised a new technology to mitigate this decline.
The investigators refer to the technology as cognitive needs and skills training, or COGNISANT, and research published in Frontiers in Aging Neuroscience shows it can provide particular benefit for older people who have low working memory capacity (WMC).
The online therapeutic exercises, developed to improve working memory, attention and vigilance, are packaged in the type of engaging interface that will be familiar to online game or App users. Brain stimulation was administered via a mobile wireless device that delivers a

small (2milliAmpère) transcranial direct current stimulation (tDCS) during training.
The study involved healthy people aged 55 to 76 years old, who were split into two groups. Both groups did the online games for 20 minutes day, over a five-day period. While one group also received tDCS, the other group wore the tDCS device, which resembles a swimming cap, but did not receive tDCS.
The researchers measured baseline working memory capacity (WMC) before the study, and two days after completion. They found that WMC improved significantly in all participants, regardless of age or whether they received tDCS.
Importantly, the combination of training games and tDCS showed particular benefit in older people with lower initial working memory. This subset included people aged 69.5 to 76 years, the advantage was evident from the first day of training and became statistically significant by the end of training.
The paper was senior authored by biomedical engineer Dr Sara Assecondi, formerly at Birmingham but now at Trento’s Centre for Mind/Brain Sciences (CIMeC), who worked on developing the technology with cognitive neuroscientist Professor Kim Shapiro from Birmingham’s
| BIOSCIENCE TODAY | 40 | cognitive decline |
School of Psychology and Centre for Human Brain Health, using online therapeutic games developed by neuropsychologist Professor Gail Eskes from Dalhousie’s departments of Psychology & Neuroscience, and Psychiatry, whose clinical practice focusses on training to improve and repair cognitive function.
The researchers are now working with technology transfer offices from the University of Birmingham and Dalhousie University, who are seeking commercial partners who wish to collaborate in developing technology using this approach and taking it to market.
Dr Sara Assecondi said: “Approaches used for hospital rehabilitation are difficult to translate to the home setting, but our approach uses online tools, and delivers brain
stimulation via a device that can be used anywhere, with the dose determined remotely by the physician.”
Professor Eskes’ clinical research, which focusses on training to improve and repair cognitive functions, designed the brain exercises. She said: “Intensive exercises at just the right difficulty are important for increasing brain capacity or efficiency. And the game-like aspects increase motivation and make it easier to stick with the challenging sessions.”
Professor Shapiro, whose research focusses on attention and memory, said: “Although cognitive decline in the elderly is an inevitability, approaches such as COGNISANT, in combination with regular physical exercise, can stem this decline and provide individuals with a higher quality of life.”
The researchers have already shown in a previous study that coupling tDCS with strategy on how to do tasks requiring working memory can help young people with low WMC improve their performance, by helping them put in place strategies that they would not otherwise be able to come up with.

Dr Assecondi added: “The effects seen in both studies were strong, with the first study indicating that the combination of stimulation and strategy instruction can improve WM performance in younger adults, and the second study showing that strategy use may be facilitated by stimulation in older participants.”
The researchers are planning further studies to examine other means of brain stimulation that may be even more effective and are currently concluding a study evaluating the benefits of COGNISANT in post-stroke patients with publication expected shortly.
REFERENCE
Older adults with lower working memory capacity benefit from tDCS when combined with working memory training: a preliminary study is published in Frontiers in Aging Neuroscience.
| BIOSCIENCE TODAY | 41 | | cognitive decline |
“Intensive exercises at just the right difficulty are important for increasing brain capacity or efficiency. And the game-like aspects increase motivation and make it easier to stick with the challenging sessions.”
Do AI models and the human brain process things in the same way?
Deep Neural Networks – part of the broader family of machine learning – have become increasingly powerful in everyday real-world applications such as automated face recognition systems and self-driving cars.
Researchers use Deep Neural Networks, or DNNs, to model the processing of information, and to investigate how this information processing matches that of humans.
While DNNs have become an increasingly popular tool to model the computations that the brain does, particularly to visually recognise real-world “things,” the ways in which DNNs do this can be very different.
Research, published in the journal Trends in Cognitive Sciences and led by the University of Glasgow’s School of Psychology and Neuroscience, presents a new approach to understand whether the human brain and its DNN models recognise things in the same way, using similar steps of computations.
Currently, Deep Neural Network technology is used in applications such as face recognition, and while it is successful in these areas, scientists still do not fully understand how these networks process information.
This opinion article outlines a new approach to better this understanding of how the process works: first, that researchers must show that both the brain and the DNNs recognise the same things – such as a face – using the same face features; and, secondly, that the brain and the DNN must process these features in the same way, with the same steps of computations.
As a current challenge of accurate AI development is understanding whether the process of machine learning matches how humans process information, it
is hoped this new work is another step forward in the creation of more accurate and reliable AI technology that will process information more like our brains do.
Prof Philippe Schyns, Dean of Research Technology at the University of Glasgow, said: “Having a better understanding of whether the human brain and its DNN models recognise things the same way would allow for more accurate real-world applications using DNNs.

“If we have a greater understanding of the mechanisms of recognition in human brains, we can then transfer that knowledge to DNNs, which in turn will help improve the way DNNs are used in applications such as facial recognition, where there are currently not always accurate.
“Creating human-like AI is about more than mimicking human behaviour – technology must also be able to process information, or ‘think’, like or better than humans if it is to be fully relied upon. We want to make sure AI models are using the same process to recognise things as a human would, so we don’t just have the illusion that the system is working.”
The study, ‘Degrees of Algorithmic Equivalence between the Brain and its DNN Models’ is published in Trends in Cognitive Sciences. The work is funded by Wellcome and Physical Sciences Research Council.
A link to the full study can be found here: www.authors. elsevier.com/sd/article/S1364-6613(22)00220-0
42 | BIOSCIENCE TODAY | | bioinformatics |
Image Credit: Univeristy of Glasgow
Stranded dolphins’ brains show common signs of Alzheimer’s disease
The brains of three different species of stranded dolphins show classic markers of human Alzheimer’s disease, according to the most extensive study into dementia in odontocetes (toothed whales).

The pan-Scotland research, a collaboration between the University of Glasgow, the Universities of St Andrews and Edinburgh and the Moredun Research Institute, studied the brains of 22 odontocetes which had all been stranded in Scottish coastal waters.
The study, which is published in the European Journal of Neuroscience, included five different species – Risso’s dolphins, long-finned pilot whales, white-beaked dolphins, harbour porpoises and bottlenose dolphins – and found that four animals from different dolphin species had some of the brain changes associated with Alzheimer’s disease in humans.
The findings may provide a possible answer to unexplained live-stranding events in some odontocete species. Study authors confirm the results could support the ‘sick-leader’ theory, whereby an otherwise healthy pod of animals find themselves in dangerously shallow waters after following a group leader who may have become confused or lost.
Whales, dolphins and porpoises are regularly stranded around the coasts of the UK. They are often found stranded in groups, or pods, in shallow waters and sometimes on beaches. While some animals can be moved to safer, deeper waters by teams of experts, other animals are less lucky and perish as a result. The underlying causes of live stranding events are not always clear, and research is ongoing to gain better insights.
For this study researchers examined stranded animals for the presence of the brain pathology that are part of the hallmarks of Alzheimer’s disease, including the formation of amyloid-beta plaques, the accumulation of phospho-tau and gliosis (a change in cell numbers in response to central nervous system damage). The results reveal that the brains of all the aged animals studied had amyloid-beta plaques.
Three animals in particular – each from a different odontocete species – had amyloid-beta plaques as well as a number of other dementia-related pathologies in their brains, showing that some odontocete species develop Alzheimer’s-like neuropathology. However, the study cannot
confirm whether any of the animals would have suffered with the same cognitive deficits associated with clinical Alzheimer’s disease in humans.
Lead researcher, Dr Mark Dagleish from the University of Glasgow, said: “These are significant findings that show, for the first time, that the brain pathology in stranded odontocetes is similar to the brains of humans affected by clinical Alzheimer’s disease. While it is tempting at this stage to speculate that the presence of these brain lesions in odontocetes indicates that they may also suffer with the cognitive deficits associated with human Alzheimer’s disease, more research must be done to better understand what is happening to these animals.”
Co-author, Professor Frank Gunn-Moore from the University of St Andrews, said: “I have always been interested in answering the question: do only humans get dementia? Our findings answer this question as it shows potential dementia associated pathology is indeed not just seen in human patients. This study is also a great example of both different research institutes, but also different branches of the Life Sciences working together.”
Professor Tara Spires-Jones, University of Edinburgh, said: “We were fascinated to see brain changes in aged dolphins similar to those in human ageing and Alzheimer’s disease. Whether these pathological changes contribute to these animals stranding is an interesting and important question for future work.”
All animals in this research were studied after a stranding event. Marine Scotland and Defra fund post-mortem examinations, via the Scottish Marine Animal Stranding Scheme (SMASS), of cetaceans (including odontocetes), pinnipeds and marine turtles that strand and die in Scottish coastal waters.
The paper, ‘Alzheimer’s disease-like neuropathology in three species of oceanic dolphin’ is published in the European Journal of Neuroscience. Ref: onlinelibrary.wiley.com/doi/ abs/10.1111/ejn.15900.
43 | BIOSCIENCE TODAY | | neurology |
Look into my eyes - and learn!
The average octopus may be shy and retiring - but they do know how to pull a party trick! Whether swimming by jet propulsion, blasting inky chemicals at enemies, or changing skin to blend in with surroundings, this clever mollusc is a biologist’s favourite.
And now a team of researchers at the University of Oregon is investigating yet another distinctive facet of these intriguing cephalopods: their outstanding visual capabilities.
Recent findings, reported in Current Biology, lay out a detailed map of the octopus’s visual system, classifying different types of neurons in a part of the brain devoted to vision. The map is a resource for other neuroscientists, giving details that could guide future experiments. And it could teach us something about the evolution of brains and visual systems more broadly, too.
Associate professor Cris Niell’s lab in the Institute of Neuroscience studies vision, mostly in mice. But a few years ago, postdoctoral researcher Judit Pungor brought a new species to the lab: the California two-spot octopus. While not traditionally used as a study subject in the lab, the cephalopod quickly captured the interest of UO neuroscientists. Unlike mice, which are not known for having good vision, “octopuses have an amazing visual system, and a large fraction of their brain is dedicated to visual processing,” Niell said. “They have an eye that’s remarkably similar to the human eye, but after that, the brain is completely different.
The last common ancestor between octopuses and humans was 500 million years ago, and the species have since evolved in very different contexts. So scientists didn’t know whether the parallels in visual systems extended beyond the eyes, or whether the octopus was instead using completely different kinds of neurons and brain circuits to achieve similar results.

“Seeing how the octopus eye convergently evolved similarly to ours, it’s cool to think about how the octopus visual system could be a model for understanding brain complexity more generally,” said Mea Songco-Casey, a graduate student in Niell’s lab and the first author on the paper. “For example, are there fundamental cell types that are required for this very intelligent, complex brain?”
Here, the team used genetic techniques to identify different types of neurons in the octopus’ optic lobe, the part of the brain that’s devoted to vision.
They picked out six major classes of neurons, distinguished based on the chemical signals they send. Looking at the activity of certain genes in those neurons then revealed further subtypes, providing clues to more specific roles. In some cases, the researchers pinpointed particular groups of
neurons in distinctive spatial arrangements, for instance, a ring of neurons around the optic lobe that all signal using a molecule called octopamine. Fruit flies use this molecule, which is similar to adrenaline, to increase visual processing when the fly is active. So it could perhaps have a similar role in octopuses.
“Now that we know there’s this very specific cell type, we can start to go in and figure out what it does,” Niell said.
About a third of the neurons in the data didn’t quite look fully developed. The octopus brain keeps growing and adding new neurons over the animal’s lifespan. These immature neurons, not yet integrated into brain circuits, were a sign of the brain in the process of expanding.
However, the map didn’t reveal sets of neurons that clearly transferred over from humans or other mammalian brains, as the researchers thought it might.
“At the obvious level, the neurons don’t map onto each other; they’re using different neurotransmitters,” Niell said. “But maybe they’re doing the same kinds of computations, just in a different way.”
Digging deeper will also require getting a better handle on cephalopod genetics. Because the octopus hasn’t traditionally been used as a lab animal, many of the tools that are used for precise genetic manipulation in fruit flies or mice don’t yet exist for the octopus, said Gabby Coffing, a graduate student in biology professor Andrew Kern’s lab who worked on the study.
“There are a lot of genes where we have no idea what their function is, because we haven’t sequenced the genomes of a lot of cephalopods,” Pungor said. Without genetic data from related species as a point of comparison, it’s harder to deduce the function of particular neurons.
Niell’s team is up for the challenge. They’re now working to map the octopus brain beyond the optic lobe, seeing how some of the genes they focused on in this study show up elsewhere in the brain. They are also recording from neurons in the optic lobe, to determine how they process the visual scene.
In time, their research might make these mysterious marine animals a little less murky — and shine a little light on our own evolution, too.
REFERENCE
“Cell types and molecular architecture of the Octopus bimaculoides visual system, Current Biology. DOI: 10.1016/j.cub.2022.10.015 Originally report: Laurel Hamers.
44 | research | | BIOSCIENCE TODAY |

Find your digital voice! Engaging your brand with audiences that matter. www.be-everywhere.co.uk E: info@be-everywhere.co.uk T: 0191 580 5990
palladium catalysts
thin lm
nickel foam
buckyballs
MOFs
Nd:YAG
perovskite crystals
europium phosphors
alternative energy
99.9999% aluminum oxide
YBCO
MOCVD
AuNPs
EuFOD
InAs wafers
III-IV semiconductors
diamond micropowder
additive manufacturing
organometallics
nanogels surface functionalized nanoparticles
nanodispersions
3D graphene foam
glassy carbon isotopes
ultralight aerospace alloys
titanium aluminum carbide
metamaterials
osmium
h-BN
Invar
GDC
NMC
CIGS
silver nanoparticles
ITO
quantum dots
niobium C103 UHP uorides
transparent ceramics
radiation shielding
sputtering targets
endohedral fullerenes
gold nanocubes
tungsten carbide
tantaloy 60
photovoltaics
graphene oxide
rare earth optical ber dopants
OLED lighting
laser crystals
scandium powder
biosynthetics
exible electronics
The Next Generation of Material Science Catalogs
mischmetal
chalcogenides
carbon nanotubes
CVD precursors
deposition slugs
platinum ink
superconductors
Over 35,000 certi ed high purity laboratory chemicals, metals, & advanced materials and a state-of-the-art Research Center. Printable GHS-compliant Safety Data Sheets. Thousands of new products. And much more. All on a secure multi-language "Mobile Responsive” platform.
ultra high purity materials
metallic glass
pyrolitic graphite
American Elements Opens a World of Possibilities..... Now Invent!
rare earth metals
mesoporus silica
www.americanelements.com
99.99999% mercury
molybdenum TZM li-ion battery materials
InGaAs
zeolites
Ti-6Al-4V
metallocenes
Now Invent. TM
© 2001-2023. American Elements is a U.S.Registered Trademark 140.116 Cerium 58 2 8 18 19 9 2 Ce 140.90765 Praseodymium 59 2 8 18 21 8 2 Pr 144.242 Neodymium 60 2 8 18 22 8 2 Nd (145) Promethium 61 2 8 18 23 8 2 Pm 150.36 Samarium 62 2 8 18 24 8 2 Sm 151.964 Europium 63 2 8 18 25 8 2 Eu 157.25 Gadolinium 64 2 8 18 25 9 2 Gd 158.92535 Terbium 65 2 8 18 27 8 2 Tb 162.5 Dysprosium 66 2 8 18 28 8 2 Dy 164.93032 Holmium 67 2 8 18 29 8 2 Ho 167.259 Erbium 68 2 8 18 30 8 2 Er 168.93421 Thulium 69 2 8 18 31 8 2 Tm 173.054 Ytterbium 70 2 8 18 32 8 2 Yb 174.9668 Lutetium 71 2 8 18 32 9 2 Lu 232.03806 Thorium 90 2 8 18 32 18 10 2 Th 231.03588 Protactinium 91 2 8 18 32 20 9 2 Pa 238.02891 Uranium 92 2 8 18 32 21 9 2 U (237) Neptunium 93 2 8 18 32 22 9 2 Np (244) Plutonium 94 2 8 18 32 24 8 2 Pu (243) Americium 95 2 8 18 32 25 8 2 Am (247) Curium 96 2 8 18 32 25 9 2 Cm (247) Berkelium 97 2 8 18 32 27 8 2 Bk (251) Californium 98 2 8 18 32 28 8 2 Cf (252) Einsteinium 99 2 8 18 32 29 8 2 Es (257) Fermium 100 2 8 18 32 30 8 2 Fm (258) Mendelevium 101 2 8 18 32 31 8 2 Md (259) Nobelium 102 2 8 18 32 32 8 2 No (262) Lawrencium 103 2 8 18 32 32 8 3 Lr 1.00794 Hydrogen 1 1 H 6.941 Lithium 3 2 1 Li 9.012182 Beryllium 4 2 2 Be 22.98976928 Sodium 11 2 8 1 Na 24.305 Magnesium 12 2 8 2 Mg 39.0983 Potassium 19 2 8 8 1 K 40.078 Calcium 20 2 8 8 2 Ca 85.4678 Rubidium 37 2 8 18 8 1 Rb 87.62 Strontium 38 2 8 18 8 2 Sr 132.9054 Cesium 55 2 8 18 8 1 Cs 137.327 Barium 56 2 8 18 8 2 Ba (223) Francium 87 2 8 18 32 18 8 Fr (226) Radium 88 2 8 18 32 18 8 Ra 44.955912 Scandium 21 2 8 9 2 Sc 47.867 Titanium 22 2 8 10 2 Ti 50.9415 Vanadium 23 2 8 11 2 V 51.9961 Chromium 24 2 8 13 1 Cr 54.938045 Manganese 25 2 8 13 2 Mn 55.845 Iron 26 2 8 14 2 Fe 58.933195 Cobalt 27 2 8 15 2 Co 58.6934 Nickel 28 2 8 16 2 Ni 63.546 Copper 29 2 8 18 1 Cu 65.38 Zinc 30 2 8 18 2 Zn 88.90585 Yttrium 39 2 8 18 9 2 Y 91.224 Zirconium 40 2 8 18 10 2 Zr 92.90638 Niobium 41 2 8 18 12 1 Nb 95.96 Molybdenum 42 2 8 18 13 1 Mo (98.0) Technetium 43 2 8 18 13 2 Tc 101.07 Ruthenium 44 2 8 18 15 1 Ru 102.9055 Rhodium 45 2 8 18 16 1 Rh 106.42 Palladium 46 2 8 18 18 Pd 107.8682 Silver 47 2 8 18 18 1 Ag 112.411 Cadmium 48 2 8 18 18 2 Cd 138.90547 Lanthanum 57 2 8 18 9 2 La 178.48 Hafnium 72 2 8 18 10 2 Hf 180.9488 Tantalum 73 2 8 18 11 2 Ta 183.84 Tungsten 74 2 8 18 12 2 W 186.207 Rhenium 75 2 8 18 13 2 Re 190.23 Osmium 76 2 8 18 14 2 Os 192.217 Iridium 77 2 8 18 15 2 Ir 195.084 Platinum 78 2 8 18 17 1 Pt 196.966569 Gold 79 2 8 18 18 1 Au 200.59 Mercury 80 2 8 18 18 2 Hg (227) Actinium 89 2 8 18 32 18 9 Ac (267) Rutherfordium 104 2 8 18 32 32 10 Rf (268) Dubnium 105 2 8 18 32 32 11 Db (271) Seaborgium 106 2 8 18 32 32 12 Sg (272) Bohrium 107 2 8 18 32 32 13 Bh (270) Hassium 108 2 8 18 32 32 14 Hs (276) Meitnerium 109 2 8 18 32 32 15 Mt (281) Darmstadtium 110 2 8 18 32 32 17 Ds (280) Roentgenium 111 2 8 18 32 32 18 Rg (285) Copernicium 112 2 8 18 32 32 18 Cn 4.002602 Helium 2 2 He 10.811 Boron 5 2 3 B 12.0107 Carbon 6 2 4 C 14.0067 Nitrogen 7 2 5 N 15.9994 Oxygen 8 2 6 O 18.9984032 Fluorine 9 2 7 F 20.1797 Neon 10 2 8 Ne 26.9815386 Aluminum 13 2 8 3 Al 28.0855 Silicon 14 2 8 4 Si 30.973762 Phosphorus 15 2 8 5 P 32.065 Sulfur 16 2 8 6 S 35.453 Chlorine 17 2 8 7 Cl 39.948 Argon 18 2 8 8 Ar 69.723 Gallium 31 2 8 18 3 Ga 72.64 Germanium 32 2 8 18 4 Ge 74.9216 Arsenic 33 2 8 18 5 As 78.96 Selenium 34 2 8 18 6 Se 79.904 Bromine 35 2 8 18 7 Br 83.798 Krypton 36 2 8 18 8 Kr 114.818 Indium 49 2 8 18 18 3 In 118.71 Tin 50 2 8 18 18 4 Sn 121.76 Antimony 51 2 8 18 18 5 Sb 127.6 Tellurium 52 2 8 18 18 6 Te 126.90447 Iodine 53 2 8 18 18 7 I 131.293 Xenon 54 2 8 18 18 8 Xe 204.3833 Thallium 81 2 8 18 18 3 Tl 207.2 Lead 82 2 8 18 18 4 Pb 208.9804 Bismuth 83 2 8 18 18 5 Bi (209) Polonium 84 2 8 18 18 6 Po (210) Astatine 85 2 8 18 18 7 At (222) Radon 86 2 8 18 18 8 Rn (284) Nihonium 113 2 8 18 32 32 18 3 (289) Flerovium 114 2 8 18 32 32 18 Fl (288) Moscovium 115 2 8 18 32 32 18 5 (293) Livermorium 116 2 8 18 32 32 18 6 Lv (294) Tennessine 117 2 8 18 32 32 18 (294) Oganesson 118 2 8 18 32 32 18 8 Nh Mc Ts Og
zircaloy
-4
SOFC
powder
borophene




 Karen Southern Editor in chief
Karen Southern Editor in chief







 Robin May
Robin May






















































